Abstract
Sex-ratio distorters are X-linked selfish genetic elements that facilitate their own transmission by subverting Mendelian segregation at the expense of the Y chromosome. Naturally occurring cases of sex-linked distorters have been reported in a variety of organisms, including several species of Drosophila; they trigger genetic conflict over the sex ratio, which is an important evolutionary force. However, with a few exceptions, the causal loci are unknown. Here, we molecularly characterize the segmental duplication involved in the Paris sex-ratio system that is still evolving in natural populations of Drosophila simulans. This 37.5 kb tandem duplication spans six genes, from the second intron of the Trf2 gene (TATA box binding protein-related factor 2) to the first intron of the org-1 gene (optomotor-blind-related-gene-1). Sequence analysis showed that the duplication arose through the production of an exact copy on the template chromosome itself. We estimated this event to be less than 500 years old. We also detected specific signatures of the duplication mechanism; these support the Duplication-Dependent Strand Annealing model. The region at the junction between the two duplicated segments contains several copies of an active transposable element, Hosim1, alternating with 687 bp repeats that are noncoding but transcribed. The almost-complete sequence identity between copies made it impossible to complete the sequencing and assembly of this region. These results form the basis for the functional dissection of Paris sex-ratio drive and will be valuable for future studies designed to better understand the dynamics and the evolutionary significance of sex chromosome drive.
Keywords: segmental duplication, meiotic drive, sex-ratio, D. simulans
Meiotic drive is a phenomenon by which one member of a pair of alleles or chromosomes of a heterozygous individual is preferentially transmitted to the next generation—a phenomenon that is in violation of Mendel’s first law (Sandler and Novitski 1957). Many examples of meiotic drive have been reported in fungi, plants, insects, worms, and mammals (Atlan et al. 1997; Lyttle 1991); sex-ratio drive specifically refers to meiotic drive in which the cheater allele is sex linked and is expressed only in heterogametic individuals, resulting in a skewed offspring sex ratio.
X chromosome drive was first observed in males of Drosophila obscura (Gershenson 1928) and has since been documented in a number of dipteran species, mainly within the Drosophila genus (Jaenike 2001). In Drosophila, the sex-ratio phenotype is usually associated with X chromosome rearrangements. Inversions of varying complexity, which presumably keep the elements contributing to the drive together, impede genetic dissection in most of the species (De Carvalho and Klaczko 1992; Dyer et al. 2007; Hauschteck-Jungen 1990; Prakash 1974; Stalker 1961; Voelker 1972). High-resolution genetic mapping has revealed gene/segmental duplication in two inversion-free sex-ratio drive systems in D. simulans: Paris (Montchamp-Moreau et al. 2006) and Winters (Tao et al. 2007a).
Mendelian alleles are favored by natural selection if they increase the fitness of their carriers. However, sex-ratio and other alleles responsible for meiotic drive are selfish genetic elements that can spread in populations as long as their preferential transmission is not offset by strong deleterious effects. The spread of a sex-linked distorter allele causes skewed population sex ratios and triggers an evolutionary arms race at the genome scale. Selective forces favor the evolution of unlinked drive suppressors to equalize the sex ratio (i.e., on the Y chromosome or the autosomes) but also favor alleles that are closely linked to the primary drive locus if they enhance distortion (Fisher 1930; Hamilton 1967). Recurrent genetic conflict over the transmission of sex chromosomes is thought to have profound evolutionary consequences, including epigenetic regulation of sex chromosomes during meiosis, genomic distribution of genes expressed in the germline, change in sex determination, and the evolution of hybrid sterility [discussed in Meiklejohn and Tao (2010) and Werren and Beukeboom (1998)]. The last hypothesis has received empirical support from studies in Drosophila (Phadnis and Orr 2009; Presgraves 2010; Tao et al. 2001). However, information about the underlying molecular mechanisms, necessary to assess the evolutionary significance of sex chromosome drive, is still critically lacking. So far, both distorter and suppressor genes together have been identified only in the Winters sex-ratio system of D. simulans, and the individual function of these genes is still elusive (Tao et al. 2007a; Tao et al. 2007b).
Here, we molecularly dissected the chromosomal region responsible for Paris sex-ratio drive—a textbook case in D. simulans (Jaenike 2008; Mercot et al. 1995). This system is particularly interesting in two ways. First, the etiology of drive is associated with a meiosis phenotype: the loss of Y-bearing sperm results from a disjunction failure of the Y chromosome sister chromatids during the second meiotic division (Cazemajor et al. 2000). Second, the emergence of Paris sex-ratio X chromosomes and the spread of these chromosomes in natural populations have triggered the evolution of autosomal and Y-linked suppressors (Atlan et al. 1997; Jutier et al. 2004). These features of the Paris system provide an opportunity to study the evolutionary impact of the emergence of sex-ratio drive and to identify a network of genes controlling segregation of the sex chromosomes.
In the Paris system, two distinct distorter elements have been fine-mapped to the cytological bands 7E-F of the sex-ratio reference chromosome XSR6: a segmental duplication and a second element located 100–150 kb away (Montchamp-Moreau et al. 2006). We used males carrying XSR6 to produce a library of bacterial artificial chromosomes (BAC). We obtained, assembled, and analyzed a sequence of about 300 kb that contains the two distorter elements. This process allowed us to identify the limits of the segmental duplication and associated repetitive elements. We were also able to shed light on the mechanism and age of the duplication event, as well as the coding potential of the different components of the duplication.
Materials and Methods
Fly stocks
Two types of males were used: (XST8)ST8 males that carry the reference standard XST8 chromosome and (XSR6)ST8 males that carry the reference sex-ratio XSR6 chromosome. Both X chromosomes are in the same ST8 genetic background (drive-suppressor free). To prevent recombination, the X chromosomes were maintained in the male lineage through repeated backcrosses with C(1)RM, y, w (ST8 background) females, as described in Montchamp-Moreau and Cazemajor (2002).
BAC construction, alignment, and annotation
DNA extraction was performed on (XSR6)ST8 males. DNA was partially digested with HindIII and separated on a 1% agarose gel by pulse field gel electrophoresis. 27,648 BACs, each about 70 kb in length, were generated according to the protocols described in Roest Crollius et al. (2000). The BACs were spotted onto nylon membranes. To screen for those covering the sex-ratio domain previously described (Montchamp-Moreau 2006), we used 32P-labeled probes consisting of gene fragments scattered along the whole domain (supporting information, Figure S1). When the clones included a part of the duplication, they were sequenced for Trf2 and/or org-1, for which a known polymorphism was used to discriminate between the two copies (Derome et al. 2008).
The BACs were sequenced by the Genoscope (Evry, France). A library was obtained for each of them after mechanical shearing of DNA and cloning of 3 kb (BAC 10c2 and 67l12) or 5 kb (BAC 58j14, 46o6, 35e19, and 24a6) fragments into a pcdna2.1 plasmid vector (Invitrogen). Additional libraries were prepared from BACs 58j14 and 46o6 by cloning 10 kb fragments into a pCNS plasmid vector (pSU18 derived). All vector DNA was purified and end-sequenced using dye terminator chemistry on ABI 3730 sequencers (Applied Biosystems, France) at ∼12× coverage. The assemblies were realized using Phred/Phrap/Consed software package (www.phrap.com; Ewing et al. 1998; Ewing and Green 1998; Gordon et al. 1998). The sequences have been deposited in the EMBL database under accession numbers FQ660547 (46o6), FQ660548 (10c2), FQ660549 (58j14), FQ660550 (35e19), FQ660551 (24a6), and FQ660552 (67l12).
The BACs were annotated using Apollo (Lewis et al. 2002). We performed BLAST analysis (Camacho et al. 2009) using the D. melanogaster genome as reference (http://flybase.org/, R5.29). When needed, the sequences were first aligned with ClustalW (Thompson et al. 2002) then manually aligned with BioEdit (Hall 1999; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The percentage of nucleotide identity between sequences was calculated using DnaSP (Librado and Rozas 2009). The repeated regions were analyzed with RepeatMasker (Chen 2004; Tarailo-Graovac and Chen 2009). Global alignment of the clones was conducted with PipMaker (Schwartz et al. 2000).
Fluorescent in situ hybridization (FISH)
The spread of chromosomes and the hybridization were performed according to the protocol described in Montchamp-Moreau et al. (2006). The probe was a fragment of Hosim1 (Figure 5 and sequence of primers in Table S1), amplified from DNA of (XSR6)ST8 males and cloned into PGEM-T Easy Vector System (Promega).
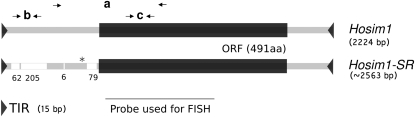
Figure 5 .
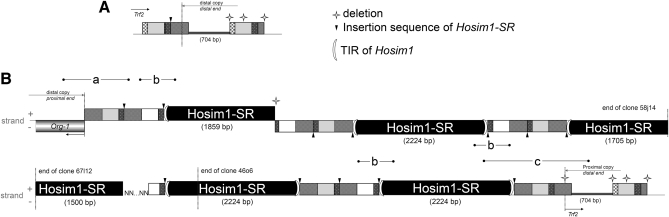
Comparison of Hosim1-SR with the canonical Hosim1: Schematic representation of the nucleotide alignment. Terminal inverted repeats (TIR): TAGTGTTGGGT. The white boxes show the position of the main deletions in Hosim1-SR, with their size below (in bp). The star shows the localization of the intron presented in Figure S7. (a) Position of primers used to amplify both Hosim1 and Hosim1-SR transcripts (Figure 7), (b) position of primers used to estimate the number of canonical Hosim1 (Figure 6A), (c) position of primers used to estimate the total number of elements (Hosim1 + Hosim1-SR, Figure 6A), and the total amount of transcripts (Figure 6B).
Southern blot
High molecular weight DNA was prepared from 300 mg of adult male (Roberts 1998). Four micrograms of each extract were digested overnight with 100 U BamHI or 100 U HindIII in 200 µl final volume, precipitated after phenol/chloroform extraction, and resuspended in 30 µl TE. Overnight electrophoresis was performed on 0.7% agarose gel in TAE 1×. The transfer onto nylon membrane (Amersham Hybon-N) was performed with a Amersham VacuGene XL Vacuum Blotting System. The probe consisted of 25 ng of Hosim1-SR PCR product (sequence of primers in Table S1) purified with Nucleospin DNA extract II (Macherey Nagel), and then labeled with α-32P dCTP using High Prime DNA Labeling Kit (Roche). After a two-hour prehybridization at 68°C, the membrane was incubated overnight with the probe in 6× SSC, 5× Denhardt's reagent, 0.5% SDS, and 100 µg/ml salmon sperm DNA, then washed twice in SSC 0.2×–SDS 0.2% and twice in SSC 0.1×–SDS 0.1%.
Quantification of DNA and cDNA by real-time PCR
The sequences of the primers are in Table S1. To estimate the copy number of Hosim1 elements per genome, we performed six independent DNA extractions from heads of 10 males for each stock under study, using a DNeasy Tissue Kit (Qiagen). The concentration of DNA was measured with a Quant-it dsDNA HS Assay kit (Invitrogen) in a Qubit fluorometer. Quantification was performed from 5 ng of DNA with a Chromo4 thermal cycler (Bio-Rad) and Bio-Rad iQ SYBR-Green kit. The reference genes were GAPDH and RPL17 (autosomal genes showing no sequence polymorphism between and within fly stocks). The efficiency of amplification was close to 100% for the six sets of primers used.
To detect and quantify Hosim1 transcripts, the testes from two-day-old males were dissected in PBS on ice and frozen in liquid nitrogen. RNA was extracted from samples of 30 testis pairs each, using a Nucleospin RNAII kit (Macherey-Nagel) following the manufacturer's protocol. For each stock, three independent RNA extracts were obtained. Two RT-PCR reactions were performed on each extract, using Bio-Rad iScript Select cDNA synthesis kit, and 2 ng of the resulting cDNAs were used for real-time PCR. The amount of transcript was standardized to the autosomal reference genes light and RPII140 that showed stable expression among samples (determined using Genorm (Vandesompele et al. 2002), Normfinder (Andersen et al. 2004), and Bestkeeper (Pfaffl et al. 2004)).
The number of DNA copies and the amount of transcript in (XSR6)ST8 males relative to (XST8)ST8 males were estimated using the ΔΔCt method (Schefe et al. 2006). For each stock, the 95% confidence interval was calculated to assess the robustness and variance of our quantifications. The values were compared with a Mann-Whitney Test, using R (http://www.r-project.org/, function wilkox.test(A,B); R Development Core Team 2010).
Results
General organization of the sex-ratio region
The sex-ratio chromosome XSR6 typically leads to 90–95% female progeny in a suppressor-free background. We sequenced four overlapping BACs, named 58j14, 46o6, 35e19, and 24a6, covering 250 kb and including the candidates regions for the two distorter elements previously mapped on XSR6 (Figure S1, Figure S2). In addition, we partially sequenced BACs 10c2 and 67l12 to check the organization of the duplication. After assembling, we aligned the resulting sequence to the genome of D. melanogaster because there are numerous gaps and assembling errors in the published D. simulans genome. The synteny of the region appeared to be conserved between the two species (Figure S2) with 92.88% sequence identity on average.
A single segmental duplication in tandem and direct orientation was detected. The duplicated fragment was found to be 37,500 bp in length (Figure 1) and contain six annotated genes. It started distally within the gene Trf2 (second intron) and ended within the gene org-1 (first intron). Of the four genes annotated in between, three had complete duplication: CG12125, CG1440, and CG12123. Analysis of their sequences did not reveal mutations that could affect their coding potential. In contrast, the distal copy of the fourth gene, CG32712, had a frameshift mutation caused by a 65 bp deletion within the second exon, which introduced an early stop codon.
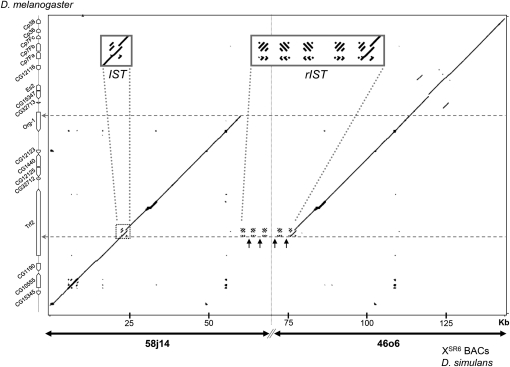
Figure 1 .
General organization of the sex-ratio duplication dot plot comparison of the duplication on the XSR6 chromosome of D. simulans (abscissa) with the homologous region in D. melanogaster (ordinate). The D. simulans sequence was obtained from BACs 58j14 and 46o6, which do not overlap (limits showed by the vertical dotted line). The black arrows show Hosim1-SR sequences (no homolog in D. melanogaster genome), separated by fragments with homologs in the second intron of Trf2 (IST). Two horizontal dotted arrows show the limits of the duplicated fragment.
Examination of the candidate region for the second element revealed the presence of an approximately 1 kb fragment between the genes spirit and CG12065 that had no homolog in the D. melanogaster genome. This insertion also exists on standard X chromosomes of D. simulans and contains a small chromodomain-containing gene (759 bp), which is annotated in the D. simulans genome as GD16106 (Figure S2). Transcripts of GD16106 have been detected in the testis of both standard and sex-ratio males (D. Ogereau, unpublished data).
Origin of the sex-ratio duplication
The two copies of the duplication had a very high sequence identity score (99.49% for exons, 98.65% for introns). Figure 2 shows that nucleotide polymorphisms were not randomly scattered along the duplication: a 10,344 bp fragment was 100% identical between the two copies. This cannot be due to an assembling error because the procedure ensured that each copy of the duplication was cloned in a different BAC (see Materials and Methods and Figure S1). This remarkable similarity between the two copies is consistent with previous direct sequencing of markers located within the genes CG12123 and CG1440, which revealed a single sequence on chromosome XSR6 (Montchamp-Moreau et al. 2006).
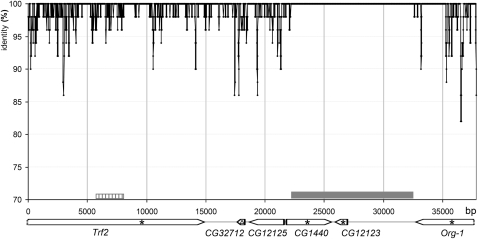
Figure 2 .
Sequence identity between the two copies of the sex-ratio duplication on the XSR6 chromosome. The analysis was performed using a 50 bp sliding window with a step size of 10 bp. The gray box represents the fragment with 100% identity; the striped box represents the region containing the DDSA traces described in Figure S3. The stars show the position of markers sequenced in the population study of Derome et al. (2008), and the triangle shows the position of the small deletion in the distal copy of CG32712.
Outside of the identical 10,344 bp fragment, the mean identity was 98.75%, similar to the value obtained for this chromosomal region in a whole-genome analysis of polymorphism among seven independent lines of D. simulans (98–99%) (Begun et al. 2007). This suggests that the duplication event occurred in the very recent past, through production of an exact copy on the donor chromosome itself. Polymorphism was later introduced by recombination. This hypothesis is consistent with experimental evidence that revealed that recombination occurs freely between the duplication and the homologous region of standard X chromosomes (Montchamp-Moreau 2006).
We therefore propose a parsimonious three-step scenario for the observed duplication pattern of XSR6. First was a tandem duplication of a fragment on the same chromosome (Figure 3A), followed by two recombination events, one affecting the proximal copy of the duplication and the other the distal copy. Assuming that the duplication originated recently and retained the ancestral sequence across a large portion, we should find signatures of the mechanism that generated the XSR6 pattern. In D. melanogaster, analysis of double-strand break repair (DSB) after P-element excision shows that DSB repair usually occurs primarily through homologous repair and, preferentially, by synthesis-dependent strand annealing (SDSA) (Engels et al. 1990; Nassif et al. 1994; Rong and Golic 2003). The template sequence is usually the allele located on the homologous chromosome or on the sister chromatid, but an ectopic site is sometimes used and thus duplicated into the DSB site (Rong and Golic 2003). The duplication-dependent strand annealing (DDSA) model is a variant of SDSA occurring after a DSB in a repeated sequence; under the DDSA model, repair uses an ectopic site that contains this repeated sequence (Fiston-Lavier et al. 2007).
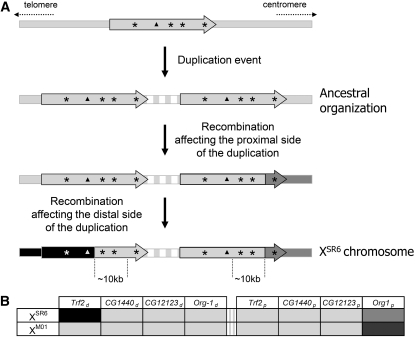
Figure 3 .
(A) Parsimonious scenario explaining the pattern of sequence variation observed between the two copies of the sex-ratio duplication carried by the chromosome XSR6. The vertical dotted lines show the limits of the 10 kb fragment with 100% sequence identity. The stars show the position of markers sequenced in Derome et al. (2008). The triangles show the position of CG32712 (the white triangle stands for the deleted allele brought by recombination). The vertical gray/white strips represent the repeated motifs of the junction region. (B) Interpretation of Figure S1 in Derome et al. (2008). XM01 is a sex-ratio chromosome from Madagascar, carrying a combination of haplotypes commonly found there. For each marker (stars in Figure 3A), the ancestral sequence is symbolized in light gray. Alleles supposed to have been brought by recombination are in medium and dark gray (proximal recombination) and in black (distal recombination).
The presence of repeated sequences at one end of the duplicated fragment suggests that the DDSA model can be applied to the sex-ratio segmental duplication. According to this model, instability of the DNA heteroduplex during repair leads to local dissociations of the nascent strand from the template. When reinvasion occurs within the same template, signatures of the repair mechanism can be found (Mcvey et al. 2004). A reinvasion upstream from the dissociation site leads to the formation of short tandem repeats within the neosynthesized copy, whereas a downstream reinvasion site that corresponds to a jump from the template causes a gap delimited by microhomology sequences within the neosynthesized copy (Fiston-Lavier et al. 2007). We analyzed the gaps in the alignment of the duplicated fragments carried by the XSR6 chromosome and, when possible, compared them to the sequences available in FlyBase (D. simulans, R1.3). We found five signatures of reinvasion in Trf2, within a fragment in which the distal copy is thought to have come from a standard chromosome via recombination (Figure 2). Three microhomologies and one tandem repeat indicated that the proximal copy was the neosynthesized sequence, whereas another tandem repeat indicated that the distal copy was the neosynthesized sequence (Figure S3). However, this latter trace can alternatively be explained by a polymerase slippage that occurred later in the proximal copy after the duplication event.
The duplication is associated with an amplified transposable element
The domain between the two copies of the segmental duplication consists of repeated modules. Each module is composed of fragments that are homologous to fragments in the second intron of the gene Trf2, which we called IST (intronic sequence of Trf2), that alternate with fragments that have no homolog on the X chromosome of D. melanogaster (Figure 1). These fragments correspond to Hosim1, a class II transposable element detected in the genome of D. simulans and D. sechellia using bioinformatics methods (de Freitas Ortiz and Loreto 2009). Hosim1 belongs to the herves transposable element family of the hAT superfamily.
In D. simulans, the gene Trf2 contains two IST motifs located 704 bp apart (Figure 4A) that show 91.3% identity without the indels and 79.8% with the indels. This organization is conserved in D. melanogaster and D. sechellia. The motifs that alternate with Hosim1 have been rearranged; we thus called them rIST (for rearranged IST). The rISTs showed more than 99% identity with each other and were always organized in direct tandems between each copy of Hosim1. The same 8 bp in the rIST sequence were duplicated at the insertion site of each Hosim1 element.
Figure 4 .
(A) Schematic representation of the canonical IST (Intronic Sequence of Trf2), found in the published D. simulans genome and in the distal copy of Trf2 on chromosome XSR6. (B) Organization of the junction region on chromosome XSR6, observed in BAC 58j14 (top) and BACs 67l12 and 46o6 (bottom). It consists of alternating Hosim1-SR elements and direct tandems of rIST. Fragments (a–c) amplified by PCR to control the organization on DNA from (XSR6)ST8 males (sequence of primers in Table S1). NNN: gap in sequence assembly
There was 100% identity among the Hosim1 copies associated with the duplication (excepted for a deletion of the 3′ part of the first element in the 58j14 clone). This finding suggests either that their amplification is very recent or that genetic conversion is frequent at this locus. We called these copies Hosim1-SR, because they were noticeably different from the four Hosim1 forms already annotated in the D. simulans genome [accession number CH986553, CH981769, CM000363, CH982471 (partial sequence)]. While the four canonical forms differ from each other by only 23 SNPs and a poly-T stretch, Hosim1-SR had four deletions in the 5′ noncoding region (Figure 5) and differed from the canonical forms by 28 SNPs. These differences, however, do not affect the size of the transposase predicted by ORF Finder (Rombel et al. 2002). Amino acid alignment with Hermes transposase showed that both Hosim1 and Hosim1-SR retained the DDE amino acids involved in the enzyme’s function (Perez et al. 2005). Furthermore, both Hosim1 and Hosim1-SR contain the LDPR sequence that is characteristic of the majority of hAT transposable elements (Handler and Gomez 1997). The terminal inverted repeats (TIR) of Hosim1 are conserved in Hosim1-SR.
While potentially active Hosim1-like elements (Hosec1) have been described in the genome of D. sechellia (de Freitas Ortiz and Loreto 2009), we found only one element, incomplete and diverging, in the D. melanogaster genome. Figure S4 shows a maximum likelihood tree obtained from the published sequences.
Checking the organization of the duplication
First, we confirmed that the presence and amplification of Hosim1-SR at the junction of the duplicated segments was not due to a cloning artifact. We performed fluorescent in situ hybridization (FISH) on polytene chromosomes with a probe targeting both Hosim1 and Hosim1-SR (Figure 5). In standard males (XST8)ST8 we detected two hybridization sites, on the chromosomal arms 3L (80A) and 2L (42C) (Figure S5). These sites correspond to the Hosim1 copies identified in the published D. simulans genome (accession number: CM000363 and CH986553, respectively). In (XSR6)ST8 males, which carry the sex-ratio XSR6 chromosome in the same autosomal background as (XST8)ST8 males, we observed an additional site in the cytological band 7E of the X chromosome. Thus, this extra signal colocalizes with the sex-ratio duplication (Montchamp-Moreau et al. 2006).
Then, because of the potential impact on the expression level of neighboring genes, we checked the gene organization at the limits between the duplicated fragments and the intervening repeated region. We extracted DNA from (XSR6)ST8 males, and we used PCR to amplify fragments that overlap between org-1 and Hosim1-SR and those that overlap between Hosim1-SR and Trf2 (Figure 4B, a–c). The sequences were found to be identical to those in the BACs.
Size and organization of the junction region
To check the organization of the junction region, we performed further screening of the BACs library and found two additional BACs (10c2 and 67l12, Figure S1) that contain a larger part of the junction region. We found no BAC that encompasses the whole region (i.e., that contains the adjacent end of the segmental duplication on both sides). The abundance of repeated motifs with very high similarity and rearrangements in clones made it impossible to completely sequence and assemble BACs 10c2 and 67l12. Nevertheless, we found only Hosim1-SR and rIST sequences in this region. The partial sequence assembly and the digestion of BACs 10c2 and 67l12 by HindIII (D. Ogereau, unpublished data) confirmed the organization proposed in Figure 4 and indicated that the junction region in 67l12 contains only Hosim1-SR/rIST/rIST modules.
In addition, we performed Southern blots using high molecular weight genomic DNA and hybridization with a Hosim1-SR probe (Figure S6). HindIII digest produced bands shared by (XST8)ST8 and (XSR6)ST8 males, which likely correspond to autosomal copies. A restriction site is present in the 5′ region of Hosim1-SR but not in IST or rIST sequences (Figure S6B). Because (XSR6)ST8 males produced a strong specific band of ∼3.6 kb, it follows that the junction region contained mainly, if not exclusively, a succession of Hosim1-SR/rIST/rIST modules in the same orientation as found in clones 46o6 and 67l12. However, a light band of ∼4.3 kb was also observed, which should correspond to a module in the opposite orientation, as found in clone 58j14 (Figure 4). This could be the signature of sporadic rearrangements, possibly favored by the repetitive structure.
Estimating the number of Hosim1 in (XSR6)ST8 males
According to the data provided by the BACs, the junction region contains at least six copies of Hosim1-SR (Figure 4). We also estimated directly on the XSR6 chromosome the total size of the repeated region using high molecular weight genomic DNA digested by BamHI. There is no BamHI restriction site in rIST and Hosim1-SR: the closest sites on either side of the junction domain are in the distal copy of org-1 (∼1.6 kb apart) and in the 5th exon of the proximal copy of Trf2 (∼10.1 kb apart). Hybridization of the Southern blot with a probe spanning the whole Hosim1-SR element revealed a large fragment estimated at 26–36 kb, which corresponds to 4.1–6.9 copies of Hosim1-SR/rIST/rIST modules of ∼3.6 kb (Figure S6).
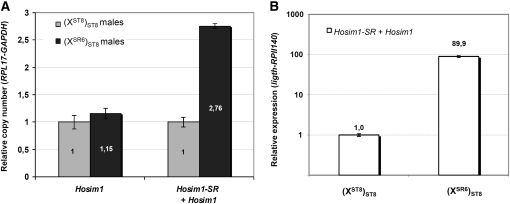
We used real-time PCR to obtain an independent estimate of the number of transposable elements in the duplication. Again we used (XSR6)ST8 and (XST8)ST8 males, which differ only by the X chromosomes. First, the published Hosim1 form (de Freitas Ortiz and Loreto 2009) was specifically amplified using primers designed within the region deleted in Hosim1-SR [Figure 5,(b)] We observed a weak difference between the two types of males (1.15 times more copies in (XSR6)ST8 than in (XST8)ST8; P = 0.04), suggesting that there is no extra canonical Hosim1 on the XSR6 chromosome (Figure 6A). We quantified the total number of elements, Hosim1 plus Hosim1-SR, by amplifying a sequence located in the coding region shared by the two forms [Figure 5,(c)]. We found 2.76 times more copies in (XSR6)ST8 than in (XST8)ST8 males (P = 2.4 × 10−12, Figure 6A). According to the results of the FISH experiment (Figure S5), there should be four copies of Hosim1 in the autosomal genome shared by (XST8)ST8 and (XSR6)ST8 males (a single and homozygous copy at each autosomal site). Under this hypothesis, about 11 copies must be present in (XSR6)ST8 males and, consequently, seven copies on the XSR6 chromosome [confidence interval 95% (6.85–7.19)].
Figure 6 .
Quantification of Hosim1 copy number (A) and Hosim1 transcripts (B) by real-time PCR. The values in (XSR6)ST8 males were estimated relative to that in (XST8)ST8 males. Vertical bars: confidence interval (95%). (A, left) “Canonical” elements (see text). (A, right) Canonical elements plus Hosim1-SR. The reference genes were RPL17 and GAPDH. (B) Total amount of testicular transcripts (= Hosim1 + Hosim1-SR). Reference genes were light and RPII140.
Expression of Hosim1 and IST in (XST8)ST8 and (XSR6)ST8 males
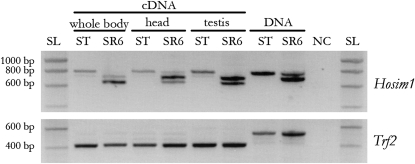
To determine whether Hosim1 and, in particular, the Hosim1-SR form are still active, we performed PCR on cDNA with a primer pair straddling the deletion characteristic of the Hosim1-SR form [Figure 5,(a)]. Transcripts were present in the whole body, in head, and in testes of both sex-ratio and standard males. We found that both forms were expressed in (XSR6)ST8 males and that the cDNA fragments were shorter than DNA fragments (Figure 7). Sequencing the shorter form revealed a 67 bp intron associated with Hosim1-SR (Figure S7). The splicing occurred for Hosim1 transcripts, too, but appeared to be less efficient [see (XST8)ST8 males in Figure 7]. In (XSR6)ST8 males, the splicing seemed to be more efficient in the testes. Real-time PCR showed that the total amount of testicular transcripts was about 90 times higher in (XSR6)ST8 males than in (XST8)ST8 males (Mann-Whitney test, P = 2.2 × 10−16, Figure 6B).
Figure 7 .
Detection of Hosim1 transcripts by RT-PCR. The primers used straddle the deletion of 79 bp specific of Hosim1-SR [Figure 5,(a)], so this element must produce a shorter band (699 bp) than the canonical Hosim1 (784 bp). Even shorter fragments were obtained from cDNAs revealing an intron of 67nt (see text). Amplification of Trf2 with primers straddling an intron was used to control the lack of DNA contamination in the cDNA samples. NC, negative control (no cDNA nor DNA); SL, SmartLadder DNA ladder (Eurogentec); SR6, (XSR6)ST8 males; ST, (XST8)ST8 males.
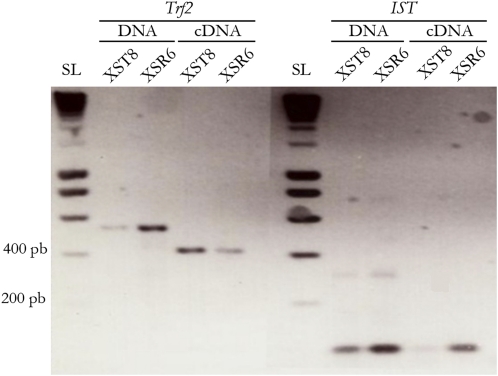
To test for the presence of transcripts that contain the IST or rIST sequences (light gray boxes in Figure 4), we designed primers that amplify both forms (Table S1); these noncoding motifs appeared to be transcribed, and more cDNAs were detected in (XSR6)ST8 males (Figure 8) than in (XST8)ST8.
Figure 8 .
Detection of IST transcripts by RT-PCR. The IST and rIST probes were designed within the region shown in light gray in Figure 4. Amplification of Trf2 gene marker was used to control the lack of DNA in the cDNA samples (see Figure 7). SL, SmartLadder DNA ladder (Eurogentec); XSR6, (XSR6)ST8 males; XST8, (XST8)ST8 males.
Discussion
Here we provide evidence that the XSR6 chromosome carries a recent tandem segmental duplication of 37.5 kb that originated through the production of an exact copy on the donor chromosome itself and that changed the copy number of six genes. By contrast, the second element required for drive is not associated with rearrangement. Yet, in the candidate region, we noticed a small gene (GD16106) that does not exist in the D. melanogaster genome. The molecular data allowed us to propose a mechanism for how the duplication was generated and to retrace its history.
Characteristics of the duplication and possible mechanisms
The two copies of the duplicated chromosome fragment are separated by repeated modules, each of which contains a Hosim1 transposable element that has small deletions but that is potentially active, and tandem motifs derived from an intronic sequence of Trf2 (rIST). Amplification of the modules may be responsible for the additional dense band revealed after DAPI coloration in the 7E section on the XSR6 polytene chromosome (Montchamp-Moreau et al. 2006), which reflects a local modification of the chromatin structure. The highly repeated nature of this region prevented its complete sequencing and assembly, but three independent methods indicated that the XSR6 chromosome carries six or seven modules. Organization like this is a potential source of instability and unequal crossovers; this instability likely produces variation in the number and organization of motifs among natural sex-ratio X chromosomes.
About 25% of the tandem duplications detected in the genome of D. melanogaster show at least one repetitive element at the breakpoint (Fiston-Lavier et al. 2007). The local sequence organization, with two repeated modules 704 bp apart within Trf2, could have favored a double-strand DNA break. Alternatively, the transposable element may have generated the break; indeed Hosim1 is a class II transposable element that is mobilized by a DNA intermediate through a ‘‘cut-and-paste’’ mechanism (de Freitas Ortiz and Loreto 2009). According to the DDSA model (Fiston-Lavier et al. 2007), a double-strand break within a repeated sequence (here IST or Hosim1) is repaired via homologous–base pairing using another copy of this repeated sequence as template. The repair leaves specific signatures that were detected on the XSR6 chromosomes and that allowed us to identify the proximal copy of the duplication as the neosynthesized sequence. The organization of the junction domain between the duplicated fragments probably resulted from secondary amplification of repeated sequences.
The duplication should induce quantitative and qualitative changes in transcripts
Testicular transcripts of the three fully duplicated genes CG12125, CG1440, and CG12123 had been detected using rtPCR in (XSR6)ST8 males, and a polymorphism in cDNA sequences led to the inference that both copies of CG12125 are active (Montchamp-Moreau et al. 2006). It was not possible to determine whether both copies of CG12123 and CG1440 are active because the distal and proximal copies are 100% identical. As none of the three genes on the XSR6 chromosome revealed any trace of frameshift or stop mutations, their duplication should result in quantitative changes in canonical transcripts. cDNA sequencing revealed that both copies of CG32712 are expressed in (XSR6)ST8 males (D. Ogereau, unpublished data). However, the 65 bp deletion in the second exon of the distal copy of CG32712 causes a nonsense mutation, so the associated mRNA cannot produce functional proteins. Other sex-ratio X chromosomes (e.g., XM01 depicted in Figure 3B) have been found to carry two 100% identical copies of the complete, likely original, proximal copy of XSR6. Thus, the deleted allele must have been introduced by recombination. As XSR6 shows strong drive ability, this suggests that CG32712 is not the distorter element in the duplication.
Although Trf2 and org-1 are not fully duplicated, transcripts produced by both copies of each of these genes were found in the testis of (XSR6)ST8 males (Montchamp-Moreau et al. 2006). In the D. melanogaster subgroup, about 78% of new genes have arisen from duplications, and of these, 32% formed chimerical structures by recruiting flanking sequences into their coding region (Zhang et al. 2008). Located on either side of the junction domain of the sex-ratio duplication, the 5′ deleted copies of the Trf2 and org-1 genes are potential actors of such a process. The distal copy of org-1 lacks its first exon, which contains the start codon and the first 54 amino acids. The nearest Hosim1-SR element in the junction domain is in the opposite orientation (Figure 4B), suggesting that a chimerical transcript cannot be produced. Trf2 is more complex because two different Trf2 transcripts have been reported. Kopytova et al. (2006) described a long Trf2 transcript (∼7.6 kb), thought to produce two proteins, one of 175 kD and one of 75 kD, that differed in their N-terminal domain; the shorter protein could have been produced via an internal ribosome entry site (IRES). Short transcripts (∼3.9 kb), initially described by Rabenstein et al. (1999), can only produce the shorter protein. In the XSR6 chromosome, the proximal copy of Trf2 lacks the two first exons of the long transcript described by Kopytova et al. (2006). However, it could potentially produce the short transcript described by Rabenstein et al. (1999), and it retains the complete coding sequence for both proteins.
The repeated sequences (Hosim1 and rIST) amplified in the junction region appeared well expressed in (XSR6)ST8 males: Hosim1 transcripts were found to be about 90 times more abundant in the testis of (XSR6)ST8 males than in (XST8)ST8 males (Figure 6B). As there are only 2.7 more Hosim1 copies in the genome of the (XSR6)ST8 males than in (XST8)ST8 males, either the Hosim1-SR form is expressed much more in the testis than the autosomal forms or there is a general overexpression of Hosim1 elements in sex-ratio males. We also detected cDNA containing IST/rIST sequences, although they are certainly noncoding (they are intronic sequences, and bioinformatics software did not detect any ORF). Such noncoding RNAs can be involved in a variety of processes, including dosage compensation, posttranscriptional gene silencing, regulation of transposable elements, and chromatin remodeling (Van Wolfswinkel and Ketting 2010). Because some rISTs and the 5′ deleted proximal copy of Trf2 are in the same orientation (Figure 4B), together they might produce chimerical transcripts.
Age of the duplication and evolutionary prospects
The 10,344 bp fragment of the sex-ratio duplication on the XSR6 chromosome with 100% identity between the copies (Figure 2) is too long to have arisen by gene conversion. We thus assumed that it represents the ancestral state, and no recombination with a standard X occurred in this region. This allows us to estimate the age of the duplication, assuming a conservative value of ∼2 × 10−8 recombination/bp/generation in the region (Derome et al. 2008; Derome et al. 2004; Montchamp-Moreau et al. 2006). As the probability of recombination or mutation is low, the number of these events follows a Poisson distribution (Sawyer and Hartl 1992). The probability of two copies of a fragment of size L remaining fully identical is given by the formula e-2(r+µ)Lt, where t = number of generations (10 per year) and µ = mutation rate/bp/generation [µ = 10−8 (Rozas et al. 2001)]. It follows that the duplication event likely took place less than 483 years ago (P = 0.05). That the duplication is so recent is well supported by previous molecular population genetics studies (Bastide et al. 2011; Derome et al. 2008). First, among the four marker loci that were surveyed, there was no fixed difference between the duplicated XSR and standard XST chromosomes sampled in the wild. In addition, these previous studies showed that most of the XSR chromosomes collected in Madagascar only 10 years ago still carried the presumed ancestral sequence with no trace of even a singleton mutation at these marker loci. The predominant variant found in this population could even have retained an ancestral fragment of larger size than that in XSR6 (Figure 3B). Thus, the structure of XSR6 is not exceptional among the present distorter X chromosomes. Note also that the region in between the identical 10,344 bp fragments may not have undergone recombination at all. If this is the case, then the duplication is younger than estimated above. Sex-ratio X chromosomes, however, can reach high frequencies in natural populations (≥50%) (Atlan et al. 1997; Jutier et al. 2004). In that case, the probability of recombination occurring between XSR and XST chromosomes would be lower than the overall recombination rate for the genome region, thus the duplication age could be more than 483 years.
Segmental duplications are frequent on the X chromosome of D. melanogaster, but only 7.21% of them are more than 10 kb long. In addition, tandem duplications are almost always shorter than other duplications (Fiston-Lavier et al. 2007). This makes the sex-ratio duplication an exceptional event with regard to both its size (more than 37 kb) and its gene content. This kind of duplication is probably deleterious most of the time and, thus, destined to disappear quickly. In the present case, the duplication has a strong, negative effect on male fertility that is a direct consequence of drive (Angelard et al. 2008; Atlan et al. 2003; Atlan et al. 2004) and should cause many other perturbations via overexpression of the six duplicated genes or rIST and Hosim1-SR activity. In this respect, meiotic drive can be understood as a process that allows genetic rearrangements, such as duplications or inversions, to be maintained in the genome in spite of associated deleterious effects, as first proposed by Hedrick (1981). These rearrangements can persist for extended evolutionary periods, as demonstrated by one of the inversions associated with the sex-ratio trait in D. pseudoobscura with an apparent divergence time of about 1 million years (Babcock and Anderson 1996; Kovacevic and Schaeffer 2000). This allows time for the genetic innovation to coevolve with the host genome and eventually lead to a neutral or advantageous form.
Now that the sequence of the reference chromosome XSR6 is known, precise study of gene expression is the next step in understanding the link between the duplication and sex-ratio meiotic drive. The duplication affects the copy number of six genes and is associated with several copies of an active transposable element and repeated modules that produce noncoding RNAs. Therefore, we must determine which of these components is involved in sex-ratio drive. In this respect, the duplicated XSR6 sequence will be a precious tool for analyzing polymorphism along this region among natural distorter chromosomes, with the goal of identifying a correlation between drive ability and duplication structure. It will also allow for the development of appropriate transgene constructs for the functional validation of candidate genes or sequences. Unraveling the molecular mechanisms that underlie the Paris sex-ratio drive should help us understand the evolutionary significance of segregation distorters.
Supplementary Material
Acknowledgments
We thank Nicole Chaminade for technical advice on FISH, Aurelie Hua-Van for help in analyzing the sequence of the transposable element, Alain Billault for production of the BAC library, and Anna-Sophie Fiston-Lavier and Pierre Gérard for their comments on the manuscript. This work was supported by the Genoscope (DNA finishing and sequencing) and the Centre National de la Recherche Scientifique. L.F. was funded by a Ph.D. scolarship from the Ecole Normale Supérieure de Lyon.
Literature Cited
- Andersen C. L., Jensen J. L., Orntoft T. F., 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245–5250 [DOI] [PubMed] [Google Scholar]
- Angelard C., Montchamp-Moreau C., Joly D., 2008. Female-driven mechanisms, ejaculate size and quality contribute to the lower fertility of sex-ratio distorter males in Drosophila simulans. BMC Evol. Biol. 8: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlan A., Capillon C., Derome N., Couvet D., Montchamp-Moreau C., 2003. The evolution of autosomal suppressors of sex-ratio drive in Drosophila simulans. Genetica 117: 47–58 [DOI] [PubMed] [Google Scholar]
- Atlan A., Joly D., Capillon C., Montchamp-Moreau C., 2004. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J. Evol. Biol. 17: 744–751 [DOI] [PubMed] [Google Scholar]
- Atlan A., Mercot H., Landre C., Montchamp-Moreau C., 1997. The sex-ratio trait in Drosophila simulans: geographical distribution if distortion and resistance. Evolution 51: 1886–1895 [DOI] [PubMed] [Google Scholar]
- Babcock C. S., Anderson W. W., 1996. Molecular evolution of the Sex-Ratio inversion complex in Drosophila pseudoobscura: analysis of the Esterase-5 gene region. Mol. Biol. Evol. 13: 297–308 [DOI] [PubMed] [Google Scholar]
- Bastide H., Cazemajor M., Ogereau D., Derome N., Hospital F., et al. , 2011. Rapid rise and fall of selfish Drosophila simulans X chromosomes in Sex-ratio: spatio-temporal analysis of phenotypic and molecular data. Mol. Biol. Evol. 28: 2461–2470 [DOI] [PubMed] [Google Scholar]
- Begun D. J., Holloway A. K., Stevens K., Hillier L. W., Poh Y. P., et al. , 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5: e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemajor M., Joly D., Montchamp-Moreau C., 2000. Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics 154: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., 2004. Using RepeatMasker to identify repetitive elements in genomic sequences, chapter 4, unit 4.10 in Current Protocols in Bioinformatics. John Wiley & Sons, New York: [DOI] [PubMed] [Google Scholar]
- de Carvalho A. B., Klaczko L. B., 1992. Age and sex-ratio expression in Drosophila mediopunctata. Genetica 87: 107–111 [DOI] [PubMed] [Google Scholar]
- de Freitas Ortiz M., Loreto E. L., 2009. Characterization of new hAT transposable elements in 12 Drosophila genomes. Genetica 135: 67–75 [DOI] [PubMed] [Google Scholar]
- Derome N., Baudry E., Ogereau D., Veuille M., Montchamp-Moreau C., 2008. Selective sweeps in a 2-locus model for sex-ratio meiotic drive in Drosophila simulans. Mol. Biol. Evol. 25: 409–416 [DOI] [PubMed] [Google Scholar]
- Derome N., Metayer K., Montchamp-Moreau C., Veuille M., 2004. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics 166: 1357–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer K. A., Charlesworth B., Jaenike J., 2007. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc. Natl. Acad. Sci. USA 104: 1587–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J., 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525 [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M., Green P., 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford, United Kingdom [Google Scholar]
- Fiston-Lavier A. S., Anxolabehere D., Quesneville H., 2007. A model of segmental duplication formation in Drosophila melanogaster. Genome Res. 17: 1458–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson S., 1928. A new sex-ratio abnormality in Drosophila obscura. Genetics 13: 488–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., 1998. Consed: a graphical tool for sequence finishing. Genome Res 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41: 95–98 [Google Scholar]
- Hamilton W. D., 1967. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science 156: 477–488 [DOI] [PubMed] [Google Scholar]
- Handler A. M., Gomez S. P., 1997. A new hobo, Ac, Tam3 transposable element, hopper, from Bactrocera dorsalis is distantly related to hobo and Ac. Gene 185: 133–135 [DOI] [PubMed] [Google Scholar]
- Hauschteck-Jungen E., 1990. Postmating reproductive isolation and modification of the 'sex ratio' trait in Drosophila subobscura induced by the sex chromosome gene arrangement A2+3+5+7. Genetica 83: 31–44 [DOI] [PubMed] [Google Scholar]
- Hedrick P., 1981. The establishment of chromosomal variants. Evolution 35: 322–332 [DOI] [PubMed] [Google Scholar]
- Jaenike J., 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32: 25–49 [Google Scholar]
- Jaenike J., 2008. X chromosome drive. Curr. Biol. 18: R508–R511 [DOI] [PubMed] [Google Scholar]
- Jutier D., Derome N., Montchamp-Moreau C., 2004. The sex-ratio trait and its evolution in Drosophila simulans: a comparative approach. Genetica 120: 87–99 [DOI] [PubMed] [Google Scholar]
- Kopytova D. V., Krasnov A. N., Kopantceva M. R., Nabirochkina E. N., Nikolenko J. V., et al. , 2006. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol. Cell. Biol. 26: 7492–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic M., Schaeffer S. W., 2000. Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics 156: 155–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. E., Searle S. M. J., Harris N., Gibson M., Iyer V., et al. , 2002. Apollo: a sequence annotation editor.Genome Biology 2002, 3(12):research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. E., Searle S. M. J., Harris N., Gibson M., Iyer V., et al. , 2002. Genome Biology, 3(12):research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lyttle T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557 [DOI] [PubMed] [Google Scholar]
- McVey M., Adams M., Staeva-Vieira E., Sekelsky J. J., 2004. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Tao Y., 2010. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 25: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercot H., Atlan A., Jacques M., Montchamp-Moreau C., 1995. Sex-ratio distortion in Drosophila simulans: co-occurence of a meiotic drive and a suppressor of drive. J. Evol. Biol. 8: 283–300 [Google Scholar]
- Montchamp-Moreau C., 2006. Sex-ratio meiotic drive in Drosophila simulans: cellular mechanism, candidate genes and evolution. Biochem. Soc. Trans. 34: 562–565 [DOI] [PubMed] [Google Scholar]
- Montchamp-Moreau C., Cazemajor M., 2002. Sex-ratio drive in Drosophila simulans: variation in segregation ratio of X chromosomes from a natural population. Genetics 162: 1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C., Ogereau D., Chaminade N., Colard A., Aulard S., 2006. Organization of the sex-ratio meiotic drive region in Drosophila simulans. Genetics 174: 1365–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Z. N., Musingarimi P., Craig N. L., Dyda F., Hickman A. B., 2005. Purification, crystallization and preliminary crystallographic analysis of the Hermes transposase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P., 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509–515 [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., 1974. Gene differences between the sex ratio and standard gene arrangements of the X chromosome and linkage disequilibrium between loci in the standard gene Drosophila pseudoobscura. Genetics 77: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing,Vienna, Austria [Google Scholar]
- Rabenstein M. D., Zhou S., Lis J. T., Tjian R., 1999. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc. Natl. Acad. Sci. USA 96: 4791–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. B. (Editor), 1998. Drosophila: A Practical Approach, Ed. 2 The Practical Approach Series Oxford University Press, New York [Google Scholar]
- Roest Crollius H., Jaillon O., Dasilva C., Ozouf-Costaz C., Fizames C., et al. , 2000. Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res. 10: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombel I. T., Sykes K. F., Rayner S., Johnston S. A., 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene 282: 33–41 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Gullaud M., Blandin G., Aguade M., 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158: 1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Novitski E., 1957. Meiotic drive as an evolutionary force. Am. Nat. 41: 105–110 [Google Scholar]
- Sawyer S. A., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe J. H., Lehmann K. E., Buschmann I. R., Unger T., Funke-Kaiser H., 2006. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CT difference” formula. J. Mol. Med. 84: 901–910 [DOI] [PubMed] [Google Scholar]
- Schwartz S., Zhang Z., Frazer K. A., Smit A., Riemer C., et al. , 2000. PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker H. D., 1961. The genetic systems modifying meiotic drive in Drosophila paramelanica. Genetics 46: 177–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Araripe L., Kingan S. B., Ke Y., Xiao H., et al. , 2007a A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol. 5: e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Hartl D. L., Laurie C. C., 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 98: 13183–13188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Masly J. P., Araripe L., Ke Y., Hartl D. L., 2007b A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. PLoS Biol. 5: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N., 2009. Using RepeatMasker to identify repetitive elements in genomic sequences, chapter 4, unit 4.10 in Current Protocols in Bioinformatics. John Wiley & Sons, New York: [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Higgins D. G., 2002. Multiple sequence alignment using ClustalW and ClustalX, chapter 2, unit 2.3 in Current Protocols in Bioinformatics. John Wiley & Sons, New York: [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Ketting R. F., 2010. The role of small non-coding RNAs in genome stability and chromatin organization. J. Cell Sci. 123: 1825–1839 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., et al. , 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R. A., 1972. Preliminary characterization of “sex ratio” and rediscovery and reinterpretation of “male sex ratio” in Drosophila affinis. Genetics 71: 597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Beukeboom L. W., 1998. Sex determination, sex ratios and genetic conflict. Annu. Rev. Ecol. Syst. 29: 233–261 [Google Scholar]
- Zhang X., Zhou J., Eickbush T. H., 2008. Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol. Biol. Evol. 25: 229–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.