Abstract
Polycomb group response elements (PRE) are cis-regulatory elements that bind Polycomb group proteins. We are studying a 181-bp PRE from the Drosophila engrailed gene. This PRE causes pairing-sensitive silencing of mini-white in transgenes. Here we show that the 181-bp PRE also represses mini-white expression in flies with only one copy of the transgene. To isolate mutations that alter the activity of the 181-bp PRE, we screened for dominant suppressors of PRE-mediated mini-white repression. Dominant suppressors of mini-white repression were rare; we recovered only nine mutations out of 68,274 progeny screened. Two of the nine mutations isolated are due to the same single amino acid change in the transcriptional activator Woc (without children). Reversion experiments show that these are dominant gain-of-function mutations in woc. We suggest that Woc can interfere with the activity of the PRE. Our data have implications for how Polycomb group proteins act to either partially repress or completely silence their target genes.
Keywords: polycomb repression, polycomb response elements, PRE, woc
Polycomb group genes (PcG) encode proteins that mediate transcriptional repression. First identified in Drosophila as genes necessary to maintain the silencing of homeotic genes, it is now evident that PcG proteins have many other targets (reviewed in Simon and Kingston 2009). Genome-wide studies show that the PcG repressive mark H3K27me3 is associated with hundreds of genes in single cell types and that targets can be cell-type specific (reviewed in Schwartz and Pirrotta 2008). Although it is evident that PcG proteins can decrease expression levels in addition to completely silencing expression, it is not clear what determines whether a gene will be completely or only partially repressed.
In Drosophila, PcG proteins are associated with Polycomb group response elements (PRE), DNA elements that recruit PcG proteins to the DNA (reviewed in Müller and Kassis 2006; Ringrose and Paro 2007). In genome-wide studies, PREs were identified as binding sites for multiple PcG proteins (Schwartz et al. 2006; Négre et al. 2006; Tolhuis et al. 2006). Two functional assays have also been used to identify PREs. In one assay, the PRE is combined in a transgene with regulatory DNA from a gene normally regulated by PcG proteins, where the PRE is required to maintain the “off” transcriptional state (Müller and Bienz 1991; Hagstrom et al. 1997). In the other assay, PREs are used to repress expression of the mini-white reporter gene in transgenic flies. Because mini-white repression is stronger in flies homozygous for the PRE-mini-white reporter, this latter assay has been called pairing-sensitive silencing (Kassis 1994).
One of the puzzles of the transgene assays for PREs is that silencing does not occur at every chromosomal insertion site. For example, for the four engrailed and invected PREs, pairing-sensitive silencing was observed at a frequency of 21–62% of insertion sites (Americo et al. 2002; Cunningham et al. 2010). PRE activity is regulated by the expression state of the gene it regulates; thus it follows that PRE activity in transgenes is dependent on the activity of regulatory elements that flank the transgene insertion site.
We have been studying a 181-bp en DNA fragment that acts as a PRE in several different assays: (1) it represses inappropriate expression in both en- and Ubx-reporter genes in embryos (Americo et al. 2002; Devido et al. 2008); (2) PcG proteins are associated with it in tissue culture cells, embryos, larvae, and adults (Strutt and Paro 1997; Négre et al. 2006; Oktaba et al. 2008); and (3) it acts as a pairing-sensitive silencing element (Kassis 1994). This fragment contains binding sites for the PRE DNA binding proteins Pho, Pho-like, GAGA factor, and Spps (Americo et al. 2002; Brown et al. 2005; Brown and Kassis 2010). Thus, the 181-bp DNA fragment is clearly a PRE. Therefore, we reasoned that conducting a genetic screen for mutations that alter the activity of this PRE might yield mutations in PcG genes.
We conducted a genetic screen for dominant suppressors of pairing-sensitive silencing by a transgene that contained the 181-bp en PRE and mini-white. These mutations were rare; we only obtained nine suppressors among 68,274 genomes screened. None of the mutations affected mini-white repression of transgenes at all chromosomal insertion sites. This suggests that none of the mutations affects PRE activity directly. Instead, we believe that these mutations affect the expression of genes flanking the transgene insertion site. Consistent with this, two of the dominant suppressors are the same gain-of-function mutation in the gene without children (woc), which encodes a transcriptional activator. Our data suggest that there is a competition between transcriptional activators and PcG repression and that certain types of activators may be better able to overcome PcG repression.
Materials and Methods
Mutagenesis
For EMS mutagenesis, adult males were fed EMS as described (Lewis and Bacher 1968; Kennison 1983), and then discarded 3–4 days following treatment to avoid pre-meiotic clusters of mutations. For the X-ray mutagenesis, males were irradiated with 30–40 Gy at 120 keV using a Faxitron Torrex 2800. The irradiated males were discarded 4–5 days following treatment.
Sequencing
DNA was isolated from homozygous or hemizygous mutant adults or larvae, and the entire woc transcription unit was sequenced.
Construction of P[L181PRE]
The 181-bp en PRE was amplified with the primers GCGGAATTCGAGATGGCATGTGGCTCT and GCGGAATTCGCATGCTGGAGCTGTCAG, cut with EcoRI, and cloned into EcoRI cut, phosphatased EK710, which contains loxP sites on both sides of the EcoRI site (Kuhn et al. 2004). A fragment of DNA containing the 181-bp PRE and flanking loxP sites was cut with NotI and cloned into NotI cut CaSpeR4. The resulting clone was sequenced to determine the orientation of the insert.
Generation and analysis of transgenic lines
P[L181PRE] was injected into homozygous Df(1)w67c23, y embryos using standard techniques. Some lines were generated by P-element mobilization by crossing to a strain with the endogenous transposase insertion P[ry+∆2, 3]99B (Robertson et al. 1988). P[L] derivative lines lacking the en181bp-PRE were obtained by crossing males with the P[L181PRE] insertion to virgin females that carried a constitutively active Cre recombinase transgene (y1w; CyO, P[Crew]/Sco) (Siegal and Hartl 1996). Progeny that contained both P[L181PRE] and CyO, P[w+Cre] were crossed to Df(1)w67c23, y flies. Two individual w+ male progeny were selected from each insertion line and crossed to the appropriate balancer chromosome. P[L] lines were established, and the deletion of the en181bp-PRE was confirmed by PCR with primers flanking the loxP sites.
qRT-PCR
Flies of the following genotypes were used: (1) w1118, (2) w1118; P[181PRE]8-10C, (3) w1118; wocD1, and (4) w1118;P[181PRE]8-10C; wocD1. Total RNA from 3rd instar larvae, 1-day-old pupae, or adult fly heads was prepared (Lorenz et al. 1989) and treated with DNase I before use. qRT-PCR was done with the QuantiTect SYBR Green RT-PCR kit (Qiagen) on the LightCycler 480 real-time PCR system (Roche Applied Sciences) using 0.2 μg total RNA/reaction. The following PCR primers were used: for the RpL32 reference gene, CGGATCGATATGCTAAGCTGT and CGACGCACTCTGTTGTCG, its amplicon is 67 bp; for CG30456, AAAATGCGCAACGATTTCC and AACTTGCCCACCAAATGCT, its amplicon is 95 bp; for GstS1, GTCAAGGACAACGATGGTCA and GGTGATGCCTGCGAAGTAG, its amplicon is 72 bp. Reverse transcription was done at 50° for 20 min, followed by incubation at 95° for 15 min to activate the PCR reaction. PCR was for 45 cycles of 94°, 10″, 60° 20″, 72°, 20″. After PCR, the reactions were heated to 95° and then cooled to 40° to analyze the melting temperatures of the PCR products.
Results
Dominant modifiers of mini-white repression
To recover mutations that affect pairing-sensitive silencing, we screened for dominant mutations that suppressed en181bp-mediated mini-white repression. We used the line P[181PRE]8-10C, which contains a P-construct with the 181-bp PRE of en DNA cloned into pCaSpeR (Construct 8 in Kassis 1994; Figure 1). pCaSpeR contains the mini-white gene; a truncated version of the white gene, which contains a promoter fragment that gives expression in the eye but no eye enhancer. The 181-bp PRE is cloned directly adjacent to the mini-white promoter. The w; P[181PRE]8-10C homozygotes have white eyes, and w; P[181PRE]8-10C heterozygotes have orange eyes (Figure 2). For the mutagenesis, we fed w; P[181PRE]8-10C males EMS and crossed to either w; P[181PRE]8-10C or w; P[181PRE]8-10C Sco/CyO females. We looked for mutations that darkened the eye color of either homozygotes or heterozygotes. We recovered nine mutations; one on the X chromosome, four on chromosome 2, and four on chromosome 3. All but two of the mutations darkened the eye color of both P[181PRE]8-10C homozygotes and heterozygotes. These mutations could identify genes involved in repression of mini-white transcription, perhaps via the PRE. One second-chromosome mutation darkened the eye color of heterozygotes only, which suggests that it is not involved in mini-white repression but might be involved in pigmentation. We did not study this mutation further. The sex-linked mutation only darkens the eye color of P[181PRE]8-10C homozygotes. The reason for this is unknown; however, it could mean that the mutation affects the interaction between PREs.
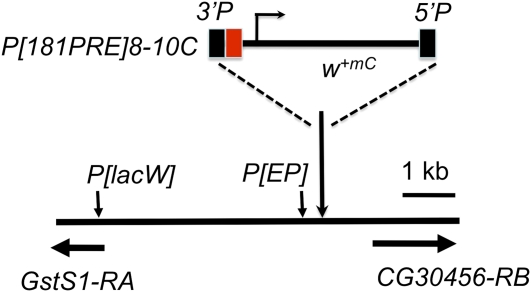
Figure 1 .
P[181PRE]8-10C construct and location in genome. 181-bp en PRE (red box) is inserted upstream of the mini-white (m+mC) gene in P[181PRE]. P[181PRE]8-10C is inserted between CG30456 and GstS1. The 5′ ends of the GstS1 and CG30456 transcription units are shown by the horizontal arrows. Vertical arrows indicate the insertion site of the EP element in P[EP]2185 and the approximate location of a cluster of six P[lacW] insertions located within 253 bp of each other near the GstS1 transcription start site. The insertion site of P[181PRE]8-10C is 2R:12989447, 925bp 5′ of the CG30456-RB transcription start site and 4512 bp from the 5′ end of GstS1-RA.
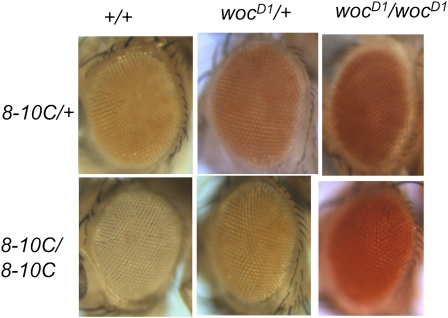
Figure 2 .
Eye colors of P[181PRE]8-10C flies in wildtype and wocD1 mutants. Pictures are of eyes of 1-day-old females. All flies were w1118/w1118, and either homozygous or heterozygous for the P[181PRE]8-10C insertion (designated by 8-10C in the figure) and wocD1 as indicated.
Two mutations cause the same single amino acid change in the transcriptional activator Woc
We mapped the mutations on chromosome 3 using the markers ru, h, th, cu, sr, es, and Pr. Two mutations mapped 2.5 map units distal to Pr. We next tried to recover recombinants between these two mutations. We found no recombinants among 448 progeny, suggesting that the two mutations are very close to each other and might be allelic. We tested whether several overlapping deletions for polytene chromosome region 96F–98B (which should include the mutations) caused a darkening of the P[181PRE]8-10C heterozygous eye color. As none did, we suspected that both mutations are gain-of-function alleles that produce proteins with altered activities. If so, then a mutation that inactivates the mutant protein should revert the dominant suppression of the P[181PRE]8-10C eye color. Therefore, we tried to revert both mutations.
We used both X-rays and EMS to generate revertants. We recovered three X-ray-induced revertants (from 24,758 progeny) and four EMS-induced revertants (from 5500 progeny). The revertants are lethal over deficiencies for the region 96F1–98A5. By crossing to overlapping deficiencies and lethals in the region, we found that all of the revertants are lethal or semilethal mutations in the gene woc.
Sequencing of the woc gene from our original suppressor mutation chromosomes showed that both of these mutations are due to the same single amino acid change in a position evolutionarily conserved throughout the Drosophila lineage, as well as in most insects (Figure 3). This amino acid change occurs in a region of the protein with no known domain or function. We named the two original suppressor mutations wocD1 and wocD2 (for wocDominant1 and wocDominant2), and we named the revertant alleles based on the allele reverted and the mutagen used (i.e., wocD1rvE8 was a revertant (rv) generated from wocD1 by EMS (E) mutagenesis). The wocD1/+ and wocD2/+ flies have no phenotypic defects. The wocD1 and wocD2 homozygotes survive, are fertile, and also show no phenotypic defects.
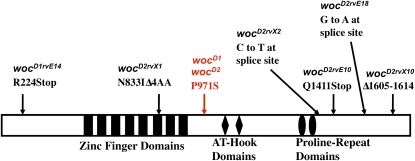
Figure 3 .
Mutations in the woc gene generated in this study. The Woc protein is depicted by the rectangle with identified domains indicated. The location of the dominant suppressors wocD1 and wocD2 is highlighted in red. The mutations present in the revertants are shown. ∆ indicates a deletion of four amino acids in wocD2rvX1. In wocD2rvX10, amino acid numbers 1605-1614 are deleted.
We also sequenced the revertants. As expected, all revertants contained the mutation present in wocD1 and wocD2, as well as an additional lesion in the woc transcription unit (Figure 3). With the exception of wocD2rvX1, all of the mutants were lethal when heterozygous with all other woc mutants. The wocD2rvX1, which contains a four amino acid deletion in the sixth zinc finger, is a hypomorphic allele. The wocD2rvX1 survives poorly in combination with the other hypomorphic woc alleles, wocrgl and woc468. Transheterozygous wocD2rvX1/wocrgl and wocD2rvX1/woc468 flies have multiple phenotypic defects, including downturned wings, lack of wing veins, slightly rough eyes, and they are sterile.
wocD suppresses the eye color in a position-specific manner
P[181PRE]8-10C is inserted in the genome between the genes GstS1 and CG30456 (Figure 1). We wanted to know whether wocD modulates the PRE directly or whether it acts through regulatory DNA flanking the insertion site of P[181PRE]8-10C. Importantly, wocD does not darken the eye color of wa, a mutation in the w gene that reduces the amount of w transcript and leads to orange eyes (Pirrotta and Bröckl 1984; Levis et al. 1984). This shows that wocD does not darken eye color indiscriminately. We examined whether the eye colors of flies heterozygous for other mini-white containing transgenes inserted near GstS1 were altered by wocD. We used a line with a P[EP] element inserted about 1.2 kb away from the insertion site of P[181PRE]8-10C and six lines with a P[lacW] inserted in the promoter region of GstS1 (Figure 1). The eye colors of P[lacW] or P[EP]/+; +/TM6C were compared with the eye colors of P[lacW] or P[EP]/+; wocD1/+ flies; no eye color differences were observed. This suggests that the effect of wocD on the eye color of P[181PRE]8-10C flies is dependent on the presence of the PRE in the transgene.
We next examined whether wocD could alter the eye color of flies with P[181PRE] inserted at different chromosomal locations. We used the transgene P[L181PRE], which contains the same 181-PRE as in P[181PRE]8-10C. In P[L181PRE], the 181-bp PRE is flanked by loxP sites (see below). Because wocD dominantly alters the eye color of P[181PRE]8-10C heterozygotes, we looked at whether wocD could dominantly alter the eye color of flies heterozygous for P[L181PRE] insertions that show mini-white repression. For 14 out of 15 P[L181PRE] lines tested, wocD does not alter the eye color. However, in P[L181PRE]-8A, the eye color was slightly darker in a wocD mutant (data not shown). P[L181PRE]-8A is inserted just upstream of the PcG-regulated gene CycA (at 3L:11826614). To determine whether the effect on the eye color of P[L181PRE]-8A flies was due to the PRE, we examined the eye color of P[L]-8A flies in which the 181-bp PRE had been removed. We found that wocD had no effect on the eye color of flies that lacked the PRE. This shows that, at least at this chromosomal location, the change in eye color mediated by wocD is dependent on the PRE. However, as wocD does not influence the eye color of most P[L181PRE] lines, we believe that wocD is not working on the PRE directly but on sequences flanking the P[181PRE] insertion sites.
wocD increases the levels of GstS1 RNA in adult heads
We examined whether the levels of GstS1 and CG30456 transcripts were altered in wocD1 mutants, both in the presence and in the absence of the P[181PRE]8-10C insertion. We examined RNA levels at three developmental stages: 3rd instar larvae, 1-day-old pupae, and adult heads. We saw no significant differences in the expression levels of GstS1 and CG30456 between wocD1 and wild-type 3rd instar larvae or one-day old pupae (data not shown). However, we saw a twofold increase in the level of GstS1-RNA in the adult heads of homozygous wocD1 mutants compared to wild-type (Figure 4). Flies with the P[181PRE]8-10C insert had about a twofold decrease in the expression levels of CG30456 at all developmental stages, suggesting that the insertion interferes with the transcription of CG30456 (Figure 4 and data not shown). However, wocD1 had no significant effect on the transcription level of CG30456 at any developmental stage either in wild-type or in P[181PRE]8-10C animals (Figure 4 and data not shown). We also examined GstS1 and CG30456 transcript levels in eye-antennal disks from third instar larvae of wild-type and wocD1 mutants with P[181PRE]8-10C and saw no significant differences (data not shown). Finally, we tested whether wocD altered the transcription level of CycA in adult heads. We saw no significant differences in CycA levels between wild-type and wocD1; P[181PRE] heads (data not shown).
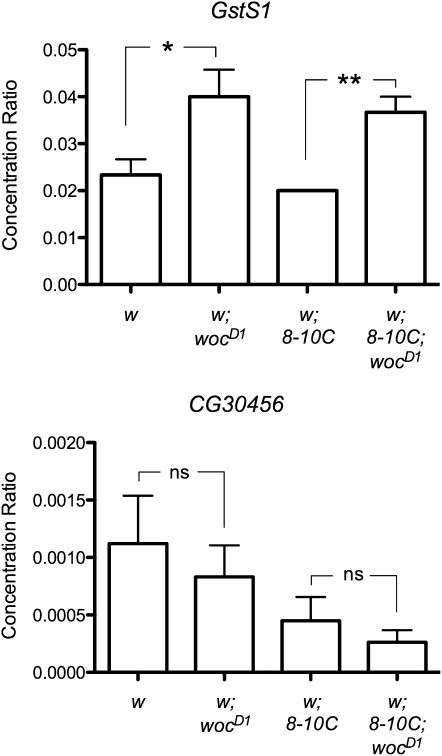
Figure 4 .
The wocD1 mutation increases the amount of GstS1 transcript in adult heads. Graph shows the concentration ratio of GstS1 and CG30456 RNA to RpL32 RNA in fly heads (by qRT-PCR). The symbol 8-10C refers to P[181PRE]8-10C. Comparisons were made between wild type and wocD1 homozygotes, in flies homozygous for P[181PRE]8-10C or lacking it entirely. wocD1 increased the amount of GstS1 RNA about 2-fold regardless of whether P[181PRE]8-10C was present. wocD1 has no significant effect on CG30456 RNA levels. Results of three independent experiments are combined and SEM is shown. *P ≤ 0.05; **P ≤ 0.01; ns = not significant as determined by unpaired t-tests.
Other dominant suppressors do not affect the expression level of GstS1 or CG30456
We tested whether four of our other dominant suppressors of pairing-sensitive silencing of P[181PRE]8-10C effect the expression levels of GstS1 or CG30465 in fly heads by qRT-PCR; we saw no effect on transcript levels of either gene (data not shown). Thus, a change in transcription level of a GstS1 is not required for suppression of the pairing-sensitive silencing of P[181PRE]8-10C. Finally, we examined the effects of three the other suppressor mutants on the eye color of P[L181PRE] at multiple insertion sites. Like wocD, none of the other suppressor mutations altered the eye color of P[L181PRE] at the insertion sites.
A single unpaired copy of the PRE reduces the eye color of mini-white transformants
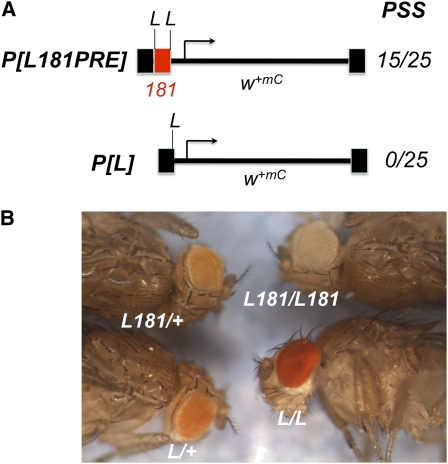
We flanked the 181-bp PRE by loxP sites and cloned it upstream of the mini-white reporter in pCaSpeR (P[L181PRE]), in the same orientation and position as in the construct P[181PRE] (Figure 5). We recovered 32 lines with insertions of P[L181PRE]. Of the 25 insertion lines that were homozygous viable, 15 exhibited pairing-sensitive silencing (60%). All lines were treated with Cre recombinase to excise the 181-bp PRE, yielding P[L]. None of the lines without the PRE showed pairing-sensitive silencing. In 7 of the 15 pairing-sensitive lines, the eye color of heterozygous flies became darker upon removal of the PRE, showing that some repression of mini-white expression occurred even in the heterozygotes (Figure 5). In contrast, in the lines that did not show pairing-sensitive silencing, the eye colors of flies heterozygous or homozygous for P[L181PRE] did not change after removal of the PRE, with one exception. In line P[L181PRE]11A, the eye color was slightly lighter after removal of the PRE, suggesting that this element was acting as a slight activator of mini-white expression at this location. This result is consistent with earlier evidence that showed that the 181-bp fragment, in the context of a reporter gene driven by the en promoter, can act as either an activator or repressor of gene expression depending on the context (Devido et al. 2008). This activation activity was weak and only occurred in 1 line out of 32. This shows that, in the mini-white assay, PRE-mediated repression is the usual situation.
Figure 5 .
P[L181PRE] construct and eye colors. (A) P[L181PRE] is identical to P[181PRE] except the 181-bp en PRE is flanked by loxP sites (L). PSS is the number of lines with pairing-sensitive silencing/the total number of viable lines obtained. (B) Eyes from 2-day-old males of lines P[L181PRE]C2A (L181) and P[L]C2A (L) are shown. Note that the P[L]/+ eyes are darker than the P[L181PRE]/+ eyes.
We determined the chromosomal insertion site for 26 of the P[L181PRE] lines (9 lines were lost prior to this part of the analysis) and examined whether the insertion occurred in or near a transcription unit (Table 1). We also examined whether the nearby gene is transcribed in the eye. We note that lines with pairing-sensitive silencing were just as likely to be inserted in or near genes transcribed in the eye as lines without it. Thus, insertion near a gene that is transcribed does not interfere with pairing-sensitive silencing.
Table 1 . P[L181PRE] insertions.
| Line Name | Locationa | Geneb | Distancec | Transcript Level in Eyed |
|---|---|---|---|---|
| Lines that show pairing-sensitive silencing | ||||
| C2A | 3R:9882721 | Foxo | +31bp | 48 |
| C5A | X:9581402 | Nej | +303bp | NID |
| C6A | X:18398636 | Wnt5 | +800bp | 18 |
| C11B | 3R:9487247 | B52 | +255bp | NID |
| 1A | X:19498690 | CG14207 | −501bp | 300 |
| 1B | 2L:11106592 | Reps | +60bp | 27 |
| 12A | 3R:21867498 | Gro | −66bp | 197 |
| 14A | 3R:7590188 | Lk6 | −9bp | 4362 |
| 24A | 2R:2051740 | Lbk | +150bp | 138 |
| 15A | 3L:19922217 | RhoGDI | +310bp | 4481 |
| 23A | 2L:10414068 | Klp31E | −34bp | 98 |
| 8A | 3L:11826614 | CycA | −81bp | NID |
| Lines that do not show pairing-sensitive silencing | ||||
| C1 | 2R:3623151 | CG18812 | −27bp | 728 |
| 2 | 3L:12074850 | Sema-5c | +77bp | 47 |
| C1-12A | 3L:13932279 | CG32137 | −73bp | 146 |
| 13A | X:14983581 | rab3-GEF | −134bp | 173 |
| 16A | 2L:2753118 | CG9894 | +319bp | 2886 |
| 17A | 2R:13435831 | MESR4 | +502bp | 41 |
| 28A | 2R:15556892 | Hrg | −325bp | 383 |
| 10 | 2L:3477289 | Thor | −1145bp | 1016 |
| 21B | 2R:8475807 | Sin3A | +793bp | NID |
Three lethal lines are included in this table: 8A, 10, and 21B. The eye color of line 8A heterozygous flies became lighter upon excision of the PRE; thus, we consider that this line undergoes mini-white repression by the PRE and classify it as having pairing-sensitive silencing. The eye colors of lines 10 and 21B did not change upon excision of the PRE and are classified as lines that do not show pairing-sensitive silencing. NID, no informative data.
Insertion site of the P[L181PRE], genome version R5.39.
Nearest gene (http://flybase.org; Tweedie et al. 2009).
Distance to the nearest transcription start site. Positive numbers indicate it is within the transcription unit. Negative numbers indicate it is upstream of the transcription unit.
Transcript level in the adult eye. Data are taken from the FlyAtlas Organ/Tissue Expression as listed on Flybase (http://flybase.org). Low (10–99.9), moderate (100–499.9), high (500–999.9), and very high expression (1000–25,000).
Discussion
To gain insight into the mechanism of pairing-sensitive silencing mediated by the 181-bp en PRE, we conducted a genetic screen for dominant mutations that affected mini-white repression by that element. Notably, we obtained only nine mutations from screening 68,274 progeny. This low frequency of mutation recovery suggests that loss-of-function alleles were not obtained in our screen. If the loss of one copy of a gene could affect mini-white repression, these mutations would have been much more frequent. Our data suggest that mini-white repression by the 181-bp PRE is not dependent on genes that are dosage sensitive.
The classical Polycomb group mutant phenotype is the presence of sex comb teeth on the second and third legs, caused by derepression of the Sex combs reduced (Scr) HOX gene (reviewed in Kennison 1995). Scr repression is sensitive to the dose of some PcG genes, as flies with only one wild-type copy have sex comb teeth on the second and third legs. In contrast, mini-white repression via the en 181bp PRE is not sensitive to a reduction in dosage of the PcG genes (Kassis 1994 and unpublished data). The 181-bp PRE is known to bind PcG proteins, and a binding site for the DNA-binding PcG group protein Pho is required for 181-bp–mediated mini-white repression (Brown et al. 1998). Thus, we believe that PcG proteins mediate en PRE mini-white repression, but that this target of PcG proteins is not dosage sensitive.
wocD1 and wocD2 are gain-of-function mutations
Heterozygosity for either wocD1 or wocD2 darkens the eye colors of P[181PRE]8-10C flies, whereas heterozygosity of woc deletions does not. This shows that wocD1 and wocD2 have acquired new activities and are gain-of-function alleles. Our results also show that wild-type Woc protein competes with WocD protein. This is suggested by the observation that P[181PRE]8-10C/P[181PRE]8-10C; wocD/woc− flies have a darker eye color than P[181PRE]8-10C/P[181PRE]8-10C; wocD/+ flies. Finally, the observation that the activity of WocD is abrogated by mutations that inactivate the Woc protein shows that the wocD mutation alters the activity of the protein.
A model for WocD
The woc gene encodes a zinc-finger transcription factor implicated in transcriptional activation (Wismar et al. 2000; Raffa et al. 2005) that acts, at least in part, through an association with HP1c (Font-Burgada et al. 2008; Able et al. 2009). There are five HP1 isoforms in Drosophila. Of these, HP1a is the best studied and is associated with heterochromatic DNA. In contrast, HP1c is excluded from centromeric heterochromatin and is associated with euchromatin.
As stated above wocD suppresses mini-white expression from P[181PRE] in a position-dependent manner. Therefore, we wanted to know whether Woc binds to the genomic regions near the P[181PRE] insertion sites it regulates. We were not able to obtain Woc antisera, and there is no published data showing where Woc binds in the genome. However, the Drosophila ModENCODE project has mapped the binding sites of HP1c in four different cell culture lines by chromatin immunoprecipitation followed by hybridization to tiling arrays (Kharchendo et al. 2010). Because Woc is often associated with HP1c, we examined whether there was a correlation between HP1c binding and the suppressor activity of wocD. HP1c is bound in a cell-type–specific manner. There is no HP1c associated with the region between GstS1 and CG30456 in S2, Kc167, or BG3 cells; however, HP1c is bound to this region in clone 8 cells. HP1c is not associated with CycA in any cell type. There is no data on HP1c localization in the pigment cells in the eye, so we cannot make any conclusion about whether WocD acts via HP1c in suppressing the PRE activity of P[181PRE]8-10C.
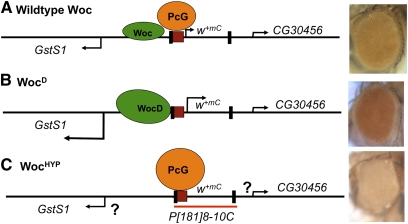
How does wocD affect the eye color of P[181PRE]8-10C? We suggest that the eye colors of P[181PRE]8-10C flies result from a competition of transcriptional repression (caused by the PRE) and transcriptional activation of GstS1 (Figure 6). In the wild-type case, we suggest there is a competition between transcriptional activation of mini-white by flanking regulatory DNA and transcriptional repression mediated by the PRE, leading to an intermediate eye color. One prediction of this model is that if Woc levels are decreased, the PRE upstream of mini-white should be able to work more strongly, and the eye color should be lighter. Consistent with this hypothesis, the eye color of P[181PRE]8-10C/+; wocrgl/wocD2X1 flies is white (Figure 6). We suggest that the PRE in line P[181PRE]8-10C modulates the levels of mini-white expression in part through a competition with flanking activators.
Figure 6 .
Interplay between Woc and PcG activity determines the eye colors of P[181PRE]8-10C flies. Genomic DNA around the P[181PRE]8-10C insertion site is denoted by the black line. The CG30456, GstS1, and w+mC promoters are designated by arrows pointing in the direction of transcription, with the height of the arrow indicating the relative level of transcription. The P-element ends (black rectangles), the 181-bp en PRE (red box), and the extent of the P[181PRE] transgene (red line at bottom) are shown. Green ovals indicate Woc activity, with WocD a bigger shape to indicate a higher activity. (Note that we have no evidence that Woc binds directly to this location.) PcG proteins are represented by orange ovals, with the level of repression indicated by the size of the oval. Eyes from flies of the genotype (A) w1118; P[181PRE]8-10C/+ (B) w1118; P[181PRE]8-10C/+; wocD1/+ (C) w1118; P[181PRE]8-10C/+; wocD2rvX1/wocrgl are shown on the right. WocHYP indicates a hypomorphic allelic combination. We were not able to obtain enough wocHYP adults to perform qRT-PCR, so we do not know the level of CG30456 and GstS1 RNA in these flies. This uncertainty is indicated by the question mark next to the transcription arrows in (C).
What determines PRE activity?
PREs have been studied for many years as silencers of homeotic gene expression in Drosophila. Recent genome-wide studies showed that PREs may play an important role in regulating gene expression levels as well. What determines whether a PRE will completely silence a gene or only decrease its expression levels? Our data suggests two things. First, the number of PREs is important. This is evident from the fact that flies homozygous for PRE-mini-white constructs, which have two PREs, repress mini-white to a much higher level than flies heterozygous. We also note that increasing the number of PREs in cis, by duplicating a P-construct with PREs, also causes an increase in mini-white repression (Kassis 1994). Second, changes in the chromatin environment, here caused by a gain-of-function mutation in the transcriptional activator Woc, can inactivate a PRE. The dependence of PRE activity in transgenes on chromosomal environment has long been recognized and is dramatically demonstrated in a recent report showing the effect of chromosome environment on the activities of the Abd-B Fab-7 PRE and the vg PRE (Okulski et al. 2011).
The relationship between transcriptional activation and PRE function is not simple. Addition of an enhancer containing three binding sites for the eye enhancer-activator protein Glass (GBS) to pCaSpeR darkens the eye color of transformants that contain an en PRE (Americo et al. 2002). If the increased transcription driven by the GBS enhancer interferes with PRE activity, one would expect to see a decrease in the number of lines with pairing-sensitive silencing in this vector. However, this did not occur. Thus, increasing the transcription of mini-white itself does not alter PRE activity. In addition, we found that insertion of P[L181PRE] into or next to genes expressed in the eye did not prevent pairing-sensitive silencing from occurring (Table 1). Therefore, we propose that it is not transcriptional activation but the actual activators present that determine whether a PRE is active or not. It has previously been suggested that PREs are general silencer elements that could act on any enhancer (Sengupta et al. 2004). The basis for this conclusion was that the Ubx PRE could act as silencers of three enhancers in reporter genes [two vestigal (vg) and one decapentaplegic (dpp) enhancer]. At that time, neither vg nor dpp was thought to be regulated by PcG proteins. However, since then, a vg PRE and a dpp PRE have been identified (Lee et al. 2005; Hauenschild et al. 2008; Okulski et al. 2011). We suggest that PREs may not be able to silence all enhancers, and in some chromosomal locations, they cannot act. It was recently reported that a human tissue–specific enhancer functions in erythroid cells by evicting PcG proteins (Vernimmen et al. 2011). Enhancers with this activity may also be present in Drosophila.
Acknowledgments
We thank Pamela Geyer and Emily Kuhn for the EK710 plasmid and the Cre recombinase flies; Mike Goldberg and Rick Padgett for woc mutants; Sarah DeVido for making the P[L181PRE] clone and for localization of many of the insertion sites; and Kristofor Langlais for statistical analysis of the qRT-PCR data. Many of the stocks used in these experiments were obtained from the Bloomington Stock Center. This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
Literature Cited
- Able J., Eskeland R., Raffa G. D., Kremmer E., Imhof A., 2009. Drosophila HP1c is regulated by an auto-regulatory feedback loop through its binding partner Woc. PLoS ONE 4: e5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Americo J., Whiteley M., Brown J. L., Fujioka M., Jaynes J. B., et al. , 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a Polycomb group response element from the Drosophila engrailed gene. Genetics 160: 1561–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Mucci D., Whiteley M., Dirksen M.-L., Kassis J. A., 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Brown J. L., Grau D. J., Devido S. K., Kassis J. A., 2005. An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res. 33: 5181–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Kassis J. A., 2010. Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development. Development 137: 2597–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M. D., Brown J. L., Kassis J. A., 2010. Characterization of the Polycomb group response elements of the Drosophila melanogaster invected locus. Mol. Cell. Biol. 30: 820–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devido S. K., Kwon D., Brown J. L., Kassis J. A., 2008. The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development 135: 669–676 [DOI] [PubMed] [Google Scholar]
- Font-Burgada J., Rossell D., Auer H., Azorin F., 2008. Drosophila HP1c isoform interacts with the zinc-finger proteins WOC and Relative-of-WOC to regulate gene expression. Genes Dev. 22: 3007–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller M., Schedl P., 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila Bithorax complex. Genetics 146: 1365–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild A., Ringrose L., Altmutter C., Paro R., Rehmsmeier M., 2008. Evolutionary plasticity of Polycomb/Trithorax response elements in Drosophila species. PLoS Biol. 6: e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., 1983. Analysis of Y-linked mutations to male-sterility in Drosophila melanogaster. Genetics 103: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29: 289–303 [DOI] [PubMed] [Google Scholar]
- Kuhn E. J., Hart C. M., Geyer P. K., 2004. Studies of the role of the Drosophila scs and scs’ insulators in defining boundaries of a chromosome puff. Mol. Cell. Biol. 24: 1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Maurange C., Ringrose L., Paro R., 2005. Suppression of Polycomb group proteins by JNK signaling induces transdetermination in Drosophila imaginal discs. Nature 438: 234–237 [DOI] [PubMed] [Google Scholar]
- Levis R., O’Hare K., Rubin G. M., 1984. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell 38: 471–481 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., Bacher F., 1968. Method of feeding ethyl methane sulfonate (EMS) to Drosophila males. Drosoph. Inf. Serv. 43: 193 [Google Scholar]
- Lorenz L. J., Hall J. C., Rosbash M., 1989. Expression of a Drosophila mRNA is under circadian clock control during pupation. Development 107: 869–880 [DOI] [PubMed] [Google Scholar]
- Müller J., Bienz M., 1991. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophla embryo. EMBO J. 10: 3147–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Kassis J. A., 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16: 476–484 [DOI] [PubMed] [Google Scholar]
- Négre N., Hennetin J., Sun L. V., Lavrov S., Bellis M., et al. , 2006. Chromosomal distribution of PcG proteins during Drosophila development. PLOS Bio. 4: 917–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K., Gutierrez L., Gagneur J., Girardot C., Sengupta A. K., et al. , 2008. Dynamic regulation of Polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Okulski H., Druck B., Bhalerao S., Ringrose L., 2011. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics and Chromatin 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Bröckl Ch., 1984. Transcription of the Drosophila white locus and some of its mutants. EMBO J. 3: 563–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa G. D., Cenci G., Siriaco G., Goldberg M. L., Gatti M., 2005. The putative Drosophila transcription factor Woc is required to prevent telomeric fusions. Mol. Cell 20: 821–831 [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R., 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232 [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz et al, 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X. Y., Bourgon R., et al. , 2006. Genome-wide analysis of Polycomb targets in Drosophila. Nat. Genet. 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V., 2008. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20: 266–273 [DOI] [PubMed] [Google Scholar]
- Sengupta A. K., Kuhrs A., Müller J., 2004. General transcriptional silencing by a Polycomb response element in Drosophila. Development 131: 1959–1965 [DOI] [PubMed] [Google Scholar]
- Siegal M. L., Hartl D. L., 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 714–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E., 2009. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Strutt H., Paro R., 1997. The Polycomb group protein complex of Drosophila has a differential composition at different target genes. Mol. Cell. Biol. 17: 6773–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Muijrers I., De Wit E., Teunissen H., Talhout W., et al. , 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38: 694–699 [DOI] [PubMed] [Google Scholar]
- Vernimmen D., Lynch M. D., De Gobbi M., Garrick D., Sharpe J. A., et al. , 2011. Polycomb eviction as a new distant enhancer function. Genes Dev. 25: 1583–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wismar J., Habtemichael N., Warren J. T., Dai J.-D., Gilbert L. I., et al. , 2000. The mutation without childrenrgl causes ecdysteroid deficiency in third-instar larvae of Drosophila melanogaster. Dev. Biol. 226: 1–17 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]