Abstract
Recent work from our group and others has argued that human induced pluripotent stem cells (hiPSCs) generated by the introduction of four viruses bearing reprogramming factors differ from human embryonic stem cells (hESCs) at the level of gene expression. Many of the differences seen were common across independent labs and, at least to some extent, are thought to be a result of residual expression of donor cell-specific genes (Chin et al., 2009; Ghosh et al., 2010; Marchetto et al., 2009). Two new reports re-analyze similar expression datasets as those used in Chin et al., (Chin et al., 2009) and come to different conclusions (Newman et al., 2010, Guenther et al., 2010). Here, we compare various approaches to perform gene expression meta-analysis that all support our original conclusions and present new data to demonstrate that polycistronic delivery of the reprogramming factors and extended culture brings hiPSCs transcriptionally much closer to hESCs than older methods.
Introduction
The recent advent of reprogramming human somatic cells to a pluripotent state has led to the examination of human induced pluripotent stem cells (hiPSCs) and their relation to human embryonic stem cells (hESCs). Our study (Chin et al., 2009) demonstrated that hiPSCs generated up until that time had statistically significant differences in gene expression compared to available hESCs. Furthermore, many of these differences, particularly those found in early passage iPSC lines, were conserved across various studies and species, suggesting that these two cell types, while very similar to each other, were nonetheless transcriptionally distinguishable. Soon after, several additional studies came to similar conclusions using hiPSCs murine iPSCs derived from fibroblasts, neural tissue, adipocytes, blood, and keratinocytes (Ghosh et al., 2010; Marchetto et al., 2009; Polo et al., 2010). These studies went further to propose that residual misexpression from the cell type of origin is responsible for the observed differences in gene expression between iPSCs and ESCs (Ghosh et al., 2010; Marchetto et al., 2009; Polo et al., 2010). Similarly, consistent differences in miRNA expression between hiPSCs and hESCs were also reported (Chin et al., 2009; Wilson et al., 2009).
hiPSCs and hESCs have also been compared at the level of the epigenome. In (Chin et al., 2009), we showed by chromatin immunoprecipitation in combination with microarrays (ChIP-chip) that histone H3K27 and H3K4 trimethylation levels at promoter regions were not distinguishable between hiPSCs and hESCs. Hawkins et al., and now Guenther et al., confirmed that trimethylation of H3K27 was very similar between hESCs and hiPSCs by ChIP in combination with massive parallel sequencing (ChIP-Seq) (Hawkins et al., 2010). On the other hand, it was shown that trimethylation on histone H3K9 is significantly different between hESCs and hiPSCs and appears to contribute to the differences in gene expression between hESCs and hiPSCs (Hawkins et al., 2010).
Along this line, two groups also demonstrated that significant differences can be detected between hESCs and hiPSCs at the level of DNA methylation (Deng et al., 2009; Doi et al., 2009), further indicating that epigenetic modifications could be differentially affected in the reprogramming process. Furthermore, when hESCs and hiPSCs from patients with Fragile X (FX) Syndrome were generated, only the FX-hESCs displayed an active FMR1 locus, whereas reprogramming failed to reactivate this locus (Urbach et al., 2010). Finally, recent work from our lab suggests that reprogramming female human fibroblasts under conventional culture conditions fails to reactivate the somatically silent × chromosome (Tchieu et al., 2010), while other work suggests that hESC lines can be generated that capture this primitive state (Lengner et al., 2010). In summary, there is considerable evidence that hiPSCs and hESCs could be epigenetically and transcriptionally different.
Given the vastly different circumstances by which hiPSCs and hESCs are generated, it is not surprising that these various analyses deem them to have distinct molecular properties. Perhaps what is most surprising is in fact the high degree to which transcription factor- induced reprogramming is able to reconstruct the pluripotent state. A key question then becomes whether any of the gene expression differences seen between hiPSCs and hESCs are functionally relevant. Interestingly, it was shown that different combinations of reprogramming factors lead to mouse (m) iPSCs with different developmental potential (Han et al., 2010). Furthermore, recent advances with mouse reprogramming suggests that miPSC lines can be derived that possess the same functional characteristics as ESCs in their capacity to generate mice via tetraploid (4N) complementation (Boland et al., 2009; Kang et al., 2009), but that most miPSC lines, even from the same starting population, do not support 4N complementation. Several new lines of evidence point to the activity of a few imprinted genes and miRNAs to be at least partially responsible for the differences in pluripotency (Liu et al., 2010; Stadtfeld et al., 2010). Specifically, cell lines that support 4N complementation express the Dlk1-Dio3 imprinted cluster, while those that fail in this approach do not (Stadtfeld et al., 2010). Thus, misexpression of just a few genes can functionally distinguish miPSCs from most miPSCs and mESCs, arguing that even very small differences can have profound consequences.
Correlating transcriptional differences between hiPSCs and hESCs with functional differences is considerably more difficult than in the mouse system because the powerful 4N complementation assay cannot be applied and quantitative assays to assess pluripotency are difficult to establish. It is conceivable that all the hiPSCs generated to date do fall into functional categories that have yet to be defined, pending the discovery of pluripotency assays that are more quantitative than teratoma formation. Given that we don’t have these defined functional sub-classifications of hiPSCs and hESCs, it is not yet possible to mine human cell reprogramming expression data for differences that correlate with functional outputs. It was also suggested that genetic background contributes significantly to gene expression changes between mESCs and miPSCs (Stadtfeld et al., 2010), but whether this also contributes to differences between hESCs and hiPSCs is still unknown. However, when comparing expression differences between early passage hiPSCs and hESCs of the reprogramming experiment from one lab with that from a different lab, we and others consistently found a significant proportion of genes to be differentially expressed between these two cell types (Chin et al., 2009; Ghosh et al., 2010; Marchetto et al., 2009).
We also reported that the overlap of expression differences decreases as more independent reprogramming experiments from different labs were compared (Chin et al., 2009). Regarding the question of consistent differences between hiPSCs and hESCs, two groups now suggest that when many lines of hiPSCs and hESCs from different labs are compared, consistent differences between them are largely lost and that most expression differences between hiPSCs and hESCs are lab-specific or stochastic in nature (Newman et al. and Guenther et al). Both groups took issue with the meta-analysis methods used in (Chin et al., 2009), and we would like to take this opportunity to discuss the meta-analyses employed to start a conversation on ‘best practice’ for meta-analysis of gene expression of hESCs and hiPSCs. We present a re-analysis of data from Chin et al. with additional statistical methods to demonstrate that the conlusions made in Chin et al were in fact appropriate, and highlight reasons for the discrepancy in interpretations between Newman et al., Guenther et al., and Chin et al.. Furthermore, we present data with new hiPSCs to demonstrate that reprogramming methods may affect the kinetics of this process.
Results and Discussion
In Chin et al. (Chin et al., 2009), we sought to determine differences in gene expression between hESCs and early passage hiPSCs and to test the repeatability of the changes seen in among different independent reprogramming experiments from several labs (Hochedlinger, Thompson, Jaenisch, and our own). We chose to analyze the reprogramming experiments from different labs independently and applied our normalization and differential gene expression discovery algorithm to each reprogramming experiment separately. Thus, our normalization strategy was different than that used in Figure 4 of Guenther et al. and Figure 2 of Newman et al., where both grouped all samples together and normalized them in a single event. In preparing (Chin et al., 2009), we also found that, because hESCs and hiPSCs are quite similar to each other, clustering all data sets from different groups together appeared to cluster them according to lab of origin, not by cell type. Because our interest was to compare the differences between the hiPSCs and hESCs in each group, we believed that independent normalization was the appropriate strategy. Since each reprogramming experiment was treated independently, any measurement error from a single reprogramming experiment should have had no influence on data obtained from another. Since we found significant overlap between the gene lists generated as differentially expressed between hESCs and hiPSCs among different reprogramming experiments, we argued that these differences were not entirely stochastic (Chin et al., 2009).
To increase the power of our analysis, we also filtered the expression data before generating lists of differentially expressed genes by collapsing the expression data for each gene. To this end, we started by only taking the “highest confidence” probesets for a given RefSeq ID and eliminating probesets that were defined by Affymetrix as potentially having more non-specific binding, then averaged expression data from remaining probe sets for a given RefSeq to further reduce false positives. Importantly, by collapsing the probe sets to characterize each RefSeq ID, we were able to include expression data from mouse experiments, which required the use of Homologene database (NCBI), a function that does not work at the probe ID level. The extent to which this original approach contributed to differences in the interpretation of results between Newman et al., Guenther et al, and Chin et al. is unclear.
To then define significant gene expression differences between hiPSCs and hESCs, we employed the combination of a simple t-test (p = 0.05) and fold change (1.5x) in our prior study (Chin et al., 2009) rather than an approach based on t-testing with false discovery correction (FDR). This approach was chosen because it was argued that the use of fold change with a P-value cutoff filtering produces lists of differentially expressed genes in a more reproducible manner (Shi et al., 2008). However, it has been proposed that multiple hypothesis testing may be the better method to eliminate noise in gene expression experiments (Chen and Sarkar, 2006; Grant et al., 2005; Shedden et al., 2005; Tsai et al., 2003). Therefore, we have now reanalyzed the same reprogramming data sets (from Hochedlinger, Thompson, and our own lab) that were used in Chin et al. (Chin et al., 2009) using a Bayesian T-test to adjust for error variance, with an FDR<0.05 and a 1.5 fold-change cutoff. This examination found a similar number of genes differentially expressed between hiPSCs and hESCs in each reprogramming experiments as described previously in Chin et al. (Fig S1A). Furthermore, the genes differentially expressed between hESCs and hiPSCs at early passage identified previously by P-value and fold-change cutoff (Chin et al., 2009) and now by FDR and fold-change were 70–98% identical, indicating that, in this case, the use of P-value and fold-change provided a similar stringency to FDR/fold-change (Fig S1A).
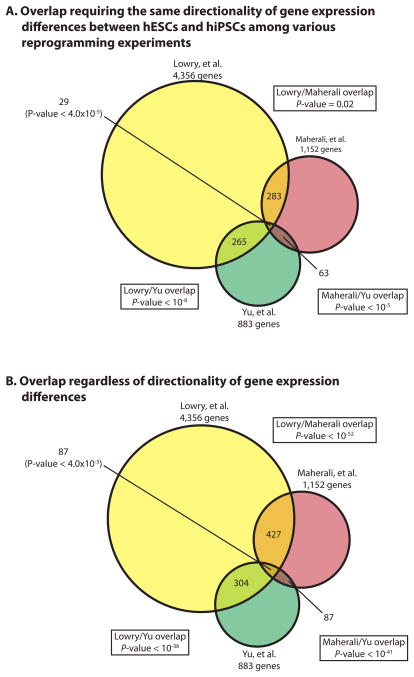
We also demonstrated in Chin et al. (Fig. S8 in (Chin et al., 2009)) that even when taking directionality of expression differences between early passage hiPSCs and hESCs into account there was still significant overlap between differential gene expression lists (hiPSCs vs hESCs) from various labs in over 80% of dual comparisons. Here we have also re-analyzed the overlap of differential gene lists generated using the Bayesian T-test with FDR correction and 1.5 fold-change cutoff between the Hochedlinger (Maherali et al., 2008), Thompson (Yu et al., 2009) and our reprogramming experiments. We found that the datasets still have a significant number of gene expression changes in common even when directionality of gene expression differences was taken into account (Fig 1A and B). When determining the gene expression differences between hESCs and hiPSCs and comparing data across different labs or from independent experiments, it is of course more likely that those differences with conserved directionality are more likely to be functionally relevant. However, until functional data suggest otherwise, it is possible that even the differences whose direction is not conserved but still found consistently found in different labs are important.
Figure 1. Overlap of differentially expressed genes between hiPSCs and hESCs from different labs, determined using the Baysian T-test with FDR correction and 1.5 fold cutoff.
Differentially expressed genes between hiPSCs and hESCs were determined for each of the three indicated reprogramming experiments (Chin et al., 2009; Lowry et al., 2008; Maherali et al., 2008; Maherali and Hochedlinger, 2008; Yu et al., 2009) using a Bayesian T-test with a FDR < 0.05 along with a greater than 1.5 fold change requirement. In (A), overlap among the differentially expressed gene lists was only considered if they were differentially expressed in the same direction between hESCs and hiPSCs. In (B), directionality was not taken into consideration for the overlap.
We have now also performed a post hoc permutation analysis to estimate the FDR for differentially expressed genes between hESCs and early passage hiPSCs from different reprogramming experiments that were determined by P-value and fold-change cutoff in Chin et al. (Chin et al., 2009). While the sample size was not large, this analysis demonstrated that the statistical stringency provided by the use of P-value and fold-change cutoff was equal to a approximated FDR of 2% for early passage hiPSCs vs hESCs (Fig S1B). Therefore, we conclude that the differentially expressed genes between early passage hiPSCs and hESCs and the overlap among different reprogramming experiments as determined in Chin et al. (Chin et al., 2009) are reproducible when using different analysis tools.
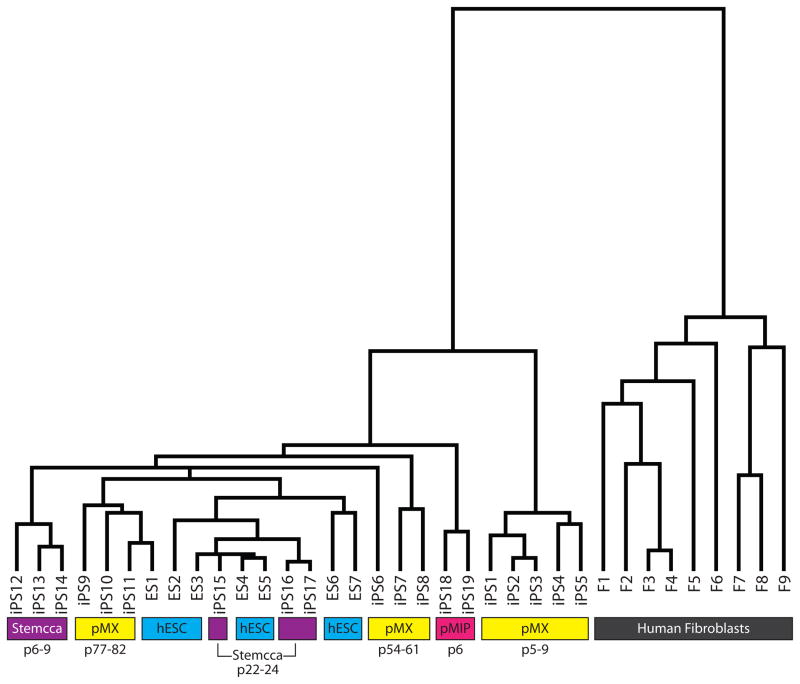
The original hiPSC lines described in Chin et al. and Lowry et al. (Chin et al., 2009; Lowry et al., 2008) were generated by the expression of the Yamanaka set of four transcription factors each from individual MMLV-based retroviruses (pMX vectors). It has since been shown that lentivirus is suitable for delivering the reprogramming factors (Brambrink et al., 2008; Maherali et al., 2008; Stadtfeld et al., 2008) and newer methods use polycistronic vectors to deliver all four reprogramming factors in one virus and were shown to be very effective at generating hiPSC lines (Carey et al., 2009; Chang et al., 2009; Gonzalez et al., 2009; Shao et al., 2009). To further define gene expression differences due to reprogramming, we have now generated new hiPSC lines with various newer methods and under improved culturing conditions during the reprogramming process. We took advantage of polycistronic vectors and employed both a MMLV-based retrovirus and a lentivirus to deliver the reprogramming factors to a variety of fibroblast lines derived from both males and females (Sommer et al., 2008; Tchieu et al., 2010) (see Supplemental Methods for a summary of the lines used and their accession numbers). As shown in Fig. 2, all early passage hiPSC lines generated with polycistronic reprogramming tools are more similar to hESCs than the hiPSCs made originally in our lab with four separate viruses each carrying one reprogramming factor. Together, these data imply that while previous methods to derive hiPSCs generated lines that could be distinguished from hESCs by gene expression as shown in three independent studies (Chin et al., 2009; Ghosh et al., 2010; Marchetto et al., 2009), newer methods and procedures appear to more faithfully reprogram fibroblasts to a pluripotent state.
Figure 2. Variation in gene expression between hiPSCs and hESCs generated in our laboratories using different technical strategies.
Hierarchical clustering of expression data obtained for hESCs (blue highlight), various hiPSCs at different passage (p) (yellow for single pMX-retroviral hiPSCs, pink for polycistronic pMIP retroviral hiPSCs, and purple for polycistronic STEMCCA lentiviral hiPSCs), and fibroblasts (F, gray). The Supplemental Methods section provides further identification for each cell line.
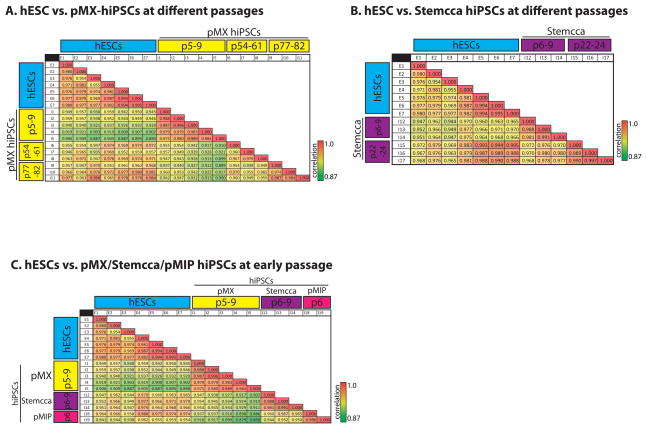
As shown in Chin et al. (Chin et al., 2009), hiPSC gene expression becomes much more similar to hESCs as a result of passaging as the gene expression profiles of our late passage pMX-hiPSCs 1,2 and 18 converged with that of hESCs. Similarly, Soldner et al. demonstrated that while gene expression differences were detected between hiPSCs and hESCs, much of this difference was eliminated upon removal of the reprogramming factors by Cre-mediated excision (Soldner et al., 2009). Whether extended passaging due to subcloning played a role in this effect is unknown. Here we show that our original pMX-hiPSC lines when passaged at least 77 times came even closer to hESCs than they were after ~55 passages (Fig 2 and 3). Furthermore, here we show that extended passaging of the polycistronic lines brought them transcriptionally even closer to hESCs (Fig 3), suggesting that passaging can play a role despite the method used to reprogram. This result was reminiscent of data with iPSCs made from various donor cell types where any residual gene expression differences were lost upon extended passaging (Polo et al., 2010).
Figure 3. Extended passaging brings the gene expression signature of hiPSCs closer to that of hESCs.
(A) Correlation table comparing normalized and filtered microarray data for hESC lines to hiPSCs generated with 4 separate pMX retroviruses, which were profiled at different passages (p) as indicated in the yellow highlight. Values present the Pearson’s correlation coefficient between two cell lines.
(B) Correlation table comparing hESC lines to hiPSCs generated with the polycistronic STEMCCA lentivirus at different passages.
(C) Correlation table comparing hESCs to hiPSCs at early passage derived from the three unique reprogramming experiments: 4-factor pMX-hiPSCs (yellow), polycistronic STEMCCA lentivirus (purple), and polycistronic pMIP retrovirus (pink).
It should be noted that the comparisons made in Figure 3 of Guenther et al. used data from late passage hiPSCs from Chin et al. but early passage hiPSC lines from Maherali et al. and Yu et al., which could shed light on the differing interpretations of Guenther et al. and Chin et al. (Chin et al., 2009). This could explain the low number of genes presented in Fig. 3c in Guenther et al. for the Chin et al. data, and why the Maherali and Yu data sets overlapped with each other but not with the Chin et al. data as they did in the original Chin et al. and in Figure 1A here.
Our new data are congruent with results from Guenther et al. with late passage hiPSC lines, where we show that newly derived lines made by polycistronic vectors and passaged significantly in our lab do not consistently cluster separately from hESCs also grown in our lab. This finding does not suggest that these newly derived hiPSCs are identical to hESCs, only that there are fewer differences between them than found in previous hiPSCs lines made with different methods. The Guenther data set shows that comparing hiPSCs and hESCs leaves just four genes with consistent gene expression differences. While this small number of differences might be irrelevant, it is worth noting that just one genetic locus containing several non-coding transcripts and miRNAs appears to distinguish different functional classes of miPSCs (Stadtfeld et al., 2010).
In light of these findings, it is important to consider passage number, reprogramming technology, and genetic background when comparing pluripotent cells from various sources. It remains unclear what drives the transition of hiPSCs closer to hESCs. Our data suggest that with improved technology one can probe deeper to find expression differences between hiPSCs and hESCs not related to lab-specific differences.
EXPERIMENTAL PROCEDURES
Tissue Culture and Reprogramming Methods
Cells were cultured and reprogrammed as described in Lowry et al. (Lowry et al., 2008) and Tchieu et al. (Tchieu et al., 2010). A summary of the cell lines used in this study is given in the Supplemental Methods. All cells were grown under a protocol approved by the Embryonic Stem Cell Research Oversight (ESCRO) committee at UCLA.
Gene Expression Analysis
The gene expression profiles of several new hiPSC and hESC as well as fibroblast lines was determined by Affymetrix Arrays as described in Chin et al. (Chin et al., 2009) and are summarized in the Supplemental Methods with their NCBI Gene Expression Omnibus (GEO) database accession numbers. The gene expression data was normalized using Robust Multichip Analysis (RMA) in R. Probeset data were then collapsed to RefSeqIDs using the highest confidence probesets as in Chin et al. (Chin et al., 2009). A Bayesian T-test was implemented to determine differentially expressed genes between hiPSC and hESC lines (Baldi and Long, 2001; Sharov et al., 2005). Genes with an FDR < 0.05 and a fold change greater than 1.5-fold were called significantly differentially expressed. Significance between any two data sets was determined using the hypergeometric test, and for three data sets the significance was measured using a simulation with replacement.
Supplementary Material
Acknowledgments
We would like to thank David Casero for his statistical advice and assistance with implementation. MHC is supported by a Ruth L. Kirschstein Institutional Training Grant (NRSA # T32CA009056), KP by the NIH Director’s Young Innovator Award (DP2OD001686) and a CIRM Young Investigator Award (RN1-00564). This work was also supported by the CIRM New Cell Line grant (Jerome Zack, PI). WEL holds the Maria Rowena Ross Chair in Cell Biology and Biochemistry and is supported by The March of Dimes.
References
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics (Oxford, England) 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen J, Sarkar SK. A Bayesian determination of threshold for identifying differentially expressed genes in microarray experiments. Statistics in medicine. 2006;25:3174–3189. doi: 10.1002/sim.2422. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nature biotechnology. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009 doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent Donor Cell Gene Expression among Human Induced Pluripotent Stem Cells Contributes to Differences with Human Embryonic Stem Cells. PLoS ONE. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Barragan Monasterio M, Tiscornia G, Montserrat Pulido N, Vassena R, Batlle Morera L, Rodriguez Piza I, Izpisua Belmonte JC. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8918–8922. doi: 10.1073/pnas.0901471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GR, Liu J, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics (Oxford, England) 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS Cells Can Support Full-Term Development of Tetraploid Blastocyst-Complemented Embryos. Cell Stem Cell. 2009 doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. The Journal of biological chemistry. 2010 doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Induced pluripotency of mouse and human somatic cells. Cold Spring Harbor symposia on quantitative biology. 2008;73:157–162. doi: 10.1101/sqb.2008.73.017. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010 doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Feng W, Sun Y, Bai H, Liu J, Currie C, Kim J, Gama R, Wang Z, Qian Z, et al. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell research. 2009;19:296–306. doi: 10.1038/cr.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics (Oxford, England) 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Shedden K, Chen W, Kuick R, Ghosh D, Macdonald J, Cho KR, Giordano TJ, Gruber SB, Fearon ER, Taylor JM, et al. Comparison of seven methods for producing Affymetrix expression scores based on False Discovery Rates in disease profiling data. BMC bioinformatics. 2005;6:26. doi: 10.1186/1471-2105-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, Guo L, Croner LJ, Boysen C, Fang H, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. iPS Cell Generation Using a Single Lentiviral Stem Cell Cassette. Stem Cells. 2008 doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010 doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman S, Aimiuwu O, Lingren A, Zack JA, Clark A, et al. Female human iPS cells retain an inactive X-chromosome. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.06.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CA, Hsueh HM, Chen JJ. Estimation of false discovery rates in multiple testing: application to gene microarray data. Biometrics. 2003;59:1071–1081. doi: 10.1111/j.0006-341x.2003.00123.x. [DOI] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. MicroRNA Profiling of Human-Induced Pluripotent Stem Cells. Stem cells and development. 2009 doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu KKS-O, Tian S, Stewart R, Slukvin I, Thomson J. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science (New York, NY. 2009 doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.