Abstract
Legius syndrome (LS) is an autosomal dominant disorder caused by germline loss-of-function mutations in the sprouty-related, EVH1 domain containing 1 (SPRED1) gene. The phenotype of LS is multiple café au lait macules (CALM) with other commonly reported manifestations, including intertriginous freckling, lipomas, macrocephaly, and learning disabilities including ADHD and developmental delays. Since the earliest signs of LS and neurofibromatosis type 1 (NF1) syndrome are pigmentary findings, the two are indistinguishable and individuals with LS may meet the National Institutes of Health diagnostic criteria for NF1 syndrome. However, individuals are not known to have an increased risk for developing tumors (compared with NF1 patients). It is therefore important to fully characterize the phenotype differences between NF1 and LS because the prognoses of these two disorders differ greatly. We have developed a mutation database that characterizes the known variants in the SPRED1 gene in an effort to facilitate this process for testing and interpreting results. This database is free to the public and will be updated quarterly.
Keywords: Legius syndrome, SPRED1, NF1, database
Legius syndrome (LS; MIM #611431) is an inherited disease caused by autosomal dominant loss-of-function (LOF) mutations in the sprouty-related, EVH1 domain containing 1 (SPRED1) gene (MIM #609291) (Brems et al. 2007). LS, also known as neurofibromatosis type 1 (NF1)-like syndrome, is a disorder with a pigmentary phenotype overlapping with NF1 syndrome. Individuals with LS may meet the diagnostic criteria set by the National Institutes of Health (NIH) for NF1 syndrome (Brems, et al., 2007; Pasmant, et al., 2009; Stevenson, et al. 2010), except people with LS have not been shown to develop tumors as found in NF1(Muram-Zborovski et al. 2010). Additional manifestations associated with LS have also been reported in individuals with NF1 (i.e., macrocephaly, lipomas, learning difficulties, Noonan syndrome-like facial features, attention deficit and hyperactivity disorder (ADHD), and pectus excavatum (Brems et al. 2007; Friedman 1998; Muram-Zborovski et al. 2010; Pasmant et al. 2009). For a clinical diagnosis of NF1, an individual must meet two or more of the following criteria: six or more café-au-lait macules (CALM); two or more neurofibromas or one plexiform neurofibroma; freckling in the axillary or inguinal regions; optic glioma; two or more Lisch nodules; distinctive osseous lesion; and/or a first-degree relative with the diagnosis of NF1. LS patients may meet the NIH diagnostic criteria for NF1 based on pigmentary findings of café-au-lait macules and distinctive freckling pattern alone. It is therefore important to fully characterize the phenotype differences between LS and NF1 because the prognoses for these two disorders differ significantly. This is especially true in children, as definitive features of NF1 are acquired with age, and the only manifestation may be CALM and axillary/inguinal freckling, making it difficult to differentiate between LS and NF1 without genetic testing.
The SPRED1 gene, located on chromosomal region 15q3.2, contains seven coding exons and produces a protein that is a negative regulator of the RAS-MAPK signaling pathway. It is believed that SPRED1 interacts directly with RAS-RAF signaling and that most germline mutations in the SPRED1 gene result in a truncated protein that is incapable of properly interacting with the RAS pathway. However, pathogenic missense mutations, as well as large deletions encompassing part of or the entire SPRED1 gene, have been described, suggesting that many different types of mutations are capable of resulting in protein products unable to properly regulate the RAS-MAPK pathway. These mutations can produce a LS phenotype and may complicate the interpretation of genetic testing results. As such, clinical and research laboratories that offer SPRED1 gene analysis often encounter rare variations and must rely on the published literature or online resources to determine the clinical relevance of the identified variant. We have developed a database devoted to LS-related mutations in the SPRED1 gene to facilitate this process.
We have recently reported gene variant archives for galactosemia (GALT), multiple endocrine neoplasia type 2 (RET), juvenile polyposis syndrome (SMAD4), and X-linked Alport syndrome (COL4A5). These clinically curated archives uniquely display genotype and associated phenotype information. Here, we introduce a new disease database for LS syndrome and SPRED1 gene variants. This database was developed to serve as a reference for all sequence variations currently reported in the SPRED1 gene as well as to provide as much phenotype information as possible (Calderon et al. 2007; Crockett et al. 2010; Margraf et al. 2009; Wooderchak et al. 2010). To date, 78 variants have been described in the SPRED1 gene, of which 49 have been classified as disease causing or pathogenic, 1 as suspected pathogenic, 7 as suspected benign, 8 as benign polymorphisms, and 13 with uncertain significance (Brems et al. 2007; Denayer et al. 2011; Messiaen et al. 2009; Muram-Zborovski et al. 2010; Pasmant et al. 2009; Spencer et al. 2011; Spurlock et al. 2009).
SPRED1 variant database objectives
The aim of the SPRED1 variant database is to record all known variants in the SPRED1 gene and their associated phenotype. This database is freely available to the public, and users are encouraged to submit newly discovered variants to the database curator using the “Database Submission” webpage. This form also can be used to update clinical information or to clarify the classification or phenotype for existing sequence variants. The submission form requests information on the sequence variation, clinical consequences, publications, and the submitter’s contact information. Those submitting new variants to the database are expected to follow their own Institutional Review Board protocols or consenting processes, as their host institutions deem necessary. To maintain the accuracy and utility of the SPRED1 database, content will be updated quarterly using new sequence variation information from literature reports, database submissions and updates, and routine clinical testing performed at ARUP Laboratories. The original start date and date of the latest update is displayed on the SPRED1 “Home” webpage.
Methods
Data sources and limitations
The SPRED1 database was constructed using gene sequence variation data published in the scientific literature since the SPRED1 gene was first identified in patients with a NF1-like phenotype but with no identifiable NF1 gene mutation in 2007 (Brems et al. 2007). To date, 78 sequence variants have been found by searching PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez) and Google (http://google.com/). Search terms included SPRED1, Legius syndrome, and NF1-like syndrome. Occasionally, laboratory-sequencing results that identify novel gene variants are also included in this archive. The database was constructed using the Human Genome Variation Society (HGVS) and Human Genome Organization (HUGO) Mutation Database Initiative recommendations for essential and optional content (http://www.hgvs.org/mdifaq.html). HGVS nomenclature recommendations for description of sequence variants were used to generate DNA and protein changes for this database. Reference sequences for this database were GenBank NC_000015.9, NM_1525940.2, and NP_689807.1; entries were verified for position and name based on these sequences. All sequence variants in the database (including future updates) are named based on their position in relation to the coding regions of the SPRED1 gene (cDNA) following recommendations listed by the HGVS (http://www.hgvs.org/mutnomen) and described in Den Dunnen and Antonarakis (2000). To comply with sequence variant nomenclature according to the guidelines of the Human Genome Variation Society, the online batch position check tool Mutalyzer 2.0 (http://www.mutalyzer.nl/) was used to confirm genomic position of all SPRED1 gene variants (Wildeman et al. 2008).
Software
SQL tables (MySQL, Inc.) were organized to represent SPRED1 mutation status, associated phenotype, literature references, and any known clinical information. PHP coding was performed in-house for dynamic HTML display to render pages and display SQL tables. Graphics were generated using FusionCharts v3 (www.fusioncharts.com). Web pages are hosted on a Mac OS X Apache server. SPRED1 mutations were added to the database and edited using phpmyadmin (www.phpmyadmin.net).
Results and Discussion
SPRED1 database website
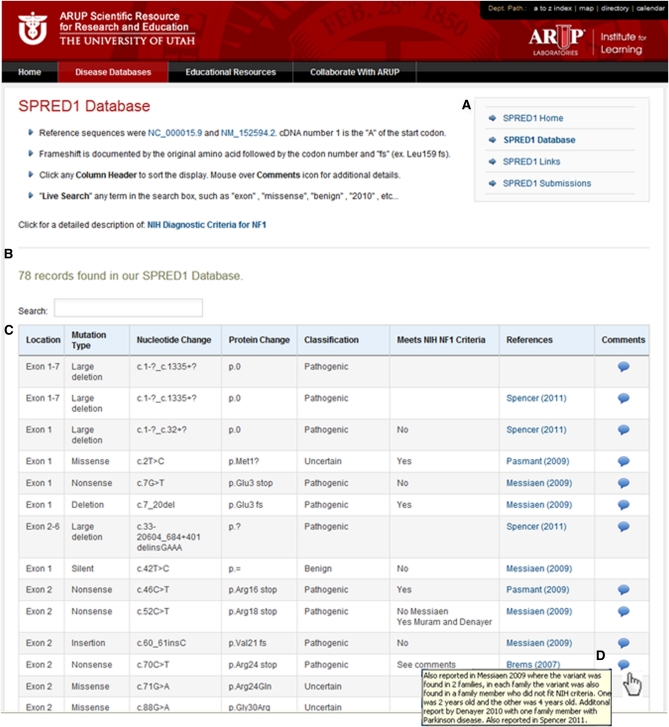
The SPRED1 database can be found under Disease Databases on the ARUP Online Scientific Resource webpage http://www.arup.utah.edu/ or accessed directly by using the URL http://www.arup.utah.edu/database/SPRED1/SPRED1_welcome.php. Navigation between the series of webpages can be done using the quick links as shown in Figure 1A. The SPRED1 “Home” webpage briefly describes Legius syndrome, phenotype, the database goals, reference sequences used, and gene sequencing tests available at ARUP Laboratories. The right frame of the page conveniently lists contact information for the appropriate experts involved in the database development and curation, including medical director, submissions, testing, and website administrator. The “Home” page also provides a graphic link to the SPRED1 gene topic hosted by the National Library of Medicine’s Genetics Home Reference (http://ghr.nlm.nih.gov/gene/SPRED1).
Figure 1 .
Legius syndrome database display. (A) Quick links for navigation within the website for SPRED1 homepage, database display, links, and submission. (B) Main database display including a “live search” function for easy filtering to gene variants of interest. (C) Clicking any display column heading will sort gene variants in ascending or descending order. (D) Mouse-over function for additional clinical “Comments” column follows the literature reference column.
The SPRED1 “Links” webpage has hyperlinks to existing online resources for Legius syndrome and SPRED1, including Databases, Gene and Protein Links, Web Reviews, and Literature Reviews. Some of the resources listed here include Gene Reviews, Genetics Home Reference, OMIM, UniProt, Human Protein Reference Database (HPRD), and the SPRED1 reference sequences used for our database curation. Lastly, recent literature reviews from experts in this disease area are posted and updated as appropriate.
The SPRED1 database can be displayed and searched in its entirety by accessing the “Database” webpage shown in Figure 1B. The database contained 78 entries as of May 2011. The default database display is by genomic position of each sequence variant (5′ to 3′) within the SPRED1 gene. Figure 1C shows the database display columns of Location, Mutation Type, Nucleotide Change, Protein Change, Classification, Meets NIH NF1 Criteria, References, and Comments. Information in the Comments column (Figure 1D) is available by placing the pointer over the comments icon. This information may further explain the data displayed for each entry. The database also has “live search” functionality, so that any term entered in the search box (e.g., exon, insertion, benign, or author name), will dynamically filter the displayed listing. Finally, clicking any column heading will sort (ascending and descending) the gene variant entries for redisplay.
The “Submissions” webpage is used for submission of novel SPRED1 sequence variants to the database or to update information for sequence variations currently archived in the database. Contact information is conveniently seen on the “Home” and “Links” pages. Requested information during submission includes gene variant location, mutation type, classification, nucleotide change, protein change, clinical significance, and contact information of the submitter. All submissions are received via autogenerated e-mail to the curators, and variant updates are added to the database quarterly.
Database display columns
SPRED1 sequence variation location and mutation type:
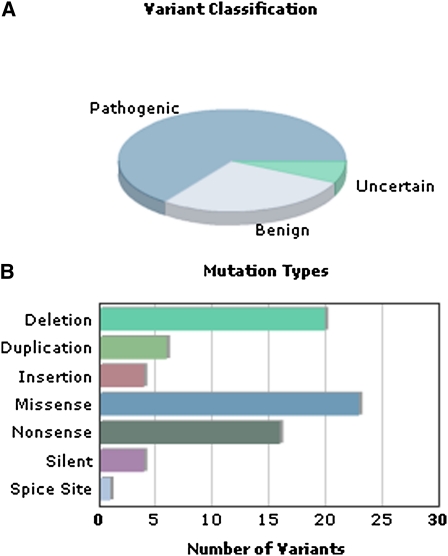
The Location column displays the exon or intron number for each SPRED1 gene variant. Mutation Type describes the biological event leading to the change in the DNA (relative to the reference sequence). Common types of variants include small deletions and insertions/duplications, missense, nonsense, splice-site, silent, and recently, large deletions (those equal to or larger than a single exon) that have been reported (Spencer et al. 2011). Large duplications have not been reported in individuals with Legius syndrome. SPRED1 gene variant mutation types are graphically summarized in Figure 2.
Figure 2 .
Dynamic graphical summary of (A) variant classification and (B) types of mutations found in the SPRED1 database.
SPRED1 genotype:
The Nucleotide Change column displays the corresponding HGVS nomenclature (Den Dunnen and Antonarakis 2000). For cDNA numbering in the Nucleotide Change column of the SPRED1 database, the first cDNA nucleotide (c.1) is the “A” of the ATG initiation (start) codon.
SPRED1 protein change:
For the Protein Change column, the single amino acid changes are listed in the database display. Where splice-site variants occur, only the genotype is listed, with the protein change indicated with a question mark.
Classification of pathogenicity:
In the Classification column, definitions for pathogenicity are Pathogenic, Suspected Pathogenic, Benign, Suspected Benign, or Uncertain. As is common practice, loss of function (LOF) mutations are assumed pathogenic, and the author’s classification for each variant was used.
Legius syndrome phenotype:
The phenotype associated with each variant is listed by author if the variant was reported more than once.
References and comments:
The final two columns of the database feature links to literature resources and genotype/phenotype findings of each individual variant. As displayed in Figure 1D, moving the cursor over the comments icon highlights a key feature and unique part of this online resource. Comments for each SPRED1 variant often contain additional information or evidence to support the classification designation and whether the patient meets NIH diagnostic criteria for a clinical diagnosis of NF1. Importantly, clinical details, such as age or other conditions, may be included. Sensitive and private patient information is never listed.
Database contents
Currently, the database displays information and classification of 78 SPRED1 gene variants. However, this number is actively increasing as additional variants are described in the scientific literature and discovered in routine clinical testing and research laboratories. Pathogenic mutations detected during routine clinical testing of patient samples at ARUP Laboratories or those submitted by other laboratories will also be added to the database. As mentioned above, for variants included from the scientific literature, the database provides a linked reference to the PubMed abstract for each variant. Because the classification of each variant is directly based on evidence from the scientific literature, the link to PubMed is a very useful feature of this database. However, in cases where a variant has not yet been reported in the literature, the database provides the clinical and/or experimental, functional data suggestive of its classification. Laboratories and database users may also contact curators via e-mail or phone regarding database entries, additional references, or clarification of information posted on the site at any time.
Figure 2 summarizes the content of the SPRED1 database by variant classification and mutation types. Analysis of the different types of mutations currently contained in the database are provided, including deletion (16), large deletion (3), insertion/deletion (1), duplication (6), insertion (4), missense (23), nonsense (16), silent (4), intronic (4) and splice site effect (1).
Clinical significance of the database
Clinical and research laboratories that offer full gene analysis often encounter rare variants and must rely on the published literature or online resources to make informed decisions about the classification of the variant when reporting their findings. We developed this database to facilitate this decision-making process and assist those who interpret results and care for patients. A key feature of this database is the ease of access with which it provides pertinent gene variant information and the associated genotype-phenotype observations. In addition, the database is curated by individuals with expertise in molecular genetics and Legius syndrome, ensuring that all of the information contained in the archive is up to date and of sufficient scientific rigor. Finally, as this database is a publicly available resource, with no login or membership requirement, it serves as an ideal centralized resource to all laboratories offering SPRED1 gene analysis, as well as to genetic counselors or physicians treating patients.
Acknowledgments
The authors thank ARUP Institute for Clinical and Experimental Pathology for financial support and the Department of Pathology, University of Utah School of Medicine for hosting the web service.
Literature Cited
- Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., et al. , 2007. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 39: 1120–1126 [DOI] [PubMed] [Google Scholar]
- Calderon F. R., Phansalkar A. R., Crockett D. K., Miller M., Mao R., 2007. Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum. Mutat. 28: 939–943 [DOI] [PubMed] [Google Scholar]
- Crockett D. K., Pont-Kingdon G., Gedge F., Sumner K., Seamons R., et al. , 2010. The Alport syndrome COL4A5 variant database. Hum. Mutat. 31: E1652–E1657 [DOI] [PubMed] [Google Scholar]
- den Dunnen J. T., Antonarakis S. E., 2000. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 15: 7–12 [DOI] [PubMed] [Google Scholar]
- Denayer E., Chmara M., Brems H., Kievit A. M., van Bever Y., et al. , 2011. Legius syndrome in fourteen families. Hum. Mutat. 32: E1985–E1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. M., 1998. Neurofibromatosis 1 in GeneReviews [Internet], bookshelf ID: NBK1109, edited by Pagon R. A., Bird T. D., Dolan C. R., Stephens K. University of Washington, Seattle, WA [Google Scholar]
- Margraf R. L., Crockett D. K., Krautscheid P. M., Seamons R., Calderon F. R., et al. , 2009. Multiple endocrine neoplasia type 2 RET protooncogene database: repository of MEN2-associated RET sequence variation and reference for genotype/phenotype correlations. Hum. Mutat. 30: 548–556 [DOI] [PubMed] [Google Scholar]
- Messiaen L., Yao S., Brems H., Callens T., Sathienkijkanchai A., et al. , 2009. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 302: 2111–2118 [DOI] [PubMed] [Google Scholar]
- Muram-Zborovski T. M., Stevenson D. A., Viskochil D. H., Dries D. C., Wilson A. R., Mao R., 2010. SPRED1 mutations in a neurofibromatosis clinic. J. Child Neurol. 25: 1203–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E., Sabbagh A., Hanna N., Masliah-Planchon J., Jolly E., et al. , 2009. SPRED1 germline mutations caused a neurofibromatosis type 1 overlapping phenotype. J. Med. Genet. 46: 425–430 [DOI] [PubMed] [Google Scholar]
- Spencer E., Davis J., Mikhail F., Fu C., Vijzelaar R., et al. , 2011. Identification of SPRED1 deletions using RT-PCR, multiplex ligation-dependent probe amplification and quantitative PCR. Am. J. Med. Genet. A. 155A: 1352–1359 [DOI] [PubMed] [Google Scholar]
- Spurlock G., Bennett E., Chuzhanova N., Thomas N., Jim H. P., et al. , 2009. SPRED1 mutations (Legius syndrome): another clinically useful genotype for dissecting the neurofibromatosis type 1 phenotype. J. Med. Genet. 46: 431–437 [DOI] [PubMed] [Google Scholar]
- Stevenson D., Viskochil D., Mao R., Muram-Zborovski T., 2010. Legius syndrome in GeneReviews [Internet], bookshelf ID: NBK47312, edited by Pagon R. A., Bird T. D., Dolan C. R., Stephens K. University of Washington, Seattle, WA [Google Scholar]
- Wildeman M., van Ophuizen E., den Dunnen J. T., Taschner P. E., 2008. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum. Mutat. 29: 6–13 [DOI] [PubMed] [Google Scholar]
- Wooderchak W., Spencer Z., Crockett D., McDonald J., Bayrak-Todemir P., 2010. Repository of SMAD4 mutations: reference for genotype/phenotype correlation. J. Data Mining in Genom. Proteomics. 1: 101. [Google Scholar]