Abstract
In Neurospora crassa, genes lacking a pairing partner during meiosis are suppressed by a process known as meiotic silencing by unpaired DNA (MSUD). To identify novel MSUD components, we have developed a high-throughput reverse-genetic screen for use with the N. crassa knockout library. Here we describe the screening method and the characterization of a gene (sad-3) subsequently discovered. SAD-3 is a putative helicase required for MSUD and sexual spore production. It exists in a complex with other known MSUD proteins in the perinuclear region, a center for meiotic silencing activity. Orthologs of SAD-3 include Schizosaccharomyces pombe Hrr1, a helicase required for RNAi-induced heterochromatin formation. Both SAD-3 and Hrr1 interact with an RNA-directed RNA polymerase and an Argonaute, suggesting that certain aspects of silencing complex formation may be conserved between the two fungal species.
Keywords: epigenetics, meiosis, MSUD, RNA interference
In the filamentous fungus Neurospora crassa, the incomplete nature of septa (cross walls) allows organelles and other cytoplasmic factors to spread throughout the entire colony (Glass et al. 2000). This lifestyle makes N. crassa especially vulnerable to attack by viruses and other repetitive elements. Perhaps for this reason, several surveillance systems have been established in this fungus. These include quelling (Romano and Macino 1992), repeat-induced point mutation (RIP; Cambareri et al. 1989), and meiotic silencing by unpaired DNA (MSUD; Shiu et al. 2001).
MSUD occurs when homologous genes are not paired during prophase I of meiosis. Such unpairing events can be caused by gene deletions, duplications, and transpositions, for example. Since its discovery, only five MSUD proteins have been identified, and all of them localize in the perinuclear region. These are SAD-1, an RNA-directed RNA polymerase (RdRP); DCL-1, a Dicer-like RNase III enzyme; SMS-2, an Argonaute-family protein; SAD-2, a protein regulating SAD-1 localization; and QIP, an exonuclease (Shiu and Metzenberg 2002; Alexander et al. 2008; Lee et al. 2003; Shiu et al. 2006; Xiao et al. 2010; Lee et al. 2010b; Kelly and Aramayo 2007). MSUD appears to involve the production of a single-stranded aberrant RNA from an unpaired segment. This RNA is exported to the perinuclear region where it acts as a template for the SAD-1–mediated double-stranded (ds)RNA synthesis. DCL-1 then processes the dsRNA into small interfering (si)RNA, which guide SMS-2 to identify and slice complementary mRNA. In our working model, SAD-2 is a protein that interacts with SAD-1 and helps transport it to the perinuclear region, whereas QIP is an exonuclease that removes the passenger strand of an siRNA duplex.
MSUD efficiency can be assayed by certain ascus (spore sac) or ascospore (meiotic spore) phenotypes in specifically designed testcrosses. Two commonly used phenotypes are ascospore shape and color, which can be manipulated through MSUD with artificial unpairing of the Round spore (r+) gene and the Ascospore maturation-1 (asm-1+) gene, respectively (Shiu et al. 2001). Expression of r+ is required for the production of American football–shaped ascospores, and its unpairing/suppression produces round ascospores. On the other hand, silencing of the asm-1+ gene results in white (inviable) ascospores instead of the normal black-pigmented ones.
In previous attempts to identify MSUD suppressors, UV and insertional mutageneses were used in forward genetic screens for mutants that suppress the silencing of unpaired r+ and asm-1+ (Shiu and Metzenberg 2002; Shiu et al. 2006). The two genes discovered through these efforts were sad-1+ and sad-2+. The mutants identified by these methods have a modified sequence of a sad gene that fails to pair with its homolog, thereby creating a self-silencing effect and compromising the silencing mechanism. The identification of SAD-1 as an RdRP pointed to the other hallmark RNAi proteins as potential MSUD players, leading to the discovery of other members of the silencing machinery (SMS-2, DCL-1, and QIP). All of the aforementioned MSUD proteins are required for sexual development, although some are needed at earlier time points than others (e.g., DCL-1 and QIP; Alexander et al. 2008; Xiao et al. 2010).
To increase the pace of our MSUD gene discovery, we have utilized an alternative screening method. This system uses a high-throughput reverse-genetic approach, which has been made possible by the construction of the N. crassa knockout library (Colot et al. 2006). This library consists of a genome-wide collection of knockout strains, each represented by a conidial (asexual spore) suspension in 1–2 wells (single or both mating types) of a 96-well microtiter plate. The screening of the knockout library for MSUD suppressors involves the transferring of conidia from a library plate to a tester strain cultured in a 96-well plate. The resulting miniature crosses can be analyzed directly with low magnification microscopy. By using this method, it is possible to systematically screen most of the genome for genes that are important for MSUD. Here we present our screening method and its utility with the identification and characterization of a novel MSUD gene sad-3.
Materials and Methods
Strains and culture media
The fungal strains used in this study are listed in Table 1. The N. crassa knockout library was obtained from the Fungal Genetics Stock Center (FGSC; McCluskey et al. 2010). N. crassa sequences can be found at http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html. MSUD tester strains were designed to unpair and silence genes that are important for the ascospore phenotype. Vegetative cultures were maintained on Vogel’s medium (Vogel 1956), and crosses were performed on synthetic crossing medium (SC) of Westergaard and Mitchell (1947). Growth media for strains with auxotrophic markers were supplemented as required (Perkins et al. 2001).
Table 1 . Strains used in this study.
| Strain | Genotype |
|---|---|
| F2-01 | fl A (FGSC 4317) |
| F2-14 | fl a |
| F2-23 | rid; fl A |
| F2-26 | rid; fl a |
| F2-27 | rid r∆::hph; fl a |
| F2-29 | rid r∆::hph; fl A |
| F2-37 | his-3+::act+; fl a |
| F2-38 | his-3+::BmlR; fl a |
| F3-23 | rid his-3+::asm-1; fl; asm-1∆::hph A |
| F3-24 | rid his-3+::asm-1; fl; asm-1∆::hph a |
| F4-31 | rid gfp-sad-3::hph; fl A |
| F5-22 | rid his-3 yfpn-sad-3::hph; fl A |
| F5-23 | fl A |
| F5-24 | sad-1Δ::hph; fl A |
| F5-25 | sad-3Δ::hph; fl A |
| F5-26 | fl; qde-1Δ::hph A |
| P3-07 | Oak Ridge wild type A (FGSC 2489) |
| P3-08 | Oak Ridge wild type a (FGSC 2490) |
| P3-25 | mep sad-1∆::hph a |
| P7-26 | sad-1∆::hph rid his-3+::sad-1-yfpc a |
| P8-01 | sad-2Δ::hph A |
| P8-02 | sad-2Δ::hph a |
| P8-18 | mep sad-1∆::hph A |
| P8-42 | rid his-3; mus-51∆::bar a |
| P8-43 | rid his-3; mus-52∆::bar A |
| P8-65 | csr-1Δ::hph a |
| P10-15 | rid his-3 A |
| P13-19 | rid his-3+::rfp-qip; qip∆::hph; mus-51∆::bar a |
| P13-59 | sad-1∆::hph rid his-3+::sad-1-rfp a |
| P14-27 | sad-3Δ::hph a (FGSC 19729) |
| P14-28 | sad-3Δ::hph A (FGSC 19730) |
| P15-06 | rid his-3+::rfp-sad-2 sad-3-gfp::hph a |
| P15-07 | rid his-3+::rfp-sad-2 sad-3-gfp::hph A |
| P15-08 | rid his-3+::rfp-sms-2 sad-3-gfp::hph a |
| P15-09 | rid his-3+::rfp-sms-2; sms-2∆::hph A |
| P15-71 | rid his-3+::sad-2-yfpc yfpn-sad-3::hph a |
| P15-72 | rid his-3+::sad-2-yfpc yfpn-sad-3::hph; inv sad-2RIP32 A |
| P16-01 | rid his-3+::yfpc yfpn-sad-3::hph; mus-51∆::bar A |
| P16-12 | rid yfpn-sad-3::hph; mus-52∆::bar; yfpc-sms-2::hph A |
| P16-13 | rid yfpn-sad-3::hph; mus-52∆::bar; yfpc-sms-2::hph a |
| P16-14 | rid yfpn-sad-3::hph; mus-52∆::bar qip-yfpc::hph A |
| P16-15 | rid yfpn-sad-3::hph; mus-52∆::bar qip-yfpc::hph a |
| P16-16 | rid his-3+::yfpc yfpn-sad-3::hph; mus-51∆::bar a |
| P16-17 | a |
| P16-18 | a |
| P16-19 | sad-3Δ::hph a |
| P16-20 | sad-3Δ::hph a |
| P16-21 | sad-1Δ::hph a |
| P16-22 | qde-1Δ::hph a |
| P16-23 | sad-3Δ::hph A |
| P16-24 | sad-3Δ::hph A |
| P16-25 | A |
| P16-26 | A |
| P16-27 | his-3 sad-3Δ::hph; mus-51Δ::bar a |
| P16-28 | his-3 sad-3Δ::hph A |
Description of genetic loci can be found in the N. crassa e-Compendium (http://bmbpcu36.leeds.ac.uk/~gen6ar/newgenelist/genes/gene_list.htm).
Library screening protocol
To prepare the female testers, 96-well V-bottom microtiter plates (Corning, Lowell, MA) were first filled with molten SC (200 µl per well). After solidification, a 96-pin replicator was used to transfer mycelia (vegetative cells) from a water suspension of the desired tester strain. The plates were incubated at room temperature, and protoperithecia (female mating structures) were allowed to develop for five days. With the aid of a replicator, protoperithecia were fertilized by transferring conidia from a slightly thawed library plate to the wells of the tester plate. Plates were then placed in humid chamber at room temperature until ascospores could be seen on the lids (∼11 days). A dissecting microscope was then used to determine the ascospore phenotype.
Quantitative analysis of MSUD suppression and ascospore production
rΔ (F2-29) and asm-1Δ (F3-23) testers, as well as other control strains, were cultured for 6 days on SC in 60 mm culture plates at room temperature for use as designated females. Conidia from each male were suspended in sterile water and adjusted to a concentration of 1000 counts per microliter. For fertilization, 3 × 33 µl aliquots of the conidial suspensions were inoculated across the surface of a female tester (in three replicas, each covering roughly the same amount of area). Ascospores were collected from the lids 21 days postfertilization (dpf) and analyzed on a hemocytometer under magnification.
N. crassa transformations
Transformations were performed by electroporation of conidia using standard techniques (Margolin et al. 1997). For localization studies, a fluorescent protein tagging protocol based on double-joint PCR was used to construct the sad-3 tagging vectors (Hammond et al. 2011). The primers used in this process are listed in supporting information, Table S1. P8-42 or P8-43 was used as a background strain to create transformants expressing SAD-3 tagged with the green-fluorescent protein (GFP), the N-terminal fragment of the yellow-fluorescent protein (YFPN), or the C-terminal fragment of the YFP (YFPC).
Genotype screening and strain confirmation
Genomic DNA was isolated with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). PCR-based confirmation of genotypes was performed with the Expand Long Range dNTPack (Roche Applied Science, Indianapolis, IN).
Photography and microscopy
A Canon Power Shot S3 IS digital camera was used to take photographs. The camera was combined with a VanGuard 1231CM or 1274ZH microscope when a sample (e.g., rosettes of asci) required magnification. For fluorescent microscopy, asci were harvested from perithecia (fruiting bodies) and prepared as previously reported (Alexander et al. 2008). Prescreening of crosses for fluorescent signal generation was typically performed with a Zeiss Universal microscope. Confocal microscopy was performed on a Zeiss LSM710 microscope. Visualization of the YFP was achieved by using a 514 nm Argon laser line for excitation, and the detector was set to collect emission at 535–600 nm. Protocols for GFP, RFP, and DAPI visualization were essentially as described (Xiao et al. 2010).
Results
Library screening links NCU09211 deletions to MSUD suppression
The high-throughput screen for gene deletions that suppress MSUD is robust, with most crosses producing ascospore levels sufficient for the analysis. Essentially, this method allows up to 96 unique strains to be assayed on a single crossing plate. As an example, Table 2 summarizes the mating success (perithecial level) and cross productivity (ascospore level) between candidates from a library plate and an rΔ tester. Most of the wells on this plate yielded high levels (∼100%) of round ascospores, typical of an r-silenced cross. However, crosses in wells E8 and E9 each produced low levels of ascospores, with a relatively high percentage (>80%) of American football–shaped progeny. Accordingly, in crosses to an asm-1Δ tester, where MSUD-proficient strains produced ∼100% white inviable ascospores, wells E8 and E9 yielded a relatively high percentage (>25%) of black ascospores. The two positive wells in this case contain opposite mating types of NCU09211Δ. On the off chance that contamination of wells E8 and E9 could have occurred, the NCU09211Δ strains were crossed to standard wild-type strains to obtain pure homokaryotic progeny. These pure cultures were subsequently used in a thorough characterization.
Table 2 . Mating and cross efficiency between knockout strains from a library plate and a rΔ tester (F2-27 or F2-29).
| 57 mat a strains crossed to the rΔ A tester | |
|---|---|
| No. of strains at a given perithecial level | No. of strains at a given ascospore level |
| P>50 = 44 | A>200 = 53 |
| P≤50 = 11 | A≤200 = 1 |
| P≤20 = 2 | A≤50 = 3 |
| P0 = 0 | A0 = 0 |
| 39 mat A strains crossed to the rΔ a tester | |
|---|---|
| No. of strains at a given perithecial level | No. of strains at a given ascospore level |
| P>50 = 37 | A>200 = 38 |
| P≤50 = 1 | A≤200 = 0 |
| P≤20 = 1 | A≤50 = 0 |
| P0 = 0 | A0 = 1 |
Perithecial and ascospore productivity were scored on a 4-level scale. For example, 44 mat a knockout strains yielded >50 perithecia when mated with the rΔ A tester.
NCU09211Δ is a suppressor of MSUD
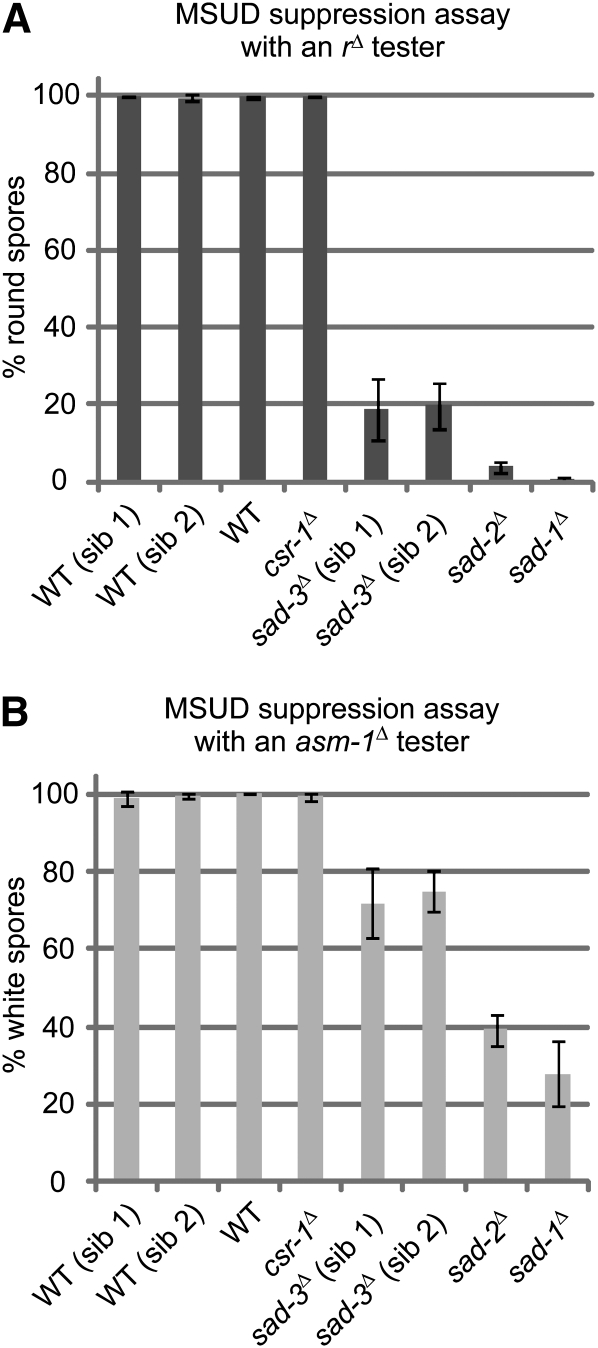
To determine if NCU09211Δ is truly a suppressor of MSUD, progeny from a cross of NCU09211Δ a × fl A were used in a quantitative assay of MSUD suppression. As expected, all MSUD-proficient (negative control) strains yielded nearly 100% round ascospores when crossed to an rΔ tester (Figure 1A). These include a standard wild-type strain, two wild-type NCU09211+ siblings, and a strain deleted for a non-MSUD gene, csr-1 (Bardiya and Shiu 2007). On the other hand, the two MSUD-deficient (positive control) strains, sad-1Δ and sad-2Δ, produced mostly American football–shaped ascospores in the same test cross. Crosses of the NCU09211Δ strains to an rΔ tester yielded ∼20% round spores, a level significantly lower than that of a negative control. Similar results were obtained for crosses of the same strains to an asm-1Δ tester (Figure 1B). Furthermore, crosses of the NCU09211Δ strain to two additional MSUD testers, ::act+ and ::bmlR (which induce the unpairing of actin and β-tubulin by an ectopic transgene, respectively), show that it can dominantly suppress their silencing (Table 3). These results demonstrate that the NCU09211Δ strains used in this study are indeed suppressors of MSUD. NCU09211 is referred to as sad-3+ (suppressor of ascus dominance-3) hereafter.
Figure 1 .
Deletions of sad-3+ (NCU09211) correlate with an MSUD-deficient phenotype. (A) An rΔ tester (F2-29) was used as the designated female in crosses with eight strains (three replicas each). The four MSUD-proficient controls include two wild-type (WT) siblings (sib) of sad-3Δ (P16-17 and P16-18), a standard wild-type strain (P3-08), and an hph-based (hygromycin-resistant) deletion strain (csr-1Δ; P8-65). The two experimental strains are sad-3Δ siblings (P16-19 and P16-20). The two MSUD-deficient controls are sad-1Δ and sad-2Δ (P3-25 and P8-02). A normal cross produces black American football (spindle)-shaped ascospores. When MSUD is active, crosses involving an rΔ tester yield nearly 100% round spores (i.e., r+ is silenced). When MSUD is suppressed (e.g., in crosses involving sad-1, 2, and 3 mutants), the percentage of round spores decreases. Three replicate crosses were performed for each male-female combination. Error bars represent the standard deviation. (B) An asm-1Δ tester (F3-23) was used as the designated female. When MSUD is active, crosses involving an asm-1Δ tester yield nearly 100% white (inviable) spores (i.e., asm-1+ is silenced). When MSUD is suppressed (e.g., in crosses involving sad-1, 2, and 3 mutants), the percentage of white spores decreases. The results demonstrate that sad-3Δ, like sad-1Δ and sad-2Δ, decreases the efficiency of MSUD.
Table 3 . Silencing by ectopic transgenes and its suppression.
| Parent 1 (mat A): mutation at sad-1 or sad-3 | Parent 2 (mat a): ectopic insertion at his-3 (::) | Unpaired gene | Ascospore count (× 103) |
|---|---|---|---|
| Wild type | No insertions | – | 415 |
| ::act+ | actin | 28.8 | |
| ::BmlR | β-tubulin | 0.612 | |
| sad-1∆ | No insertions | - | 209 |
| ::act+ | actin | 358 | |
| ::BmlR | β-tubulin | 399 | |
| sad-3∆ | No insertions | - | 29.9 |
| ::act+ | actin | 74.3 | |
| ::BmlR | β-tubulin | 59.7 |
Meiotic silencing of unpaired actin or β-tubulin leads to the reduced production of ascospores. Such reduction is not observed in crosses carrying sad-1∆ or sad-3∆. Strains used in this experiment include F2-14, F2-37, F2-38, P3-07, P8-18, and P14-28.
sad-3 encodes a putative eukaryotic helicase
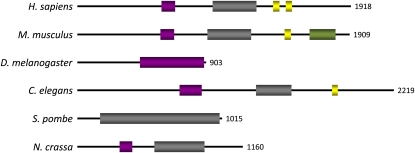
Three of the previously discovered MSUD proteins (DCL-1, SAD-1, and SMS-2) have orthologs in other eukaryotes (e.g., Caenorhabditis elegans and Schizosaccharomyces pombe), possibly because they are core components of RNA silencing. Like the aforementioned, SAD-3 also has related proteins in a wide range of organisms (Figure 2). However, except for S. pombe Hrr1 (Motamedi et al., 2004), none of the indicated orthologs has been extensively characterized. A search of NCBI's conserved domain database with the predicted SAD-3 sequence finds two helicase domains, suggesting that SAD-3 may possess the ability to unwind nucleic-acid strands.
Figure 2 .
SAD-3 is a putative eukaryotic helicase. GenBank identification numbers and BLASTP E-values for each protein (relative to Neurospora crassa SAD-3) are as follows: Homo sapiens, GI:28626521, 1e-46; Mus musculus, GI:124487311, 3e-49; Drosophila melanogaster, GI:24649577, 4e-09; Caenorhabditis elegans, GI:17538027, 5e-21; Schizosaccharomyces pombe, GI:19075911, 6e-68. Purple and gray, helicase domains; yellow, zinc finger; green, SMC (structural maintenance of chromosomes) domain.
SAD-3 is required for sexual development
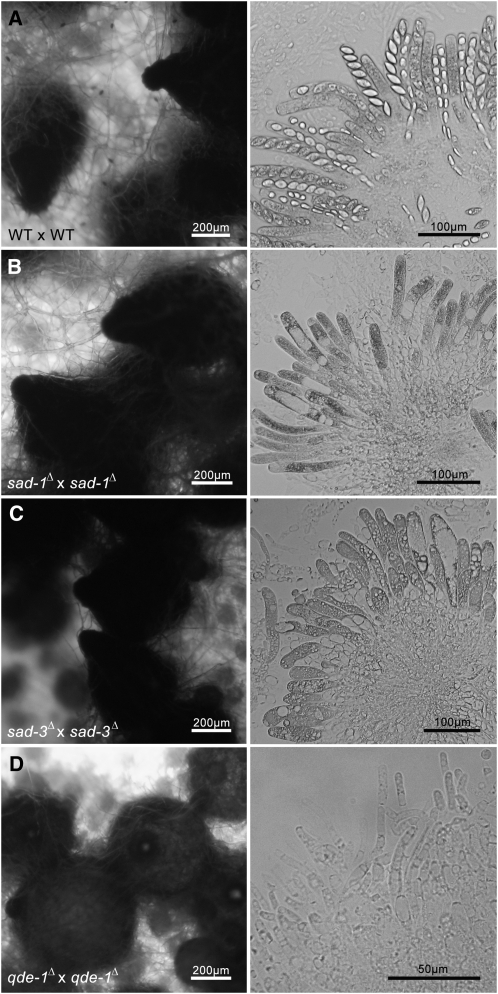
The previously characterized MSUD genes are all required for sexual development. For example, when both parents of a cross carry either sad-1Δ or sad-2Δ, perithecia with normal beaks and rosettes with elongated asci are produced, but no ascospores are made (Shiu et al. 2001, 2006). Crosses lacking a functional dcl-1 or qip gene are also barren, but the resulting perithecia have underdeveloped beaks and no recognizable asci (Alexander et al. 2008; Xiao et al. 2010). Our analysis of sad-3Δ suggests that it is also required for sexual development. Crosses homozygous for sad-3Δ produce perithecia with normal beaks and elongated asci but no ascospores (Figure 3). In other words, SAD-3 plays an important role in ascospore development.
Figure 3 .
sad-3Δ × sad-3Δ crosses produce perithecia with normal beaks and pigmentation, but no ascospores. Perithecia (left) and their contents (right) are shown for various homozygous crosses (sad-3Δ and three controls). (A) A WT cross (F5-23 × P16-17) produces normal perithecia and rosettes of ascospores. (B) A sad-1Δ cross (F5-24 × P16-21) produces elongated asci with no ascospores. (C) A sad-3Δ cross (F5-25 × P16-20) also produces elongated asci with no ascospores. (D) A qde-1Δ cross (F5-26 × P16-22) produces perithecia with underdeveloped beaks and no recognizable asci (right: contents scraped from the inside of the perithecial walls are shown.) qde-1 encodes an RNA polymerase required for vegetative silencing (Quelling; Lee et al. 2010a).
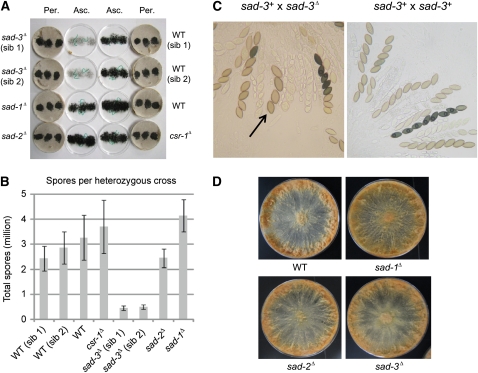
To determine the effect of sad-3Δ on sexual development in heterozygous crosses, ascospore levels were quantified in crosses between sad-3Δ and wild-type strains. As expected, wild-type crosses yielded abundant ascospores, all within a similar range of 2.4–3.3 million ascospores per cross (Figure 4A, B). In contrast, a sad-3Δ × WT (wild type) cross produced far fewer progeny (∼0.5 million or 18% of WT level). As a low number of recoverable ascospores in a cross could be due to a defect in spore ejection (and not spore production), rosettes of sad-3Δ × WT asci were analyzed.
Figure 4 .
SAD-3 is required for sexual development. (A) Crosses heterozygous for sad-3Δ produce a low level of shot progeny. The lids [containing ascospores (Asc.)] and crossing plates [containing perithecia (Per.)] from various crosses are shown. A standard female (F2-23) and eight males (same as Figure 1) were used. (B) Quantitative analysis of shot ascospores from crosses depicted in A. Error bars represents the standard deviation among three replicates. (C) Pictures of rosettes from sad-3+ (F2-01) × sad-3Δ (P16-19) and from sad-3+ (F2-01) × sad-3+ (P3-08). Four-spored asci are often observed in a cross heterozygous for sad-3Δ (arrow). See Figure S1 for additional pictures. (D) Conidia (2 × 104) from WT (P3-07), sad-1Δ (P8-18), sad-2Δ (P8-01), and sad-3Δ (P16-24) were each inoculated onto the center of a 150-mm Vogel’s plate and incubated for seven days at room temperature. There are no obvious morphological differences between WT and any of the MSUD-suppressing (sad) strains in the vegetative phase.
Compared with wild-type crosses, rosettes heterozygous for sad-3Δ produce mostly aborted asci. Asci that are not aborted sometimes contain four large or otherwise deformed ascospores, instead of the normal eight (Figure 4C). This indicates that the low ascospore count in heterozygous crosses is due to a combination of ascus abortion and a reduction in the number of ascospores per ascus. This aspect of sad-3Δ is interesting in light of the fact that the other characterized MSUD mutant alleles do not appear to affect sexual development dramatically in heterozygous crosses. To ensure these unique results were not due to an unknown (and closely linked) mutation in the sad-3Δ strains, sad-3+ was deleted independently in two additional strains. Analysis of the new deletion strains showed similar results, confirming that the aberrant sexual phenotypes were due to sad-3Δ and not to a closely linked mutation (Figure S1). sad-3Δ strains are normal during the vegetative phase. Linear growth and vegetative morphology assays revealed no significant differences between sad-3Δ and wild-type strains (Figure 4D and Figure S2).
SAD-3 colocalizes with other members of the MSUD machinery
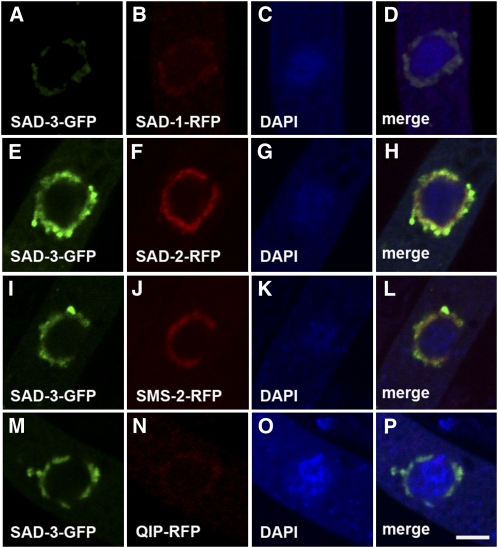
To determine the subcellular localization of its protein product, sad-3 was tagged with the GFP sequence at its native locus. The sad-3-gfp strains are fertile, suggesting the tag does not affect the normal function of SAD-3. Microscopic analysis revealed that SAD-3 is localized in the perinuclear region, similar to the previously characterized MSUD proteins (Figure 5A). To determine if SAD-3 colocalizes with these proteins, an analysis was performed by expressing sad-3-gfp with rfp-tagged versions of sad-1, sad-2, sms-2, and qip in a cross. The results demonstrate that SAD-3 indeed colocalizes with other components of the MSUD machinery (Figure 5). It is conceivable that SAD-3 physically interacts with these proteins and is part of a meiotic silencing complex.
Figure 5 .
SAD-3 colocalizes with SAD-1, SAD-2, SMS-2, and QIP in the perinuclear region. Micrographs illustrate prophase asci expressing (A–D) sad-3-gfp and sad-1-rfp (F4-31 × P13-59), (E–H) sad-3-gfp and sad-2-rfp (P15-06 × P15-07), (I–L) sad-3-gfp and sms-2-rfp (P15-08 × P15-09), and (M–P) sad-3-gfp and qip-rfp (F4-31 × P13-19). The chromatin was stained with DAPI. Bar, 5 μm.
Interaction study suggests that SAD-3 is part of an MSUD protein complex
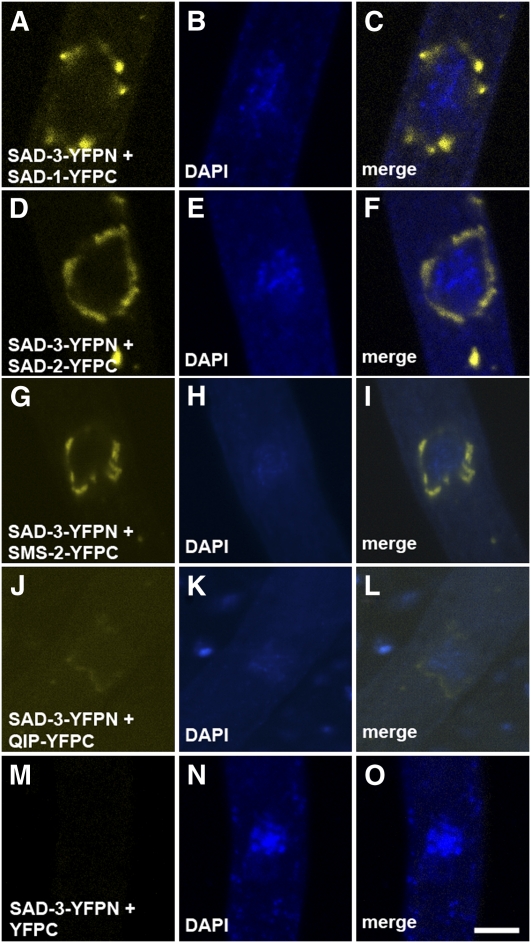
Bimolecular fluorescence complementation (BiFC) analysis is an in vivo assay of protein-protein interactions (Hu et al. 2002). Essentially, two proteins of interest are tagged with nonfunctional fragments of a fluorescent protein (e.g., YFP). If the two tagged proteins are in close proximity (e.g., in the same complex), a functional fluorophore may be reconstituted. Using BiFC, we demonstrated the direct interaction between SAD-1 and SAD-2 (Bardiya et al. 2008), and between QIP and SMS-2 (Hammond et al. 2011). In this study, we have found that SAD-3 interacts with SAD-1, SAD-2, SMS-2, and QIP (Figure 6), giving credence to the notion that these proteins form an RNA-processing complex in the perinuclear region.
Figure 6 .
SAD-3 interacts with SAD-1, SAD-2, SMS-2, and QIP in the perinuclear region. BiFC experiments were performed between SAD-3 (tagged with YFPN, the N-terminal fragment of YFP) and other MSUD proteins (tagged with YFPC). In each case, fluorescent signal was observed in the perinuclear region, suggesting that the tagged MSUD proteins have intimate interaction in vivo that allows the formation of a functional fluorophore. SAD-3-YFPN does not yield a fluorescent signal when YFPC is not attached to a SAD-3–interacting protein. Micrographs illustrate prophase asci expressing (A–C) sad-3-yfpn and sad-1-yfpc (F5-22 × P7-26), (D–F) sad-3-yfpn and sad-2-yfpc (P15-71 × P15-72), (G–I) sad-3-yfpn and sms-2-yfpc (P16-12 × P16-13), (J–L) sad-3-yfpn and qip-yfpc (P16-14 × P16-15), and (M–O) sad-3-yfpn and yfpc (P16-01 × P16-16). The chromatin was stained with DAPI. Bar, 5 μm.
Discussion
In this study, we have developed a high-throughput reverse-genetic screen to identify suppressors of MSUD. The methodical gene-by-gene nature of this approach has several advantages over a forward genetic screen (e.g., in time and labor costs). However, this screening method does come with several caveats. First, only annotated genes are included in the knockout library, limiting what can be discovered by this method. Second, some deletions may not suppress MSUD dominantly to a level that is sufficient for detection. Finally, not every compatible fertilization on each plate results in the production of enough ascospores for the visual assay (Table 2). Poor mating or low ascospore count can sometimes result from random technical problems, such as a low medium level in (or poor conidial transfer to) a given well. These potential problems suggest that there may be considerable value in performing multiple screens for each library plate.
SAD-3 is a helicase-domain protein required for MSUD. Loss of sad-3+ by gene deletion is sufficient to induce a dominant MSUD suppression phenotype in crosses to four different MSUD tester strains (each containing a rearranged sexual development gene). This dominant suppression can be explained by the “silencing-the-silencer” model first proposed by Shiu et al. (2001). In this case, a sad-3Δ × sad-3+ cross results in the unpairing of sad-3+ during meiosis, which makes it a target for silencing by the MSUD machinery. Because sad-3+ is part of the MSUD machinery, its suppression leads to a negative feedback and the eventual defeat of the silencing mechanism. The finding of sad-3Δ being an overall weaker dominant suppressor of MSUD (compared with sad-1Δ and sad-2Δ) may suggest that sad-3Δ is a less capable “self-silencer” than the others are. However, suppression strength in the “silencing-the-silencer” assay may be related to the expression profile of each MSUD gene and its product, including protein/mRNA turnover rates and expression timing. It is not necessarily an indicator of the importance of a SAD protein to the mechanism. Interestingly, despite the lower strength of MSUD suppression, dramatic sexual defects were observed in heterozygous sad-3Δ × sad-3+ crosses, an observation unique among all reported MSUD genes to date.
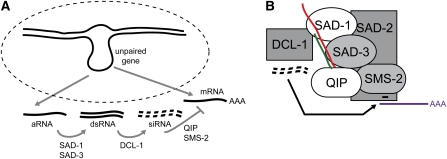
All of the MSUD proteins characterized thus far, including SAD-3, show a perinuclear localization pattern. It seems possible that they form a silencing complex in the perinuclear region, which inspects (and if necessary, processes) RNA molecules as they exit the nucleus. This hypothesis is consistent with the colocalization and BiFC results demonstrated in this and previous studies. Based on the identity of the proteins involved and what we have learned to date, a simple model for MSUD is depicted as follows (Figure 7A). Briefly, recognition of unpaired DNA leads to the production of an aberrant RNA, which is made double-stranded (by SAD-1) and processed into siRNA (by DCL-1) capable of directing mRNA degradation (by SMS-2). SAD-3, a putative helicase, may assist SAD-1 in dsRNA synthesis by increasing its processivity on RNA templates. QIP functions to degrade the passenger strand of an siRNA duplex, whereas SAD-2 may serve as a scaffolding protein and assemble the necessary components in the perinuclear region.
Figure 7 .
A working model for MSUD (see text for details). (A) An unpaired gene triggers the production of an aberrant (a)RNA molecule that is destined for the perinuclear region, a region surrounding the nucleus (dotted line). There, the aRNA is made double-stranded (SAD-1/SAD-3) and processed into siRNA (DCL-1), which are subsequently used to destroy complementary mRNA (QIP/SMS-2). (B) An MSUD complex (as suggested by our BiFC results) is shown interacting with different RNA molecules. SAD-2 is responsible for recruiting SAD-1 (and possibly others) to the perinuclear region. Red, aRNA; green, newly synthesized RNA; black, siRNA; purple, mRNA.
S. pombe Hrr1 is a SAD-3 ortholog required for RNAi-mediated heterochromatin formation (Motamedi et al. 2004). It is a component of the RNA-directed RNA polymerase complex (RDRC) that also includes Rdp1 (an RdRP) and Cid12 (a polyA polymerase family member). RDRC interacts with the RNA-induced transcriptional silencing complex (RITS), which is comprised of the Ago1 Argonaute (an SMS-2 ortholog) and two other proteins. The model for RDRC/RITS assembly involves an RdRP-helicase-Argonaute interaction. This mirrors our BiFC results, which suggest an interaction between SAD-1 and SAD-3, and between SAD-3 and SMS-2. It seems plausible that the complex formations that mediate MSUD and RNAi-induced heterochromatin assembly are related to some degree.
Supplementary Material
Acknowledgments
This work would not have been possible without the efforts of the Neurospora Functional Genomics group and the Fungal Genetics Stock Center. The authors thank James Birchler, members of the Shiu Laboratory, and colleagues from our community for their indispensable help. T.M.H. was supported by a Life Science fellowship from the University of Missouri and a Ruth L. Kirschstein National Research Service Award from the National Institute of General Medical Sciences. This work was supported by National Science Foundation grant MCB0918937 (to P.K.T.S.).
Literature Cited
- Alexander W. G., Raju N. B., Xiao H., Hammond T. M., Perdue T. D., et al. , 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45: 719–727 [DOI] [PubMed] [Google Scholar]
- Bardiya N., Alexander W. G., Perdue T. D., Barry E. G., Metzenberg R. L., et al. , 2008. Characterization of interactions between and among components of the MSUD machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178: 593–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya N., Shiu P. K. T., 2007. Cyclosporin A resistance–based gene placement system for Neurospora crassa. Fungal Genet. Biol. 44: 307–314 [DOI] [PubMed] [Google Scholar]
- Cambareri E. B., Jensen B. C., Schabtach E., Selker E. U., 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244: 1571–1575 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Jacobson D. J., Shiu P. K. T., 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34: 165–186 [DOI] [PubMed] [Google Scholar]
- Hammond T. M., Xiao H., Rehard D. G., Boone E. C., Perdue T. D., et al. , 2011. Fluorescent and bimolecular-fluorescent protein tagging of genes at their native loci in Neurospora crassa using specialized double-joint PCR plasmids. Fungal Genet. Biol. 48: 866–873 [DOI] [PubMed] [Google Scholar]
- Hu C. D., Chinenov Y., Kerppola T. K., 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Aramayo R., 2007. Meiotic silencing and the epigenetics of sex. Chromosome Res. 15: 633–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Pratt R. J., McLaughlin M., Aramayo R., 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C., Aalto A. P., Yang Q., Chang S. S., Huang G., et al. , 2010a The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 8: e1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. W., Millimaki R., Aramayo R., 2010b QIP, a component of the vegetative RNA silencing pathway, is essential for meiosis and suppresses meiotic silencing in Neurospora crassa. Genetics 186: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U., 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36 [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126 [DOI] [PubMed] [Google Scholar]
- Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., et al. , 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Sachs M. S., 2001. The Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego [Google Scholar]
- Romano N., Macino G., 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6: 3343–3353 [DOI] [PubMed] [Google Scholar]
- Shiu P. K. T., Metzenberg R. L., 2002. Meiotic silencing by unpaired DNA: properties, regulation, and suppression. Genetics 161: 1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P. K. T., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916 [DOI] [PubMed] [Google Scholar]
- Shiu P. K. T., Zickler D., Raju N. B., Ruprich-Robert G., Metzenberg R. L., 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103: 2243–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora (Medium N). Microbial Genet. Bull. 13: 42–43 [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577 [Google Scholar]
- Xiao H., Alexander W. G., Hammond T. M., Boone E. C., Perdue T. D., et al. , 2010. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 186: 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.