Abstract
Ischemic stroke (IS) is among the leading causes of death in Western countries. There is a significant genetic component to IS susceptibility, especially among young adults. To date, research to identify genetic loci predisposing to stroke has met only with limited success. We performed a genome-wide association (GWA) analysis of early-onset IS to identify potential stroke susceptibility loci. The GWA analysis was conducted by genotyping 1 million SNPs in a biracial population of 889 IS cases and 927 controls, ages 15–49 years. Genotypes were imputed using the HapMap3 reference panel to provide 1.4 million SNPs for analysis. Logistic regression models adjusting for age, recruitment stages, and population structure were used to determine the association of IS with individual SNPs. Although no single SNP reached genome-wide significance (P < 5 × 10−8), we identified two SNPs in chromosome 2q23.3, rs2304556 (in FMNL2; P = 1.2 × 10−7) and rs1986743 (in ARL6IP6; P = 2.7 × 10−7), strongly associated with early-onset stroke. These data suggest that a novel locus on human chromosome 2q23.3 may be associated with IS susceptibility among young adults.
Keywords: epidemiology, genetics, brain infarction, FMNL2
Age-related stroke is a common debilitating disease that ranks among the leading causes of death in the United States (Rosamond et al. 2008). Ischemic stroke (IS) comprises approximately 85% of all strokes. While hypertension, cigarette smoking, and diabetes are well-established risk factors for IS, family and twin studies provide evidence that IS aggregates within families (Flossmann et al. 2004), thus implicating a genetic contribution to stroke susceptibility.
Efforts to identify the genetic determinants of IS have had limited success. One strategy to enhance the chances of identifying stroke susceptibility genes is to study stroke cases that are more likely to have a strong genetic predisposition. Such cases might include those with a strong family history of stroke or those experiencing a stroke at a relatively young age. Such strategies have been successful in identifying genes associated with early-onset forms of other complex diseases, including breast cancer (Walsh et al. 2006), Parkinson’s disease (Lesage et al. 2008), obesity (Hinney et al. 2007; Scherag et al. 2010), inflammatory bowel disease (Imielinski et al. 2009), and myocardial infarction (Kathiresan et al. 2009). Evidence from twin (Brass et al. 1992; Brass et al. 1998; Hassan and Markus 2000) and familial aggregation studies (Schulz et al. 2004), in fact, supports a stronger genetic contribution to early-onset than later-onset stroke. We have previously reported in our own study of young-onset stroke that the proportion of cases reporting a family history of stroke increases with decreasing age of the proband (MacClellan et al. 2006).
We have performed a case-control study of IS in subjects ages 15–49 years. In this report, we present results of a genome-wide association (GWA) analysis based on 1.4 million SNPs genotyped and/or imputed across the genome for their association with risk of IS.
Materials and Methods
Study population
The Genetics of Early Onset Stroke (GEOS) Study is a population-based, case-control study designed to identify genes associated with early-onset ischemic stroke and to characterize interactions of identified stroke genes and/or SNPs with environmental risk factors. Participants were recruited from the greater Baltimore-Washington area in four different periods: Stroke Prevention in Young Women-1 (SPYW-1) conducted from 1992 to 1996, Stroke Prevention in Young Women-2 (SPYW-2) conducted from 2001 to 2003, Stroke Prevention in Young Men (SPYM) conducted from 2003 to 2007, and Stroke Prevention in Young Adults (SPYA) conducted in 2008. From these samples, we identified a total of 921 cases and 941 controls that consented to having their DNA used for genetic studies of stroke. This study was conducted with the consent of all study participants and was approved by the University of Maryland at Baltimore Institutional Review Board.
Definitions of cases and controls
“Case participants” were hospitalized with a first cerebral infarction identified by discharge surveillance from one of the 59 hospitals in the greater Baltimore-Washington area and direct referral from regional neurologists. IS with the following characteristics were excluded from participation: stroke occurring as an immediate consequence of trauma; stroke within 48 hr after a hospital procedure, stroke within 60 days after the onset of a nontraumatic subarachnoid hemorrhage, and cerebral venous thrombosis. Additional exclusions for these genetic analyses are listed in Table 1. All cases had neuroimaging that was consistent with cerebral infarction, although neuroimaging was not used for case ascertainment. The abstracted hospital records of cases were reviewed and adjudicated for IS subtype by a pair of neurologists according to previously published procedures (Johnson et al. 1995; Kittner et al. 1998), with disagreements resolved by a third neurologist. The IS subtype classification system retains information on all probable and possible causes, and it is reducible to the more widely used TOAST system (Adams et al. 1993) that assigns each case to a single category. All cases had age of first stroke between 15 and 49 years and were recruited within three years of stroke.
Table 1 . Subject exclusion criteria for genetic analysis in GEOS study.
| Exclusion Criteria |
|---|
| 1. Known single-gene or mitochondrial disorders recognized by a distinctive phenotype; e.g., cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), mitochondrial encephalopathy with lactic acidosis and stroke-like episodes (MELAS), homocystinuria, Fabry disease, or sickle cell anemia |
| 2. Mechanical aortic or mitral valve at the time of index stroke |
| 3. Untreated or actively treated bacterial endocarditis at the time of the index stroke |
| 4. Neurosyphilis or other CNS infections |
| 5. Neurosarcoidosis |
| 6. Severe sepsis with hypotension at the time of the index stroke |
| 7. Cerebral vasculitis by angiogram and clinical criteria |
| 8. Post-radiation arteriopathy |
| 9. Left atrial myxoma |
| 10. Major congenital heart disease |
| 11. Cocaine use in the 48 hr prior to the index stroke |
“Control participants” without a history of stroke were identified by random-digit dialing. Controls were balanced to cases by age and region of residence in each study and were additionally balanced for race in SPYW-2 and SPYM. Traditional stroke risk factors and other study variables, including age, race/ethnicity, history of hypertension, diabetes, myocardial infarction (MI), and current smoking status (defined as use within one month prior to event for cases and at a comparable reference time for controls), were also collected during a standardized interview.
Genotyping
Genomic DNA was isolated from a variety of sample types, including cell line (55.2%), whole blood (43.1%), mouth wash (0.4%), and buccal swab (0.05%). Whole-genome amplification (Qiagen REPLI-g kit, Valencia, CA) was used to obtain sufficient DNA for genotyping in 1.3% of samples. The distribution of sample types did not differ significantly between cases and controls (56.3% cell lines in cases vs. 54.1% in controls; 41.5% whole blood in cases vs. 44.9% in controls). Samples were genotyped at the Johns Hopkins Center for Inherited Disease Research (CIDR), and genotyping was performed using the Illumina HumanOmni1-Quad_v1-0_B BeadChip (Illumina, San Diego, CA). Case and control samples were balanced across the plates, and self-identified whites and African Americans were placed on different plates. Samples of 50 self-reported whites were also placed on African American plates for quality control. All study samples, including 39 blind duplicates (19 whites and 20 African Americans), were plated and genotyped together with 42 HapMap control samples, including 26 Utah residents with ancestry from northern and Western Europe (CEU) and 16 Yoruba (YRI) samples, and all samples were processed together in the lab. Allele cluster definitions for each SNP were determined using Illumina BeadStudio Genotyping Module version 3.3.7, Gentrain version 1.0, and the combined intensity data from all released samples. Genotypes were not called if the quality threshold (Gencall score) was below 0.15.
Genotypes of a total of 1827 study individuals (99% of attempted samples) were released by CIDR, and all had a genotype call rate greater than 98%. Genotyping concordance rate was 99.996% based on study duplicates. We excluded 11 individuals from analysis due to unexpected duplicates, gender discrepancy, or unexpected relatedness, leaving a total of 1816 individuals (889 cases and 927 controls) in the final analysis. A total of 1,014,719 SNPs were released by CIDR (99.83% of attempted). Genotypes were not released for SNPs that had call rates less than 85%, a cluster separation value of less than 0.2, more than 1 HapMap replicate error, more than a 5% (autosomal) or 6% (X) difference in call rate between sexes, more than 0.3% male AB frequency (X), or more than a 11.3% (autosomal) or 10% (XY) difference in AB frequency. Individual SNPs were excluded postanalysis if they had excessive deviation from Hardy-Weinberg equilibrium (HWE) proportions (P < 1.0 × 10−7) or genotype call rates less than 95%. Departure from HWE was assessed by χ2 test among controls only and among each ethnic group separately. For this report, only SNPs having minor allele frequencies (MAF) greater than 1% and SNPs passing HWE filtering in both genetically defined (see Statistical Analysis below) European ancestry (EA) and African ancestry (AA) populations were included (N = 784,766 SNPs).
Imputation
Imputation was performed for genetically defined EA cohort (N = 946) and AA cohort (N = 733) separately using BEAGLE v3.3 software (Browning and Browning 2009). Genetically defined EA participants were those within two standard deviations (SD) of the mean of the first and second principal components for subjects self-identified as “white,” and similarly, genetically defined AA participants were those within two SD of the mean of the first and second principal components for those self-identified as “African American.” The combined samples of CEU and Tuscans from Italy (TSI) from the HapMap Phase 3 populations were used as the reference panel for EA cohort imputation. A combination of five HapMap Phase 3 populations, CEU, TSI, YRI, Luhya in Webuye, Kenya (LWK), and African ancestry in southwest United States (ASW), was used as reference panel for AA cohort imputation, in accordance with the recommendation that a mixture of populations be used as the reference panel for African Americans (Hao et al. 2009). All SNPs failing the composite quality filter were removed [i.e. SNPs with missing call rate ≥ 5%, SNPs with Mendelian errors in three HapMap trios > 0, HWE P < 1 × 10−4, and SNPs with more than one discordant genotype call across the 89 duplicated study samples (39 blind duplicates and 50 white duplicates plated on African American plates]. All remaining SNPs having a MAF ≥ 1% in each respective cohort were used as input SNPs for imputation. A total of 1,387,466 SNPs were obtained via imputation, which included 532,595 and 582,186 input SNPs for the EA and AA cohorts, respectively. Imputed SNPs that have an estimated squared correlation between the estimated allele dosage and the true allele dosage (dosage r2) < 0.3 were removed postanalysis. The dosage r2 can be derived from arguments found in Appendix 1 of Browning and Browning (2009). For this study, we supplemented 784,766 SNPs genotyped by the Illumina Omni1-Quad chip with 637,233 imputed SNPs with r2 > 0.3 for a total of 1,421,999 SNPs in the final genome-wide association report.
To assess the performance of imputation, analysis was performed of a masked set of SNPs on chromosome 1. We first masked a randomly selected set of 20% of all Omni1 array SNPs on chromosome 1 that passed the preimputation filters. The masked SNPs were then removed from input files for imputation, and then imputation was performed using the same procedures as described previously. The imputed genotypes at the “masked” SNPs were then compared with the experimental (typed) genotypes from Omni1 GWAS data to assess the accuracy of imputation. The SNP concordance rates, which were defined as fraction of identical genotypes between the most likely imputed and observed, were 0.9858 for EA and 0.9805 for AA (filtering imputed genotypes at posterior probability ≥ 0.9). The correlations between observed vs. expected allele frequencies for all imputed SNPs were also very high (0.99967 for EA and 0.99937 for AA). The plot of the observed allelic frequency vs. expected allelic frequency from imputation is shown in supporting information, Figure S1.
Statistical analysis
Summary statistics of study characteristics were calculated and expressed as unadjusted means ± SDs (SD) or percentages, stratified by case status. We compared cases and controls for difference in means using t-tests (for continuous variables) or difference in proportions by χ2 tests (for categorical variables) to obtain the unadjusted two-sided P values. Logistic regression with case/control status as the outcome and baseline characteristics as predictors was used to obtain the age- and sex-adjusted P values. All summary statistics of study characteristics were obtained using the STATA statistical package (version 9.2; StataCorp, College Station, TX).
GWA analysis was performed using a logistic regression model to test for associations of each SNP with IS under an additive model (for imputed SNPs, estimated allelic dosage was used). A t-score was used to assess the significance of the beta coefficient (β) for the SNP, and the odds ratio (OR) was derived by exponentiation of the beta coefficient. All estimates were obtained after adjusting for the effect of age, recruitment periods (three indicator variables), and population structure. Population structure was estimated using multidimensional scaling (MDS) analysis on the matrix of genome-wide identical-by-state (IBS) pairwise distances, which were estimated based on 127,901 typed (not imputed) SNPs that were selected for having MAF > 5%, SNP call rates > 95%, and not being in linkage disequilibrium with other selected SNPs (i.e. pair-wise LD between any two SNPs was constrained to be r2 < 0.2). The first 10 dimensions of population structure were extracted using MDS, but only the first dimension was significantly associated with case status and included as a covariate in the association analysis. Similar to imputation, study subjects were divided into genetically defined EA and AA groups for association analyses based on population structure analysis. EA participants were defined as those failing within two SD of the mean of the first and second components for self-reported “white” subjects, and AA participants were defined as falling within two SD of the mean of the first and second components for self-reported “African American” subjects. Association analyses were conducted in the total population, adjusting for the first MDS component, as well as for genetically defined EA and AA groups separately. Genome-wide significance level was set at α = 5.0 × 10−8 after Bonferroni correction for multiple testing. All analyses were performed using existing programs implemented in PLINK version 1.07 (Purcell et al. 2007). Power calculations (using Quanto v1.2.4) indicated that our sample size of 889 cases and 927 controls provided 80% power to detect ORs ranging 1.55–2.30 for allele frequencies ranging 10–50% at an α = 5.0 × 10−8.
We also reviewed previous genome-wide association studies on IS. We identified one gene, NINJ2, reported previously to be genome-wide significantly associated with stroke risk in a large-scale meta-analysis (CHARGE consortium) (Ikram et al. 2009). To replicate the reported associations with NINJ2, a logistic regression model was conducted under an additive genetic assumption to obtain beta coefficients (and odds ratios) and 95% confidence intervals after adjusting the same set of covariates described above. We first tested whether the two SNPs previously reported by CHARGE (rs12425791 and rs11833579) were associated with young-onset stroke in our sample. Because of our a priori hypothesis that each SNP would be associated with young-onset IS, we did not adjust for multiple comparisons in this analysis (α = 0.05). We also conducted an agnostic screening of all genotyped/imputed SNPs located within NINJ2 to identify the most-significant SNP from the gene based on empirical P values to control for the number of SNPs tested. Empirical P values for the most-significant SNPs were obtained by permuting the case/control status 1000 times to obtain the null distribution of the maximum T statistics (over all SNPs within the gene) and calculating the number of times the maximum test statistics exceeded the observed test statistic. Analyses were conducted using a set-based permutation procedure implemented in PLINK 1.07 (Purcell et al. 2007).
Results
Characteristics of study population
A total of 1816 GEOS study participants, including 889 cases and 927 controls, were included in the analysis. Characteristics of study participants are summarized in Table 2. The mean age was 41.3 years for cases and 39.6 years for controls (P < 0.001). The population is primarily composed of two self-reported race groups, white (54.5%) and African American (40.4%), with the remaining 5.1% of individuals comprising other races, including Chinese, Japanese, other Asians, and other unspecified. There were more males than females among both cases and controls. Cases were more likely than controls to report having prevalent hypertension, diabetes, and myocardial infarction and to being current smokers.
Table 2 . Population characteristics by case-control status.
| Characteristic | Case (n = 889) | Control (n= 927) | Pa |
|---|---|---|---|
| Age (mean ± SD, years) | 41.3 ± 6.9 | 39.6 ± 6.8 | <0.001 |
| Female (%) | 41.5 | 43.6 | 0.37 |
| Self-reported ethnicity (%) | |||
| White | 52.42 | 56.42 | 0.22 |
| African American | 42.41 | 38.51 | |
| Other | 5.17 | 5.07 | |
| Subtype (%) | |||
| Cardioembolic | 20.0 | — | — |
| Large artery | 7.1 | ||
| Lacunar | 16.1 | ||
| Other known cause | 6.5 | ||
| Undetermined cause | 50.3 | ||
| Hypertension (%) | 42.7 | 19.2 | <0.001 |
| Diabetes mellitus (%) | 16.7 | 5.1 | <0.001 |
| Angina/myocardial infarction (%) | 5.3 | 0.7 | <0.001 |
| Current smoker (%) | 42.5 | 28.6 | <0.001 |
Unadjusted P values for age, sex, and race; age and sex-adjusted P values for other characteristics.
Genome-wide association of early-onset IS
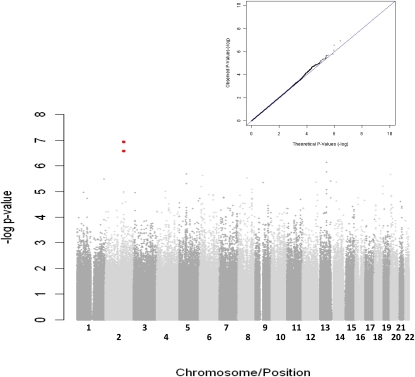
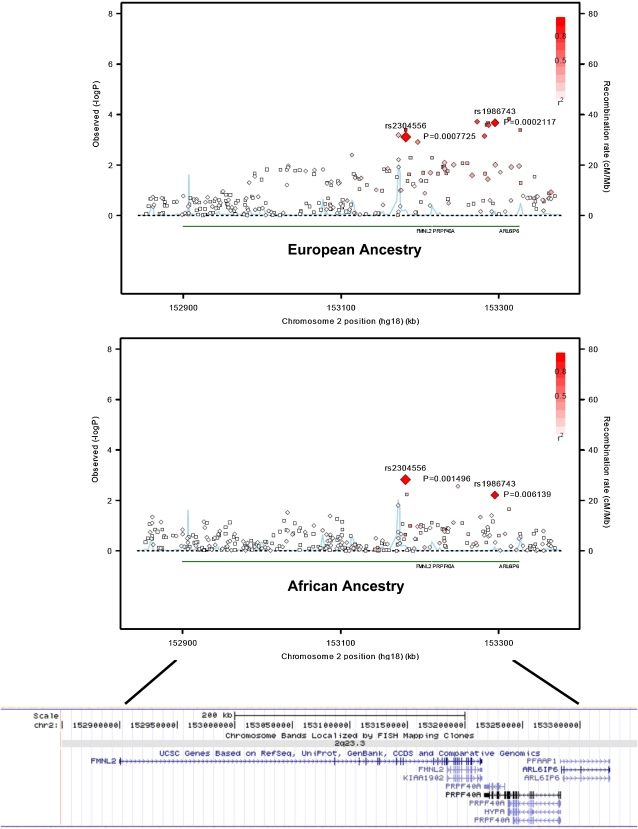
Results from the GWA analysis are summarized in Figure 1. The genomic inflation factor was 1.01, indicating that the distribution of observed P values across most of the genome was consistent with that expected under the null hypothesis. The 10 most strongly associated SNPs are listed in Table 3. No single SNP reached genome-wide significance (α = 5.0 × 10−8). However, we found SNPs, rs2304556 (P = 1.2 × 10−7) and rs1986743 (P = 2.7 × 10−7) located on chromosome 2q23.3, that were associated with IS but not at genome-wide significance (P < 5 × 10−8). These two SNPs are separated by 113 kb and are in strong linkage disequilibrium (LD) in EA (D′ = 0.90, r2 = 0.75) although not in AA participants (D′ = 0.35, r2 = 0.08) in our data [see Figure 2 for regional association plots for EA and AA obtained with the SNAP program (Johnson et al. 2008)]. The estimated OR for both SNPs was 0.69 (95% CI = 0.60–0.79) per copy of the minor allele (Table 3). The minor alleles of the two SNPs were more common in controls than cases and more common in AA than EA populations. The effects of the minor allele on risk of IS are similar between EA and AA populations for rs2304556 (OR= 0.71, P = 0.00077 in EA and OR = 0.71, P = 0.0015 in AA, allele G) and for rs1986743 (OR= 0.68, P = 0.0002 in EA and OR = 0.73, P = 0.006 in AA, allele A). After adjusting for the effect of rs2304556, the effect of rs1986743 became nonsignificant in EA (OR = 0.72, P = 0.11) and borderline significant in AA (OR = 0.79, P = 0.06). Additionally, we reexamined the associations of the two SNPs by further adjustment for prior history of MI, hypertension, diabetes and for current smoking, and the effects of the two SNPs remained similar (rs2304556: OR = 0.68, 95% CI = 0.59∼0.79; P = 1.9 × 10−7; rs1986743: OR = 0.70, 95% CI = 0.61∼0.82; P = 3.6 × 10−6). rs2304556 is in the intron of FMNL2, which encodes a formin-related protein, and rs1986743 is in the intron of ARL6IP6, which encodes ADP-ribosylation-like factor 6 interacting protein 6. The cluster plots of both SNPs (rs2304556 and rs1986743) also showed clear separation of the three genotypes, indicating good clustering of genotype calls for the two SNPs (see Figure S2). Most of the other top-associated SNPs were located in intergenic regions, except rs9465922 in an intron of CDKAL1 and rs16834810 in an intron of RYR2.
Figure 1 .
Negative log10 of genome-wide P values from logistic regression model and a quantile-quantile (Q-Q) plot (upper right) of the distribution of these observed test statistics against expected distribution in overall samples. Red dots represent the two most significant SNPs in chromosome 2q (rs2304556 and rs1986743).
Table 3 . Association results of the top 10 SNPs based on results of overall population, ranked by P values.
| Effect Allele Frequency | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European Ancestrya | African Ancestrya | Combined | |||||||||||
| SNP | Chr | Positionb | Impc | Effect/Noneffect Alleled | Case | Control | P | Case | Control | P | OR (95% CI)e | P | Closest Genef |
| rs2304556 | 2 | 153182040 | No | G/T | 0.28 | 0.36 | 7.7E-04 | 0.40 | 0.48 | 1.5E-03 | 0.69 (0.60, 0.79) | 1.2E-07 | FMNL2 (intron) |
| rs1986743 | 2 | 153295145 | No | A/G | 0.30 | 0.38 | 2.1E-04 | 0.31 | 0.37 | 6.1E-03 | 0.69 (0.60, 0.79) | 2.7E-07 | ARL6IP6 (intron) |
| rs3909263 | 13 | 71753222 | Yes | C/G | 0.26 | 0.31 | 1.4E-02 | 0.35 | 0.46 | 2.9E-06 | 0.68 (0.59, 0.79) | 7.3E-07 | DACH1 (413.9 kb) |
| rs1146849 | 13 | 71770207 | No | A/G | 0.24 | 0.29 | 2.2E-02 | 0.36 | 0.47 | 3.3E-06 | 0.7 (0.61, 0.81) | 1.7E-06 | DACH1 (430.9 kb) |
| rs17366217 | 5 | 61530870 | No | T/C | 0.08 | 0.11 | 5.5E-03 | 0.15 | 0.22 | 5.0E-04 | 0.61 (0.5, 0.75) | 2.0E-06 | KIF2A (147.8 kb) |
| rs879012 | 20 | 957788 | Yes | C/T | 0.38 | 0.29 | 1.7E-05 | 0.48 | 0.41 | 2.3E-02 | 1.43 (1.23, 1.65) | 2.2E-06 | RSPO4 (26.9 kb) |
| rs9465922 | 6 | 20973576 | No | C/A | 0.03 | 0.07 | 4.9E-04 | 0.14 | 0.20 | 1.8E-03 | 0.57 (0.45, 0.72) | 2.3E-06 | CDKAL1 (intron) |
| rs2605877 | 8 | 74309320 | No | A/G | 0.37 | 0.27 | 5.8E-06 | 0.37 | 0.32 | 4.0E-02 | 1.41 (1.22, 1.64) | 3.0E-06 | RPL7 (56.1 kb) |
| rs16834810 | 1 | 235325422 | Yes | A/G | 0.01 | 0.03 | 3.4E-03 | 0.02 | 0.06 | 2.6E-04 | 0.3 (0.18, 0.5) | 3.3E-06 | RYR2 (intron) |
| rs9604365 | 13 | 111625198 | No | G/A | 0.06 | 0.05 | 5.0E-01 | 0.52 | 0.39 | 8.1E-07 | 1.54 (1.28, 1.86) | 3.9E-06 | SOX1 (144.7 kb) |
Abbreviations: Chr, chromosome; CI, confidence interval; Imp, Imputation; OR, odds ratio; P, association P value.
European ancestry and African ancestry are defined based on multidimensional scaling (MDS) analysis.
Physical position is based on National Center for Biotechnology Information (NCBI) build 36.
Flag indicating whether the SNP was imputed (Yes) or directly genotyped (No). The total number of individuals in the combined analysis is 1816 for directly genotyped SNPs and 1679 for imputed SNPs.
Polymorphism is reported based on genome assembly plus strand.
Odds ratio per copy of effect allele.
The closest gene to the SNP and the location/distance of the SNP to the gene.
Figure 2 .
Regional association plots showing associations of FMNL2-ARL6IP6I region in European ancestry (top) and in African ancestry populations (middle), and the position of the genes in this region (bottom). Each diamond represents a single typed SNP, and each square represents a single imputed SNP. The color within each square/diamond represents the extent of LD correlation, r2, between the SNP and rs2304556. The blue line represents the recombination rate. Recombination rate and r2 were estimated based on HapMap3 CEU and YRI samples, respectively. Regional association plots were drawn by modifying the R program codes from the SNAP program, and the position of the gene were obtained from the University of California, Santa Cruz (UCSC) genome browser.
No SNPs achieved genome-wide significance in ethnic group–specific analyses (data not shown). In the EA population, the SNP most strongly associated with IS was rs2242878 at RUNX1 (OR = 1.81, 95% CI = 1.42–2.31, P = 1.8 × 10−6, T allele), but this SNP was not significantly associated with IS in the AA population (OR = 0.95, 95% CI = 0.55–1.65, P = 0.87). Similarly, the SNP most strongly associated with IS in AAs was rs9604365 near SOX1 (OR = 1.74, 95% CI = 1.40–2.17, P = 8.1 × 10−7, allele G), but this SNP was not associated with IS in EA (OR= 1.15, 95% CI = 0.77–1.72, P = 0.5). It should be noted that rs9604365 was the tenth most significant SNP based on the overall population results (see Table 3).
Genome-wide association of stroke subtypes
We further analyzed the genome-wide associations stratified by each stroke subtype defined by TOAST criteria, including cardioembolic, large artery, lacunar subtypes, other known causes, and other undetermined causes. No SNPs were associated with any of the stroke subtypes at genome-wide levels of statistical significance, although the power to detect associations for subtypes was very modest. Summary results showing all SNPs associated with any stroke subtype at P < 1.0 × 10−5 in combined analyses of EA and AA are provided in Table S1. Neither rs2304556 nor rs1986743, the two SNPs most strongly associated with overall stroke in our sample, showed subtype-specific patterns of association; i.e., the associations for both SNPs were approximately consistent across all stroke subtypes (ORs ranged 0.52–0.76 for rs2304556 and 0.57–0.73 for rs1986743) (Table S2).
Replications of chromosome 2 SNP associations in other Caucasian cohorts
There are currently no other large cohorts of young-onset stroke for which to attempt replication. We did, however, assess the association of rs2304556, our peak SNP, with IS in two independent Caucasian cohorts with older-onset stroke: the Ischemic Stroke Genetics Study and Siblings with Ischemic Stroke Studies (ISGS/SWISS; n = 1070 cases and 1488 controls) and the Genes Affecting Stroke Risk and Outcomes Study (GASROS; n = 516 cases and 1202 controls). There was no evidence for association with IS in either cohort (OR = 1.10, P = 0.12 in ISGS/SWISS and OR = 0.95, P = 0.63 in GASROS for minor allele G). These cohorts had 80% power to detect odds ratios as low as 1.21 (ISGS/SWISS) and 1.26 (GASROS) for this SNP.
Associations of NINJ2 variants candidate genes with early-onset IS
We additionally tested whether SNPs from NINJ2, which was identified in a prior large GWAS study to be associated with IS, were associated with young-onset stroke in our study (Table 4). Of the two SNPs near NINJ2 that were previously reported to be associated with IS (Ikram et al. 2009), neither was associated with early-onset IS in our samples (rs11833579, combined P = 0.54, EA P = 0.99, AA P = 0.76; and rs12425791, combined P = 0.49, EA P = 0.74, AA P = 0.91). None of the other SNPs from NINJ2 was significantly associated with stroke risk in EA and AA populations after controlling for the number of SNPs analyzed within the gene. When further investigating the NINJ2 associations by TOAST subtypes, none of the SNPs was significantly associated with any of the stroke subtype, either (data not shown).
Table 4 . Association results of NINJ2 in the GEOS Study.
| Previously Reported Variant | Best SNP within the Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of SNPsa | Variant | Effect/Noneffect Allele | OR (95% CI) | Pb | Variant | Effect/Noneffect Allele | OR (95% CI) | Pb | Empirical Pc | |
| EAd | 70 | rs12425791 | A/G | 0.96 (0.76, 1.21) | 0.74 | rs12229103 | A/C | 1.34 (0.90, 2.00) | 0.15 | 0.95 |
| rs11833579 | A/G | 1.00 (0.81, 1.24) | 0.99 | |||||||
| AAd | 70 | rs12425791 | A/G | 1.02 (0.73, 1.42) | 0.91 | rs11063749 | T/C | 0.74 (0.60, 0.93) | 0.008 | 0.27 |
| rs11833579 | A/G | 1.04 (0.80, 1.35) | 0.76 | |||||||
Abbreviations: CI, confidence interval; OR, odds ratio; P, association P value.
Number of SNPs within the gene that were either directly genotyped (SNP call rate > 95% and HWE P > 1.0 × 10−7 in either European or African ancestry) or imputed (imputation dosage r2 > 0.3).
Nominal P was obtained by t-statistics in the logistic regression.
Empirical P was obtained by permutations as described in Materials and Methods.
European ancestry (EA) and African ancestry (AA) are defined based on multidimensional scaling (MDS) analysis.
Discussion
Suggestive associations between chromosome 2q23.3 and early-onset IS
The GEOS Study is the largest genetics study of young-onset stroke carried out to date. Despite having 80% power to detect odds ratios of 1.55–2.30 (at allele frequencies 10–50%), we had limited power (<70%) to detect odds ratio less than 1.5 and failed to identify associations with SNPs meeting conventional genome-wide levels of significance. However, we did detect associations with two SNPs on chromosome 2q associated with overall IS that achieved nearly genome-wide levels of significance (P < 2.65 × 10−7). The most strongly associated SNPs were near FMNL2 on chromosome 2. The minor allele of this SNP (rs2304556), which is located in the intron of FMNL2, is associated with an approximately 31% decrease in risk per copy of minor allele in both EA and AA populations. A similar association with stroke risk was observed for a nearby SNP, rs1986743, located on ARL6IP6, although the strength of association for this SNP diminished substantially after adjusting for rs2304556, suggesting the two SNPs may not represent independent signals. As there are no other large studies of young strokes currently available for replication, we sought replication in two other cohorts with older strokes (ISGS and GASROS). Although we were unable to replicate the associations of the chromosome 2 SNPs in other Caucasian cohorts, the lack of replications may be due to the genetic heterogeneity possibility between younger vs. older strokes.
FMNL2, which encodes formin-like 2 protein, is involved in actin-dependent processes and is implicated in cell motility and invasion (Kitzing et al. 2010). Although the function of this gene is largely unknown, the gene is highly expressed in the brain and other central/peripheral nervous system tissues (http://www.cgl.ucsf.edu/Research/genentech/genehub-gepis/index.html) (Gardberg et al. 2010) and is conserved in different species (http://www.ncbi.nlm.nih.gov/sites/homologene). A microdeletion on chromosome 2q region involving FMNL2 has previously been implicated in mental retardation (Lybæk et al. 2009), and association and linkage studies have implicated this same chromosomal region with susceptibility to autism (Imgsac. 2001; Maestrini et al. 2009). The function of ARL6IP6 is currently unknown. Replication studies, particularly in young stroke cases, are required to assess the role of rs2304556 and/or other variants in and around FMNL2 with young-onset stroke.
Interestingly, our GWAS also showed CDKAL1, a gene previously shown to be associated with type 2 diabetes (T2D) (Scott et al. 2007; Zeggini et al. 2007), to be marginally associated with IS in our data. T2D is a strong risk factor for stroke. However, the association between rs9465922 and IS remained highly significant after adjusting for self-reported diabetes status in our population (OR = 0.55, 95% = 0.44–0.70, P = 5.5 × 10−7), suggesting the effect of this gene on IS may be independent of the effect of T2D. The function of CDKAL1 is not currently known, although the association of this gene to T2D may be related to an effect of the associated SNP on insulin response (Steinthorsdottir et al. 2007).
Failure to replicate previously reported associations with NINJ2
We were unable to replicate the previous GWA associations with two SNPs in NINJ2 as reported by the CHARGE consortium (Ikram et al. 2009), even though power was 98% in our sample to detect associations of the magnitude of that observed in CHARGE (OR = 1.39–1.41 for SNPs with these allele frequencies). Notably, the association with NINJ2 reported in CHARGE could also not be replicated in a recent very large meta-analysis by the International Stroke Genetics Consortium and Wellcome Trust Case-Control Consortium 2 (2010).
Strengths and limitations of the study
We used the strategy of studying the early-onset form of IS to identify genetic variants associated with this complex disease because the early-onset form may be less susceptible to the influence of environmental exposures, and genetic components could potentially have a greater contribution to disease risk. Although it is possible that genetic variants associated with early-onset stroke may have little relevance to late-onset stroke, it is also possible that studying early-onset form of diseases may reveal important insights about stroke pathogenesis, even implicating key genes and pathways relevant to late-onset disease even though the responsible variants may differ.
Although we did identify a region on chromosome 2 with suggestive evidence for association in the GEOS population, we did not identify any variants reaching conventional thresholds for statistical significance at a genome-wide level. The absence of robust associations may be due to the limited sample size that provided insufficient power to detect causal variants with small effect sizes (e.g. less than 80% power to detect genetic risk effect less than 1.55). Alternatively, one can argue that the early-onset form of IS may be more likely to be caused by rare variants with larger effects (and higher penetrance). Such rare variants are generally not well covered on currently available genotyping platforms and, thus, would be difficult to detect through a GWA approach. Exome and/or whole-genome sequencing may be a promising research approach to pursue the roles of these infrequent and highly penetrant variants on IS risk. Notably, our ability to identify IS-associated variants could be further limited by the stroke phenotype itself, which is composed of several heterogeneous subtypes with different underlying causes and potentially different genetic dispositions (Marnane et al. 2010). Thus, the genetic variants contributing to stroke risk may differ based upon stroke subtype. Although we analyzed according to TOAST subtype, in doing so we incurred further losses in power due to the smaller number of cases available in each subtype. As IS occurs infrequently before age 50, it is difficult for a single study to obtain the sufficient number of strokes necessary to power subtype-specific analyses. Therefore, combining genetic data among multiple early-onset stroke studies to increase the number of cases for overall stroke and stroke subtype analyses will be essential to ensure sufficient power to detect causal variants with moderate effects.
Conclusions
Our early-onset IS GWA study identified a novel IS susceptibility region around FMNL2 on chromosome 2q. Further studies in other young-stroke populations are needed to replicate these findings and to evaluate the associations between these SNPs and the more common older-onset form of stroke. Meta-analysis of early-onset stroke ensuring sufficient statistical power for moderate genetic effects and sequencing efforts to explore the effect of rare variants are also required.
Supplementary Material
Acknowledgments
GEOS was supported by the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) Grant U01 HG-004436, as part of the GENEVA consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK-072488); and by the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR) and funded by GEI Grant U01 HG-004438-01. Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG-004446; Bruce S. Weir, P.I.). Study recruitment and assembly of datasets were supported by a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control, and by National Institute of Neurological Disorders and Stroke (NINDS) and NIH Office of Research on Women's Health Grants R01 NS-45012 and U01 NS-069208-01. The ISGS/SWISS study was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the National Institute on Aging, NIH project Z01 AG-000015-50, human subjects protocol number 2003-078.The ISGS study was funded by NIH-NINDS Grant R01 NS-42733 (J. F. Meschia, P.I.). The SWISS study was funded by NIH-NINDS Grant R01 NS-39987 (J. F. Meschia, P.I.). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov). GASROS was supported by American Heart Association/Bugher Foundation Centers for Stroke Prevention Research Grant 0775010N; NIH-NINDS Grants K23 NS-064052 (N. S. Rost), R01 NS-063925 (J. Rosand), and P50 NS-051343 (J. Rosand and K. L. Furie); the Deane Institute for Integrative Study of Atrial Fibrillation and Stroke; and the Keane Stroke Genetics Research Fund. Drs. A. Biffi, N. S. Rost, and C. D. Anderson were supported in part by the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research.
Literature Cited
- Adams H. P., Jr, Bendixen B. H., Kappelle L. J., Biller J., Love B. B., et al. , 1993. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24: 35–41 [DOI] [PubMed] [Google Scholar]
- Brass L. M., Isaacsohn J. L., Merikangas K. R., Robinette C. D., 1992. A study of twins and stroke. Stroke 23: 221–223 [DOI] [PubMed] [Google Scholar]
- Brass L. M., Page W. F., Lichtman J. H., 1998. Stroke in twins III: a follow-up study. Stroke 29(Suppl): 256 [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flossmann E., Schulz U. G., Rothwell P. M., 2004. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke 35: 212–227 [DOI] [PubMed] [Google Scholar]
- Gardberg M., Talvinen K., Kaipio K., Iljin K., Kampf C., et al. , 2010. Characterization of Diaphanous-related formin FMNL2 in human tissues. BMC Cell Biol. 11: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K., Chudin E., McElwee J., Schadt E., 2009. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A., Markus H. S., 2000. Genetics and ischaemic stroke. Brain 123(Pt 9): 1784–1812 [DOI] [PubMed] [Google Scholar]
- Hinney A., Nguyen T. T., Scherag A., Friedel S., Bronner G., et al. , 2007. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE 2: e1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram M. A., Seshadri S., Bis J. C., Fornage M., DeStefano A. L., et al. , 2009. Genomewide association studies of stroke. N. Engl. J. Med. 360: 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M., Baldassano R. N., Griffiths A., Russell R. K., Annese V., et al. , 2009. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 41: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC), 2001. A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am. J. Hum. Genet. 69: 570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Stroke Genetics Consortium and Wellcome Trust Case-Control Consortium 2, 2010. Failure to validate association between 12p13 variants and ischemic stroke. N. Engl. J. Med. 362: 1547–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Handsaker R. E., Pulit S. L., Nizzari M. M., O’Donnell C. J., et al. , 2008. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. J., Kittner S. J., McCarter R. J., Sloan M. A., Stern B. J., et al. , 1995. Interrater reliability of an etiologic classification of ischemic stroke. Stroke 26: 46–51 [DOI] [PubMed] [Google Scholar]
- Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., et al. , 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittner S. J., Stern B. J., Wozniak M., Buchholz D. W., Earley C. J., et al. , 1998. Cerebral infarction in young adults: the Baltimore-Washington Cooperative Young Stroke Study. Neurology 50: 890–894 [DOI] [PubMed] [Google Scholar]
- Kitzing T. M., Wang Y., Pertz O., Copeland J. W., Grosse R., 2010. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 29: 2441–2448 [DOI] [PubMed] [Google Scholar]
- Lesage S., Lohmann E., Tison F., Durif F., Durr A., et al. , 2008. Rare heterozygous parkin variants in French early-onset Parkinson disease patients and controls. J. Med. Genet. 45: 43–46 [DOI] [PubMed] [Google Scholar]
- Lybæk H., Ørstavik K. H., Prescott T., Hovland R., Breilid H., et al. , 2009. An 8.9 Mb 19p13 duplication associated with precocious puberty and a sporadic 3.9 Mb 2q23.3q24.1 deletion containing NR4A2 in mentally retarded members of a family with an intrachromosomal 19p-into-19q between-arm insertion. Eur. J. Hum. Genet. 17: 904–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacClellan L. R., Mitchell B. D., Cole J. W., Wozniak M. A., Stern B. J., et al. , 2006. Familial aggregation of ischemic stroke in young women: the Stroke Prevention in Young Women Study. Genet. Epidemiol. 30: 602–608 [DOI] [PubMed] [Google Scholar]
- Maestrini E., Pagnamenta A. T., Lamb J. A., Bacchelli E., Sykes N. H., et al. , 2009. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol. Psychiatry. 15: 954–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnane M., Duggan C. A., Sheehan O. C., Merwick A., Hannon N., et al. , 2010. Stroke subtype classification to mechanism-specific and undetermined categories by TOAST, A-S-C-O, and causative classification system: direct comparison in the North Dublin Population Stroke Study. Stroke 41: 1579–1586 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosamond W., Flegal K., Furie K., Go A., Greenlund K., et al. , 2008. Heart disease and stroke statistics–2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 117: e25–e146 [DOI] [PubMed] [Google Scholar]
- Scherag A., Dina C., Hinney A., Vatin V., Scherag S., et al. , 2010. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 6: e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz U. G., Flossmann E., Rothwell P. M., 2004. Heritability of ischemic stroke in relation to age, vascular risk factors, and subtypes of incident stroke in population-based studies. Stroke 35: 819–824 [DOI] [PubMed] [Google Scholar]
- Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., et al. , 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., et al. , 2007. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 39: 770–775 [DOI] [PubMed] [Google Scholar]
- Walsh T., Casadei S., Coats K. H., Swisher E., Stray S. M., et al. , 2006. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295: 1379–1388 [DOI] [PubMed] [Google Scholar]
- Zeggini E., Weedon M. N., Lindgren C. M., Frayling T. M., Elliott K. S., et al. , 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.