Abstract
Genetically homogeneous populations, such as inbred strains, are powerful experimental tools that are ideally suited for studying immunology, cancer, and genetics of complex traits. The zebrafish, Danio rerio, has been underutilized in these research areas because homogeneous strains of experimental fish have not been available in tractable condition. Here, we attempted to inbreed two zebrafish wild-type strains, Tübingen and India, through full sib-pair mating. Although the inbred Tübingen strain failed to thrive and was lost after 13 generations, an inbred India strain (IM) has been maintained successfully. The IM strain has endured 16 generations of inbreeding and has maintained a healthy condition. Two additional strains, IM12m and IM14m, were established as closed colonies from the branches of the IM strain. Genotype analyses using genetic markers revealed a dramatic decrease in polymorphisms (62% dropped to 5%) in both IM (generation 14) and the two closed colonies. This indicates a high level of homogeneity in these strains. Furthermore, scale transplantations between individuals within each strain were successful. These data suggest that extremely homogeneous zebrafish strains have been established, thereby creating a valuable resource for practical application.
Keywords: Danio rerio, sib-pair mating, inbreeding depression, loss of heterogeneity
Genetically uniform strains of animals are valuable tools in biomedical research. Multiple laboratories can use these strains without concern that genetic background variability will confound the experimental results. Indeed, genetic background often affects phenotypes associated with a particular mutant or experimental manipulation in several organisms (Anzai et al. 2010; Nojima et al. 2004; Nomura 1991; Yamamura et al. 2001). In such cases, genetically isogenic strains that can be maintained for generations play an important role in providing highly reproducible results (even several or more years later), and enable us to directly compare and evaluate results obtained from multiple laboratories. Genetically homogeneous strains also provide essential tools for the identification of quantitative loci that affect evolutionarily and biomedically important traits (Frankel 1995; Kimura et al. 2007; Klingenberg et al. 2004; Klingenberg et al. 2001; Tomida et al. 2009; Xiao et al. 2010). Extremely reliable and reproducible data obtained from genetically homogeneous model organisms enable the dissection of both gene-gene and gene-environmental interaction patterns. Furthermore, to identify the susceptible genes, it is often necessary to generate congenic strains; strains that differ from their inbred partner strain by only a short chromosomal segment. The stably maintained homogeneous strain makes it possible to backcross continuously to generate congenic strains (Markel et al. 1997; Morel et al. 1996; Yui et al. 1996).
Numerous attributes in the zebrafish, Danio rerio, including large numbers of eggs per clutch, a short generation time, and relatively low costs associated with maintenance, enable the application of a wide variety of biological techniques to the organism, such as embryonic manipulations, forward and reverse genetics, and molecular biology. Consequently, over the past two decades, the zebrafish has become an important vertebrate model organism in developmental biology, neuroscience, and cancer research. However, the zebrafish system has one major weakness; namely, the lack of highly homogeneous strains that can be stably maintained for many generations. For example, in both mice and medaka, over 10 inbred strains have been maintained by full sib-pair mating for more than 20 generations; these strains have expanded the use of these animals as vertebrate model systems (Naruse et al. 2004; Peters et al. 2007). Comparable zebrafish inbred strains have not been available to the scientific community.
Streisinger et al. (1981) demonstrated that gynogenesis could be used to produce homozygous diploid fish in which only the maternal genome is represented in the offspring. Two zebrafish strains, C32 and SJD, were generated in this manner and were subsequently inbred for a number of generations (Johnson et al. 1995; Nechiporuk et al. 1999; Streisinger et al. 1981). Genetic analysis of single nucleotide polymorphisms (SNPs) revealed that 7% and 11% of tested loci were polymorphic in the SJD and C32 strains, respectively (Guryev et al. 2006). As 14.1% of these SNPs were polymorphic in an outbred strain, WIK, it is difficult to conclude that SJD and C32 truly represent homogeneous lines (Guryev et al. 2006).
Recently, several additional clonal zebrafish strains have been generated by gynogenesis. Successful transplantations between adult fish have been performed with these strains without severe immune reactions (Mizgirev and Revskoy 2010). It is too early to know, however, whether these strains can be maintained and expanded over many generations. Finally, there is an additional inbred strain, SJA, which is listed in the Zebrafish Model Organism Database (ZFIN, Bradford et al. 2011). It is unclear how SJA was created, however, and only 85% of its genome is guaranteed to be monomorphic.
It is clear that some logistical difficulties (i.e., inbreeding depressions), including high mortality levels of embryos and larvae, biased sex ratio, and few eggs from natural crosses, have made it hard to inbreed zebrafish. However, it is not clear whether these difficulties are too severe to establish and maintain homogeneous strains in zebrafish. In this study, we have inbred two zebrafish wild-type strains, attempting to know the severity of inbreeding depression in zebrafish and to establish highly homogeneous zebrafish strains through continuous full sib-pair mating.
Materials and Methods
Strains and fish maintenance
Two outbred wild-type strains, India and Tübingen, were used to generate genetically homogeneous strains of zebrafish. The India strain is a strain obtained from expedition to Darjeeling (see ZFIN at http://zfin.org/action/genotype/genotype-detail?zdbID=ZDB-GENO-980210-28). The Tübingen strain originated from a local pet shop and was maintained for many generations in the laboratory at Tübingen (Haffter et al. 1996). This strain has been used by the Sanger Institute for the Danio rerio Sequencing Project. The two inbreeding strains were named IM (India-Mishima) and TM (Tübingen-Mishima). For the IM strain, male and female fish of generation n were denoted In-M# and In-F#, respectively (# represents the serial number). An IM pair (consisting of one male and one female) within generation n was denoted In-#. For example, the IM founder pair (I0-1) consisted of one male (I0-M1) and one female (I0-F1). The same nomenclature was applied to TM fish, with a “T” replacing the “I.” The method for fish breeding is described in supporting information, File S1.
Generation and selection of pairs
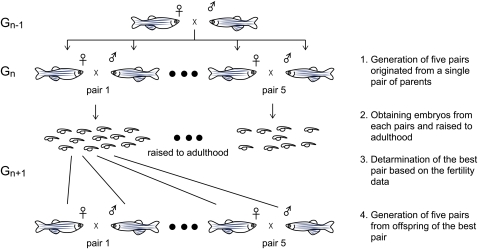
To generate genetically homogeneous zebrafish strains, sister-brother mating was performed for many consecutive generations. A basic strategy for inbreeding is schematically represented in Figure 1. Typically, five males and five females were selected from offspring obtained from a single pair of parents. Five single-pair matings were established from them, and embryos from each pair were raised to adulthood. To determine which pair’s offspring was intercrossed to generate the next generation, the following criteria were used: (1) average number of eggs laid; (2) fertilization efficiency; (3) average number of embryos surviving until three days postfertilization (dpf); (4) survival rate of embryos at three dpf; (5) normal development in the most embryos; (6) normal growth into healthy adult fish in the most larvae; and (7) sex ratio. If most pairs of a given generation revealed problems in any of these parameters, additional pairs were generated, and the process was repeated. Paired fish of generation n were fixed in 100% ethanol and placed at –30° after confirmation that their offspring were producing viable progeny (generation n + 2). If a paired fish was found dead before this confirmation was possible, the deceased fish was fixed in 100% ethanol and immediately placed at –30°. Closed colonies from two distinct branches of the IM family were maintained through mass-mating of healthy-looking females and males (three to eight each). Significant differences between the fertility data at generation n and those at generation 0 were tested by t-test.
Figure 1.
Schematic representation of typical inbreeding strategy. (1) Five pairs at generation n (Gn) were generated from offspring originated from a single pair of fish at generation n-1 (Gn-1). (2) Embryos (Gn+1) from each pairs were obtained and raised to adulthood. Data corresponding to the fertility of each pair were recorded. (3) The best pair at generation n (pair 1 in this figure) was determined based on the fertility data (see Materials and Methods for detailed criteria). (4) Inbreeding was repeated from (1) for the next generation; i.e., five pairs at generation n + 1 (Gn+1) were generated from the offspring of the best pair at generation n.
Genetic monitoring
During the course of the experiment, the fish were genotyped to monitor genetic homogeneity and to guard against contamination. Genotyping was performed using the simple sequence length polymorphisms (SSLP) listed on the ZFIN web site. Positional information for each marker was obtained from ZFIN and the Massachusetts General Hospital Zebrafish server (Shimoda et al. 1999). The allele size of each SSLP marker was determined for both TM and IM founder pairs (generation 0), and markers that were polymorphic in the original pair were identified. Furthermore, using four progeny from each strain (generation 1), codominant inheritance of the selected markers was confirmed. Genomic DNA was extracted from fin-clips of living fish or one-quarter of an ethanol-fixed fish using the Maxwell 16 Automated Purification System (Promega, Tokyo, Japan). Each fish’s genotype was determined using methods described in Kimura et al. (2005), with some modifications.
Scale transplantation
Two fish were anesthetized using 0.01% ethyl 4-aminobenzoate (w/v) and placed on a wet Kimwipe with the lateral side up. For autotransplantation, one dorsal and one ventral scale were removed using forceps. The dorsal scale then was inserted into the ventral position (where the ventral scale had just been removed). For transplantations between individuals, one dorsal scale from the donor fish was inserted into the ventral region of the recipient fish. Dorsal scales contain melanophores and, therefore, are easily identified within the recipient’s nonpigmented ventral region. The day after transplantation, the presence of the donor scale was confirmed. If the transplanted scale was missing at this time, the operation was deemed a failure. Between posttransplantation days 3 and 24, the transplanted scale was checked frequently (posttransplantation days 3 to 9, every two days; posttransplantation days 10–24, every three days) to monitor for allograft rejection. Significant differences between autotransplantation and transplantations between individuals were tested by Fisher’s exact test.
Results
TM inbreeding
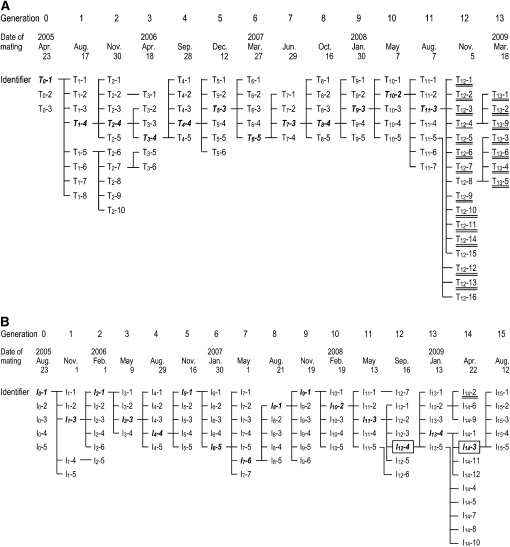
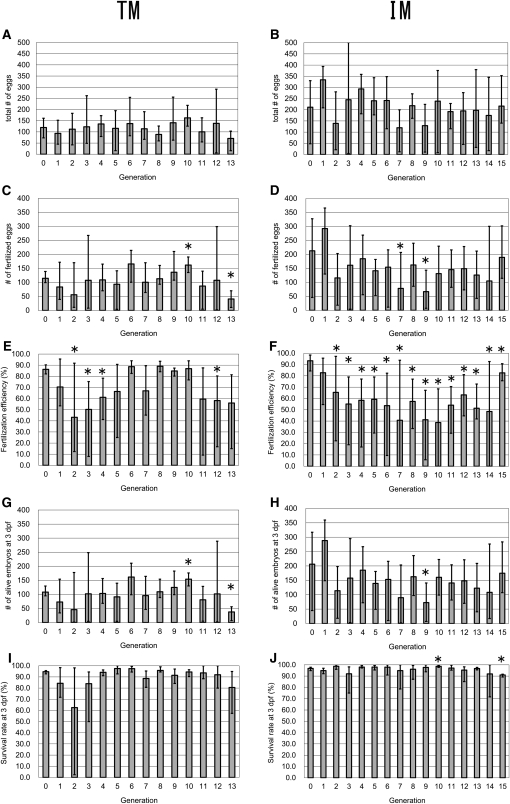
Tübingen is a heterogeneous, wild-type strain of zebrafish, and in April 2005, inbreeding was initiated with three independent single-pair matings (generation 0) (Figure 2A). Complete fertility records for each pair in each generation are shown in Table S1. The first obstacle to inbreeding arose in generation 2 (Figure 3C, E, G, I). At this point, the average number of fertilized eggs per clutch was 56 (the lowest value measured during the study, except for the value in generation 13), and the fertilization efficiency was only 43.2%, significantly lower than the value at generation 0. At three dpf, the number of living embryos and the survival rate (each measured on a per clutch basis) also were quite low (46 and 62.5%, respectively), although the differences were not significant compared to the data at the beginning of the inbreeding. In this generation, 8 of 10 pairs yielded less than 50 living embryos at three dpf per clutch. Because of generation 2’s fertility problems, additional pair matings were established from generation 1 fish, and two subfamily branches (one originating from T1-4 and the other from T1-5) were maintained (Figure 2A). Because of these difficulties, it took approximately five months to establish the next generation successfully, whereas the typical zebrafish generation time is three months (Laale 1977) (Figure 2A). After generation 4, no additional major problems arose, and the TM strain was successfully propagated until generation 11 (Figures 2A and 3). The body size and lifespan seemed the same as that of the original strain, although most TM fish were usually disposed of after around one year to save space in the fish facility. However, a biased sex ratio was observed in pair matings of generation 2 (T2-2, T2-3, and T2-8), generation 5 (T5-1, T5-4), and generation 6 (T6-2). In each case, the progeny from the crosses were predominantly male.
Figure 2.
Pedigree of the highly homogeneous strains. (A) Pedigree of the TM strain. (B) Pedigree of the IM strain. Bolded paired identifiers represent the pair whose offspring were kept inbreeding. Single-underlined paired identifiers represent pairs that produced some embryos with the TM phenotype, whereas double-underlined paired identifiers represent pairs that only produced embryos with the TM phenotype. Two closed colonies (IM12m and IM14m) were established from the pairs in boxes.
Figure 3.
Fertility of the TM and IM strains at each generation. Graphed data from TM (A, C, E, G, and I) and IM (B, D, F, H, and J) are shown. (A, B) Number of eggs per clutch. (C, D) Number of fertilized eggs per clutch. (E, F) Percentage of fertilized eggs per clutch (fertilization efficiency). (G, H) Number of embryos that survived until three dpf per clutch. (I, J) Percentage of embryos that survived until three dpf per clutch. Bars indicate mean values; error bars indicate range of values. Both maximum (up) and minimum (down) values are indicated. Asterisks indicate the significant differences (P < 0.05) from the values observed in generation 0.
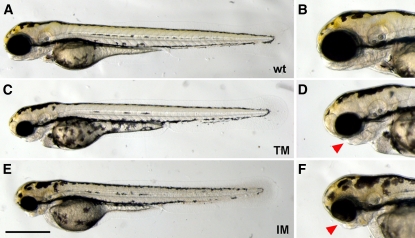
The second major difficulty arose in generation 12 of the TM strain. Most pairs of generation 12 produced embryos with phenotypes that included a small head, an underdeveloped lower jaw, and poor circulation (Figure 4). These phenotypes were collectively designated the “TM phenotype.” Embryos with the TM phenotype died within 10 dpf from cell necrosis within the brain or starvation or both. As a result, additional single-pair matings were established from generation 12 fish. Pairs T12-8, T12-15, and T12-16 produced phenotypically normal embryos, whereas T12-4 produced both normal embryos and embryos with the TM phenotype (Figure 2A, Table S1). The remaining 12 single-pair matings, however, only produced embryos with the TM phenotype. Normal embryos obtained from the above single-pair matings were raised to adulthood. Unfortunately, the T12-15 and T12-16 progeny were all male. In generation 13, therefore, only fish from T12-4 and T12-8 were available for sib-pair mating (Figure 2A). All progeny from generation 13, however, exhibited the TM phenotype. In a final attempt to overcome this problem, additional single-pair matings were established from generation 11 (T11-6 and T11-7). Embryos from these crosses appeared normal and healthy; however, all T11-7 progeny were male, and the T11-6 progeny did not produce any fertilized eggs. Therefore, despite much effort, inbreeding of the TM strain ended at generation 13.
Figure 4.
The TM phenotype. Embryos with the TM phenotype had small heads and underdeveloped jaws. Lateral views of the embryos are shown. (A, B) Wild-type embryo. (C, D) TM embryo with the TM phenotype. (E, F) IM embryo with the TM phenotype. Arrowheads mark the underdeveloped jaws. Scale bar is 0.7 mm.
IM inbreeding
Inbreeding of the IM strain began in August 2005 (Figure 2B). India is a wild-type strain that has been used often as the reference line for mapping mutations (Geisler 2002). Inbreeding of India fish was initiated with five single-pair matings (generation 0). Complete fertility records for each pair in each generation are shown in Table S2. Successful progression from one generation to the next took between two-and-one-half and four months for IM (Figure 2B), indicating a relatively smooth inbreeding process.
As with TM, the fertilization efficiency of the IM strain decreased soon after the inbreeding process was initiated. IM efficiency remained significantly low (approximately 50%, Figure 3F). In contrast, TM fertilization efficiency had recovered by generation 6. However, because IM females tended to lay more eggs than TM females (Figure 3A, B), the number of fertilized eggs and surviving embryos at three dpf were mostly higher in IM compared with TM (Figure 3C, D, G, H). Furthermore, although the value was still significantly lower than that of generation 0, the fertilization efficiency of IM suddenly increased to ∼80% at generation 15, providing hope that this parameter may improve in subsequent generations. The survival rate at three dpf did not vary much during the course of the experiment (Figure 3J). Surprisingly, one pair (I14-2) produced embryos with a phenotype indistinguishable from the TM phenotype (Figure 2B, compare Figure 4E, F with Figure 4C, D). Therefore, we established additional single-pair matings using IM fish from some pairs in generation 13 (I13-2, I13-4, and I13-5). To be safe, generation 15 single-pair matings were prepared from I14-3, which had different parents than I14-2 (Figure 2B). Phenotypically wild-type embryos were obtained from all generation 15 pairs, and the progeny, the IM fish of generation 16, have been growing well. During earlier stages of the IM inbreeding process, there were examples of unbalanced sex ratios. I3-1, I3-2, and I7-1 produced primarily male progeny; however, this did not develop into a persistent problem. In addition, generation 15 and generation 16 fish generally looked a little bit smaller than fish of their original strain, but they appeared healthy in all other ways and consistently produced fertilized eggs without artificial insemination. Just like fish in the India strain, their original strain, most of IM fish were usually in good condition until they were disposed of to surrender space (about one year, normally). We will continue to propagate the IM strain through sib-pair matings.
As was learned from the TM strain, continuous sib-pair mating carried the risk of strain extermination. To avoid complete loss of the genetically homogeneous IM strain, therefore, we also established two IM branch families and maintained them as closed colonies. I12-4 and I14-3 served as founder pairs of these colonies (Figure 2B), and they were named IM12m and IM14m, respectively. These strains have been successfully propagated for two to four generations by mass mating.
Genetic assessment of the strains inbred
To monitor inbreeding progress and to avoid strain contamination, the pairs whose progeny was crossed in the next generation were frequently used in a genetic analysis. Each fish was genotyped using two genetic markers per chromosome (50 markers in all). These 50 markers were prepared for the TM and IM strains as listed in Table S3 and Table S4, respectively. Polymorphic markers in founder fish were included in the marker sets as much as possible. In generation 0 of the TM and IM strains, polymorphisms were detected at 52% and 84% of the examined loci, respectively (Table 1). By generation 9, the number of polymorphic makers had decreased to 12% (TM) and 8% (IM), indicating a dramatic loss of genetic diversity. In the TM strain, 8% of the markers were polymorphic at generation 11, whereas in IM, only 2% of the markers were polymorphic by generation 13. No unexpected alleles were detected in this analysis, suggesting a lack of contamination during the inbreeding process.
Table 1. Number of polymorphic markers within the TM and IM strains.
| Generation | ||||||||
|---|---|---|---|---|---|---|---|---|
| Marker Set | Strain | 0 | 9 | 11 | 13 | 14 | 15 | |
| TM marker set (n = 50) | TM | n | 26 | 6 | 4 | ND | ND | ND |
| (%) | 52.0 | 12.0 | 8.0 | ND | ND | ND | ||
| IM marker set (n = 50) | IM | n | 42 | 4 | ND | 1 | 1 | 1 |
| (%) | 84.0 | 8.0 | ND | 2 | 2.0 | 2.0 | ||
| IM marker set + additional marker set (n = 100) | IM | n | 62 | ND | ND | ND | 5 | ND |
| (%) | 62.0 | ND | ND | ND | 5.0 | ND | ||
ND, not determined.
To determine more precisely which genomic regions remained polymorphic within the IM strain, an additional 50 genetic markers were analyzed. This “additional marker set” contained two more markers per chromosome than the original IM marker set. The I14-3 pair, which consisted of I14-M3 and I14-F3, was chosen for this analysis. In addition, three fish from IM14m that had been mass-mated for two generations (IM14m2), three fish from IM12m that had been mass-mated for one generation (IM12m1), and three fish from IM12m that had been mass-mated for three generations (IM12m3) were included in the analysis. The 100 markers covered the zebrafish genome with an average interval of 19.4 ± 12.3 cM. The alleles found in these fish are shown in Table 2. At generation 0, 62% of the markers were polymorphic, whereas only 5% were polymorphic in I14-3 (Table 1) and IM12m1 (data not shown). Alleles that remained polymorphic at late stages of inbreeding included: Z6802 at LG1; Z191 at LG7; Z6867 at LG8; Z6895 at LG15; and Z4003 at LG23. Four of the five markers (except for Z6895 at LG15) also were polymorphic in IM12m3. In addition, four of the five markers (except for Z6867 at LG8) were polymorphic in IM14m2. For each of these IM strains, therefore, ∼95% of the genome was homozygous. As such, we have successfully generated highly inbred and genetically homogeneous strains of zebrafish by sequential sib-pair mating.
Table 2. Alleles present in IM (generation 14), IM14m2, and IM12m3.
| LG | Markera | Position (cM) | Allele | LG | Markera | Position (cM) | Allele |
|---|---|---|---|---|---|---|---|
| 1 | Z9394 | 8.5 | 279 | 14 | Z9857 | 10.5 | 215 |
| Z5508 | 24.2 | 208 | Z5435 | 34.6 | 110 | ||
| Z6802 | 61.3 | 187, 191 | Z8801 | 55.5 | 229 | ||
| Z6223 | 85.1 | 210 | Z11837 | 91.9 | 133 | ||
| 2 | Z27170 | 12.9 | 282 | 15 | Z6312 | 9.7 | 188 |
| Z9361 | 43.1 | 211 | Z6895 | 49.3 | 128, 132c | ||
| Z10838 | 57.0 | 205 | Z9773 | 85.2 | 212 | ||
| Z9037 | 86.2 | 119 | Z10193 | 97 | 151 | ||
| 3 | Z11244 | 14.0 | 230 | 16 | Z14570 | 3.6 | 184 |
| Z15457 | 40.9 | 128 | Z9881 | 27.7 | 195 | ||
| Z22516 | 64.9 | 155 | Z9685 | 43.4 | 192 | ||
| Z7486 | 100.6 | 181 | Z9269 | 78.9 | 251 | ||
| 4 | Z1366 | 10.9 | 106 | 17 | Z6010 | 2.3 | 138 |
| Z20533 | 26.0 | 206 | Z22674 | 33.5 | 162 | ||
| Z11250 | 44.9 | 147 | Z9692 | 64.5 | 251 | ||
| Z7524 | 61.6 | 96 | Z21144 | 81.1 | 167 | ||
| 5 | Z15414 | 3.6 | 190 | 18 | Z11685 | 4.3 | 223 |
| Z8921 | 28.2 | 255 | Z9231 | 26.2 | 267 | ||
| Z9871 | 54.0 | 181 | Z8343 | 49.4 | 197 | ||
| Z9185 | 103.0 | 86 | Z7961 | 75.4 | 182 | ||
| 6 | Z1265 | 3.5 | 104 | 19 | Z4009 | 12.7 | 242 |
| Z880 | 25.2 | 170 | Z4825 | 28.1 | 243 | ||
| Z10183 | 51.3 | 240 | Z7686 | 48.8 | 162 | ||
| Z9230 | 81.0 | 186 | Z9512 | 80.8 | 153 | ||
| 7 | Z191 | 1.9 | 120, 122 | 20 | Z6804 | 7.9 | 184 |
| Z9133 | 35.5 | 132 | Z10056 | 38.9 | 259 | ||
| Z1239 | 70.6 | 186 | Z21067 | 64.9 | 188 | ||
| Z13936 | 85.8 | 190 | Z21485 | 115.8 | 146 | ||
| 8 | Z1637 | 4.9 | 101 | 21 | Z58858 | 1.4 | 127 |
| Z11001 | 39.9 | 144 | Z6869 | 49.4 | 129 | ||
| Z6867 | 62.3 | 220, 236b | Z1497 | 119 | 251 | ||
| Z9279 | 81.5 | 151 | Z6537 | 125.2 | 110 | ||
| 9 | Z8348 | 8.3 | 263 | 22 | Z9516 | 6.8 | 177 |
| Z11785 | 48.8 | 190 | Z13794 | 26.7 | 225 | ||
| Z5564 | 74.5 | 236 | Z3286 | 59.2 | 89 | ||
| Z7564 | 86.7 | 151 | Z4682 | 68.7 | 292 | ||
| 10 | Z10251 | 8.6 | 221 | 23 | Z1668 | 0 | 149 |
| Z8146 | 27.9 | 147 | Z4003 | 16.6 | 243, 247 | ||
| Z3835 | 48.7 | 153 | Z5141 | 36.6 | 88 | ||
| Z7558 | 77.3 | 150 | Z4120 | 60.1 | 137 | ||
| 11 | Z22552 | 0.0 | 137 | 24 | Z10961 | 1.2 | 132 |
| Z13411 | 22.6 | 236 | Z23011 | 36 | 220 | ||
| Z13666 | 51.4 | 273 | Z13229 | 53.5 | 159 | ||
| Z11809 | 69.5 | 131 | Z6923 | 75.7 | 100 | ||
| 12 | Z7135 | 6.3 | 170 | 25 | Z21302 | 4.3 | 158 |
| Z9891 | 32.0 | 186 | Z1197 | 10.9 | 210 | ||
| Z4830 | 55.0 | 158 | Z21055 | 42.5 | 133 | ||
| Z1312 | 78.7 | 147 | Z1431 | 67.8 | 172 | ||
| 13 | Z9878 | 4.7 | 268 | ||||
| Z6104 | 25.2 | 179 | |||||
| Z17223 | 51.5 | 125 | |||||
| Z6007 | 80.7 | 128 | |||||
LG, linkage group.
Markers belonging to the IM marker set are in bold.
The allele in IM14m2 was 220.
The allele in IM12m3 was 132.
Tolerance for tissue transplantation within the IM strain
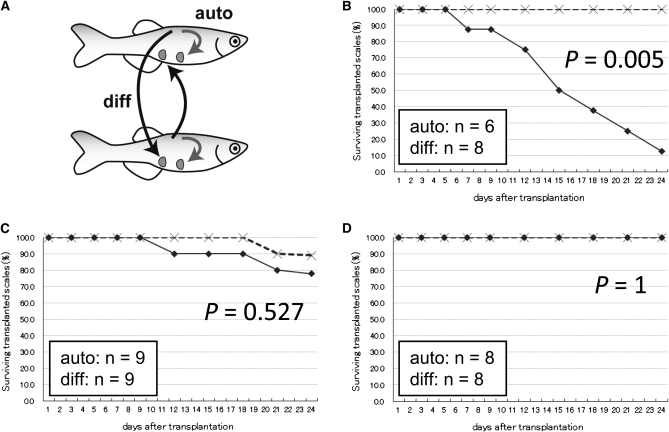
As with other animals, adult zebrafish that are part of an outbred colony reject transplanted tissue from another individual (Figure 5B). In mice, however, successful tissue transplantation between individuals occurs if the two mice originated from the same inbred strain (Medina 2010). Similar results have been seen in medaka after 8 generations of sib-pair mating (Hyodo-Taguchi and Egami 1985). To test whether tissue transplantation between adult individuals was possible in zebrafish strains inbred for many generations, scale transplantation experiments were performed (Figure 5A). IM animals from generations 14 and 16, as well as IM12m1, were chosen for analysis. In autograft experiments (in which a dorsal scale is place in a ventral region of the same individual), 86–100% of the autografted scales remained at the transplanted region 24 days after transplantation (Figure 5B–D). In contrast, most grafts between individuals of the outbred India strain were rejected and, therefore, not detected except for one scale at 24 days after transplantation (Figure 5B, P = 0.005 by Fisher’s exact test). When scales were transplanted between IM individuals (generation 14), 78% of the scales survived 24 days (Figure 5C); this did not represent a significant difference from autograft experiments (P = 0.527 by Fisher’s exact test). For both I16 (Figure 5D) and IM12m1 (data not shown), no grafted scales were rejected (P = 1 by Fisher’s exact test). These strains of zebrafish inbred for 12 or more generations are, therefore, sufficiently homogeneous to support transplantation of cells between adult individuals.
Figure 5.
Scale transplantation experiments. (A) Schematic representation of scale transplantation. A dorsal scale was always transplanted to the ventral side. The “diff” (black arrows) indicates transplantation between individuals, and “auto” (gray arrows) indicates transplantation within the same fish. (B–D) The percentage of surviving transplanted scales during the 24 days after transplantation. Crosses indicate autotransplantation, and diamonds indicate transplantation between individuals. Transplantation data from the India outbred strain (B), IM fish at generation 14 (C), and IM fish at generation 16 (D) are shown. P-values were calculated from the data at 24 days after transplantation.
Discussion
In this study, we inbred two wild-type zebrafish strains, TM and IM, by full sib-pair mating. Although the TM strain was lost after 13 generations, the IM strain successfully produced offspring from pairs at generation 15; that is, the IM strain endured 16 generations of sib-pair mating. Furthermore, two additional strains (IM12m and IM14m) were established as closed colonies. Genotype analysis revealed that polymorphisms associated with the IM founder pair (62% of the interrogated genetic markers) were dramatically reduced (5%) in IM, IM12m, and IM14m. These data suggest a high level of genetic homogeneity within each strain. In fact, allograft rejection was not observed between individuals within these strains. Together, the IM, IM12m, and IM14m strains represent important resources for the zebrafish scientific community, particularly in the fields of complex trait analysis. Furthermore, the inbreeding strategy described here will be a good reference to establish more strains of zebrafish that are highly homogeneous. Those strains are available from M. Shinya by request.
Highly homogeneous zebrafish strains: IM, IM12m, and IM14m
The genetic analysis of SSLP markers showed that the genomes of the IM, IM12m, and IM14m strains were ∼95% nonpolymorphic (Tables 1 and 2). The coefficient of inbreeding is the probability that two alleles at a randomly chosen locus in an individual are identical by descents, and it is theoretically calculated as 0.951 for an individual generated by full sib-pair mating for 14 generations (Falconer 1989). Therefore, our estimation that ∼95% of the genomes were homozygous in the IM and its related strains is consistent with the theoretical value, even though we examined only 100 loci. To determine more precisely the degree of polymorphism within these strains, it will be necessary to genotype more genetic markers, including SNPs. Alternatively, direct sequencing of these strains will provide the most accurate assessment of genetic homogeneity. In this regard, we have begun sequencing two genomes from I14-3 (I14-M3 and I14-F3), which will identify polymorphisms within the IM strain.
Highly homogeneous strains of zebrafish are typically difficult to manipulate and maintain. Problems associated with these strains include physiological weakness, biased sex ratios, and the inability to perform natural crosses. Homozygous diploid zebrafish lines C32 and SJD (Johnson et al. 1995; Streisinger et al. 1981) were shown to be homozygous in over 90% of genomes through SSLP marker analyses (Nechiporuk et al. 1999). Unfortunately, C32 lacked vigor, and SJD progeny were predominantly male. Reciprocal introgression of genes to overcome these phenotypes was attempted by crossing C32 and SJD (Rawls et al. 2003). The IM strain and two related closed colonies were shown to be ∼95% homozygous. As a result, they are somewhat weaker physiologically than heterogeneous populations (e.g., lower tolerance for sudden changes to pH or temperature). These weaknesses, however, have not hindered their propagation or maintenance. Furthermore, all three strains are capable of laying fertilized eggs under natural-cross conditions. While inbreeding IM, we occasionally observed biased sex ratios. In generations 3 and 7, some pairs predominantly yielded males (see Results), but after generation 8, no further gender bias was detected. No sex ratio bias has been seen for either IM12m or IM14m. Taken together, IM strains do not exhibit the phenotypic weaknesses that typically plague homogeneous populations of zebrafish. Careful selection for those phenotypes in each generation may have contributed to this successful outcome.
The genetic homogeneity and health condition of IM, IM12m, and IM14m make them ideally suited for experimental manipulation. Testicular cell grafting experiments have already been performed using these strains (Kawasaki et al. 2010). To effectively utilize these strains in genetic analyses of quantitative traits, however, it will be necessary to generate at least one additional inbred strain from a different genetic background (i.e., not India). We have begun additional inbreeding by following the inbreeding strategy described here with some modifications to attempt to generate more homogeneous lines from other zebrafish wild-type strains.
TM phenotype
TM inbreeding failed at generation 13 because of the TM phenotype (Figure 4). This phenotype arose so suddenly and with such a high frequency (almost every embryo from generation 12 breeding pairs was affected) (Figure 2A) that propagation of the line was not possible. Although TM and IM were raised under essentially identical conditions, the TM phenotype primarily affected only one line. It is likely, therefore, that a genetic factor(s) caused this condition. Phenotypes associated with maternal contributions often arise quickly, but the TM phenotype does not seem to be inherited maternally. One female (T12-F13) produced embryos with the TM phenotype when crossed with T12-M13 but yielded wild-type embryos when crossed with T12-M14 (Table S1). The pattern of phenotypic appearance within the TM family (rapid and essentially ubiquitous) makes it difficult to explain this phenomenon with a single factor. Many factors (perhaps including environmental factors) may have been involved in the phenotypic expression. Genomic comparisons between zebrafish parents that produce embryos with either the wild-type or TM phenotype may lead to identify the causative loci. As we have sampled most pairs for DNA preparation, we are now planning to perform the genomic analyses using those samples to elucidate the genetic basis of the TM phenotype.
The TM phenotype was also seen in a single cross of the IM strain, which was derived from a completely different genetic background. Furthermore, similar phenotypes have been observed in another wild-type strain, AB, which was established long ago in Oregon (H. Yokoi, personal communication). These observations suggest that the TM phenotype is generally associated with increased homozygosity of the zebrafish genome. “Inbreeding depression” is a term used to describe the reduced fitness of a given population that results from breeding related individuals (Charlesworth and Willis 2009). The TM phenotype exemplifies inbreeding depression. Then, in addition to the inbreeding problems described above, how to cope with the TM phenotype is an important issue to establish inbred strains in zebrafish. One way to avoid extinction caused by TM phenotype is the way taken in the IM inbreeding at generation 14: generating many branches and for the next generation, selecting a single-mating pair whose parents are different from those of pairs with TM phenotype (Figure 2B). Alternatively, we can avoid the phenotype, if the causative loci are identified. In this sense, the genomic analyses proposed above will provide a big help to establish inbred strains in zebrafish more easily.
Level of inbreeding depression in zebrafish
Inbreeding depression presents a major difficulty in generating and maintaining highly homogeneous strains. The degree of inbreeding depression varies from species to species. Quail, for example, exhibit remarkably strong inbreeding depression (Sato 1986). The lethal equivalent (LE) is an index that attempts to quantify inbreeding depression. An LE is defined as a set of alleles that would cumulatively cause an individual to fail to produce its offspring, if all alleles in the set are homozygous in the individual (Morton et al. 1956). The number of LEs in quail has been estimated at 8.7 (Sittmann et al. 1966), whereas values of 1.4 to 3.6 LEs have been reported for zebrafish (McCune et al. 2002; McCune et al. 2004). It seems clear, therefore, that zebrafish exhibit less inbreeding depression than birds, a fact that contributed to the success of our IM project.
In both mice and medaka, inbreeding depression is weak enough to allow 20 or more generations of continuous sib-pair mating. However, there is a clear difference between these two species. Inbred strains of medaka are generated quite easily, as only two to three single-pair matings are typically established and tested per generation (Taguchi 1980). In contrast, sibling crosses in mice generate many sterile animals and require numerous single-pair matings at each generation, especially the first generation (Silver 1995). This inbreeding depression typically begins to diminish by generation 8. Therefore, inbreeding depression is stronger in mice than in medaka. In this study, we typically established five single-pair matings per generation. Although we lost one strain, IM remains maintained by continuous sib-pair mating. These data suggest that inbreeding depression in zebrafish is greater than in medaka but likely less than in mice.
Inbreeding depression is caused by increased homozygosity for a single locus with heterozygous advantages and/or a single or multiple deleterious mutations with recessive effects (reviewed in Charlesworth and Willis 2009). It is interesting and important to identify the genomic factors involved in inbreeding depression and in its variation among species. Although some genetic analyses have been performed for the traits related to fitness (Remington and O’Malley 2000), they have not identified those factor(s) yet. Together with the quantitative trait loci (QTL) analyses for the fitness traits, genomic sequencing analyses using TM and IM fish in some generations might give some insights into the factor(s) related to inbreeding depression. Those kinds of data from several organisms will provide information about the genomic factor(s) that contribute to the variation of inbreeding depression among those species.
Generation of zebrafish inbred strain by full sib-pair mating
In mice and rats, an inbred strain is defined as a population that was established with a single ancestral pair and has been inbred (brother × sister) for 20 or more consecutive generations (Guidelines for Nomenclature of Mouse and Rat Strains). This definition also is used by the medaka community (Taguchi 1990). Although homogeneous lines have been generated in zebrafish through gynogenesis (Johnson et al. 1995; Streisinger et al. 1981), no zebrafish strains have been established that meet the inbred strain definition used by the mice and medaka scientific communities. To our knowledge, the 16 generations of continuous sib-pair mating that characterize the IM strain represent the longest such effort to date. With 4 more generations of careful inbreeding (for a total of 20 generations), the IM strain will become the first inbred strain of zebrafish that meets the definition of inbred strain used for mice and medaka.
Supplementary Material
Acknowledgments
We thank Ms. Kimiko Saka, Ms. Tomoko Hoshikawa, and Ms. Asami Takagi for their valuable assistance with this study; Ms. Michiyo Kojima for fish maintenance; and all other laboratory members for their help feeding fish over the holidays. We also thank Drs. Atsuko Shimada, Kiyoshi Naruse, and Koichi Kawakami for their helpful discussions. The work presented here was supported by Grant-in-Aid for Young Scientists (B) from Japan Society for the Promotion of Science and, in part, by the Center for the Promotion of Integrated Sciences (CPIS) of Sokendai.
Literature Cited
- Anzai T., Ullmann L. G., Hayashi D., Satoh T., Kumazawa T., et al. , 2010. Effects of strain differences and vehicles on results of local lymph node assays. Exp. Anim. 59: 245–249 [DOI] [PubMed] [Google Scholar]
- Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., et al. , 2011. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 39: 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Willis J. H., 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10: 783–796 [DOI] [PubMed] [Google Scholar]
- Falconer D. S., 1989. Introduction to Quantitative Genetics. Longman, Essex, UK [Google Scholar]
- Frankel W. N., 1995. Taking stock of complex trait genetics in mice. Trends Genet. 11: 471–477 [DOI] [PubMed] [Google Scholar]
- Geisler R., 2002. Mapping and cloning, pp. 175–212 in Zebrafish, edited by Nusslein-Volhard C., Dahm R. Oxford University Press, New York [Google Scholar]
- Guryev V., Koudijs M. J., Berezikov E., Johnson S. L., Plasterk R. H., et al. , 2006. Genetic variation in the zebrafish. Genome Res. 16: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Granato M., Brand M., Mullins M. C., Hammerschmidt M., et al. , 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1–36 [DOI] [PubMed] [Google Scholar]
- Hyodo-Taguchi Y., Egami N., 1985. Establishment of inbred strains of the medaka Oryzias latipes and the usefulness of the strains for biomedical research. Zoolog. Sci. 2: 305–316 [Google Scholar]
- Johnson S. L., Africa D., Horne S., Postlethwait J. H., 1995. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics 139: 1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Yoshida K., Shimada A., Jindo T., Sakaizumi M., et al. , 2005. Genetic linkage map of medaka with polymerase chain reaction length polymorphisms. Gene 363: 24–31 [DOI] [PubMed] [Google Scholar]
- Kimura T., Shimada A., Sakai N., Mitani H., Naruse K., et al. , 2007. Genetic analysis of craniofacial traits in the medaka. Genetics 177: 2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Saito K., Shinya M., Olsen L. C., Sakai N., 2010. Regeneration of spermatogenesis and production of functional sperm by grafting of testicular cell aggregates in Zebrafish (Danio rerio). Biol. Reprod. 83: 533–539 [DOI] [PubMed] [Google Scholar]
- Klingenberg C. P., Leamy L. J., Routman E. J., Cheverud J. M., 2001. Genetic architecture of mandible shape in mice: effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157: 785–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg C. P., Leamy L. J., Cheverud J. M., 2004. Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics 166: 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laale H. W., 1977. The biology and use of zebrafish, Brachydanio rerio in fisheries research: a literature review. J. Fish Biol. 10: 121–173 [Google Scholar]
- Markel P., Shu P., Ebeling C., Carlson G. A., Nagle D. L., et al. , 1997. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17: 280–284 [DOI] [PubMed] [Google Scholar]
- McCune A. R., Fuller R. C., Aquilina A. A., Dawley R. M., Fadool J. M., et al. , 2002. A low genomic number of recessive lethals in natural populations of bluefin killifish and zebrafish. Science 296: 2398–2401 [DOI] [PubMed] [Google Scholar]
- McCune A. R., Houle D., Mcmillan K., Annable R., Kondrashov A. S., 2004. Two classes of deleterious recessive alleles in a natural population of zebrafish, Danio rerio. Proc. Biol. Sci. 271: 2025–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D., 2010. Of mice and women: a short history of mouse mammary cancer research with an emphasis on the paradigms inspired by the transplantation method. Cold Spring Harb. Perspect. Biol. 2: a004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgirev I., Revskoy S., 2010. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat. Protoc. 5: 383–394 [DOI] [PubMed] [Google Scholar]
- Morel L., Yu Y., Blenman K. R., Caldwell R. A., Wakeland E. K., 1996. Production of congenic mouse strains carrying genomic intervals containing SLE-susceptibility genes derived from the SLE-prone NZM2410 strain. Mamm. Genome 7: 335–339 [DOI] [PubMed] [Google Scholar]
- Morton N. E., Crow J. F., Muller H. J., 1956. An estimate of the mutational damage in man from data on consanguineous marriages. Proc. Natl. Acad. Sci. USA 42: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K., Hori H., Shimizu N., Kohara Y., Takeda H., 2004. Medaka genomics: a bridge between mutant phenotype and gene function. Mech. Dev. 121: 619–628 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A., Finney J. E., Keating M. T., Johnson S. L., 1999. Assessment of polymorphism in zebrafish mapping strains. Genome Res. 9: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H., Shimizu T., Kim C. H., Yabe T., Bae Y. K., et al. , 2004. Genetic evidence for involvement of maternally derived Wnt canonical signaling in dorsal determination in zebrafish. Mech. Dev. 121: 371–386 [DOI] [PubMed] [Google Scholar]
- Nomura T., 1991. Paternal exposure to radiation and offspring cancer in mice: reanalysis and new evidence. J. Radiat. Res. (Tokyo) 32(Suppl. 2): 64–72 [DOI] [PubMed] [Google Scholar]
- Peters L. L., Robledo R. F., Bult C. J., Churchill G. A., Paigen B. J., et al. , 2007. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8: 58–69 [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Frieda M. R., Mcadow A. R., Gross J. P., Clayton C. M., et al. , 2003. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics 163: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington D. L., O'Malley D. M., 2000. Whole-genome characterization of embryonic stage inbreeding depression in a selfed loblolly pine family. Genetics 155: 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., 1986. Inbreeding depression in Japanese quail. Proceedings of Okayama Association for Laboratory Animal Science, Okayama, Japan. 4: 18–23 [Google Scholar]
- Shimoda N., Knapik E. W., Ziniti J., Sim C., Yamada E., 1999. Zebrafish genetic map with 2000 microsatellite markers. Genomics 58: 291–232 [DOI] [PubMed] [Google Scholar]
- Silver L. M., 1995. Mouse Genetics. Oxford University Press, New York [Google Scholar]
- Sittmann K., Abplanalp H., Fraser R. A., 1966. Inbreeding depression in Japanese quail. Genetics 54: 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Walker C., Dower N., Knauber D., Singer F., 1981. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291: 293–296 [DOI] [PubMed] [Google Scholar]
- Taguchi Y., 1980. Establishment of inbred strains of the Medaka. Zoological Magazine 89: 283–301 [Google Scholar]
- Taguchi Y., 1990. Inbred strains and its characteristics, pp. 129–142 in Biology of Medaka, edited by Egami N., Yamagami K., Shima A. University of Tokyo Press, Tokyo, Japan [Google Scholar]
- Tomida S., Mamiya T., Sakamaki H., Miura M., Aosaki T., et al. , 2009. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 41: 688–695 [DOI] [PubMed] [Google Scholar]
- Xiao J., Liang Y., Li K., Zhou Y., Cai W., et al. , 2010. A novel strategy for genetic dissection of complex traits: the population of specific chromosome substitution strains from laboratory and wild mice. Mamm. Genome 21: 370–376 [DOI] [PubMed] [Google Scholar]
- Yamamura K., Tani M., Hasegawa H., Gen W., 2001. Very low dose of the Na(+)/Ca(2+) exchange inhibitor, KB-R7943, protects ischemic reperfused aged Fischer 344 rat hearts: considerable strain difference in the sensitivity to KB-R7943. Cardiovasc. Res. 52: 397–406 [DOI] [PubMed] [Google Scholar]
- Yui M. A., Muralidharan K., Moreno-Altamirano B., Perrin G., Chestnut K., et al. , 1996. Production of congenic mouse strains carrying NOD-derived diabetogenic genetic intervals: an approach for the genetic dissection of complex traits. Mamm. Genome 7: 331–334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.