Abstract
Chrysanthemum is a typical short-day (SD) plant that responds to shortening daylength during the transition from the vegetative to the reproductive phase. FLOWERING LOCUS T (FT)/Heading date 3a (Hd3a) plays a pivotal role in the induction of phase transition and is proposed to encode a florigen. Three FT-like genes were isolated from Chrysanthemum seticuspe (Maxim.) Hand.-Mazz. f. boreale (Makino) H. Ohashi & Yonek, a wild diploid chrysanthemum: CsFTL1, CsFTL2, and CsFTL3. The organ-specific expression patterns of the three genes were similar: they were all expressed mainly in the leaves. However, their response to daylength differed in that under SD (floral-inductive) conditions, the expression of CsFTL1 and CsFTL2 was down-regulated, whereas that of CsFTL3 was up-regulated. CsFTL3 had the potential to induce early flowering since its overexpression in chrysanthemum could induce flowering under non-inductive conditions. CsFTL3-dependent graft-transmissible signals partially substituted for SD stimuli in chrysanthemum. The CsFTL3 expression levels in the two C. seticuspe accessions that differed in their critical daylengths for flowering closely coincided with the flowering response. The CsFTL3 expression levels in the leaves were higher under floral-inductive photoperiods than under non-inductive conditions in both the accessions, with the induction of floral integrator and/or floral meristem identity genes occurring in the shoot apexes. Taken together, these results indicate that the gene product of CsFTL3 is a key regulator of photoperiodic flowering in chrysanthemums.
Keywords: Chrysanthemum, floral transition, flowering, FT, short day
Introduction
Daylength plays an important role in floral transitions such as the shift from vegetative to inflorescence meristem identity in plants. In their pioneering work, Garner and Allard (1920) classified plants according to their responses to daylength. They showed that long-day (LD) plants take less time to flower when light exposure exceeds a certain critical daylength, while short-day (SD) plants flower earlier when light exposure is shorter than a certain critical daylength. The main site for the perception of daylength is recognized to be the leaf. Through tests with chrysanthemums, in 1936 Chailakhyan determined the leaf to be the site where a plant hormone called florigen is produced (for a review, see Chailakhyan and Krikorian, 1975). After a florigen is produced in the leaf, it travels through the phloem and into the shoot apical meristem (SAM), which is where flowering is initiated. This approach allows the process to be subdivided into two successive steps: (i) ‘induction’ mechanisms, which occur in the leaf, and (ii) floral ‘evocation’, which consists of the events occurring in the SAM that commit it to producing flowers (Evans, 1969).
Recent advances in molecular biology and traditional grafting studies have revealed that the gene product of FLOWERING LOCUS T (FT) in Arabidopsis, an LD plant, and that of Heading date 3a (Hd3a) in rice (Oryza sativa L.), an SD plant, can function as florigens to regulate flowering (Corbesier et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007; Notaguchi et al., 2008). Light plays important roles in the regulation of flowering in Arabidopsis by regulating CONSTANS (CO) and FT (Kobayashi and Weigel, 2007). The expression of CO mRNA is under the control of the circadian clock. Post-translational regulation of the stability of the CO protein by light plays an important role in the regulation of FT expression (Suarez-Lopez et al., 2001; Valverde et al., 2004). Induction of flowering by FT-like proteins seems to be broadly conserved across plants. In tomato, a photoperiod-insensitive plant, SINGLE FLOWER TRUSS (SFT/SP3D), an orthologue of FT, is induced in mature leaves and triggers flowering (Lifschitz et al., 2006). Grafting of shoots from SFT-overexpressing tomato plants to sft mutants rescued the late-flowering phenotype of the mutants, indicating that SFT-dependent graft-transmissible signals substituted for the developmental defects in the mutants; in addition, these signals substituted for LD stimuli in Arabidopsis and SD stimuli in Maryland Mammoth tobacco (Lifschitz et al., 2006). This indicates that a systemic graft-transmissible floral signal is transmitted from the donor to the receptor and broadly conserved across plants (Shalit et al., 2009). The fact that homologues of the FT protein are normally present in the phloem sap of cucurbits (Lin et al., 2007) and rice (Aki et al., 2008) supports the idea that FT-like proteins are universal signalling molecules travelling from the leaves to the meristems via phloem.
FT/Hd3a belongs to a small protein family whose members show homology to the mammalian phosphatidylethanolamine-binding protein (PEBP; Kardailsky et al., 1999; Kobayashi et al., 1999; Kojima et al., 2002). In Arabidopsis, in addition to FT, five PEBP family genes are found: TERMINAL FLOWER 1 (TFL1), TWIN SISTER OF FT (TSF), BROTHER OF FT AND TFL1 (BFT), Arabidopsis thaliana CENTRORADIALIS homologue (ATC), and MOTHER OF FT AND TFL1 (MFT) (Bradley et al., 1997; Mimida et al., 2001). Phylogenetic analysis has resolved three major clades within this family, corresponding to FT-like, TFL1-like, and MFT-like genes. FT-like and TFL1-like genes function in controlling flowering time (Bradley et al., 1997). FT-like genes promote flowering, whereas TFL1-like genes delay flowering and prevent conversion of the SAM into a floral meristem. In Arabidopsis, FT and TFL1 have antagonistic effects on flowering time, and their functions have been related to the presence of critical amino acid residues—Tyr85/Gln140 in FT and His88/Asp144 in TFL1 (Hanzawa et al., 2005; Ahn et al., 2006). In Arabidopsis, FT and its closest paralogue, TSF, play a similar role in inducing flowering and show similar patterns of regulation of responses to photoperiod and vernalization (Yamaguchi et al., 2005). Rice is classified as a facultative SD plant; its flowering is not completely suppressed under LD conditions. It uses two florigen genes, Hd3a and its closest paralogue, Rice flowering locus T1 (RFT1), depending on daylength (Komiya et al., 2008, 2009). Hd3a is the major floral activator under SD conditions, while RFT1 is the major floral activator under LD conditions. Recently, a negative effect of FT-like genes on flowering has been postulated in sunflower and sugar beet. In sunflower, the in-frame allele of Helianthus annuus FT1 (HaFT1) accelerates flowering and the frameshift allele represses flowering by interfering with the function of an activating paralogue, HaFT4 (Blackman et al., 2010). In sugar beet, Beta vulgaris FT2 (BvFT2) accelerates flowering, and its paralogue, BvFT1, represses flowering. A comparison analysis of BvFT1 and BvFT2 revealed that substitutions in the fourth exon, encoding an external loop of PEBP, are essential for the antagonistic functions (Pin et al., 2010). MFT-like genes may play different roles in plant development, compared with FT-like and TFL1-like genes. MFT regulates seed germination via the modulation of abscisic acid (ABA) and gibberellin (GA) signalling in Arabidopsis (Xi et al., 2010).
FT is transported from phloem companion cells to shoot apexes via the phloem and interacts with a basic-leucine zipper (bZIP) transcription factor, FLOWERING LOCUS D (FD) (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007). Unlike FT, FD is only expressed at the shoot apexes and is independent of photoperiodic induction (Abe et al., 2005; Wigge et al., 2005). At the shoot apexes in Arabidopsis, FT and FD probably activate important regulators of floral fate, such as APETALA1 (AP1) and FRUITFULL (FUL) (Kobayashi and Weigel, 2007). FLORICAULA (FLO) in snapdragon and its Arabidopsis orthologue, LEAFY (LFY), appear to play pivotal roles in specifying floral meristem identity (Weigel and Nilsson, 1995). FLO/LFY plays an important role in reproductive transition and regulates flower development by establishing the expression of floral organ identity genes (Benlloch et al., 2007). The MADS-box genes CAULIFLOWER (CAL), AP1, and FUL act in a redundant manner to control meristem identity in Arabidopsis. FUL is expressed in inflorescence meristems and leaves, while AP1 and CAL are preferentially expressed in inflorescences and floral meristems (Mandel et al., 1992; Kempin et al., 1995; Mandel and Yanofsky, 1995; Ferrándiz et al., 2000). In addition to promoting floral identity, FUL also promotes floral transition (Ferrándiz et al., 2000). From chrysanthemum, CmSOC1, CmAFL1, CmFL, and CDM111 have been isolated as homologues of SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), AP1/FUL, FLO/LFY, and AP1, respectively (Shchennikova et al., 2004; Li et al., 2009). The expression of CmSOC1, CmAFL1, CmFL, and CDM111 is induced in differentiating floral shoot apexes, suggesting that preserved mechanisms trigger reproductive transition in chrysanthemums (Shchennikova et al., 2004; Li et al., 2009; Sumitomo et al., 2009).
Chrysanthemum (Chrysanthemum morifolium Ramat.) is a typical SD plant widely cultivated worldwide; variations in the flowering time of chrysanthemums (from early summer to winter) under natural conditions mainly reflect differences in their critical daylengths for flowering (Langton, 1977; Kawata, 1987). Seasonal changes in the extension growth and flowering of chrysanthemums are an adaptation to temperate climates at middle latitudes. Growth and flowering depend on the combination of growth temperature, daylength, and the environment in the previous season (for a review, see Cathey, 1969). Researchers have been trying to reveal how environmental conditions such as daylength and temperature regulate flowering, in order to achieve stable year-round flower production (Link, 1936; Post and Kamemoto, 1950; for a review, see Cathey, 1969). Chrysanthemum growers are currently trying to produce constantly throughout the year. However, in the case of year-round production, unexpected developments such as premature flower budding occasionally occur due to deviations from normal environmental conditions in greenhouses. Despite the efforts of researchers to understand the mechanisms underlying flowering in chrysanthemums, very little is known about floral transition and further development of floral meristems at the molecular level. Chrysanthemum morifolium is a complex hybrid derived from several species that grow in the wild in China and Japan (for a review, see Cathey, 1969), which makes its genetic analysis difficult. A wild diploid chrysanthemum, Chrysanthemum seticuspe (Maxim.) Hand.-Mazz. f. boreale (Makino) H. Ohashi & Yonek (C. seticuspe hereafter; 2n=18), probably exhibits similar seasonal growth and flowering responses to other chrysanthemum cultivars. To overcome the issue of complex hybridity and polyploidy, C. seticuspe was used as an alternative model of chrysanthemum cultivars in this study.
The present study reports the isolation and functional analysis of homologues of FT/Hd3a from C. seticuspe. Ectopic expression of the FT-like gene CsFTL3 promoted flowering in chrysanthemum under non-inductive conditions. The CsFTL3 expression levels in the leaves of the two C. seticuspe accessions that differed in their critical daylengths for flowering closely coincided with the induction of the floral identity genes at the shoot apexes and capitulum development.
Materials and methods
Plant material and growth conditions
Chrysanthemum seticuspe accessions NIFS-3 and Matsukawa were used for the experiments. Stock plants were grown in a greenhouse maintained at an air temperature >18 °C and ventilated when the temperature increased above 25 °C, under a natural photoperiod with a 4 h night break (NB; 23:00–03:00 h) provided by incandescent lamps (K-RD100V60W; Matsushita Electric Industrial Co. Ltd, Osaka, Japan). Rooted cuttings from the stock plants were planted into 7.5 cm plastic pots containing a commercial horticultural soil (Kureha-Engei-Baido; Kureha Chemical Industry Co. Ltd, Tochigi, Japan) and grown in the greenhouse. When the plants developed four or five expanded leaves, they were transferred to a growth chamber maintained at 20 °C with a 16 h photoperiod (LD conditions). Light was supplied with fluorescent tubes (FL40SW; Mitsubishi Co. Ltd, Tokyo, Japan) at a photosynthetic photon flux density (PPFD) of 200 μmol m−2 s−1. After 7 d of growth under these conditions, the plants were transferred to a growth chamber maintained at 20 °C with an 8 h photoperiod (SD conditions), a 16 h photoperiod (LD conditions), or an 8 h SD+an NB (NB conditions). Depending on the experiments, two kinds of NBs were given: a 4 h NB was supplied by fluorescent tubes at 200 μmol m−2 s−1 and a 15 min NB was supplied by red light-emitting diodes (LEDs) (LED-R; 660 nm; EYELA Co. Ltd, Tokyo, Japan) at 30 μmol m−2 s−1.
CsFTL1 cloning
Total RNA was extracted from young fully expanded leaves and shoot apexes of C. seticuspe NIFS-3 by using the RNeasy Plant Mini Kit (QIAGEN K.K., Tokyo, Japan) and treated with RNase-free DNase (QIAGEN K.K.) according to the manufacturer’s instructions. cDNAs were synthesized from 500 ng of total RNA by using the TaKaRa RNA PCR Kit (AMV) version 2.1 (TaKaRa BIO Inc., Shiga, Japan) according to the manufacturer’s instructions; these cDNAs were used as templates in subsequent PCR experiments. PCR amplification was performed with an oligo(dT)-M13M4 adaptor primer (5′-GTTTTCCCAGTCACGAC-3′) and the degenerate primers (FT-F2: 5′-TAYACIYTIGTIATGGTIGAYCC-3′; FT-R2: 5′-CCISWYTCICKYTGRCARTT-3′). PCR products ∼320 bp long were cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) as CsFTL fragments and sequenced. Internal gene-specific primers were designed to isolate full-length cDNAs of CsFTL1. The open reading frames (ORFs) of the genes were determined by 3′- and 5′-rapid amplification of cDNA ends-PCR (RACE-PCR), using the TaKaRa RNA PCR Kit (AMV) version 2.1 (TaKaRa BIO Inc.) and the Roche 5′/3′-RACE second-generation Kit (Roche Diagnostics K.K., Tokyo, Japan) according to the manufacturers’ instructions. The full-length cDNAs of CsFTL1 were amplified using the cDNA synthesized from the total RNA extracted from young fully expanded leaves as the template and cloned into the pGEM-T Easy Vector (Promega).
CsFTL2 and CsFTL3 cloning
Genomic DNA was extracted from the leaves of C. seticuspe NIFS-3 by using the DNeasy Plant Mini Kit (QIAGEN K.K.) according to the manufacturer’s instructions. For CsFTL2, PCR amplification was performed with CsFT-F (5′-ATGCCGAGGGAAAGGGATCC-3′) and CsFT-1R (5′-AGCTCCTGTAGTCTCTGGAA-3′) primers based on the CsFTL1 cDNA sequence. PCR products ∼1500 bp long were cloned into the pGEM-T Easy Vector (Promega) and sequenced. Clones that were even slightly different from CsFTL1 were designated CsFTL2.
For CsFTL3, PCR amplification was performed with CsFT-F (5′-ATGCCGAGGGAAAGGGATCC-3′) and CsFT-2R (5′-CCCAATTGCCGGAATAGCAC-3′) primers. PCR products ∼380 bp long were cloned into the pGEM-T Easy Vector (Promega) and sequenced. Clones that were even slightly different from both CsFTL1 and CsFTL2 were designated CsFTL3.
For full-length cDNAs of CsFTL2 and CsFTL3, internal gene-specific primers, CsFTL2-F (5′-TGGGTGCGATCTCAAACCCTCTCAGA-3′) and CsFTL3-F (5′-ACTTACACTGGTTGGTTACC-3′), were used for 3′-RACE-PCR. The products were cloned into the pGEM-T Easy Vector (Promega) and sequenced.
Comparison of CsFTL genes and phylogenetic analysis
The sequences of the CsFTL genes were aligned with ClustalX, and Ks values were calculated with Molecular Evolutionary Genetics Analysis version 5 (MEGA 5) (http://www.megasoftware.net). The amino acid sequences of the PEBP family were assembled with ClustalX. A Neighbor–Joining phylogenetic tree was generated with MEGA 5, using the Poisson model with gamma-distributed rates and 1000 bootstrap replicates.
Expression analysis by quantitative real-time PCR
The abundance of transcripts was investigated by quantitative real-time PCR (QRT-PCR). Total RNA was extracted from each tissue by using the RNeasy Plant Mini Kit (QIAGEN K.K.) and treated with RNase-free DNase (QIAGEN K.K.) according to the manufacturer’s instructions. For each sample, 500 ng of total RNA was reverse-transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics K.K.) according to the manufacturer’s instructions. The cDNA was diluted to 20% of its original concentration, and 5 μl of cDNA solution was used in a QRT-PCR (reaction mixture, 15 μl) with SYBR Premix Ex Taq (TaKaRa BIO Inc.) on a LightCycler system (Roche Diagnostics K.K.). The QRT-PCR products of CsFTL1, CsFTL2, and CsFTL3 were cloned into the pGEM-T Easy Vector (Promega), and 10 clones were sequenced to verify the contributing genes. All the sequences were identical to those of CsFTL1, CsFTL2, and CsFTL3. The transcription levels of CsFTL1, CsFTL2, CsFTL3, CsAFL1, CsSOC1, CsM111, and CsFL in the samples were directly compared after normalization against the CsACTIN (AB679277) or CsEF1α (AB679278) loading standard. The ‘calibrator sample’ was designated as the most highly expressed time point for each gene of interest and therefore showed the highest relative expression level of 1.0 in each QRT-PCR run. The experiments were performed with three independently isolated RNA samples from the tissue. The data represent the mean ±standard error (SE) of at least three biological replicates. Primer sequences and PCR conditions are listed in Supplementary Table S1 (available at JXB online).
Arabidopsis and chrysanthemum transformation
A full-length coding sequence of CsFTL3 was cloned into the vector pENTR221 (Invitrogen Carlsbad, CA, USA) via the BP reaction by using Gateway BP Clonase II enzyme mix (Invitrogen). The sequence was then subcloned into the binary vector pGWB2 (Nakagawa et al., 2007) via the LR reaction by using Gateway LR Clonase II enzyme mix (Invitrogen) as described in the manufacturer’s instructions. Arabidopsis (ecotype Columbia) plants were transformed using the floral dip method (Clough and Bent, 1998), and the flowering time of the T3 line was evaluated. Transgenic lines of C. morifolium ‘Jimba’ were obtained by an Agrobacterium-mediated transformation system, as described by Aida et al. (2004). Candidate transformants were selected on the basis of paromomycin resistance. To confirm the presence of the transgene in T0 plants, genomic DNA was extracted from the leaves and the NPTII region was amplified using NPTII-F (5′-GAGAGGCTATTCGGCTATGA-3′) and NPTII-R (5′-GATGCTCTTCGTCCAGATCA-3′) primers. Each T0 line was propagated in vitro. The stem segments with axillary buds were used as explants. The emerged shoots were transplanted and used for further analysis.
Grafting of chrysanthemum plants
CsFTL3-overexpressing chrysanthemum plants (#20-66) and wild-type Jimba plants were used as stock. Wild-type Jimba plants (for experiment 1) and Nagano-queen plants (for experiment 2) were used as the scion, respectively. A wedge-shaped/slit grafting technique was applied, with the site of union wrapped with Parafilm. After grafting, the plants were kept for 2 weeks under high-humidity conditions, with a plastic film covering. In experiment 1, the plants were kept in a growth chamber at 20 °C with a 16 h photoperiod during the experiment (for 18 weeks). In experiment 2, the plants were kept in a closed greenhouse (maintained at an air temperature >18 °C, and ventilated when it rose above 25 °C) under a natural photoperiod plus a 6 h NB during the experiment (for 9 weeks).
Anatomical observations
For microscopic observation, tissue samples were fixed in formalin–acetic acid–alcohol (FAA) [70% ethanol:formalin:acetic acid, 90:5:5 (v/v/v)] and stored at room temperature. For observation by scanning electron microscopy (SEM), the fixed floral buds were dehydrated in an ethanol series [50, 70, 90, 95, and 99.5% (v/v)], following which ethanol was replaced with t-butanol. The samples were then freeze-dried and observed by SEM (VE-7000; Keyence Co., Osaka, Japan).
Results
Flowering response of C. seticuspe and identification of FT-like genes
Flowering in C. seticuspe NIFS-3 was induced under 8 h SD conditions and efficiently inhibited under 16 h LD and 8 h SD+4 h NB conditions (Table 1). The number of days to visible flower bud formation was much less under floral-inductive SD conditions compared with that under non-inductive LD and NB conditions (Table 1).
Table 1.
Effect of daylength on flowering in C. seticuspe NIFS-3
| Flowering (%) | Days to visible flower buds | |
| SD | 100 | 20.8±0.38 |
| NB | 0 | >30 |
| LD | 0 | >30 |
SD, short day (8 h photoperiod); NB, night break (8 h SD+4 h NB); LD, long day (16 h photoperiod).
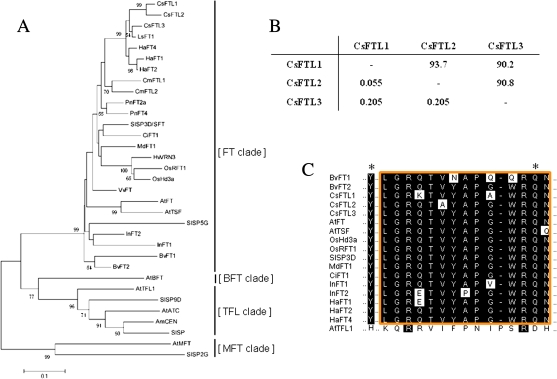
Three FT-like genes, CsFTL1 (AB679270), CsFTL2 (AB679271), and CsFTL3 (AB679272), which show significant homology to FT/Hd3a, were isolated from C. seticuspe NIFS-3. Sequence analysis of the deduced amino acid sequences showed that all the three CsFTL genes shared high identities (>90% at the amino acid sequence level) with each other (Fig. 1; Supplementary Fig. S1 at JXB online). The synonymous substitution rate between two sequences (i.e. Ks) provided a measure of time since divergence (Fig. 1). Ks for the comparison of CsFTL3 with the other two CsFTL genes was 0.205, while that for the comparison of CsFTL1 with CsFTL2 was 0.055, indicating that duplication probably occurred within the genus Chrysanthemum. A phylogenetic tree was constructed using the amino acid sequences of several plant FT/Hd3a- and TFL1-like proteins (Fig. 1). The tree was divided into four major clades represented by FT, TFL1, BFT, and MFT. According to the phylogenetic tree analysis, all the three CsFTLs were clustered into the FT-like protein family and separated from the TFL1-, BFT-, and MFT-like protein families (Fig. 1). A single amino acid substitution of Tyr85 to His88 and of Gln140 to Asp144 is the most critical residue change for distinguishing FT and TFL1, respectively, in Arabidopsis (Hanzawa et al., 2005; Ahn et al., 2006). All the CsFTLs had Tyr84 and Gln139 in the positions corresponding to Tyr85 and Gln140, respectively, in Arabidopsis FT (Fig. 1; Supplementary Fig. S1). The segment B in the fourth exon, encoding an external loop of PEBP, is also important for the antagonistic functions of FT and TFL1 in Arabidopsis (Ahn et al., 2006). In sugar beet, an antagonistic pair of FT paralogues, BvFT1 and BvFT2, have three substitutions in 14 amino acid residues in the external loop of PEBP, which is the major cause of their antagonistic functions (Pin et al., 2010). The deduced amino acid sequence of CsFTL3 in the external loop of PEBP was identical to those of most other flowering-promoting FT-like proteins, including FT and BvFT2, whereas CsFTL1 and CsFTL2 had double and single amino acid substitutions, respectively, in the region (Fig. 1). CsFTL1 had the charge-changing substitution (lysine at the position Glu135 in BvFT2) and the replacement with a non-polar amino acid (alanine at the position Gly141 in BvFT2). These substitutions might have a profound effect on the activity of the FT-like proteins.
Fig. 1.
Cloning of FT-like genes from C. seticuspe. (A) Phylogenetic tree based on the deduced amino acid sequences of the PEBP gene family, including those of CsFTL1, CsFTL2, and CsFTL3, from several plant species. The tree was constructed by the Neighbor–Joining method. Bootstrap percentages >50% are shown along the branches. Species abbreviations: Antirrhinum majus, Am; Arabidopsis thaliana, At; Beta vulgaris, Bv; Citrus unshiu, Ci; Cucurbita maxima, Cm; Chrysanthemum seticuspe, Cs; Helianthus annuus, Ha; Hordeum vulgare, Hv; Ipomoea nil, In; Lactuca sativa, Ls; Malus×domestica, Md; Oryza sativa, Os; Populus nigra, Pn; Solanum lycopersicum, Sl; and Vitis vinifera, Vv. (B) Comparisons of CsFT paralogues. Ks and amino acid sequence identity (%) values for pairwise comparisons of CsFT paralogues are shown: Ks values are shown below the diagonal, while amino acid sequence identity (%) values are shown above the diagonal. (C) Partial amino acid sequence alignment of the PEBP family members. Asterisks indicate the residues Tyr85(Y)/Gln140(Q) and His88(H)/Asp144(D) contributing to FT and TFL1 functioning, respectively (Hanzawa et al., 2005; Ahn et al., 2006). The conserved segment region B in the fourth exon, corresponding to the external loop of the PEBP family proteins, is boxed. (This figure is available in colour at JXB online.)
Organ-specific expression patterns of CsFTL1, CsFTL2, and CsFTL3 in chrysanthemum
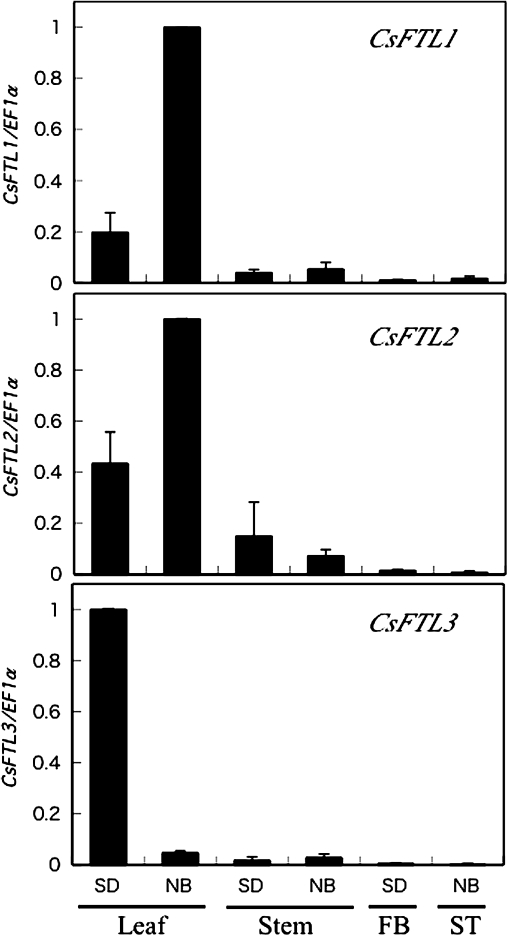
To examine the organ-specific expression patterns of the three CsFTL genes in C. seticuspe at dawn, QRT-PCR analysis was performed. CsFTL mRNAs were detected in all the investigated tissues with huge differences in their expression levels (Fig. 2). The expression levels of all the three CsFTL genes were higher in the leaves than in the other tested tissues. Interestingly, the CsFTL1 and CsFTL2 expression levels were higher under NB (non-inductive) conditions with NB provided with red light than under SD (floral-inductive) conditions (Fig. 2). In contrast, the CsFTL3 expression levels were higher under SD (floral-inductive) conditions than under non-inductive conditions (Fig. 2).
Fig. 2.
Expression patterns of CsFTLs in different tissues of C. seticuspe under SD or NB conditions. Chrysanthemum seticuspe NIFS-3 plants were grown under LD (16 h) conditions and then transferred to SD (8 h) or NB (SD+15 min exposure to red light around the middle of the night) conditions. The tissues were harvested 4 weeks after the plants were transferred to SD or NB conditions and subjected to QRT-PCR analysis. FB, flower bud (∼3 mm in diameter); ST, shoot tip (∼3 mm long). CsFTL1, CsFTL2, and CsFTL3 expression in the leaves was normalized against CsEF1α expression.
Expression patterns of CsFTL1, CsFTL2, and CsFTL3 in chrysanthemum under different light conditions
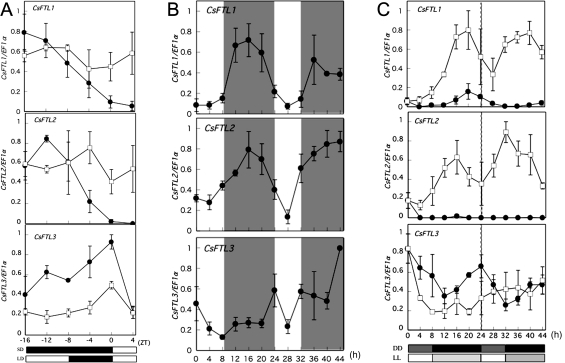
To investigate the effect of light on CsFTL expression in C. seticuspe, the relationship between the gene expression patterns in the leaves and floral-inductive photoperiod conditions was investigated. After 7 d of growth under LD conditions, the plants were shifted to SD conditions. The CsFTL1 and CsFTL2 transcript levels decreased after transfer to SD conditions (Fig. 3A), although CsFTL2 expression was transiently increased 4 h after transfer to SD conditions under the experimental conditions. In contrast, the CsFTL3 transcript levels rapidly increased after transfer to SD conditions; they were higher under SD conditions than under LD conditions in the dark period (Fig. 3A). Since FT-like genes, which accelerate flowering, are up-regulated under floral-inductive conditions (Kojima et al., 2002; King et al., 2006; Hayama et al., 2007; Kikuchi et al., 2009; Pin et al., 2010), CsFTL3 is selected as an FT-like gene candidate to regulate flowering in C. seticuspe. In addition, the counterparts of CsFTL1, CsFTL2, and CsFTL3 were identified in a chrysanthemum cultivar, Reagan, and it was observed that their expression profiles in the Reagan plants exposed to LD or SD conditions for 7 d coincided with those in C. seticuspe under both LD and SD conditions (Supplementary Fig. S2 at JXB online). This suggests that the alternative model system of using C. seticuspe for chrysanthemum cultivars is applicable for basic phenomena in chrysanthemums, such as flowering.
Fig. 3.
Expression patterns of CsFTLs in leaves of C. seticuspe under different light conditions. (A) Diurnal expression patterns of CsFTL genes in C. seticuspe under LD conditions and after transfer to SD conditions. Chrysanthemum seticuspe NIFS-3 plants were grown under 16 h LD conditions and then transferred to 8 h SD (filled circles) or maintained under 16 h LD (open squares) conditions. White and black horizontal bars represent light and dark periods, respectively. (B) Diurnal expression patterns of CsFTL genes in C. seticuspe under repeated SD conditions. Chrysanthemum seticuspe NIFS-3 plants were transferred to 8 h SD conditions after growth under 16 h LD conditions. Transcription levels were monitored from the beginning of the second SD cycle for the subsequent 44 h. Grey vertical areas represent dark periods. (C) Circadian expression patterns of CsFTL genes in C. seticuspe under continuous dark and continuous light conditions. Chrysanthemum seticuspe NIFS-3 plants exposed to an 8 h SD cycle after growth under 16 h LD conditions were transferred to continuous dark (DD; filled circles) or continuous light (LL; open squares) conditions. Transcription levels were monitored for the subsequent 44 h. All expression values of CsFTL1, CsFTL2, and CsFTL3 were normalized against CsEF1α expression.
Under repeated SD conditions, the expression of CsFTL genes in C. seticuspe plants showed diurnal oscillations (Fig. 3B). CsFTL1 and CsFTL2 transcripts accumulated during the dark period of the day, peaking around the middle of the dark period (Fig. 3B). Further, CsFTL3 transcripts accumulated during the dark period of the day, peaking at dawn (Fig. 3B). Under repeated SD conditions, the CsFTL3 transcript levels increased with SD cycles. The CsFTL1 and CsFTL2 transcript levels showed a clear diurnal oscillation as compared with the CsFTL3 transcript levels. The diurnal oscillations suggested that the genes were under the control of the circadian clock. In fact, the CsFTL1 transcript levels showed a circadian rhythm under both continuous light and continuous dark conditions, although the peak amplitude under continuous dark conditions was much lower than that under continuous light conditions (Fig. 3C). CsFTL2 transcripts were barely detectable under continuous dark conditions but showed a similar pattern to CsFTL1 transcripts under continuous light conditions (Fig. 3C). The CsFTL3 transcript levels also showed a circadian rhythm under continuous dark conditions, peaking at subjective dawn; on the other hand, they showed an arrhythmic pattern under continuous light conditions (Fig. 3C). The transcript levels under the two different conditions (continuous light versus continuous darkness) were almost the same.
Comparison of CsFTL3 expression and induction of floral integrator and/or floral identity genes, CsAFL1, CsFL, and CsM111, in the two accessions with different critical daylengths for flowering
Chrysanthemum cultivars have different critical daylengths for flowering. The flowering response of two C. seticuspe accessions, NIFS-3 and Matsukawa, to different photoperiods (11, 12, 13, and 14 h) was examined. All NIFS-3 plants flowered under 11 h and 12 h photoperiods; photoperiods longer than 13 h completely suppressed flowering until the end of the experiment (Table 2). Leaf initiation increased dramatically and slightly before floral transition under 13 h and 12 h photoperiods, respectively, compared with that under an 11 h photoperiod (Table 2). All Matsukawa plants flowered under 11, 12, and 13 h photoperiods; a 14 h photoperiod completely suppressed flowering until the end of the experiment (Table 2). Leaf initiation increased dramatically and slightly before floral transition under 14 h and 13 h photoperiods, respectively, compared with that under 11 h and 12 h photoperiods (Table 2). Therefore, NIFS-3 and Matsukawa are estimated to have 12 h and 13 h photoperiods as their critical daylengths for flowering.
Table 2.
Flowering response of C. seticuspe NIFS-3 and Matsukawa under different daylengths
| NIFS-3 |
Matsukawa |
|||
| Daylength (h) | Flowering (%) | No. of leaves | Flowering (%) | No. of leaves |
| 11 | 100 | 21.4±1.43 | 100 | 20.4±1.17 |
| 12 | 100 | 23.9±2.18 | 100 | 21.8±0.79 |
| 13 | 0 | >30 | 100 | 24.7±0.82 |
| 14 | 0 | >30 | 0 | >30 |
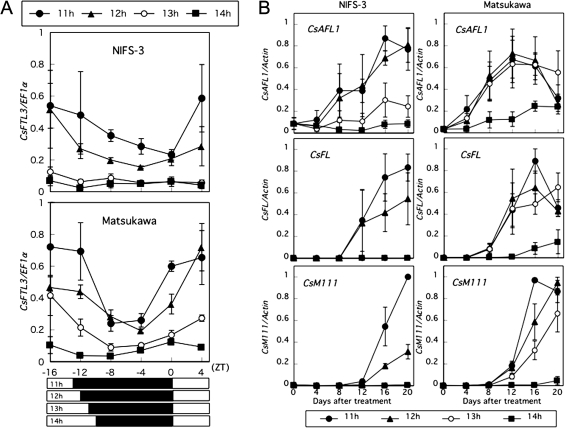
The CsFTL3 expression levels were investigated in both NIFS-3 and Matsukawa plants grown under the different photoperiod conditions described above. Leaves from the plants already exposed to seven cycles of each photoperiod were collected every 4 h over a 20 h period. The CsFTL3 expression levels increased as the photoperiod shortened (Fig. 4A). The CsFTL3 expression levels in NIFS-3 remained low under photoperiods >13 h, compared with those under 11 h and 12 h photoperiods; however, the expression levels in Matsukawa remained low only under a 14 h photoperiod (Fig. 4A). These CsFTL3 expression patterns are correlated with their estimated critical daylengths for flowering. The CsFTL3 mRNA expression levels clearly exhibited a diurnal rhythm in plants grown under shorter photoperiods; they started to increase towards the end of the night and peaked by midday (Fig. 4A).
Fig. 4.
Analysis of CsFTL3 in leaves and floral integrator and/or floral identity gene expression in shoot tips in two different C. seticuspe accessions under different daylengths. Two different C. seticuspe accessions, NIFS-3 and Matsukawa, were grown under 16 h LD conditions and then transferred to 11 h (filled circles), 12 h (filled triangles), 13 h (open circles), or 14 h (filled squares) photoperiod conditions. (A) Diurnal expression patterns of CsFTL3 in leaves. Seven days after transfer, leaves were harvested every 4 h and subjected to QRT-PCR analysis. The CsFTL3 expression was normalized against CsEF1α expression. White and black horizontal bars represent light and dark periods, respectively. (B) Analysis of floral integrator and/or floral identity gene expression in shoot tips. Shoot apexes were harvested every 4 d over a 20 d period and subjected to QRT-PCR analysis. The expression of each gene of interest was normalized against CsACTIN expression.
The expression of floral integrator and/or floral identity genes in shoot apexes under the different daylength conditions described above was also investigated. The C. seticuspe counterparts of CmAFL1, CmFL, and CDM111 (Shchennikova et al., 2004; Li et al., 2009) were cloned and named CsAFL1 (AB679273), CsFL (AB679274), and CsM111 (AB679275), respectively. The coding sequences of CsAFL1, CsFL, and CsM111 cDNAs exhibited 99.4, 99.8, and 99.7% identity, respectively, to those of their counterparts at the nucleotide level. The expression of these floral integrator and/or floral identity genes was examined in shoot apexes every 4 d over a 20 d period. CsAFL1, CsFL, and CsM111 were involved in the photoperiodic regulation of floral transition (Fig. 4B). In NIFS-3, the CsAFL1 expression levels started to increase 4 d after the plants were transferred to 11 h and 12 h photoperiod conditions; they increased more rapidly after day 4 (Fig. 4B). Although the CsAFL1 transcript levels under a 13 h photoperiod were lower than those under 11 h and 12 h photoperiods, CsAFL1 expression increased slightly over the 20 d period under the 13 h photoperiod. The CsAFL1 expression levels remained low under a 14 h photoperiod. CsFL and CsM111 transcripts were first detected on day 12 under 11 h and 12 h photoperiods; the increase in the CsFL and CsM111 expression levels was higher under an 11 h photoperiod than under a 12 h photoperiod (Fig. 4B). In Matsukawa, the CsAFL1 expression levels started to increase 4 d after the plants were transferred to 11, 12, and 13 h photoperiod conditions; they increased more rapidly after day 4 (Fig. 4B). CsAFL1 expression decreased slightly on day 20 under 11 h and 12 h photoperiods. On day 20, the CsAFL1 transcript levels under 11 h and 12 h photoperiods were the same as those under a 14 h photoperiod. CsFL transcripts were first detected on day 8 under 11, 12, and 13 h photoperiods and on day 16 under a 14 h photoperiod (Fig. 4B). Under 11, 12, and 13 h photoperiods, CsM111 transcripts were first detected on day 12; their levels increased more rapidly after day 16 under shorter photoperiods (Fig. 4B). Under a 14 h photoperiod, the CsM111 expression levels remained relatively low. Thus, CsAFL1, CsFL, and CsM111 were coordinately activated after floral transition in shoot apexes and their induction closely coincided with the accessions’ critical daylengths for flowering (Table 2).
Early flowering due to the ectopic expression of CsFTL3 in chrysanthemum and Arabidopsis
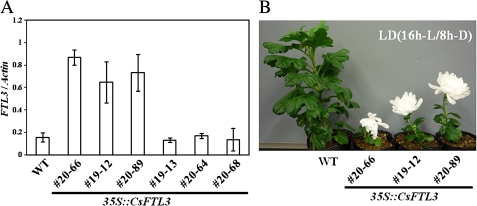
To investigate the potential function of CsFTL3 as a floral promoter, transgenic experiments with chrysanthemum were performed. Transgenic chrysanthemum plants with CsFTL3 expressed under the control of the Cauliflower mosaic virus (CaMV) 35S promoter were generated. As a result, 30 independent T0 lines were obtained and their FTL3 (CsFTL3 and CmFTL3) expression levels were investigated. Of these, 14 transgenic lines of in vitro plants developed flower buds under non-inductive LD conditions, with high FTL3 transcript levels (Supplementary Fig. S3 at JXB online). Six transgenic lines were selected on the basis of the preliminary analysis of exogenous CsFTL3 mRNA levels. Depending on soil conditions, three out of the six transgenic lines (# 20-66, #19-12, and #20-89) flowered under non-inductive LD conditions (16 h), with 5–7 times higher FTL3 mRNA levels than those in the wild-type plants (Fig. 5A, B). The other three out of the six transgenic lines (#19-13, #20-64, and #20-68) did not show any visible flower buds under LD conditions or any alterations in their FTL3 mRNA levels compared with those in the wild-type plants (Fig. 5A). Furthermore, transgenic Arabidopsis, which ectopically expressed CsFTL3, flowered earlier than the wild-type plants under SD conditions (Supplementary Fig. S4 at JXB online).
Fig. 5.
Effect of CsFTL3 overexpression on flowering in Chrysanthemum morifolium under LD conditions. (A) Transcript levels of FTL3 (CsFTL3 and CmFTL3) in the leaves were analysed by QRT-PCR in six independent transgenic lines and wild-type (WT) plants. (B) Three independent transgenic lines showed high expression levels of FTL3 and flowered under LD conditions, while the wild-type plants did not. (This figure is available in colour at JXB online.)
To demonstrate the potential function of the gene product of CsFTL3 as a graft-transmissible floral promoter, florigen, in chrysanthemum, grafting experiments were performed with CsFTL3-overexpressing transgenic plants (#20-66). The experiments showed the translocation of CsFTL3 from the transgenic stock to the wild-type scion under non-inductive LD or NB conditions. Four out of six plants with Jimba (experiment 1) and all the plants with Nagano-queen (experiment 2) resulting from grafting between the wild-type and 35S::CsFTL3 transgenic plants produced a crown bud, a flower bud that initiates capitulum primordia with involucral bracts but in which the development is arrested before floret initiation (Fig. 6; Supplementary Table S2 at JXB online). It is a sign of the transition to the reproductive phase in chrysanthemums. All the plants resulting from grafting between the wild-type stock and scion remained vegetative. To induce transfer of the floral stimulus from the donor to the receptor in grafting experiments, it is often necessary to defoliate the receptor plants (Thomas and Vince-Prue, 1997). In this study, defoliating the receptor plants increased the number of plants producing a crown bud (Supplementary Table S2).
Fig. 6.
Grafting experiment with Chrysanthemum morifolium plants overexpressing CsFTL3 under NB conditions. Plants resulting from grafting between the Nagano-queen wild-type plants (WT) and 35S::CsFTL3 transgenic plants (#20-66) produced a crown bud, a flower bud that initiates the capitulum primordia with involucral bracts. Arrows indicate the graft junctions. Photograph was taken at 9 weeks after the grafting. (This figure is available in colour at JXB online.)
Number of SD cycles required for the initiation and development of capitulum in chrysanthemums
The NIFS-3 plants maintained under NB conditions (8 h SD+4 h NB with fluorescent tubes) continued their vegetative growth for 70 d, whereas those maintained under SD conditions flowered by 50 d from the initiation of the SD cycle (Fig. 7). The plants required four SD cycles to determine the node number below the terminal capitulum. The first morphological signs of the initiation of capitulum primordia and involucral scales (bracts or modified leaves) appeared at the periphery of the dome on day 8; floret primordia differentiated acropetally from the bottom of the capitulum meristem on day 12. About 50% of the dome of the enlarged capitulum meristem was covered with floret primordia (Fig. 7). The development changed on transfer to NB conditions. The plants transferred to NB conditions after exposure to SD conditions for 4 d produced a crown bud, which initiated involucral bracts on the edge of the apical receptacle; however, the plants did not initiate florets on the apical receptacle until the end of the experiment (day 70). The plants transferred to NB conditions after exposure to SD conditions for 8 d initiated florets on the apical receptacle, but capitulum development was strongly suppressed by NB conditions.
Fig. 7.
Alternative pathways of capitulum development in C. seticuspe. Chrysanthemum seticuspe NIFS-3 plants were grown under NB conditions (8 h SD+4 h NB) and then subjected to different treatments: maintained under NB conditions (0 d of SD exposure); transferred to 8 h SD conditions (4 d or 8 d) and then returned to NB conditions; or maintained under SD conditions. (A) SEM images illustrating the developmental stages of the capitulum in C. seticuspe under 8 h SD conditions and the change in development on transfer to NB conditions. (B) The photograph was taken at 50 d after the initiation of the SD cycle. (This figure is available in colour at JXB online.)
Alteration in the photoperiodic cycle (i.e. transfer of plants from SD to NB conditions) down-regulated FTL3 expression in the leaves (Supplementary Fig. S5 at JXB online). The expression of the floral integrator and/or floral identity genes CsAFL1, CsSOC1 (AB679276), CsFL, and CsM111 was examined in shoot apexes every 4 d. Under NB conditions, the expression levels of all genes of interest remained low or undetectable (Fig. 8). The expression patterns of CsAFL1, CsFL, and CsM111 in the plants maintained under 8 h SD conditions were identical to those in the plants grown under an 11 h photoperiod (Figs 4B, 8). CsSOC1 expression in the plants maintained under SD conditions increased, peaking on day 8; after day 8, the expression levels started to decrease gradually, almost reaching the levels observed initially (Fig. 8). In the plants exposed to SD conditions for 4 d, the expression levels of all genes of interest increased slightly as compared with those in the plants maintained under NB conditions, but remained low at all tested time points (Fig. 8). In the plants exposed to SD conditions for 8 d, CsAFL1 and CsSOC1 expression was suppressed after transfer to NB conditions (Fig. 8). On the other hand, the expression levels of CsFL and CsM111 were identical to those in the plants maintained under SD conditions until day 12, but were suppressed thereafter (Fig. 8).
Fig. 8.
Effect of transfer from SD to NB conditions on the floral integrator and/or floral identity gene expression in C. seticuspe. Chrysanthemum seticuspe NIFS-3 plants were grown as described in Fig. 7. The plants were maintained under NB conditions (0 d of SD exposure; open squares); exposed to SD conditions for 4 d (filled triangles) or 8 d (filled squares) and then transferred to NB conditions; or maintained under SD conditions (filled circles). Shoot apexes were harvested every 4 d over a 20 d period and subjected to QRT-PCR analysis. The expression of each gene of interest was normalized against CsACTIN expression.
Discussion
Chrysanthemum has been used in classical physiological experiments on flowering. These experiments have demonstrated the presence of a kind of hormonal substance, called a florigen, produced in the leaves (for a review, see Chailakhyan and Krikorian, 1975). The present study used C. seticuspe, a wild diploid chrysanthemum. Under the present experimental conditions, two accessions of C. seticuspe, NIFS-3 and Matsukawa, showed typical photoperiodic flowering responses as obligate SD plants (Tables 1, 2). The FT-like genes CsFTL1, CsFTL2, and CsFTL3 and the floral integrator and/or floral identity genes CsAFL1, CsSOC1, CsFL, and CsM111 isolated from C. seticuspe showed high sequence identity (>98.8%) to their C. morifolium counterparts. The expression profiles of the FT-like and floral integrator and/or floral identity genes in C. morifolium coincided with those in C. seticuspe under both SD and LD conditions (Figs 3A, 4A; Li et al., 2009). These observations suggest that to understand the genetic and molecular mechanisms of the reproductive process in chrysanthemums and overcome the issue of complex hybridity and polyploidy, C. seticuspe can be a useful alternative model of chrysanthemum cultivars.
Identification of FT-like genes in chrysanthemum
In this study, CsFTL1, CsFTL2 and CsFTL3 were isolated as FT-like genes from C. seticuspe. These chrysanthemum paralogues of Arabidopsis FT are more closely related to each other than to homologues from other species (Fig. 1). The synonymous substitution rate between two sequences (i.e. Ks) and phylogenetic analysis indicated that CsFTL3 is separated from CsFTL1 and CsFTL2 (Fig. 1). Amino acid sequence comparison of the three encoded proteins showed that all the three CsFTLs carried the functionally important signatures Tyr84 and Gln139 in the positions corresponding to Tyr85 and Gln140, respectively, in Arabidopsis FT (Fig. 1; Supplementary Fig. S1 at JXB online), which are important for the antagonistic functions of FT and TFL1, respectively, in Arabidopsis (Hanzawa et al., 2005; Ahn et al., 2006). Unlike two FT family genes in Arabidopsis, FT and TSF, which exhibit similar expression patterns, the FT-like genes in chrysanthemum showed distinctly different expression patterns. All the three FT-like genes were expressed mainly in the leaves, as expected (Fig. 2). CsFTL3 was up-regulated under floral-inductive SD conditions, whereas CsFTL1 and CsFTL2 were down-regulated, showing an antagonistic expression pattern to that of CsFTL3; CsFTL1 and CsFTL2 were preferentially expressed under non-inductive conditions (Figs 2, 3A). The study showed a particularly strong correlation between flowering and CsFTL3 expression under various photoperiodic conditions in the two C. seticuspe accessions that differed in their critical daylengths for flowering (Table 2; Fig. 4A). Recently, a negative effect of FT-like genes on flowering has been postulated in sugar beet (Pin et al., 2010) and sunflower (Blackman et al., 2010). In sugar beet, amino acid changes within the region encoding an external loop of PEBP determine the ability of BvFT2 and BvFT1 to promote and repress flowering, respectively (Pin et al., 2010). CsFTL3 has the amino acids in the external loop of PEBP which are conserved in most other FT-like proteins, whereas CsFTL1 and CsFTL2 had substituted amino acids in the region (Fig. 1). Correlation between flowering and the expression patterns of the three CsFTL genes together with sequence comparison indicated that at least CsFTL3 might have a hypothetical function as a mobile signal for flower induction under SD conditions, and the other closely related FT-like genes in C. seticuspe (i.e. CsFTL1 and CsFTL2) might have distinct roles as transmissible signals, possibly antagonistic functions as postulated in sugar beet and sunflower.
Early flowering due to the ectopic expression of CsFTL3 in Arabidopsis and chrysanthemum
Transgenic Arabidopsis plants overexpressing CsFTL3 flowered earlier than the wild-type plants (Supplementary Fig. S4 at JXB online). Early flowering phenotypes of transgenic Arabidopsis are reported to occur upon ectopic expression of FT-like genes from heterologous plant species such as tomatoes (Lifschitz et al., 2006) and poplars (Igasaki et al., 2008). This shows that CsFTL3 has the potential to induce early flowering in Arabidopsis in a similar manner to that of the FT-like genes in other plant species. In functional complementation analyses using Arabidopsis, transgenic plants overexpressing FT-like genes isolated from lettuce and sunflower, and which showed close correlation with their chrysanthemum paralogues in phylogenetic analysis, did not flower as early as the plants overexpressing Arabidopsis FT (Blackman et al., 2010; Fukuda et al., 2011). The extremely early flowering phenotype of CsFTL3-overexpressing Arabidopsis plants could not be observed in this study (Supplementary Fig. S4). This suggests that some functional difference exists between FT in Arabidopsis and that in Compositae species such as chrysanthemum, lettuce, and sunflower.
To investigate the potential function of CsFTL3 as a floral promoter in chrysanthemum, transgenic experiments with chrysanthemum were performed. The transgenic chrysanthemum plants with high CsFTL3 expression levels (#20-66, #19-12, and #20-89) flowered in vitro and in soil under non-inductive LD conditions (Fig. 5; Supplementary Fig. S3 at JXB online). These results strongly indicated that the gene product of CsFTL3 could function as a floral activator to regulate flowering under non-inductive LD conditions. Post-transcriptional regulation might have altered the level of protein production by CsFTL3 in the experiment. Aida et al. (2008) reported that the β-glucuronidase (GUS) activity levels in their study did not quantitatively correspond to the GUS mRNA expression levels, but depended on the translational efficiency of the transgenes in a GUS assay of chrysanthemum transformants. This is a potential reason why some of the transgenic plants with high CsFTL3 expression levels did not show the early flowering phenotype (Supplementary Fig. S3).
The grafting experiment with CsFTL3-overexpressing chrysanthemum plants (#20-66) may reflect the translocation of CsFTL3 from the transgenic stock to the wild-type scion under non-inductive LD conditions, although the plants failed to flower (Fig. 6). The reason might be that not enough graft-transmissible signals were transmitted to the wild-type scion for complete development of the capitulum. This small effect may be explained in terms of the sink–source relationship between the receptor shoot and the donor stock (Zeevaart, 2006). There is another argument that transmissible inhibitors may be produced in the leaves when the plants are exposed to non-inductive photoperiodic cycles (Lang et al., 1977; Thomas and Vince-Prue, 1997). The result suggested that the gene product of CsFTL3 could function as a graft-transmissible signal, florigen, in chrysanthemum, and that there possibly exists an antagonistic inhibitor in the wild-type scion under non-inductive LD conditions.
Correlation between CsFTL3 expression in leaves and critical daylengths for flowering
Elucidation of the mechanisms underlying the determination of critical daylengths for flowering will offer the advantage of year-round chrysanthemum production. In Pharbitis (Ipomoea) nil (L.) Choisy (Hayama et al., 2007) and rice (Itoh et al., 2010), orthologues of the FT gene are highly expressed when the daylength is shorter than their respective critical daylengths for flowering; conversely, the expression levels decrease markedly as the photoperiod increases. In this study, differences were observed in critical daylengths for flowering and CsFTL3 expression between the C. seticuspe accessions NIFS-3 (<12 h) and Matsukawa (<13 h) (Fig. 4A; Table 2). Consistent with the increased CsFTL3 mRNA expression, the expression of the floral activator genes CsAFL1, CsFL, and CsM111 was up-regulated in shoot tips under shorter photoperiods (Fig. 4B). Together with the function of CsFTL3 as a floral inducer, increased CsFTL3 expression in leaves seems to be a key step in the gene activation cascades involved in the induction of flowering in C. seticuspe under SD conditions. The mRNA expression of orthologues of the FT gene, namely PnFT in P. nil and Hd3a in rice, is suppressed to undetectable levels when daylength is longer than the critical daylengths for flowering (Hayama et al., 2007; Itoh et al., 2010). In contrast, detectable expression levels of CsFTL3 transcripts were observed when daylength was longer than the critical daylengths in both the C. seticuspe accessions; they were relatively low compared with those under floral-inductive conditions (Figs 3A, 4A). This might indicate that chrysanthemums require a threshold expression level of CsFTL3 mRNA in leaves for flowering. As argued, perhaps, the fact also indicates the possibility of the existence of an antagonistic inhibitor in unfavourable photoperiodic cycles, which suppresses the function of the gene product of CsFTL3 as a floral inducer.
Effect of light period on and circadian regulation of the expression of FT-like genes
It has been demonstrated that a period of light preceding an inductive dark period is essential for flowering in chrysanthemums: a minimum of 3–5 h of light per day is required (for a review, see Cathey, 1969). In the present study, the expression of CsFTL3, which is involved in flowering, increased during repeated 8 h SD cycles (Fig. 3B). However, the expression did not increase under continuous dark conditions (Fig. 3C). This indicates that a period of light preceding an inductive dark period is important for CsFTL3 expression, and, thus, flowering. In addition, there is a clear relationship between the daily light integral during SDs and the time to both flower initiation and flower development in chrysanthemums (for a review, see Carvalho and Heuvelink, 2001). Low light intensity suppresses floral transition under floral-inductive SD conditions. Although it is not clear whether the daily light integral regulates CsFTL3 expression, in Arabidopsis LD promotion of flowering with FT expression up-regulated by ‘photosynthetically active radiation’—possibly mediated by sucrose—has been highlighted (King et al., 2008).
Recent advances in molecular biology have elucidated the mechanisms underlying the determination of the accurate critical daylength threshold. In the external coincidence model (Pittendrigh and Minis, 1964), a signal is produced when an environmental signal (light) coincides with the sensitive phase of an endogenous circadian rhythm of photoresponsiveness. Two distinct gating mechanisms in rice could enable the manipulation of slight differences in daylength to control Hd3a transcription with a critical daylength threshold (Itoh et al., 2010). In the present study, expression of CsFTL genes showed diurnal oscillations (Fig. 3B) and a circadian rhythm after the plants were exposed to an SD cycle (Fig. 3C). A key challenge now is to determine how the divergent photoperiodic flowering responses are regulated in chrysanthemums via the control of the genes regulated by both light and the circadian clock. CsFTL genes will provide important information with recent advances in the understanding of the mechanisms underlying the determination of the accurate critical daylength threshold.
Gene activation cascade for flowering under SD conditions in chrysanthemums
The SAM generates leaves and shoots during the vegetative phase; in the reproductive phase, after floral transition, it becomes transformed into an inflorescence meristem, and the new lateral primordia produced develop as floral meristems. The chrysanthemum flower is a capitulum, a head type of inflorescence, which apparently mimics a large single flower, with the elongated ray florets located on the edge of the receptacle and the disc florets located at the centre of the receptacle. The photoperiodicity for the development of the capitulum as a large single flower in chrysanthemums differs from that for floral initiation and developmental processes in other model plants. Chrysanthemum seticuspe plants require SD cycles for capitulum development (Fig. 7). In Arabidopsis, FT activation in leaves induces the transition to the reproductive phase and flowering. The FT protein translocates into the SAM and interacts with FD, forming the FT–FD complex, which regulates the downstream target genes such as AP1 and FUL (Abe et al., 2005; Wigge et al., 2005). It has been indicated that AP1 expression is a relatively late event during floral induction. In Arabidopsis, LFY and FUL [AGAMOUS-LIKE 8 (AGL8)] are induced 24 h after plants are transferred to floral-inductive LD conditions, while AP1 is induced 72 h after transfer (Hempel et al., 1997). It has been suggested that SOC1 mainly regulates LFY for floral initiation in the SAM (Lee et al., 2008). SOC1 and LFY at the SAM also influence phase transition through the regulation of the GA pathway (Lee and Lee, 2010). LFY and AP1 are the key regulators of floral meristem identity and the main activators of the cascade of genes initiating floral development. In the two C. seticuspe accessions that differed in their critical daylengths for flowering, the expression levels of CsFTL3 in the leaves closely coincided with the induction of floral integrator and/or floral identity genes and the accessions’ flowering responses (Figs 3A, 4A; Tables 1, 2). In Arabidopsis, FUL is expressed throughout the shoot apex and the expression is up-regulated in inflorescence meristems, consistent with floral transition and identity (Ferrandiz et al., 2000). The expression patterns of CsAFL1, a homologue of FUL, suggest that CsAFL1 functions around the time of the transition to the reproductive phase and during early capitulum development in C. seticuspe, similar to FUL in Arabidopsis. It has been suggested that CO activates SOC1 mainly through the regulation of FT, and up-regulation of SOC1 in the SAM is one of the earliest events in floral transition (Lee and Lee, 2010). In the SAM, interaction of SOC1 with AGL24 induces LFY expression (Lee et al., 2008). In C. seticuspe, CsSOC1 expression in the shoot apexes was up-regulated at the time of the first morphological signs of capitulum primordia initiation (day 8; Figs 7, 8). After the up-regulation of CsSOC1 expression, CsFL, an LFY homologue, was induced in the shoot apexes (day 12; Fig. 8). CsSOC1 expression was not up-regulated and CsFL was not induced in the plants maintained under NB conditions (Fig. 8). No florets were initiated and CsFL was only weakly expressed with slightly up-regulated CsSOC1 expression in the plants exposed to SD conditions for 4 d (Figs 7, 8). It seems that CsFL induction by CsSOC1, possibly like LFY induction by SOC1 through interaction with AGL24, is conserved in the process of floral meristem induction in C. seticuspe. CsFL and CsM111, an AP1 homologue, were induced—CsFL slightly earlier than CsM111—at the time of the first morphological signs of floret primordia initiation (day 12; Figs 7, 8). The expression patterns of CsFL and CsM111 suggest that they function around the time of floral meristem (floret) differentiation rather than at the time of the transition to the reproductive phase. In Arabidopsis, shortly after the onset of floret differentiation, SOC1 is down-regulated by AP1 to initiate the downstream pathways required for inducing B- and C-class floral organ identity genes (Liu et al., 2008). As in Arabidopsis, it was observed in C. seticuspe plants maintained under SD conditions that temporally increased CsSOC1 expression started to decrease with CsM111 induction and floret initiation (Figs 7, 8). The present study results suggest that floral integrator and/or floral identity gene activation cascades are conserved in the floral transition and developmental processes regulated by photoperiodic cycles in C. seticuspe, like in Arabidopsis.
The approach of reciprocal transfer experiments was useful for further investigation of the activation cascades involved in floral transition and development regulated by photoperiodic cycles in chrysanthemums. In plants that have been studied in detail as physiological models of SD or LD plants, such as P. nil ( Imamura, 1967), Sinapis alba L. (Bernier, 1963), Lolium temulentum L. (Evans, 1958), and Arabidopsis (Hempel and Feldman, 1994), floral transition can be induced after exposure to a single inductive photoperiod. When floral transition is induced by a single inductive photoperiod, the floral primordia proceed to develop further under non-inductive conditions. However, chrysanthemums require SD cycles for capitulum and floret development; non-inductive conditions inhibit this development (Cockshull and Horridge, 1980; Adams et al., 1998). In the SD plant P. nil, PnFT abundance induced by a single floral-inductive dark period is sufficient for flower bud initiation and development (Hayama et al., 2007; Higuchi et al., 2011). In the LD plant S. alba, induction of flowering by a single photoperiodic cycle is fine-tuned by SaFT in the leaves (D’Aloia et al., 2009). SaFT abundance induced by a single floral-inductive light period is sufficient for flowering. In the SAM, SaSOC1, SaLFY, and SaAP1 activation occurs after SaFT activation in the leaves. SaAP1 activation is considered the final step in floral transition in the SAM. A similar activation pattern for FT in leaves is observed on the induction of flowering by a single photoperiodic cycle in Arabidopsis (King et al., 2008). Unlike the above-mentioned model plants, C. seticuspe plants require SD cycles for flowering (i.e. capitulum development; Fig. 7). In the C. seticuspe plants exposed to SD conditions for 4 d, the node number below the terminal capitulum was determined and a crown bud was produced (Fig. 7). This response indicated that a slight increase in the CsAFL1 and CsSOC1 transcript levels could induce the transition to the reproductive phase (Fig. 8). In the C. seticuspe plants exposed to SD conditions for 8 d, the initiated florets failed to develop and the expression of all the tested genes was remarkably suppressed after transfer to NB conditions (Figs 7, 8). This indicated that the activities of CsFL and CsM111 in the plants were not sufficient for floret development. In Arabidopsis, LFY directly activates AP1 (Mandel and Yanofsky, 1995). AP1 also promotes LFY transcription as part of a positive feedback loop (Kaufmann et al., 2010). In addition, the FT–FD complex plays a role in up-regulating FUL and AP1 expression in floral primordia (Wigge et al., 2005). Since FTL3 expression in the leaves was down-regulated by alteration in the photoperiod conditions [i.e. transfer from SD to NB conditions (Supplementary Fig. S5 at JXB online)], the down-regulation may be associated with CsAFL1 and CsM111 expression in the shoot apexes of C. seticuspe (Fig. 8). The weak CsM111 expression related to the down-regulation of CsFTL3 expression might modify CsFL up-regulation during floret development. This suggests that a similar signal network of the genes for flowering is conserved in chrysanthemums, as in Arabidopsis, and that there exists a possibility that in chrysanthemums, the photoperiodicity for capitulum primordia initiation and further development is tuned by CsFTL3 in the leaves. This is supported by the fact that the 35S::CsFTL3 transgenic lines could overcome the inhibition of floral organ development under non-inductive LD conditions (Fig. 5).
In conclusion, the present study provides strong evidence that CsFTL3 promotes floral transition in a similar manner to FT-like genes in other plant species. CsFTL3 induction occurred in the leaves and its up-regulation was correlated with the events occurring in the SAM (i.e. floral evocation) that commit it to producing flowers (Evans, 1969) with the activation of floral integrator and/or floral identity genes. Further, CsFTL3-dependent graft-transmissible signals partially substituted for SD stimuli in chrysanthemum. Taken together, these observations indicate that the product of the CsFTL3 gene could function as a florigen to regulate flowering, which has been previously demonstrated in studies on flowering physiology in chrysanthemum (Chailakhyan and Krikorian, 1975). It seems likely that the photoperiodic control of flowering may depend on the balance of promoters and inhibitors from leaves rather than on the accumulation of a threshold amount of a single floral stimulus at the shoot apex (Thomas and Vince-Prue, 1997). The observations suggest an argument that existence of inhibitors cannot be excluded and they might play an important role in the photoperiodic control of flowering. Further investigations to determine the role of CsFTL genes in the growth and flowering of chrysanthemums will provide clues for better understanding of the suggested argument.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Comparison of deduced amino acid sequences of CsFTL1, CsFTL2, CsFTL3, FT, and TFL1.
Figure S2. Expression of CmFTL genes in the leaves of Chrysanthemum morifolium ‘Reagan’ under SD and LD conditions.
Figure S3. Relative expression levels of exogenous CsFTL3 mRNA in in vitro transgenic (35S::CsFTL3) Jimba plants.
Figure S4. Effect of CsFTL3 overexpression on flowering in Arabidopsis under SD conditions.
Figure S5. Effect of transfer from SD to NB conditions on CsFTL3 expression in C. seticuspe.
Table S1. Primer sequences and PCR conditions used in the study.
Table S2. Effect of defoliation on the transition from the vegetative to the reproductive phase in the plants resulting from grafting between wild-type (WT) and 35S::CsFTL3 transgenic plants.
Acknowledgments
We thank Professor Tsuyoshi Nakagawa for providing the binary vector pGWB2 for Arabidopsis and chrysanthemum transformation experiments, and Dr. Tsuyoshi Tanaka for performing the phylogenetic analysis. This work was supported by a grant-in-aid from the Ministry of Agriculture, Forestry and Fisheries of Japan (Project: Elucidation of biological mechanisms of photoresponse and development of advanced technologies utilising light).
Glossary
Abbreviations
- ABA
abscisic acid
- bZIP
basic-leucine zipper
- CaMV
Cauliflower mosaic virus
- FAA
formalin–acetic acid–alcohol
- GA
gibberellin
- GUS
β-glucuronidase
- LD
long day
- LED
light-emitting diode
- NB
night break
- ORF
open reading frame
- PEBP
phosphatidylethanolamine-binding protein
- PPFD
photosynthetic photon flux density
- QRT-PCR
quantitative real-time PCR
- RACE
rapid amplification of cDNA ends
- SAM
shoot apical meristem
- SD
short day
- SEM
scanning electron microscopy
- ZT
Zeitgeber time
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Adams SR, Pearson S, Hadley P. An appraisal of the use of reciprocal transfer experiments: assessing the stages of photoperiod sensitivity in chrysanthemum cv. Snowdon (Chrysanthemum morifolium Ramat.) Journal of Experimental Botany. 1998;49:1405–1411. [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO Journal. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida R, Ohira K, Tanaka Y, Yoshida K, Kishimoto S, Shibata M, Ohmiya A. Efficient transgene expression in chrysanthemum, Dendranthema grandiflorum (Ramat.) Kitamura, by using the promoter of a gene for chrysanthemum chlorophyll-a/b-binding protein. Breeding Science. 2004;54:51–58. [Google Scholar]
- Aida R, Narumi T, Ohtsubo N, Yamaguchi H, Kato K, Shinmyo A, Shibata M. Improved translation efficiency in chrysanthemum and torenia with a translational enhancer derived from the tobacco alcohol dehydrogenase gene. Plant Biotechnology. 2008;25:69–75. [Google Scholar]
- Aki T, Shigyo M, Nakano R, Yoneyama T, Yanagisawa S. Nano scale proteomics revealed the presence of regulatory proteins including three FT-like proteins in phloem and xylem saps from rice. Plant and Cell Physiology. 2008;49:767–790. doi: 10.1093/pcp/pcn049. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G. Sinapis alba L., a new long-day plant requiring a single photoinductive cycle. Naturwissenschaften. 1963;50:101–104. [Google Scholar]
- Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH. The role of recently derived FT paralogs in sunflower domestication. Current Biology. 2010;20:629–635. doi: 10.1016/j.cub.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Carvalho SMP, Heuvelink E. Influence of greenhouse climate and plant density on external quality of chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura): first steps towards a quality model. Journal of Horticultural Science and Biotechnology. 2001;76:249–258. [Google Scholar]
- Cathey HM. Chrysanthemum morifolium (Ramat.) Hemsl. In: Evans LT, editor. The induction of flowering: some case histories. Melbourne, VIC: Macmillan Company of Australia; 1969. pp. 268–290. [Google Scholar]
- Chailakhyan MK, Krikorian AD. Forty years of research on the hormonal basis of plant development—some personal reflections. Botanical Review. 1975;41:1–29. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cockshull KE, Horridge JS. Further evidence of a relationship between size of the Chrysanthemum shoot apex and inflorescence development. Annals of Botany. 1980;46:125–127. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- D’Aloia M, Tamseddak K, Bonhomme D, Bonhomme F, Bernier G, Périlleux C. Gene activation cascade triggered by a single photoperiodic cycle inducing flowering in Sinapis alba. The Plant Journal. 2009;59:962–973. doi: 10.1111/j.1365-313X.2009.03927.x. [DOI] [PubMed] [Google Scholar]
- Evans LT. Lolium temulentum L., a long-day plant requiring only one inductive photocycle. Nature. 1958;182:197–198. [Google Scholar]
- Evans LT. The nature of flower induction. In: Evans LT, editor. The induction of flowering: some case histories. Melbourne, VIC: Macmillan Company of Australia; 1969. pp. 457–480. [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Matsuo S, Kikuchi K, Kawazu Y, Fujiyama R, Honda I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. Journal of Plant Physiology. 2011;168:1602–1607. doi: 10.1016/j.jplph.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Journal of Agricultural Research. 1920;18:553–606. [Google Scholar]
- Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Sciences, USA. 2005;102:7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Agashe B, Luley E, King R, Coupland G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. The Plant Cell. 2007;19:2988–3000. doi: 10.1105/tpc.107.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ. Bi-directional inflorescence development in Arabidopsis thaliana: acropetal initiation of flowers and basipetal initiation of paraclades. Planta. 1994;192:276–286. [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Higuchi Y, Sage-Ono K, Sasaki R, Ohtsuki N, Hoshino A, Iida S, Kamada H, Ono M. Constitutive expression of the GIGANTEA ortholog affects circadian rhythms and suppresses one-shot induction of flowering in Pharbitis nil, a typical short-day plant. Plant and Cell Physiology. 2011;52:638–650. doi: 10.1093/pcp/pcr023. [DOI] [PubMed] [Google Scholar]
- Igasaki T, Watanabe Y, Nishiguchi M, Kotoda N. The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in Lombardy poplar. Plant and Cell Physiology. 2008;49:291–300. doi: 10.1093/pcp/pcn010. [DOI] [PubMed] [Google Scholar]
- Imamura S. Physiology of flowering in Pharbitis nil. Tokyo: Japanese Society of Plant Physiologists; 1967. [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nature Genetics. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- Kawata J. The phasic development of chrysanthemum as a basis for the regulation of vegetative growth and flowering in Japan. Acta Horticulturae. 1987;197:280–287. [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular-basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiology. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Hisamatsu T, Goldschmidt EE, Blundell C. The nature of floral signals in Arabidopsis. I. Photosynthesis and a far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT ) Journal of Experimental Botany. 2008;59:3811–3820. doi: 10.1093/jxb/ern231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Moritz T, Evans LT, Martin J, Andersen CH, Blundell C, Kardailsky I, Chandler PM. Regulation of flowering in the long day grass, Lolium temulentum L., by gibberellins and the gene, FLOWERING LOCUS T (FT ) Plant Physiology. 2006;141:498–507. doi: 10.1104/pp.106.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes and Development. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Lang A, Chailakhyan MK, Frolova IA. Promotion and inhibition of flower formation in a dayneutral plant in grafts with a short-day plant and a long-day plant. Proceedings of the National Academy of Sciences, USA. 1977;74:2412–2416. doi: 10.1073/pnas.74.6.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton FA. The responses of early-flowering chrysanthemums to daylength. Scientia Horticulturae. 1977;7:277–289. [Google Scholar]
- Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany. 2010;61:2247–2254. doi: 10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. The Plant Journal. 2008;55:832–843. doi: 10.1111/j.1365-313X.2008.03552.x. [DOI] [PubMed] [Google Scholar]
- Li T, Niki T, Nishijima T, Douzono M, Koshioka M, Hisamatsu T. Roles of CmFL, CmAFL1, and CmSOC1 in the transition from vegetative to reproductive growth in Chrysanthemum morifolium Ramat. Journal of Horticultural Science and Biotechnology. 2009;84:447–453. [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proceedings of the National Academy of Sciences, USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Belanger H, Lee YJ, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CB. Preliminary studies on flower bud differentiation in relation to photoperiodic response. Proceedings of the American Society for Horticultural Science. 1936;34:621–623. [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135:1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Kuttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W. Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes to Cells. 2001;6:327–336. doi: 10.1046/j.1365-2443.2001.00425.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T. Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant and Cell Physiology. 2008;49:1645–1658. doi: 10.1093/pcp/pcn154. [DOI] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL, Nilsson O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. American Naturalist. 1964;98:261–294. [Google Scholar]
- Post K, Kamemoto H. A study on the number of short photoperiods required for flower bud initiation and the effect of interrupted treatment on flower spray formation in two commercial varieties of chrysanthemums. Proceedings of the American Society for Horticultural Science. 1950;55:477–482. [Google Scholar]
- Shalit A, Rozman A, Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y, Lifschitz E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences, USA. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchennikova AV, Shulga OA, Immink R, Skryabin KG, Angenent GC. Identification and characterization of four chrysanthemum MADS-box genes, belonging to the APETALA1/FRUITFULL and SEPALLATA3 subfamilies. Plant Physiology. 2004;134:1632–1641. doi: 10.1104/pp.103.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Sumitomo K, Li T, Hisamatsu T. Gibberellin promotes flowering of chrysanthemum by upregulating CmFL, a chrysanthemum FLORICAULA/LEAFY homologous gene. Plant Science. 2009;176:643–649. [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. London: Academic Press; 1997. [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. The Plant Cell. 2010;22:1733–1748. doi: 10.1105/tpc.109.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant and Cell Physiology. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD. Florigen coming of age after 70 years. The Plant Cell. 2006;18:1783–1789. doi: 10.1105/tpc.106.043513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.