Abstract
We have previously characterized an EMS-induced allele of the bubR1 gene (bubR1D1326N) that separates the two functions of BubR1, causing meiotic nondisjunction but retaining spindle assembly checkpoint activity during somatic cell division in Drosophila melanogaster. Using this allele, we demonstrate that bubR1 meiotic nondisjunction is dosage sensitive, occurs for both exchange and nonexchange homologous chromosomes, and is associated with decreased maintenance of sister chromatid cohesion and of the synaptonemal complex during prophase I progression. We took advantage of these features to perform a genetic screen designed to identify third chromosome deficiencies having a dominant effect on bubR1D1326N/bubR1rev1 meiotic phenotypes. We tested 65 deficiencies covering 60% of the third chromosome euchromatin. Among them, we characterized 24 deficiencies having a dominant effect on bubR1D1326N/bubR1rev1 meiotic phenotypes that we classified in two groups: (1) suppressor of nondisjunction and (2) enhancer of nondisjunction. Among these 24 deficiencies, our results show that deficiencies uncovering the polo locus act as suppressor of bubR1 nondisjunction by delaying meiotic prophase I progression and restoring chiasmata formation as observed by the loading of the condensin subunit SMC2. Furthermore, we identified two deficiencies inducing a lethal phenotype during embryonic development and thus affecting BubR1 kinase activity in somatic cells and one deficiency causing female sterility. Overall, our genetic screening strategy proved to be highly sensitive for the identification of modifiers of BubR1 kinase activity in both meiosis and mitosis.
Mitosis is a process that results in the production of two identical daughter cells from a single cell. At metaphase-anaphase transition, the accuracy of chromosome segregation is ensured by the spindle assembly checkpoint (SAC) that monitors microtubule-kinetochore attachment and prevents mitotic exit until all chromosomes are attached to the bipolar spindle and under tension. The Mad (mitotic arrest deficient) and Bub (budding uninhibited by benomyl) SAC components were first identified in budding yeast through genetic screens designed to isolate mutations which override the mitotic arrest in the presence of microtubule depolymerizing drugs (Hoyt et al. 1991; Li and Murray 1991). Immunolocalization studies have shown that these conserved proteins localize to kinetochores that are unattached or under reduced tension (Chen et al. 1998; Taylor et al. 1998; Logarinho et al. 2004). The SAC proteins impose a mitotic arrest by inhibiting the activity of the anaphase-promoting complex/cyclosome (APC/C) that is essential for sister chromatid separation and mitotic exit (Li and Benezra 1996; Taylor and Mckeon 1997; Bernard et al. 1998; Gorbsky et al. 1998; Basu et al. 1999). However, the Bub1-related kinase (BubR1), which displays N-terminal homology with the yeast Mad3 protein and C-terminal homology with the Bub1 kinase domain, is found only in higher eukaryotes (Taylor et al. 1998). In addition to its involvement in the SAC, BubR1 is also required for proper mitotic timing, capture, and stabilization of kinetochore-microtubule attachment at prometaphase-metaphase transition (Basu et al. 1999; Ditchfield et al. 2003; Logarinho et al. 2004; Harris et al. 2005; Lampson and Kapoor 2005) and for mitotic arrest in the presence of DNA damage (Fang et al. 2006). Furthermore, BubR1 is essential to prevent early aging and infertility in mice (Baker et al. 2006; Hartman et al. 2007; Matsumoto et al. 2007).
In contrast to mitosis, meiosis results in the production of haploid gametes from a diploid parental cell. Meiosis involves one single round of DNA replication followed by two sequential rounds of cell division (meiosis I and meiosis II). During meiotic prophase I, homologous chromosomes undergo a complex series of modifications through pairing, synapsis, and exchange to ensure chromosome reduction and sister chromatid separation during meiosis. Nondisjunction (NDJ), the failure to properly segregate the genome during meiosis, produces haploid cells that have unbalanced genetic composition. The vast majority of meiotic segregation errors occur during meiosis in females, and the error rate increases with advancing maternal age. The frequency of missegregation in human oocytes is remarkably high (about 10% of meiosis), and this is thought to be one reason for the high rate of miscarriages (spontaneous abortions) in early stages of pregnancy (Alberts et al. 2002).
In contrast to mitosis, where BubR1 has an important role in controlling the metaphase-anaphase transition, it was shown that MAD3/BubR1 has an essential and conserved function during prophase I progression in meiosis. MAD3/BubR1 is required to delay prophase I in response to nonexchange chromosomes in budding yeast (Cheslock et al. 2005). In D. melanogaster, BubR1 was shown to play an essential role during meiotic progression in both sexes and to prevent missegregation of both chiasmate and achiasmate homologous chromosomes. During oogenesis, BubR1 is also required to maintain the synaptonemal complex (SC) (Malmanche et al. 2007). In mice, BubR1-depleted oocytes have a reduced capacity to trigger essential meiotic arrest through destabilization of the APC/C inhibitor Cdh1 (Homer et al. 2009). It was also shown that overexpression of exogenous BubR1 arrests oocyte maturation during meiosis I, while dominant-negative BubR1 expression accelerates meiotic progression (Wei et al. 2010). These studies indicate an essential requirement for BubR1 function in timing the complex events taking place during meiotic prophase I progression.
In D. melanogaster, we have previously characterized a bubR1 EMS allele with a point mutation in a conserved amino acid essential for its kinase activity, bubR1D1326N (Malmanche et al. 2007). Using this allele, we showed that individuals with bubR1D1326N in trans with two previously characterized alleles, bubR11 (Basu et al. 1999) and bubR1rev1 (Perez-Mongiovi et al. 2005), and with Df(2R)nap9, a deficiency uncovering bubR1, lack any of the visible phenotypes that are characteristic of a defect in mitosis and have a functional SAC. However, during meiosis, bubR1D1326N trans-heterozygotes show meiotic NDJ for the autosome and sex chromosomes, with the frequency being BubR1 dosage-dependent. We took advantage of these sensitized genetic backgrounds (Malmanche et al. 2007) to perform a genetic screening to identify third chromosome deficiencies displaying suppression or enhancement of X NDJ frequency. For this, we used the third chromosome D. melanogaster deficiency kit from the Bloomington Drosophila Stock Center and we tested 60% of the third chromosome euchromatin. Our results identified 18 deficiencies that suppress and 6 deficiencies that enhance X NDJ. Among the candidate genes tested, we identified Polo kinase as a strong suppressor of the NDJ phenotype and demonstrate that this effect is related to the maintenance of SC during prophase I progression. We identified one deficiency that causes female sterility and two deficiencies affecting BubR1 kinase activity during embryonic development and somatic cell cycle progression, indicating that other pathways can complement BubR1 kinase activity in an otherwise wild-type genetic background.
Materials and Methods
Drosophila melanogaster stocks
The third chromosome deficiency kit, as well as some of the mutant alleles, were obtained from the Bloomington Drosophila Stock Center.
Genetic screen
From the 96 deficiency stocks of the third chromosome, we were able to generate and maintain 65 deficiencies in the bubR1D1326N genetic background. To test their effect on female X NDJ, bubR1rev1/bubR1D1326N;Df/+ females were crossed to C(1;Y)1, v f B/O males, and the resulting X NDJ frequency was calculated from the progeny. From this cross, regular female progeny are Bar (X/C(1;Y)1, v f B) and regular male progeny are non-Bar (X/O), exceptional female progeny are non-Bar (XX/O) and exceptional male progeny are vermilion, forked, and Bar (C(1;Y)1, v f B/O). The X NDJ frequency was calculated using the following formula: X NDJ = (2 × males and females exceptional progeny)/Adjusted total progeny, with the Adjusted total progeny = (2 × males and females exceptional progeny) + normal progeny. The exceptional progeny was multiplied by 2 due to the lethality associated with half of the exceptional progeny (triplo-X and nullo-X).
Immunofluorescence on ovaries
Ovaries were dissected in cold 1× PBS. Fixation was performed for 20 min in 2% EM grade formaldehyde as described in Page and Hawley (2001), followed by 2 hr permeabilization in 1× PBS - 0,5% Triton X-100–10% calf serum. Primary antibodies used were: guinea pig anti-C(3)G (Page and Hawley 2001), rat anti-SMC1 (Malmanche et al. 2007), rabbit anti-SMC2 (Savvidou et al. 2005), mouse anti-Orb (Lantz et al. 1994). The following secondary antibodies were used at 1:1000: anti-rabbit Alexa Fluor 488, anti-mouse Alexa Fluor 568 and anti-guinea pig Alexa Fluor 647. DNA was detected in 1X PBS containing 1 µg/ml of DAPI. Images were obtained using a Leica TCS SP2 AOBS Confocal Microscope (Leica Microsystems, Heidelberg), deconvolved using Huygens Essential (version 3.0.2pl) and processed with Adobe Photoshop 7.0.
Immunofluorescence on embryos
Wild-type and bubR1 mutant embryos were collected and aged at 25°C. Immunodetection on embryos was performed as described in Sullivan et al. (2000). Primary antibody used was: anti-phospho H3 rabbit polyclonal, used at 1:500 (Upstate Biotechnology). Secondary antibody used was: anti-rabbit Alexa Fluor 488 used at 1:2000 (Molecular Probes). DNA was detected in 1× PBS containing 1 µg/ml of DAPI. For colchicine treatment, embryos were permeabilized in n-heptane, containing 250 µM colchicine in 1× PBS for 30 min before fixation. Images were obtained using a Leica TCS SP2 AOBS Confocal Microscope (Leica Microsystems, Heidelberg), deconvolved using Huygens Essential (version 3.0.2pl) and processed with Adobe Photoshop 7.0.
Results
Analysis of X chromosome NDJ in bubR1D1326N/bubR1rev1 females
To determine whether a specific third chromosome deficiency has an effect on bubR1 X NDJ, first we analyzed the overall variations observed in the control genotype, X/X;bubR1D1326N/bubR1rev1. During the screening procedure, we performed 16 rounds of X NDJ experiments, using for each group a control with the X/X;bubR1D1326N/bubR1rev1 genotype. Within these 16 controls, the X NDJ frequency ranges from 16.60% to 35.20%, allowing us to calculate an average frequency of 25.82% with a standard deviation of 4.87% (Table 1). These results indicate an inherent variability in the experimental procedure that reflects uncontrollable environmental effects, as well as sampling errors. Thus, in a first approach, we consider that a given third chromosome deficiency has an effect on bubR1 X NDJ if the frequency is below 20.95% or above 30.69% of the control values, and if the given deficiency shows the same effect when compared to the control for its specific experimental round. For that, we performed a statistical analysis using the multinomial-Poisson hierarchy model (Zeng et al. 2010) to take into account variations in sample size and sampling errors. Using these parameters, we identified 24 deficiencies that affect bubR1 X NDJ with a 95% of confidence interval.
Table 1 . NDJ of X/X ; bubR1D1326N/bubR1rev1 females of the 16 control experiments.
| Experiment | Normal Progeny | Exceptional Progeny | Total Adjusted Progeny | X NDJ | |
|---|---|---|---|---|---|
| X/XY and X/O | XX/O | O/XY | |||
| 1 | 1869 | 122 | 64 | 2241 | 16.60% |
| 2 | 2641 | 280 | 40 | 3281 | 19.51% |
| 3 | 1452 | 149 | 48 | 1846 | 21.34% |
| 4 | 2885 | 272 | 153 | 3735 | 22.76% |
| 5 | 1782 | 233 | 33 | 2314 | 22.99% |
| 6 | 1286 | 111 | 85 | 1678 | 23.36% |
| 7 | 1138 | 157 | 28 | 1508 | 24.54% |
| 8 | 972 | 130 | 32 | 1296 | 25.00% |
| 9 | 1545 | 170 | 107 | 2099 | 26.39% |
| 10 | 737 | 102 | 34 | 1009 | 26.96% |
| 11 | 1324 | 178 | 72 | 1824 | 27.41% |
| 12 | 282 | 45 | 10 | 392 | 28.06% |
| 13 | 631 | 79 | 49 | 887 | 28.86% |
| 14 | 1116 | 210 | 44 | 1624 | 31.28% |
| 15 | 775 | 167 | 22 | 1153 | 32.78% |
| 16 | 681 | 166 | 19 | 1051 | 35.20% |
| Average | 25.82% ± 4.87% | ||||
Third chromosome deficiencies modifying bubR1 X chromosome NDJ
The third chromosome deficiency kit that we originally used included 96 deficiencies. From these, we were able to generate 65 stocks in the appropriate bubR1D1326N genetic background that covers 60% of the third chromosome euchromatin. As stated above, we performed 16 rounds of experiments to screen these 65 deficiencies (Table 2 and supporting information, Table S1). Using the appropriate parameters for comparison with controls (see above), we identified 6 deficiencies that enhance and 18 deficiencies that suppress the bubR1 X NDJ. We also identified one deficiency (Df(3L)ED4674) causing sterility in bubR1D1326N/bubR1rev1 females and two deficiencies (Df(3L)BSC10 and Df(3R)BSC56) inducing a zygotic developmental arrest. Following the first round of screening, we performed a second round of genetic screens using overlapping deficiencies and mutant alleles of candidate genes for three deficiencies that enhance and two that suppress bubR1 X NDJ, for the deficiency that causes female sterility, and for the two deficiencies that induce an embryonic lethal phenotype. We also performed complementation tests using a number of overlapping deficiencies for the remaining enhancers and suppressors to confirm the breakpoints of the deficiencies in the stocks we established and used to test bubR1 X NDJ (Table S2).
Table 2 . Deficiencies that enhance or suppress X NDJ of X/X;bubR1D1326N/bubR1rev1 females.
| Cytogenetic Breakpoints | Normal ProgenyX/XY and X/O | Exceptional Progeny | Total Adjusted Progeny | Δ With Matched Controla | Δ With Average Controlb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Deficiency Name | XX/O | O/XY | X NDJ | Candidate | |||||
| Enhancer deficiencies | |||||||||
| Df(3L)BSC23 | 62E8;63B5-6 | 550 | 127 | 16 | 836 | 34.21%c | 14.70% | 8.39% | Mrtf; aly |
| Df(3L)BSC13 | 66B12-C1;66D2-4 | 841 | 281 | 14 | 1431 | 41.23%c | 18.24% | 15.41% | mtrm |
| Df(3L)Pc-2q | 78C5-6;78E3-79A1 | 323 | 109 | 15 | 571 | 43.43%c | 18.43% | 17.61% | pzg; Pc; SAK |
| Df(3R)ED5177 | 83B4;83B6 | 462 | 105 | 16 | 704 | 34.38%c | 9.84% | 8.56% | asl |
| Df(3R)ED5780 | 89E11;90C1 | 61 | 22 | 4 | 113 | 46.02%c | 17.16% | 20.20% | cal1; ald |
| Df(3R)D605 | 97E2;98A3-4 | 274 | 85 | 15 | 474 | 42.19%c | 17.19% | 16.37% | Klp98A |
| Suppressor deficiencies | |||||||||
| Df(3L)pbl-X1 | 65F6;66B7-8 | 780 | 30 | 19 | 878 | 11.16%c | −11.83% | −14.66% | Pdp1;pbl; Arp66B |
| Df(3L)ZP1 | 66A17-20;66C1-5 | 968 | 81 | 21 | 1172 | 17.41%c | −5.58% | −8.41% | pbl; Arp66B |
| Df(3L)BSC8 | 74D3-75A1;75B2-5 | 687 | 56 | 17 | 833 | 17.53%c | −7.01% | −8.29% | CycT |
| Df(3L)ED4858 | 76D3;77C1 | 566 | 23 | 13 | 638 | 11.29%c | −17.58% | −14.53% | polo |
| Df(3L)rdgC-co2 | 77A1;77D1 | 686 | 37 | 11 | 782 | 12.28%c | −14.68% | −13.54% | polo |
| Df(3L)ED4978 | 78D5;79A2 | 1712 | 69 | 27 | 1904 | 10.08%c | −6.52% | −15.74% | |
| Df(3L)Ten-m-AL29 | 79C1-3;79E3-8 | 1079 | 64 | 19 | 1245 | 13.33%c | −6.17% | −12.49% | |
| Df(3R)Tpl10 | 83C1-2;84B1-2, 83D4-5;84A4-5;98F1-2 | 647 | 20 | 22 | 731 | 11.49%c | −8.02% | −14.33% | bcd |
| Df(3R)GB104 | 85D12;85E10 | 189 | 2 | 1 | 195 | 3.08%c | −13.52% | −22.74% | βTub85D; hyd; αTub85E; topi |
| Df(3R)ED5559 | 86E11;87B11 | 270 | 0 | 0 | 270 | 0.00%c | −21.34% | −25.82% | aur; ssp5 |
| Df(3R)sbd105 | 88F9-89A1;89B9-10 | 1022 | 106 | 23 | 1280 | 20.16%c | −15.05% | −5.66% | c(3)G; msps |
| Df(3R)ED5942 | 91F12;92B3 | 725 | 78 | 18 | 917 | 20.94%c | −6.47% | −4.88% | |
| Df(3R)BSC55 | 94D2-10;94E1-6 | 613 | 43 | 20 | 739 | 17.05%c | −10.36% | −8.77% | sav |
| Df(3R)mbc-30 | 95A5-7;95C10-11 | 181 | 8 | 1 | 199 | 9.05%c | −18.37% | −16.77% | Rpn9; eIF4G2; Pros26.4; CG13599; SMC1 |
| Df(3R)mbc-R1 | 95A5-7;95D6-11 | 1302 | 32 | 49 | 1464 | 11.07%c | −20.22% | −14.75% | Rpn9; eIF4G2; Pros26.4; CG13599; SMC1 |
| Df(3R)Exel6202 | 96D1;96D1 | 837 | 64 | 29 | 1023 | 18.18%c | −10.68% | −7.64% | |
| Df(3R)BSC42 | 98B1-2;98B3-5 | 1849 | 126 | 35 | 2171 | 14.83%c | −16.45% | −10.99% | Sce; btz |
| Df(3R)3450 | 98E3;99A6-8 | 1003 | 42 | 28 | 1143 | 12.25%c | −7.26% | −13.57% | Doa; Slu7; yemaα; dgt6; Slbp; stg |
Difference between X NDJ of the deficiency-bearing flies vs. matched control.
Difference between X NDJ of the deficiency-bearing flies vs. average controls.
The percentage of X NDJ is significantly higher/lower than in X/X;bubR1D1326N/bubR1rev1 females (multinomial-Poisson hierarchy model, P < 0.05).
Enhancers of bubR1 X chromosome NDJ
In the group of deficiencies that enhance the frequency of bubR1 X NDJ, we narrowed down to a smaller genomic region or identified candidate genes for the following deficiencies: Df(3R)D605 (97E2;98A3-4), Df(3L)Pc-2q (78C5-6;78E3-79A1), and Df(3L)BSC13 (66B12-C1;66D2-4). Because the breakpoints of some of these deficiencies have not been defined molecularly, it is likely that these regions and the genes they contain cannot be accurately defined.
Df(3R)D605.
Df(3R)D605 (97E2;98A3-4) enhances bubR1 X NDJ by 17.19% when compared with its own matched control (Table 2). To identify a smaller genomic region, we tested four overlapping deficiencies [Df(3R)ED6255 (97D2;97F1); Df(3R)Exel6206 (97E1;97E5); Df(3R)ED6237 (97E4;97E11) and Df(3R)IR16 (97F1-2;98A)] and one deficiency covering completely Df(3R)D605 [Df(3R)ED6265 (97E2;98A7)]. Among these five deficiencies, only Df(3R)ED6265 has a similar effect as Df(3R)D605 on bubR1 X NDJ, indicating that the gene responsible for the observed enhancement lies within 98A1;98A4 genomic region (Table S3).
Df(3L)Pc-2q.
Df(3L)Pc-2q (78C5-6;78E3-79A1) enhances bubR1 X NDJ by 18.43% when compared to its own matched control (Table 2). Within the third chromosome deficiency kit, we also tested Df(3L)ED4978 (78D5;79A2) which partially overlaps with Df(3L)Pc-2q. However, Df(3L)ED4978 suppresses bubR1 X NDJ by 6.52% when compared to its own matched control (Table 2). Thus, according to the cytogenetic map coordinates for both deficiencies, we can conclude that the enhancer gene is located in the genomic region 78C5-6;78D4, while the suppressor gene is located in the genomic region 79A1;79A2.
Df(3L)BSC13.
Df(3L)BSC13 (66B12-C1;66D2-4) enhances bubR1 X NDJ by 18.24% when compared with its own matched control (Table 2). Within the third chromosome deficiency kit, we also tested Df(3L)ZP1 (66A17-20;66C1-5), which partially overlaps with Df(3L)BSC13. However, Df(3L)ZP1 suppresses bubR1 X NDJ (Table 2), suggesting that the enhancer gene should be in the region 66C6-66D4. Within this region, we identified matrimony (mtrm – 66C11) as a candidate gene. mtrm was previously identified in a screen of the major autosomes as being haploinsufficient for achiasmate segregation in Drosophila oocytes (Harris et al. 2003). We tested a null allele of mtrm, mtrm126 (Xiang et al. 2007) in a bubR1D1326N/bubR1rev1 mutant female background and observed a 13.40% increase of bubR1 X NDJ. However, mtrm126/+ females have a frequency of X NDJ of 9.45%. Thus, it is likely that the enhancement of bubR1 X NDJ observed in bubR1D1326N/bubR1rev1; mtrm126/+ females is only due to an additive effect of the frequency of NDJ displayed by mtrm and bubR1 mutants alone.
Suppressors of bubR1 X NDJ
In the group of deficiencies that suppress the frequency of bubR1 X NDJ, we narrowed down to a smaller genomic region or identified candidate genes for the following deficiencies: Df(3R)ED5559 (86E11;87B11) and Df(3L)rdgC-co2 (77A1;77D1).
Df(3R)ED5559.
Df(3R)ED5559 (86E11;87B11) completely suppresses bubR1-induced X NDJ (Table 2). To uncover the gene responsible for the full phenotypic rescue, we tested four deficiencies that overlap Df(3R)ED5559 [Df(3R)ED5516 (86D8;86E13); Df(3R)Exel8154 (86E13;86E18); Df(3R)Exel7310 (86E18;87A1) and Df(3R)ED5577 (86F9;87B13)]. Our results show that only Df(3R)ED5577 has a similar effect, suggesting that the candidate gene is within the genomic region 87A2;87B11. In this region, we identified aurora (aur) as a potential candidate gene. Aur is essential for several aspects of the cell cycle and mitotic progression (Berdnik and Knoblich 2002; Marumoto et al. 2003; Portier et al. 2007). We tested two EMS aur alleles, aur1 (hypomorphic allele) and aur87Ac-3 (null allele) (Glover et al. 1995). However, none of these alleles showed a full rescue of bubR1 X NDJ, suggesting that a different gene within the region is responsible for this effect (Table S3).
Df(3L)rdgC-co2.
Df(3L)rdgC-co2 (77A1;77D1) suppresses bubR1 X NDJ by 14.68% when compared with its own matched control (Table 2). We identified two other overlapping deficiencies that also suppress bubR1 X NDJ: Df(3L)ED4858 (76D3;77C1) and Df(3L)Exel6136 (77B2;77C6), suggesting that the gene responsible is within the genomic region 77B2;77C1 (Table 3). In this genomic region, we identified polo (77B2-77B3) as a candidate gene, so we tested three polo mutant alleles (polo1, polo2, and polo9). Our results show that all three alleles suppress bubR1 X NDJ and the level of suppression increases with stronger polo alleles (Table 3). polo1 is a weak hypomorph allele with an EMS-induced point mutation in the kinase domain and is hemizygous viable (Sunkel and Glover 1988). polo9 allele has a P-element insertion that reduces Polo levels, and is homozygous lethal at third-instar larvae (Donaldson et al. 2001). polo2 was induced by P-M hybrid dysgenesis and is consider as the strongest polo allele (C.E. Sunkel, R.E. Karess, and D.M. Glover, unpublished results).
Table 3 . NDJ of Df(3L)rdgC-co2, its overlapping deficiencies, and mutant alleles.
| Maternal Genotype | Cytogenetic Breakpoints | Normal ProgenyX/XY and X/O | Exceptional Progeny | Total Adjusted Progeny | X NDJ | |
|---|---|---|---|---|---|---|
| XX/O | O/XY | |||||
| bubR1D1326N/bubR1rev1 | 737 | 102 | 34 | 1009 | 26.96% | |
| bubR1D1326N/bubR1rev1;Df(3L)rdgC-co2/+ | 77A1;77D1 | 686 | 37 | 11 | 782 | 12.28%a |
| bubR1D1326N/bubR1rev1;Df(3L)ED4858/+ | 76D3;77C1 | 566 | 23 | 13 | 638 | 11.29%a |
| bubR1D1326N/bubR1rev1;Df(3L)Exel6136/+ | 77B2;77C6 | 1996 | 9 | 5 | 2024 | 1.38%a |
| bubR1D1326N/bubR1rev1;polo1/+ | 438 | 35 | 17 | 542 | 19.19%a | |
| bubR1D1326N/bubR1rev1;polo2/+ | 1612 | 32 | 25 | 1726 | 6.60%a | |
| bubR1D1326N/bubR1rev1;polo9/+ | 979 | 36 | 21 | 1093 | 10.43%a | |
The percentage of X NDJ is significantly lower than in X/X;bubR1D1326N/bubR1rev1 females (multinomial-Poisson hierarchy model, P < 0.05).
During meiosis, CDC5, the yeast polo homolog, is required to phosphorylate and remove meiotic cohesin from chromosome arms (Lee and Amon 2003), to form chiasmata (Clyne et al. 2003), to co-orient sister kinetochores and to cosegregate sister centromeres at meiosis I (Clyne et al. 2003; Lee and Amon 2003). In D. melanogaster meiosis, Polo is involved in the timing of meiotic prophase I entry, in the restriction of meiosis to the oocyte and in the initiation/maintenance of SC (Mirouse et al. 2006). At later stage, Polo is required to activate Twine, a germline-specific form of the Cdc25 phosphatase, that initiates the chain of events leading to GVBD and prometaphase I progression (Xiang et al. 2007). At meiosis II, Polo is essential to phosphorylate and remove MEI-S332, the Shugoshin homolog, and to allow sister chromatid segregation (Clarke et al. 2005).
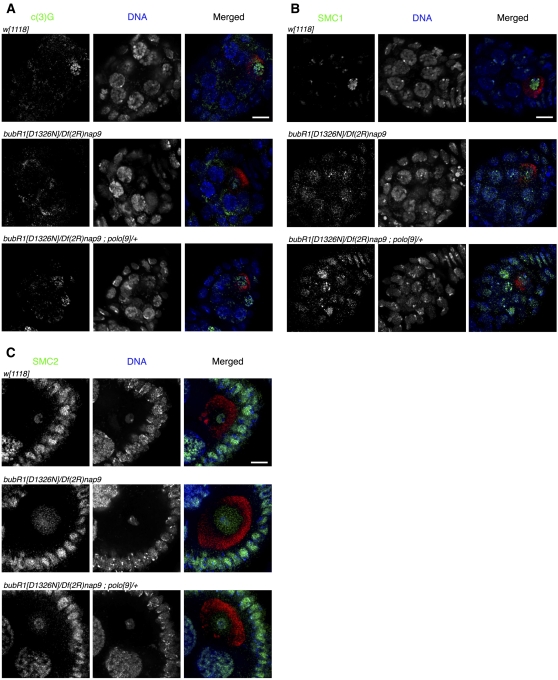
To further investigate at the molecular level the nature of the decrease in X NDJ, we first analyzed whether reducing Polo dosage in bubR1 mutant females restores in part, the meiotic prophase I phenotypes associated with the bubR1 mutation. Because BubR1 and Polo appear to have opposite effects during early stages of meiotic prophase I, with Polo controlling meiotic entry and SC assembly and BubR1 controlling meiotic prophase I progression and SC disassembly, we analyzed the nature of the SC and sister chromatid cohesion in bubR1D1326N/Df(2R)nap9;polo9/+ mutant combinations in region 3 of the germarium (Figure 1). Interestingly, while mutations in bubR1 result in premature disassembly of the SC, in the bubR1D1326N/Df(2R)nap9;polo9/+ we observed the maintenance of the SC protein, C(3)G, and the sister chromatid cohesin subunit, SMC1 (Figure 1, A and B). This suggests that decreasing Polo dosage in bubR1 mutant ovarioles delays sufficiently prophase I entry and/or progression to allow a more accurate prophase I. In addition, we investigated if a reduction of Polo dosage in bubR1 mutant females allows a higher efficiency in the specific replacement of cohesin complex by condensin complex during chiasmata formation (Yu and Koshland 2005) and SC disassembly (Ivanovska et al. 2005; Resnick et al. 2009). Thus, we analyzed the localization of SMC2 condensin subunit at stage 5-7, which corresponds to the pachytene-diplotene transition. In wild-type oocytes, condensin subunit SMC2 is gradually loaded on the bivalent to fully cover the karyosome by stage 5-7 (Figure 1C). However, in bubR1 mutant oocytes, SMC2 localizes within the nuclear space rather than being bound to the karyosome (Figure 1C). Thus, in addition to the decrease in the maintenance of the SC and of the sister chromatid cohesion, mutations in bubR1 also affect the changes in bivalent configuration at diplotene. Interestingly, decreasing Polo dosage restores condensin loading and bivalent modifications in bubR1 mutant oocytes. Taken together, these results suggest that the decreased X NDJ induced by a reduction of Polo dosage in bubR1 mutant oocytes occurs through a Polo-dependent mechanism during the initial stages of meiosis.
Figure 1 .
Polo is a suppressor of bubR1 X NDJ. (A) Structure of SC in wild-type, in bubR1 mutant and in bubR1;polo mutant in region 3 of the germarium. C(3)G is in green, DNA is in blue, and Orb (as oocyte marker) is in red. Decreasing polo dosage in a bubR1 genetic background rescues the maintenance of the SC in the oocyte nucleus comparing to bubR1 mutant. (B) Sister chromatid cohesin subunit SMC1 in wild-type, in bubR1 mutant and in bubR1;polo mutant in region 3 of the germarium. SMC1 is in green, DNA is in blue, and Orb is in red. Decreasing polo dosage in bubR1 genetic background rescues the maintenance of the sister chromatid cohesion in the oocyte nucleus comparing to bubR1 mutant. (C) Condensin subunit SMC2 in wild-type, in bubR1 mutant and in bubR1;polo mutant at the pachytene/diplotene transition (stages 5-7). SMC2 is in green, DNA is in blue, and Orb is in red. Decreasing polo dosage in bubR1 genetic background rescues the loading of condensin subunit at stages 5–7 in the oocyte nucleus during chiasmata formation comparing to bubR1 mutant oocyte. Scale bars = 10 μm.
Deficiency causing sterility in bubR1D1326N/bubR1rev1 females
Df(3L)ED4674 (73B5;73E5) causes sterility in bubR1D1326N/bubR1rev1 mutant females, suggesting a strong enhancement of the bubR1 meiotic phenotype. To narrow down the genomic region of interest, we tested four overlapping deficiencies [Df(3L)BSC561 (73A2;73C1); Df(3L)Exel9004 (73D1;73D5); Df(3L)Exel7253 (73D5;73E4) and Df(3L)BSC414 (73E1;74C3)]. Among these four deficiencies, our results show that only Df(3L)BSC561 causes sterility in bubR1D1326N/bubR1rev1 mutant females, indicating the presence of a dosage-sensitive gene within the genomic region 73B5-73C1. Within this region, we found two potential candidate genes, Baldspot and Lasp, which have an essential function during spermatogenesis (Jung et al. 2007; Lee et al. 2008).
Identification of two deficiencies affecting somatic BubR1 kinase activity
It was previously shown that different domains of BubR1 have different functions, with the KEN box being essential for SAC activity but the kinase domain being required only for spindle formation during prometaphase (Elowe et al. 2010). Mutations in BubR1 kinase domain give rise to viable adults lacking any visible phenotype, and mutant cells are able to properly segregate their genetic material during somatic cell division (Malmanche et al. 2007; Rahmani et al. 2009). Interestingly, we identified two deficiencies that induce an embryonic lethality in the bubR1 genetic background indicating that a decrease in some components of the cell cycle network can impair cell cycle progression and zygote development in a BubR1 kinase dead context.
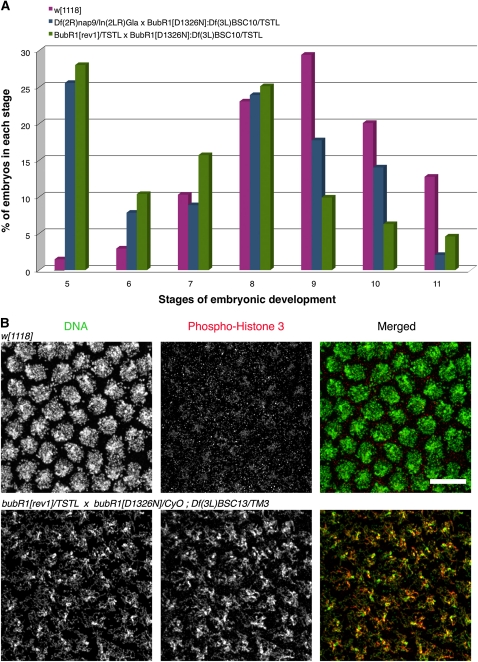
Df(3L)BSC10.
Df(3L)BSC10 deletes the genomic region 69D4-5;69F5-7. First, we characterized the embryonic stage at which the development of bubR1D1326N/bubR1rev1;Df(3L)BSC10/+ embryos fail to proceed. For this, we analyzed staged embryonic collections from 0-2 hr that were aged for 3 hr before fixation. Our results showed a developmental arrest at stage 5 that corresponds to the point when the syncytial cell cycle 13 ends and cellularization and gastrulation initiates (Figure 2A). Thus, we asked if the mutant embryos failed to undergo gastrulation due to a modified cytoskeleton network. For this, embryos were collected as before, but they were treated with colchicine prior to fixation, to depolymerize microtubules. In wild-type embryos, colchicine treatment does not affect the overall nuclear structure, and the replicated chromosomes remain decondensed (Figure 2B). However, in the mutant embryos, we observed the condensation of interphase chromatin after colchicine treatment. Moreover, chromosome condensation is associated with the presence of the phospho-H3 epitope that is normally only detected in dividing nuclei (Figure 2B). To identify candidate genes within this deficiency, we tested overlapping deficiencies covering the Df(3L)BSC10 deleted region [Df(3L)iro-2 (69B1-5;69D1-6); Df(3L)Exel6117 (69D1;69E2), Df(3L)E44 (69D2;69E3-5) and Df(3L)ED4486 (69C4;69F6)]. However, none of the deficiencies tested induce an embryonic lethal phenotype, suggesting that the candidate gene is within the genomic region 69F6;69F7. Within this small region, we found RpS4 (Ribosomal protein S4), a gene involved in mitotic spindle elongation and organization (Goshima et al. 2007). However, we cannot exclude the possibility of Df(3L)BSC10 having another unknown chromosomal aberration.
Figure 2 .
BubR1 kinase activity is essential during somatic cell cycle (A) The developmental arrest induced by Df(3L)BSC10 in BubR1 kinase dead embryos takes place after the syncitial division and prevents the initialization of gastrulation. (B) Embryonic phenotype at stage 5 after 250 μM colchicine treatment in wild-type and progeny from a cross made between bubR1rev1/TSTL females to bubR1D1326N/CyO;Df(3L)BSC10/TM3 males. Phospho-Histone 3 is in red and DAPI is in green. In contrast to wild-type embryos, replicated chromosomes in mutant embryos after colchicine treatment appear condensed and the presence of phospho-histone 3 epitope is detected. Scale bars = 10μm.
Df(3R)BSC56.
Df(3R)BSC56 (94E1-2;94F1-2) also induces a developmental arrest of bubR1D1326N/bubR1rev1;Df(3R)BSC56/+ mutant embryos. We used five overlapping deficiencies covering the Df(3R)BSC56 deleted region to identify candidate genes [Df(3R)BSC55 (94D2-10;94E1-6), Df(3R)Exel6193 (94D3;94E4), Df(3R)ED6103 (94D3;94E9), Df(3R)Exel6274 (94E4;94E11) and Df(3R)Exel6194 (94F1;95A4)]. Among these deficiencies, only Df(3R)Exel6274 induces an embryonic lethality, suggesting the presence of the candidate gene within the genomic region 94E9;94E11, defined between the breakpoint of Df(3R)ED6103 and Df(3R)Exel6274. Within this region, we identified cdc16 (94E9) as a potential candidate gene. Cdc16/Apc6 is a tetratricopeptide repeat (TPR) subunit of APC/C, essential for G2 progression. cdc16RNAi animals die as P5(i) early pupae and cdc16RNAi cells show mitotic phenotypes, including high mitotic index and overcondensed chromosomes in a metaphase-like arrest and depletion of Cdc16 also affects cyclin B degradation (Pal et al. 2007). We therefore tested a cdc16 allele (cdc16MB09129) that has a transposable element insertion in an intron of cdc16. However, this allele does not induce the lethality observed with Df(3R)BSC56, suggesting that either cdc16 is not the relevant gene or the allele we tested, which has not been extensively characterized, still produces sufficient active protein that allow embryonic development.
Discussion
We have shown previously that BubR1 kinase activity is essential during meiotic prophase I progression to ensure the correct timing of SC disassemble and chromosome NDJ in D. melanogaster female (Malmanche et al. 2007). Given the frequency of chromosome NDJ observed in a dose-sensitive manner in bubR1D1326N trans-heterozygotes, we decided to perform a genetic screen to identify third-chromosome haploinsufficient synthetic modifiers of the NDJ phenotype. We could test 65 of the 96 deficiencies that are part of the third chromosome deficiency kit generated by the Bloomington Drosophila Stock Center. We could not test all of them due to the presence of haplolethal and haplosterile loci (Marygold et al. 2007) in some deficiencies. Within the 60% of the euchromatin screened, we identified enhancers and suppressors of bubR1 X NDJ. Furthermore, we show that Polo kinase acts as a strong suppressor of female bubR1 X NDJ by delaying meiotic prophase I progression. Interestingly, we also found deficiencies that cause synthetic lethality or affect the fertility of the mutant females. Thus, this screen proved to be highly sensitive for the identification of modifiers of the BubR1 kinase activity.
Ideally, this genetic screen should test simultaneously all mutant genetic backgrounds to minimize variables, such as temperature, humidity and food. However, because the genotype used to test NDJ only accounts for 1/6 of the total female F2 generation, all the third-chromosome deficiencies could not be tested at the same time. Therefore, we performed 16 different experiments. To classify the deficiencies as enhancer, suppressor, or having no effect, we took into account the variation in X NDJ observed in the control genotype and then performed a statistical analysis using the multinomial-Poisson hierarchy model (Zeng et al. 2010) with a 95% confidence interval.
From the 65 deficiencies tested, we identified six deficiencies that enhance and 18 deficiencies that suppress female bubR1 X NDJ. From these 24 positive hits, we narrowed down the genomic region of interest using overlapping deficiencies for three enhancers [Df(3R)D605, Df(3L)Pc-2q and Df(3L)BSC13], and two suppressors [Df(3R)ED5559 and Df(3L)rdgC-co2] to identify the genes responsible for the modification of the X NDJ frequency. We identified the gene responsible for modification of X NDJ for two deficiencies: Df(3L)BSC13 and Df(3L)rdgC-co2. Further studies are needed to identify among the subset of candidate genes, those responsible for the modification of the female X NDJ for the other deficiencies.
Despite the difficulties in identifying the gene involved in the process of X NDJ for every positive interaction, our genetic strategy allowed us to identify Polo kinase as a suppressor of bubR1 X NDJ. Our previous results (Malmanche et al. 2007) illustrate an essential requirement for BubR1 kinase activity in the timing of the early stages of meiotic prophase I, suggesting that BubR1 and Polo have opposite functions in early prophase I. BubR1 appears to slow down progression by maintaining the SC in place, whereas Polo accelerates the process by driving prophase I forward. Through indirect immunofluorescence, we confirmed that Polo dosage antagonizes BubR1 function during the early stages of meiotic prophase I, first by increasing SC and sister chromatin cohesion maintenance, and second by allowing a higher efficiency of bivalent reorganization during karyosome formation, as observed by the loading of the condensin subunit SMC2 at pachytene-diplotene transition.
Our genetic screen also allowed the identification of two deficiencies that impair BubR1 kinase activity in somatic cells and zygotic development. It has been recently shown that BubR1 kinase activity is not essential for accurate somatic chromosome segregation and animal development, because BubR1 kinase dead homozygous adult flies lack any obvious phenotype and are recovered in a normal Mendelian proportion (Malmanche et al. 2007; Elowe et al. 2010). Nonetheless, although these results strongly suggest that BubR1 kinase activity is not essential for somatic development, our findings indicate that subtle modifications in the dosage of other key components of the cell cycle machinery can trigger an essential requirement for BubR1 kinase activity.
Taken together, our results suggest that the genetic screen strategy presented above is highly efficient in recovering genes that interact with BubR1 kinase function. Our results allowed us to identify deficiencies showing genetic interactions, spanning from full recovery of X NDJ to sterility, the strongest effect expected for genes positively involved in BubR1 kinase activity. In addition, the screen also revealed an unexpected result, because it identified deficiencies that impaired zygotic development of BubR1 kinase dead embryos, suggesting a somatic phenotype. Further genetic and cytological experiments will allow the identification of genes responsible for the effect observed in the characterized deficiencies, allowing the build up of a genetic map of essential genes involved in BubR1 kinase function.
Supplementary Material
Acknowledgments
We thank R. Scott Hawley for the mutant mtrm126 fly stock and anti-C(3)G antibody. N.M. was supported by a postdoctoral grant (SFRH/BDP/20888/2004). S.S.G. was supported by a Ph.D. studentship (SFRH/BD/39865/2007) from Fundação para a Ciência e a Tecnologia (FCT). The laboratory of C. E. Sunkel was funded by grants from the FCT in Portugal.
Literature Cited
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., 2002. Molecular Biology of the Cell. Garland, New York [Google Scholar]
- Baker D. J., Jeganathan K. B., Malureanu L., Perez-Terzic C., Terzic A., et al. , 2006. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B. C., et al. , 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146: 13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D., Knoblich J. A., 2002. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 12: 640–647 [DOI] [PubMed] [Google Scholar]
- Bernard P., Hardwick K., Javerzat J. P., 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. H., Shevchenko A., Mann M., Murray A. W., 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheslock P. S., Kemp B. J., Boumil R. M., Dawson D. S., 2005. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat. Genet. 37: 756–760 [DOI] [PubMed] [Google Scholar]
- Clarke A. S., Tang T. T., Ooi D. L., Orr-Weaver T. L., 2005. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev. Cell 8: 53–64 [DOI] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., et al. , 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., et al. , 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson M. M., Tavares A. A., Ohkura H., Deak P., Glover D. M., 2001. Metaphase arrest with centromere separation in polo mutants of Drosophila. J. Cell Biol. 153: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Dulla K., Uldschmid A., Li X., Dou Z., et al. , 2010. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J. Cell Sci. 123: 84–94 [DOI] [PubMed] [Google Scholar]
- Fang Y., Liu T., Wang X., Yang Y. M., Deng H., et al. , 2006. BubR1 is involved in regulation of DNA damage responses. Oncogene 25: 3598–3605 [DOI] [PubMed] [Google Scholar]
- Glover D. M., Leibowitz M. H., McLean D. A., Parry H., 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81: 95–105 [DOI] [PubMed] [Google Scholar]
- Gorbsky G. J., Chen R. H., Murray A. W., 1998. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 141: 1193–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., et al. , 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D., Orme C., Kramer J., Namba L., Champion M., et al. , 2003. A deficiency screen of the major autosomes identifies a gene (matrimony) that is haplo-insufficient for achiasmate segregation in Drosophila oocytes. Genetics 165: 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L., Davenport J., Neale G., Goorha R., 2005. The mitotic checkpoint gene BubR1 has two distinct functions in mitosis. Exp. Cell Res. 308: 85–100 [DOI] [PubMed] [Google Scholar]
- Hartman T. K., Wengenack T. M., Poduslo J. F., van Deursen J. M., 2007. Mutant mice with small amounts of BubR1 display accelerated age-related gliosis. Neurobiol. Aging. 28: 921–927 [DOI] [PubMed] [Google Scholar]
- Homer H., Gui L., Carroll J., 2009. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science 326: 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T., 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517 [DOI] [PubMed] [Google Scholar]
- Ivanovska I., Khandan T., Ito T., Orr-Weaver T. L., 2005. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 19: 2571–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A., Hollmann M., Schafer M. A., 2007. The fatty acid elongase NOA is necessary for viability and has a somatic role in Drosophila sperm development. J. Cell Sci. 120: 2924–2934 [DOI] [PubMed] [Google Scholar]
- Lampson M. A., Kapoor T. M., 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7: 93–98 [DOI] [PubMed] [Google Scholar]
- Lantz V., Chang J. S., Horabin J. I., Bopp D., Schedl P., 1994. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8: 598–613 [DOI] [PubMed] [Google Scholar]
- Lee B. H., Amon A., 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300: 482–486 [DOI] [PubMed] [Google Scholar]
- Lee S., Zhou L., Kim J., Kalbfleisch S., Schock F., 2008. Lasp anchors the Drosophila male stem cell niche and mediates spermatid individualization. Mech. Dev. 125: 768–776 [DOI] [PubMed] [Google Scholar]
- Li R., Murray A. W., 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531 [DOI] [PubMed] [Google Scholar]
- Li Y., Benezra R., 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science 274: 246–248 [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J. M., Lopes C., Amorim I., et al. , 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 117: 1757–1771 [DOI] [PubMed] [Google Scholar]
- Malmanche N., Owen S., Gegick S., Steffensen S., Tomkiel J. E., 2007. Drosophila BubR1 is essential for meiotic sister-chromatid cohesion and maintenance of synaptonemal complex. Curr. Biol. 17: 1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., Honda S., Hara T., Nitta M., Hirota T., et al. , 2003. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278: 51786–51795 [DOI] [PubMed] [Google Scholar]
- Marygold S. J., Roote J., Reuter G., Lambertsson A., Ashburner M., et al. , 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8: R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Baker D. J., d'Uscio L. V., Mozammel G., Katusic Z. S., et al. , 2007. Aging-associated vascular phenotype in mutant mice with low levels of BubR1. Stroke 38: 1050–1056 [DOI] [PubMed] [Google Scholar]
- Mirouse V., Formstecher E., Couderc J. L., 2006. Interaction between Polo and BicD proteins links oocyte determination and meiosis control in Drosophila. Development 133: 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., Hawley R. S., 2001. (3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Nagy O., Menesi D., Udvardy A., Deak P., 2007. Structurally related TPR subunits contribute differently to the function of the anaphase-promoting complex in Drosophila melanogaster. J. Cell Sci. 120: 3238–3248 [DOI] [PubMed] [Google Scholar]
- Perez-Mongiovi D., Malmanche N., Bousbaa H., Sunkel C., 2005. Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 132: 4509–4520 [DOI] [PubMed] [Google Scholar]
- Portier N., Audhya A., Maddox P. S., Green R. A., Dammermann A., et al. , 2007. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev. Cell 12: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani Z., Gagou M. E., Lefebvre C., Emre D., Karess R. E., 2009. Separating the spindle, checkpoint, and timer functions of BubR1. J. Cell Biol. 187: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick T. D., Dej K. J., Xiang Y., Hawley R. S., Ahn C., et al. , 2009. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics 181: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidou E., Cobbe N., Steffensen S., Cotterill S., Heck M. M., 2005. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell Sci. 118: 2529–2543 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., Hawley R. S., 2000. Drosophila Protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sunkel C. E., Glover D. M., 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89(Pt 1): 25–38 [DOI] [PubMed] [Google Scholar]
- Taylor S. S., Ha E., McKeon F., 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., McKeon F., 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89: 727–735 [DOI] [PubMed] [Google Scholar]
- Wei L., Liang X. W., Zhang Q. H., Li M., Yuan J., et al. , 2010. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle 9: 1112–1121 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Takeo S., Florens L., Hughes S. E., Huo L. J., et al. , 2007. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol. 5: e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. G., Koshland D., 2005. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123: 397–407 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Li H., Schweppe N. M., Hawley R. S., Gilliland W. D., 2010. Statistical analysis of nondisjunction assays in Drosophila. Genetics 186: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.