Abstract
Vernalization genes determine winter/spring growth habit in temperate cereals and play important roles in plant development and environmental adaptation. In wheat (Triticum L. sp.), it was previously shown that allelic variation in the vernalization gene VRN1 was due to deletions or insertions either in the promoter or in the first intron. Here, we report a novel Vrn-B1 allele that has a retrotransposon in its promoter conferring spring growth habit. The VRN-B1 gene was mapped in a doubled haploid population that segregated for winter-spring growth habit but was derived from two spring tetraploid wheat genotypes, the durum wheat (T. turgidum subsp. durum) variety ‘Lebsock’ and T. turgidum subsp. carthlicum accession PI 94749. Genetic analysis revealed that Lebsock carried the dominant Vrn-A1 and recessive vrn-B1 alleles, whereas PI 94749 had the recessive vrn-A1 and dominant Vrn-B1 alleles. The Vrn-A1 allele in Lebsock was the same as the Vrn-A1c allele previously reported in hexaploid wheat. No differences existed between the vrn-B1 and Vrn-B1 alleles, except that a 5463-bp insertion was detected in the 5′-UTR region of the Vrn-B1 allele. This insertion was a novel retrotransposon (designated as retrotrans_VRN), which was flanked by a 5-bp target site duplication and contained primer binding site and polypurine tract motifs, a 325-bp long terminal repeat, and an open reading frame encoding 1231 amino acids. The insertion of retrotrans_VRN resulted in expression of Vrn-B1 without vernalization. Retrotrans_VRN is prevalent among T. turgidum subsp. carthlicum accessions, less prevalent among T. turgidum subsp. dicoccum accessions, and rarely found in other tetraploid wheat subspecies.
Keywords: Triticum turgidum, tetraploid wheat, vernalization, Vrn-B1, retrotransposon
Vernalization genes determine winter/spring growth habit in temperate cereals and are one of the major genetic factors that affect plant life cycle and enable the crop plant to adapt to a wide range of environments (reviewed by Distelfeld et al. 2009; Rousset et al. 2011; Golovnina et al. 2010). On the basis of vernalization requirement, wheat (Triticum L. sp.) crops are traditionally divided into winter and spring wheat. Winter wheat requires an exposure to a period of low temperatures to induce flowering, whereas spring wheat precludes the requirement for low temperatures to flower (Stelmakh 1987). Increasing our understanding of allelic variation in vernalization genes will be useful for more effective development of wheat cultivars adapted to various environments.
In diploid wheat (T. monococcum L., 2n = 2x = 14, AmAm) and barley (Hordeum vulgare L., 2n = 2x = 14, HH), vernalization is controlled by at least three genes, VRN1, VRN2, and VRN3 (Takahashi and Yasuda 1971; Tranquilli and Dubcovsky 2000; Yan et al. 2004b, 2006; Hemming et al. 2009). In hexaploid wheat (T. aestivum L, 2n = 6x = 42, AABBDD), the three homoeologues of the VRN1 gene, VRN-A1, VRN-B1, and VRN-D1 on the long arms of chromosomes 5A, 5B, and 5D, respectively, are major genes determining growth habit (Pugsley 1971; Law et al. 1976; Worland 1996; Dubcovsky et al. 1998; Snape et al. 2001; Barrett et al. 2002; Danyluk et al. 2003; Leonova et al. 2003; Trevaskis et al. 2003; Yan et al. 2003, 2004a; Fu et al. 2005). A dominant allele for any one of the three genes leads to spring growth habit regardless of the allelic state of the other vernalization genes, but the presence of recessive alleles for all three genes leads to winter growth habit (Tranquilli and Dubcovsky 2000; Yan et al. 2006; Zhang et al. 2008). VRN1 is an ortholog of the Arabidopsis meristem identity gene AP1 (Danyluk et al. 2003; Murai et al. 2003; Trevaskis et al. 2003; Yan et al. 2003), which encodes a MADS-box protein and is responsible for the initiation of the transition from vegetative to reproductive apices in Arabidopsis (Mandel et al. 1992).
As one of a few genes that have been successfully cloned in wheat, VRN1 and its allelic variants have been extensively studied. The dominant Vrn-A1 allele for spring growth habit originated from mutations either in the promoter or in the first intron of a recessive vrn-A1 allele for wild-type winter growth habit in diploid, tetraploid (T. turgidum L., 2n = 4x = 28, AABB), and hexaploid wheat (Yan et al. 2004a; Fu et al. 2005; Dubcovsky et al. 2006; Pidal et al. 2009). In the promoter region, different lengths of deletions (Vrn-Am1a, Vrn-Am1b, Vrn-Am1g) and a 1-bp deletion (Vrn-Am1f) in the CArG-box were identified in Vrn-Am1 of T. monococcum (Yan et al. 2003; Dubcovsky et al. 2006; Pidal et al. 2009). In addition to similar deletions (Vrn-A1d and Vrn-A1e) in the CArG-box region, a deletion in the VRN-box (Vrn-A1b) was found in tetraploid wheat (Yan et al. 2004a; Pidal et al. 2009). In hexaploid wheat, most spring cultivars carry a dominant Vrn-A1a allele that has a duplicated MITE insertion in the VRN-box (Yan et al. 2004a). On the contrary, the dominant Vrn-B1 and Vrn-D1 alleles do not harbor variation in their promoter regions, but they contain large deletions in the first intron in tetraploid wheat and hexaploid wheat (Yan et al. 2004a; Fu et al. 2005).
In this study, we discovered a novel allele of the dominant Vrn-B1 gene in tetraploid wheat as a result of mapping genetic loci associated with growth habit in a doubled haploid (DH) population derived from two spring tetraploid wheat genotypes, a cultivar ‘Lebsock’ of durum wheat [T. turgidum subsp. durum (Desf.) Husnot, 2n = 4x = 28, AABB] and an accession (PI 94749) of T. turgidum subsp. carthlicum (Nevski in Kom.) Á. Löve & D. Löve (2n = 4x = 28, AABB). Variation between the Vrn-B1 and vrn-B1 alleles that segregate in this population was exclusive to a retrotransposon insertion in the 5′-untranslated region (UTR) of the Vrn-B1 allele in PI 94749. Molecular markers were developed to determine the frequencies of the dominant Vrn-B1 and recessive vrn-B1 alleles in a global collection of tetraploid wheat. The evolutionary mechanism that induced the allelic variation and resulted in a conversion of winter to spring growth habit in tetraploid wheat is discussed.
Materials and Methods
Plant materials
A DH population (hereafter referred as to LP749), consisting of 146 lines derived from a cross between Lebsock and PI 94749 (Chu et al. 2010), was used to identify genes responsible for growth habit within the population (supporting information, Table S1). The F1 and F2 plants from the cross between Lebsock and PI 94749 and two backcross populations (Lebsock/PI 94749//Lebsock and Lebsock/2*PI 94749) were then generated to evaluate the genetic effects of each identified gene in the DH population.
Evaluation of winter/spring growth habit
Growth habit was evaluated following the procedures described in Yan et al. (2003). Plants of LP749, two parental lines, F1 hybrids, and F2 and backcross populations were grown in a greenhouse at 20–25° without vernalization and under a 16 h photoperiod by providing additional artificial light. Under this temperature-photoperiod–controlled regime, plants that flowered within two months after planting were classified as spring types, and plants that were still in vegetative growth one month after the spring plants flowered were characterized as winter types. The winter-type plants were then vernalized to induce flowering and seed production. More than 20 plants for each of the two parental lines and their F1 hybrids were tested in three greenhouse seasons. Evaluation included 200 plants for the F2 population and 100 plants for each of the two BC1F2 populations. For the DH lines, growth habit was evaluated in two greenhouse seasons.
QTL analysis
The linkage maps previously developed for the LP749 population (Chu et al. 2010) were used in this study. For QTL analysis, a subset of 188 markers that were spaced greater than 2 cM apart and gave the most complete genome coverage was used, and values of 0 and 1 were assigned to spring and winter growth habits, respectively. Composite interval-regression mapping (CIM) was performed using the computer program Map Manager QTX (Manly et al. 2001) to evaluate marker intervals associated with growth habit. A permutation test with 5000 permutations indicated that a logarithm of the odds (LOD) threshold of 2.91 in this population yielded an experiment-wise significance level of 0.05. Markers with significant (P < 0.001) main effects were tested against all other markers (Manly et al. 2001) in the dataset to identify significant (P < 10−6) interactions among QTL.
Gene-specific marker development and sequence analysis
Allelic variation in the VRN-A1 and VRN-B1 genes is mostly confined to the promoter regions and the first introns of the genes (Fu et al. 2005; Yan et al. 2004a; Pidal et al. 2009), and 10 pairs of primers that were designed from those regions were used in this study (Table 1). In addition, several primers were designed to amplify the VRN-A1 and VRN-B1 genes based on the conserved sequence between Vrn-Am1 (BAC 231A16 of T. monococcum accession DV92, GenBank ID AY188331) (Yan et al. 2003) or VRN-A1 (BAC 1226C17, GenBank ID AY616452) and VRN-B1 (BAC 1225D16, GenBank ID AY616453) of durum wheat ‘Langdon’ (Yan et al. 2004a). In addition to these, several primers specific to VRN-B1 were designed to identify allelic variation in VRN-B1 (Table 1).
Table 1 . Primers used for detecting allelic variation in VRN-A1 and VRN-B1 in tetraploid wheat.
| Primer | Sequence (5′–3′) | Position | Reference | |
|---|---|---|---|---|
| Loci | Gene Region | |||
| VRN1AF | GAAAGGAAAAATTCTGCTCG | VRN-A1 | Promoter | Yan et al. (2004a) |
| VRN1AR | TGCACCTTCCC(C/G)CGCCCCAT | VRN-A1 | Promoter | |
| Ex1/C/F | GTTCTCCACCGAGTCATGGT | VRN-A1 | Intron 1 | Fu et al. (2005) |
| Intr1/A/R3 | AAGTAAGACAACACGAATGTGAGA | VRN-A1 | Intron 1 | |
| Intr1/C/F | GCACTCCTAACCCACTAACC | VRN-A1 | Intron 1 | |
| Intr1/AB/R | TCATCCATCATCAAGGCAAA | VRN-A1 | Intron 1 | |
| Intr1/B/F | CAAGTGGAACGGTTAGGACA | VRN-B1 | Intron 1 | |
| Intr1/B/R3 | CTCATGCCAAAAATTGAAGATGA | VRN-B1 | Intron 1 | |
| Intr1/B/F | CAAGTGGAACGGTTAGGACA | VRN-B1 | Intron 1 | |
| Intr1/B/R4 | CAAATGAAAAGGAATGAGAGCA | VRN-B1 | Intron 1 | |
| VRNBPF1 | CCCCTGCTACCAGTGCCTACTA | VRN-B1 | 960 bp upstream from start codon | Newly developed |
| VRNBPF2 | GGCTTGGGGTGTAGGGTTGG | VRN-B1 | 18 bp upstream from start codon | |
| VRNBPF3 | CCCTCTCTTCCGCCTCACCCAAC | VRN-B1 | 116 bp upstream from start codon | |
| VRNBPR1 | GCCCCATCTCCGCTGGAGAACG | VRN-B1 | Contained start codon | |
| VRNBPR2 | CAGGTGGTTGGGTGAGGCGGAAG | VRN-B1 | 110 bp upstream from start codon | |
| VRNBPR3 | CGTAAATATCCCCAGCCAGA | VRN-B1 | 586 bp downstream from start codon | |
| VBINSR1 | CCCGCTTGTTGGCTGGTGAG | VRN-B1 | 42 bp downstream from VRNBPF3 in PI 94749 | |
Total genomic DNA was isolated from leaves (Faris et al. 2000), and the DNA concentration was adjusted to 50–100 ng/µl for PCR reactions. For DNA fragments of less than 2 kb, a total volume of 15 µl per PCR reaction was conducted, which contained 200 nM of each primer, 0.2 mM of each dNTP, 1.5 mM MgCl2, 1 unit Taq polymerase (Qiagen Sciences Inc., Germantown, MD), and 100–200 ng of template DNA. The amplification was performed at 94° for 4 min, followed by 40 cycles, each consisting of 30 sec at 94°, 45 sec at 50–60° (depending on the annealing temperatures of primers), 45–90 sec at 72° (depending on the size of the products with a rate of 1 min per kb), and a final extension step of 10 min at 72°. For PCR products greater than 2 kb, a total volume of 25 µl per reaction was performed using the LongAmp Taq polymerase kit (New England Biolabs Inc., Ipswich, MA), and 200–400 ng of template DNA was applied in each reaction. PCR amplification was performed at 94° for 30 sec, followed by 30 cycles, each consisting of 20 sec at 94°, 20 sec at 55–58°, 5–10 min at 65° (time estimated based on the rate of 50 sec/kb), and a final extension step of 10 min at 65°. PCR products were mixed with 3 µl loading buffer (40% sucrose, 0.2% bromophenol blue) for electrophoresis, which was carried out on 0.8–1% agarose gels in 1× TAE buffer at 80 W for 1–1.5 h. Gels were stained using 0.001% GelRed (Biotium Inc., Hayward, CA) for 10 min and visualized using a Gel Logic 200 imaging system (Kodak Inc., New Haven, CT). PCR products of the VRN-B1 gene were purified with the Qiagen spin miniprep kit (Qiagen Sciences Inc.), cloned into the pCR2.1 TOPO vector (Invitrogen Inc., Carlsbad, CA), and then sequenced by the DNA Facility at Iowa State University (Ames, Iowa).
PCR markers for VRN-A1 and VRN-B1 were developed to analyze the LP749 population. The new markers were evaluated for linkage with the previously mapped SSR markers (Chu et al. 2010) using the computer program MAPMAKER (V2.0) for Macintosh (Lander et al. 1987) with a minimum LOD threshold of 3.0 and the Kosambi mapping function (Kosambi 1944).
Frequency of the Vrn-B1 allele carrying the retrotransposon insertion in tetraploid wheat
The newly developed VRN-B1 markers were used to investigate the frequency of the dominant and recessive alleles in 154 spring accessions/lines of six domesticated tetraploid wheat subspecies, including durum wheat, T. turgidum subsp. carthlicum, T. turgidum subsp. dicoccum (Schrank ex Schübler) Thell., T. turgidum subsp. polonicum (L.) Thell., T. turgidum subsp. turanicum (Jakubz.) Á. Löve & D. Löve, and T. turgidum subsp. turgidum (Table S2). For accessions that carried the retrotransposon insertion as indicated by the Vrn-B1 marker, the 5′ and 3′ ends of the insertion were sequenced. The 5′ end of the insertion was amplified using primers VBINS2F (5′-CTCACCCAACCACCTGACAGC-3′) and VBINS2R (5′-ATCCGCCCCACTTGGGATT-3′), and the 3′ end of the insertion was amplified using primers VBINS5F (5′-CAACCAGAGGCAATTCTGGACAC-3′) and VRNBPR1 (5′-GCCCCATCTCCGCTGGAGAACG-3′).
Sequence analysis of the retrotransposon insertion in Vrn-B1
The web-based computer program GenScan at GeniusNet (http://genome.dkfz-heidelberg.de/cgi-bin/GENSCAN/genscan.cgi) was used to predict the open reading frame and deduce the encoded amino acid sequences. The primer binding site (PBS) was identified by aligning the sequence downstream of the 5′ long terminal repeat (LTR) to that of 3′ regions of tRNAs. The polypurine tract (PPT) was identified by examining the purine-rich region upstream of the 3′ LTR. The web-based computer program RPS-BLAST (reverse position-specific BLAST) (http://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html) was used to identify conserved domains in the deduced amino acid sequences.
Cloning and sequencing of the Vrn-B1 and vrn-B1 alleles
To determine whether the insertion in the PI 94749 Vrn-B1 was the only difference compared with the Lebsock vrn-B1 allele, the complete gene, including the 27-bp UTL upstream from the start codon, the 44-bp UTL downstream from the stop codon, and 12,343 bp between the start codon and the stop codon, was cloned for sequence analysis. For convenience of cloning, this large gene was divided into three fragments. The first fragment accounted for the 5′-UTR region of 27 bp, exon 1, and 4778 bp of intron 1, and was amplified by primers VRN-A1F1 (5′-TAGGGTTGGCCCGGTTCTC-3′) and VRNB1-C1-R7 (5′-AGGCCGACGCCTTGTGAC-3′). The second fragment, which spanned most of intron 1, consisted of 4927 bp that overlapped 917 bp with the first fragment and was amplified by primers VRNB1-C1-F6 (5′-ACCGCACTCCTAACCCACTA-3′) and VRNB1-C2-R2 (5′-ATGCCTAGATAAGTAGGACGATACGA-3′). The third fragment spanned 3069 bp of intron 1, which overlapped 2192 bp with the second fragment, and the remaining region, including 7 exons and 6 introns. The third fragment was amplified by primers VRNB1-INT1-F4 (5′-TCCCTAAAGAAAGATCGGAAGAC-3′) and VRNB1-3end-R3 (5′-GTTACTCTCTACTGAATAGTACGCTTA-3′). These primers specific to VRN-B1 were designed by comparison of VRN-A1 (AY616452) and VRN-B1 (AY616453) of Langdon (Yan et al. 2004a), and the sizes of PCR products were estimated based on the sequence of the VRN-B1 gene.
Gene expression analysis of VRN-A1 and VRN-B1
For gene expression experiments, the two parents along with 16 spring DH lines carrying either Vrn-A1 or Vrn-B1 or both and 8 winter DH lines carrying recessive vrn-A1 and vrn-B1 alleles were grown in the greenhouse under constant temperature (20–25°) and long day length (16 h light). For each of the winter DH lines, five plants were vernalized by treating them for 6 weeks at 4–6° and 16 h day length, and another five plants were not vernalized and kept in the greenhouse as controls.
For detecting transcripts of VRN-A1 and VRN-B1, leaf samples from all tested lines were collected at 9:00 am to avoid potential effect of the VRN-1 diurnal expression pattern reported by Shimada et al. (2009). Total RNA was extracted using the TRIZOL method (Invitrogen Inc.) (Yan et al. 2003) from leaves at the third leaf stage, and the two conserved primers, Ex4-5_F1 and Ex8_R1 specific for VRN-A1 and VRN-B1, respectively, were used to simultaneously amplify their transcripts using the protocol described previously (Loukoianov et al. 2005). The mixed VRN-A1 and VRN-B1 transcripts were separated and distinguished by digesting the PCR products with restriction enzyme HinfI, which resulted in undigested PCR products for VRN-A1 (313 bp) and digested PCR products for VRN-B1 (173 bp + 110 bp + 30 bp) (Loukoianov et al. 2005). Because no transcripts were observed in the unvernalized winter DH lines at the third leaf stage, RNA samples were also collected at the seventh and ninth leaf stages for these lines. The VRN-A1 and VRN-B1 transcripts were further investigated in the winter DH lines that were vernalized for six weeks. In addition, the primer pair specific for actin (ACTIN, forward: TGTGGATATCAGGAAGGA and reverse: CTCATACGGTCAGCAATAC) (Yan et al. 2003), which produced a cDNA fragment of 85 bp, was used as an endogenous control for detecting transcripts.
Results
Restoration of winter growth habit in the DH population derived from two spring parents
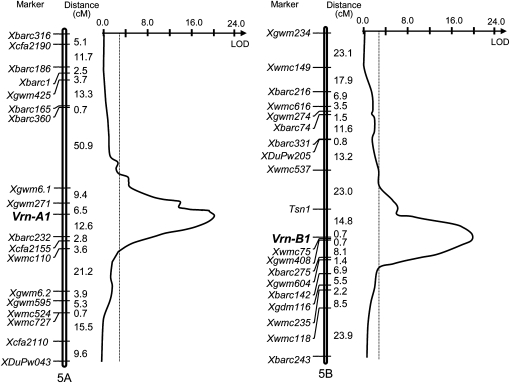
Of 146 DH lines generated from two spring parents, 101 were spring type and 45 were winter type (Table 2 and Table S1), indicating winter growth habit was restored due to homoeoallelic recombination of genes controlling growth habit. The observed segregation ratio was not significantly different from a 3:1 ratio (χ2df = 1 = 2.64, P = 0.10) and fit a two-gene model. CIM revealed that two QTL were significantly associated with growth habit and centered at the VRN-A1 and VRN-B1 loci on chromosome arms 5AL and 5BL, respectively (Figure 1). The VRN-A1 and VRN-B1 QTL had LOD values of 19.67 and 19.45, respectively, and they each explained 30% of the total phenotypic variation. No other major loci for growth habit were detected in this population.
Table 2 . Characterization of winter/spring growth habit.
| Lines/Plants | Number of Plants/Lines | ||
|---|---|---|---|
| Total | Winter Type | Spring Type | |
| Parent | |||
| Lebsock | > 20 | 0 | > 20 |
| PI 94749 | > 20 | 0 | > 20 |
| F1 | > 20 | 0 | > 20 |
| F2 | 200 | 13 | 187 |
| DH population | 146 | 45 | 101 |
| Backcross population | |||
| Lebsock/PI 94749//Lebsock | 100 | 0 | 100 |
| Lebsock/2*PI 94749 | 100 | 0 | 100 |
Winter/spring growth habit of parental lines Lebsock and PI 94749 and their F1 plants, F2 plants, doubled haploid (DH), and two BC1F2 populations.
Figure 1 .
QTL mapping for growth habit segregation in the Lebsock × PI 94749–derived DH population. QTL analysis was performed with composite interval mapping using the computer program Map Manager QTX (Manly et al. 2001). The positions of marker loci are shown to the left of each linkage group and centiMorgan (cM) distances between loci are shown along the right. The vertical dotted line indicates the logarithm of the odds (LOD) significance threshold of 2.91. R2 and LOD values are 0.30 and 19.67 for the QTL on chromosome 5AL (left), and 0.30 and 19.45 for the QTL on chromosome 5BL (right), respectively.
Analysis of genetic effects of VRN-A1 and VRN-B1 on winter/spring growth habit in F1 plants and in F2 and BC1F2 populations
The parental lines and their F1 hybrids were all spring type (Table 2), indicating that spring type was dominant and that the two parents each carried only one, but different, dominant allele of either Vrn-A1 or Vrn-B1. Among 200 F2 plants, 187 were spring type and 13 were winter type (Table 2), and the segregation fit a 15:1 ratio (χ2df = 1 = 0.02, P = 0.88). In the two BC1F2 populations, each consisting of 100 plants, no winter-type plants were observed (Table 2). Therefore, the results of the QTL analysis in the LP749 population combined with the results from the F1 plants and the F2 and BC1F2 populations demonstrated that the genotypes of Lebsock and PI 94749 were Vrn-A1Vrn-A1vrn-B1vrn-B1 and vrn-A1vrn-A1Vrn-B1Vrn-B1, respectively.
Allelic variation in VRN-A1 between Lebsock and PI 94749
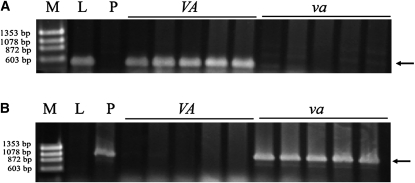
The paired primers VRN1AF and VRN1AR (Table 1) were used to detect allelic variation in the promoter region of VRN-A1 between Lebsock and PI 94749. A 484-bp fragment was amplified in both parental lines. Sequencing results confirmed that they had the same DNA sequences as the recessive vrn-A1 allele (Figure S1). However, the paired primers Ex1/C/F and Intrl/A/R3 (Table 1) produced an expected 522-bp fragment in Lebsock (Figure 2A), which demonstrated the presence of the large deletion within the first intron of the Vrn-A1 allele, whereas primer pair Intrl/C/F and Intrl/AB/R (Table 1) generated the expected 1068-bp fragment in PI 94749 (Figure 2B), which demonstrated the absence of this large deletion in the vrn-A1 allele. Both primer pairs were then used to genotype all DH lines in LP749, and VRN-A1 mapped to a location that defined the peak of the QTL on chromosome 5AL as expected (Figure 1).
Figure 2 .
Markers for allelic variation in the first intron of VRN-A1. PCR products were amplified by primer pairs Ex1/C/F and Intr1/A/R3 (A) and Intr1/C/F and Intr1/AB/R (B) for detecting presence and absence of the large deletion in the first intron of VRN-A1, respectively. Lanes: M = size standard; L = Lebsock; P = PI 94749; VA and va are DH lines carrying dominant and recessive VRN-A1, respectively. Arrows indicate the expected size of the products.
Allelic variation in VRN-B1 between Lebsock and PI 94749
The two primer pairs Intr1/B/F and Intr1/B/R3 and Intr1/B/F and Intr1/B/R4 (Table 1), which had been used to detect the presence/absence of a large deletion in intron 1 of the VRN-B1 allele (Fu et al. 2005), were used to analyze Lebsock and PI 94749, but no polymorphic products were observed. Sequencing results confirmed that both Lebsock and PI 94749 had the same 1149-bp fragment (Figure S2) of the vrn-B1 allele as in Langdon (GenBank ID AY747602) (Fu et al. 2005). Therefore, no allelic variation within intron 1 was found between the Vrn-B1 allele in PI 94749 and the vrn-B1 allele in Lebsock.
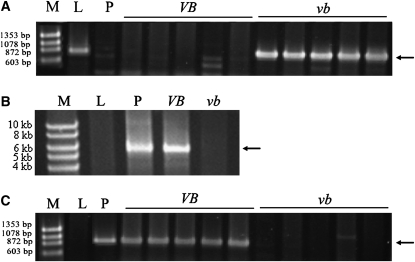
To determine whether allelic variation existed between the promoter regions of Vrn-B1 and vrn-B1, PCR analysis was conducted using the primers pair VRNBPF1 and VRNBPR1 (Table 1). However, only Lebsock yielded an amplified product, which was 989 bp in size (Figure 3A). Sequencing results indicated that the fragment from Lebsock had the same sequence as the reported VRN-B1 in Langdon (GenBank ID AY616453) (Yan et al. 2004a) (Figure S3). This primer pair was then used to genotype all DH lines, and the 989-bp fragment was amplified only in the lines carrying the vrn-B1 allele (Figure 3A), which facilitated mapping of the VRN-B1 gene in the DH population (Figure 1).
Figure 3 .
Markers for allelic variation in VRN-B1. PCR products were amplified by primer pairs VRNBPF1 and VRNBPR1 (A), VRNBPF3 and VRNBPR1 (B), and VRNBPF1 and VBINSR1 (C). Lanes: M = size standard; L = Lebsock; P = PI 94749; VB and vb are DH lines carrying dominant and recessive VRN-B1, respectively. Arrows indicate the expected polymorphic PCR products.
To understand why PI 94749 did not yield a product when amplified with VRNBPF1/VRNBPR1, a new primer (VRNBPR2) (Table 1) located 95 bp upstream of VRNBPR1, was combined with the primer VRNBPF1 for PCR. This primer pair produced the same expected 872-bp fragment in both Lebsock and PI 94749 (Figure S4), indicating that no differences existed in the region between VRNBPF1 and VRNBPR2 of the VRN-B1 gene. PCR results using the primer pair VRNBPF2 and VRNBPR3 (Table 1), which are located 3 bp upstream and 582 bp downstream of VRNBPR1, respectively, also indicated that Lebsock and PI 94749 had the same expected 647-bp amplicon. Primers VRNBPR2 and VRNBPF2 were 72 bp apart in vrn-B1 in Lebsock, but they failed to amplify the expected PCR product in PI 94749. We hypothesized that a large insertion might be located between VRNBPF1 and VRNBPR2 in the Vrn-B1 allele in PI 94749. To test this hypothesis, a long-range PCR reaction, including primers VRNBPF3 (with sequence overlapped with VRNBPR2) and VRNBPR1, was conducted. A 5.5-kb fragment was amplified in PI 94749 and a DH line carrying the dominant Vrn-B1 allele but not in Lebsock or a DH line carrying the recessive vrn-B1 allele (Figure 3B).
Sequence analysis of the 5.5-kb insertion
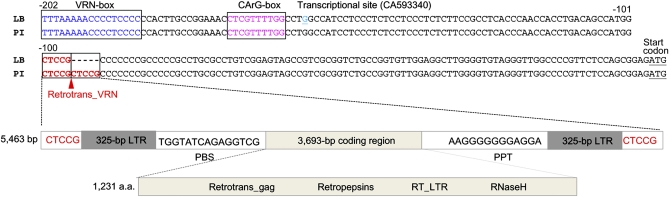
Sequencing of the 5.5-kb fragment from PI 94749 verified that the dominant Vrn-B1 allele had an insertion of 5463 bp in the 5′-UTR region (Figure 4). PI 94749 had duplicated CTCCG motifs flanking the insertion, whereas Lebsock had only one copy of this motif (Figure 4 and Figure S5). The 5-bp sequence CTCCG is a typical target site duplication (TSD) sequence for a retrotransposable element (retrotransposon) to insert in a new site of the genome (Han et al. 2010). A 325-bp LTR was found within the insertion, each having a short inverted repeat 5′-TG...CA-3′ at both ends (Figure S5). Another striking feature of this retrotransposon is the presence of PBS and PPT motifs (Figure 4 and Figure S5).
Figure 4 .
Position and structure of retrotrans_VRN identified in the dominant Vrn-B1 allele of PI 94749. The aligned sequences were from positions at VRN-box to ATG start codon in PI 94749 (PI) and Lebsock (LB). The VRN-box and CArG-box are framed and shown in blue and pink, respectively. The ATG start codon is underlined. The duplicated target site CTCCG is shown in red with the position of retrotrans_VRN indicated by a red triangle. Positions of the primer binding site (PBS) and polypurine tract (PPT), long terminal repeat (LTR), and the coding region are all indicated. The numbers “−202” and “−101” indicate the start base positions in each line relative to start codon, and “−100” indicates the end base position in each line relative to the start codon. The DNA sequence of the 5.5-kb insertion in the dominant Vrn-B1 allele in PI 94749 is deposited in the EMBL/GenBank Data Libraries under accession no. HQ186251.
An open reading frame in the 5463-bp insertion that encoded a total of 1231 amino acids was predicted (Figure S5 and Figure S6). Searches of the deduced amino acid sequence against the NCBI nonredundant protein database (nr) (ftp://ftp.ncbi.nlm.nih.gov/blast/db) showed that it had 63% similarity to a rice retrotransposon (CAD40278) along the whole predicted protein sequence, which included multiple conserved domains, such as retropepsin, retrotrans_gag, and a Gypsy family of RNase H (Figure 4). No other wheat or grass sequences had significant homology to the nucleic acid or deduced amino acid sequences associated with this 5.5-kb insertion. The new retrotransposon in the Vrn-B1 allele is designated retrotrans_VRN (GenBank ID HQ186251).
To determine whether the retrotrans_VRN insertion was the only difference between Vrn-B1 in PI 94749 and vrn-B1 in Lebsock, the complete gene from each parent was sequenced. A total of 13,347 bp, including 960 bp upstream from the start codon, 12,343 bp from the start codon to the stop codon, and 44 bp downstream from the stop codon, was determined for the recessive vrn-B1 allele (GenBank ID JN817431). The dominant Vrn-B1 allele (GenBank ID JN817430) was exactly the same as the recessive vrn-B1 allele in the sequenced region. Therefore, the retrotrans_VRN insertion was the only difference between the PI 94749 Vrn-B1 allele and the Lebsock vrn-B1 allele.
Frequency of the Vrn-B1 allele containing retrotrans_VRN in tetraploid wheat
The primer VBINSR1 (Table 1), which was close to the 5′ end of the retrotrans_VRN insertion but 42 bp downstream from VRNBPF3, was paired with VRNBPF1 to genotype all DH lines. The expected 928-bp fragment was produced only in PI 94749 and those DH lines carrying the Vrn-B1 allele (Figure 3C). Therefore, this primer pair was used to observe the presence of retrotrans_VRN in Vrn-B1, whereas the primer pair VRNBPF1 and VRNBPR1 was used to detect the absence of retrotrans_VRN in vrn-B1.
The above two primer pairs were used to determine the frequency of the Vrn-B1 allele containing retrotrans_VRN in 154 spring-type accessions/lines from six T. turgidum subspecies (Table 2). No accessions/lines from subspecies durum, polonicum, turanicum, or turgidum were found to carry retrotrans_VRN. However, 19 of 22 T. turgidum subsp. carthlicum accessions and 3 of 30 T. turgidum subsp. dicoccum accessions had retrotrans_VRN (Table 3 and Table S2). These results indicated that the dominant Vrn-B1 allele containing retrotrans_VRN is frequent among T. turgidum subsp. carthlicum accessions, is much less frequent among T. turgidum subsp. dicoccum accessions, and is rare among other tetraploid wheat subspecies. The accessions carrying retrotrans_VRN had no specific geographical distribution, suggesting that the insertion of retrotrans_VRN in Vrn-B1 occurred before the accessions spread worldwide. Sequencing results showed that these 22 accessions carrying retrotrans_VRN had the same sequences as PI 94749 at the 5′ and 3′ LTRs of retrotrans_VRN (Figure S7).
Table 3 . Presence and absence of the 5.5-kb retrotrans_VRN in 154 spring accessions/lines from six T. turgidum subspecies.
| Subspecies | Number of Accessions/Lines | ||
|---|---|---|---|
| Total | Presence of retrotrans_VRN | Absence of retrotrans_VRN | |
| T. turgidum subsp. durum | 33 | 0 | 33 |
| T. turgidum subsp. carthlicum | 22 | 19 | 3 |
| T. turgidum subsp. dicoccum | 30 | 3 | 27 |
| T. turgidum subsp. polonicum | 20 | 0 | 20 |
| T. turgidum subsp. turanicum | 30 | 0 | 30 |
| T. turgidum subsp. turgidum | 19 | 0 | 19 |
Differential expression of VRN1 genes between the dominant and recessive alleles
Under the same greenhouse conditions without vernalization, 75 plants from 16 spring DH lines carrying the dominant allele(s) of Vrn-A1, or Vrn-B1, or both, flowered at 43–64 days with an average of 54 days after planting, whereas 32 plants from 8 winter DH lines carrying both vrn-A1 and vrn-B1 alleles flowered at 89–145 days with an average of 123 days after planting. Several unvernalized plants did not flower even 145 days after planting when the experiment was terminated (Table S3). On average, vernalization decreased flowering time by 35 days compared with plants of the same lines that did not undergo vernalization (Table S3).
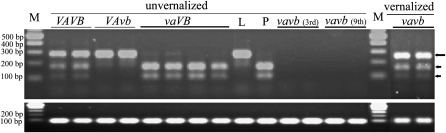
In spring DH lines carrying both dominant Vrn-A1 and Vrn-B1 (retrotrans_VRN) alleles, transcripts from both Vrn-A1 and Vrn-B1 were observed in leaves at the third leaf stage (Figure 5), whereas in Lebsock and the spring DH lines carrying the dominant Vrn-A1 and the recessive vrn-B1 alleles, transcripts were only observed from Vrn-A1 but not from vrn-B1 (Figure 5). Similarly, in PI 94749 and the spring DH lines carrying the recessive vrn-A1 allele and the dominant Vrn-B1 alleles, transcripts were observed from Vrn-B1 but not from vrn-A1 (Figure 5). No transcripts were detected from the recessive allele of either vrn-A1 or vrn-B1 in the winter DH lines in leaves at the third, seventh, or ninth leaf stages (Figure 5). However, after the winter lines were vernalized for six weeks, transcripts from both vrn-A1 and vrn-B1 were detectable (Figure 5).
Figure 5 .
Expression of VRN-1 in DH lines derived from the cross between Lebsock and PI 94749. (Upper gel) Image of HinfI-digested cDNA fragments amplified by primers Ex4-5_F1 and Ex8_R1 for separating transcripts of VRN-A1 (313 bp) (indicated by long arrow) and the VRN-B1 (173 bp + 110 bp + 30 bp) (indicated by short arrows). (Lower gel) Actin was expressed at the same level in each of the lines carrying different VRN-1 alleles. Lanes: M = size standard; L = Lebsock; P = PI 94749; vernalized and unvernalized indicates plants with or without cold treatment at 4–6° for six weeks, respectively. VAVB, VAvb, vaVB, and vavb indicate DH lines having genotype of Vrn-A1Vrn-A1Vrn-B1Vrn-B1, Vrn-A1Vrn-A1vrn-B1vrn-B1, vrn-A1vrn-A1Vrn-B1Vrn-B1, and vrn-A1vrn-A1vrn-B1vrn-B1, accordingly. The third and ninth mean leaf samples were taken at the third and ninth leaf stages, respectively.
Discussion
In the present study, we analyzed a DH population derived from two tetraploid wheat lines with spring growth habit and subsequent F2 and BC1F2 populations, and we found that the VRN-A1 and VRN-B1 genes controlled the segregation of growth habit. The durum cultivar Lebsock carried the dominant Vrn-A1 and the recessive vrn-B1 alleles, whereas PI 94749 had the recessive vrn-A1 and the dominant Vrn-B1 alleles. Because one dominant gene, either Vrn-A1 or Vrn-B1, was sufficient to confer spring growth habit in the parental lines, the appearance of winter lines in the LP749 population was due to the homoeoallelic recombination. All of the DH lines with the double-recessive genotype (vrn-A1vrn-B1) had winter growth habit, demonstrating the decisive role of the VRN1 gene in determining winter/spring growth habit in tetraploid wheat.
Alleles with variation in the first intron of VRN-A1 are the same for tetraploid and hexaploid wheat (Fu et al. 2005). However, the allelic variant of VRN-B1 identified in tetraploid wheat in this study is different from that previously reported in hexaploid wheat. Allelic variation in VRN-B1 was reported to occur in intron 1 due to a large deletion in the dominant allele in extensive spring cultivars (Fu et al. 2005; Zhang et al. 2008), and no allelic variation in this gene was reported in its promoter or any other region. In this study, no variation was observed within the first intron of dominant and recessive VRN-B1 alleles. Instead, the variation in VRN-B1 was due to the insertion of the retrotrans_VRN element within the 5′ UTR of Vrn-B1 in a position near the corresponding CArG-box and VRN-box regulatory sites of VRN-A1 in diploid and hexaploid wheat (Yan et al. 2003, 2004a; Dubcovsky et al. 2006; Pidal et al. 2009).
Kane et al. (2007) reported that the MADS-box transcription factor TaVRT2 binds the CArG-box in the promoter region of the recessive vrn1 allele. Li and Dubcovsky (2008) showed that TaFDL2, a functional homolog of Arabidopsis FD, can also interact with the promoter of the recessive vrn1 allele. In both cases vrn1 cannot be transcribed due to the binding of these transcriptional repressor(s) in the promoter region. Therefore, homozygous recessive vrn1 cannot flower without vernalization. Deletions in the promoter region disrupt the ability of repressors to bind, allowing Vrn1 to be transcribed and function in a dominant fashion, which results in spring growth habit (Yan et al. 2003; Dubcovsky et al. 2006; Pidal et al. 2009). In the same way, the insertion of retrotrans_VRN in the critical promoter region of the recessive vrn-B1 allele may disrupt repressor binding, allowing the gene to be transcribed. The gene expression experiment proved that the Vrn-B1 allele containing retrotrans_VRN was transcribed and the spring lines carrying the Vrn-B1 allele flowered without vernalization. In the winter DH lines carrying the recessive vrn-B1 allele, vrn-B1 transcripts were detected only after plants were vernalized, and the vernalized plants flowered earlier than those without vernalization treatment did. Most insertions have deleterious effects on the expression of nearby genes in plants (Hollister et al. 2011). On the contrary, this study provides an example of an insertion that resulted in the liberation of gene expression. Mutation of the VRN-B1 gene is the mechanism by which tetraploid wheat evolved from the winter wild type to spring type.
A survey of 154 spring-type accessions/lines of six T. turgidum subspecies indicated that none of the accessions from the subspecies durum, polonicum, turanicum, or turgidum had retrotrans_VRN in VRN-B1, but the insertion was present in some accessions of T. turgidum subsp. dicoccum and in most accessions of T. turgidum subsp. carthlicum. This suggested that the insertion event in the dominant Vrn-B1 allele might have occurred during the evolution of tetraploid wheat and may have contributed to adaptation to certain environments. Identification of the VRN-B1 allele in a global collection of tetraploid wheat could provide important molecular information for wheat breeding. Both T. turgidum subsp. carthlicum and T. turgidum subsp. dicoccum are potentially important sources for improving resistance to tan spot and Stagonospora nodorum blotch (Chu et al. 2008a), Fusarium head blight (Oliver et al. 2008), and stem rust (Olivera Firpo et al. 2010) for durum and bread wheat grown in North America. Molecular identification of the dominant gene for spring growth habit will facilitate selection of the proper wheat parents to breed cultivars for specific environments or to develop spring populations that do not have a vernalization requirement.
Effects of different VRN1 genes on heading date have been studied extensively in spring wheat (Law et al. 1976; Stelmakh 1993; Kato et al. 1999; Tóth et al. 2003; Hanocq et al. 2004; Chu et al. 2008b). The dominant Vrn-A1 allele normally provides complete insensitivity to vernalization; thus, it is considered the most potent for spring growth habit, whereas a dominant Vrn-B1 or Vrn-D1 allele may result in the partial elimination of the vernalization requirement (Pugsley 1971, 1972; Stelmakh 1993). Vrn-A1 is indeed expressed at earlier and higher levels than Vrn-B1 and Vrn-D1 in Triple Dirk isogenic lines (Loukoianov et al. 2005). The Triple Dirk A (TDA) line carrying the dominant Vrn-A1 allele flowered several days earlier than the Triple Dirk B (TDB) line carrying the dominant Vrn-B1 allele and the Triple Dirk E (TDE) line carrying the dominant Vrn-D1 allele (Trevaskis et al. 2003; Loukoianov et al. 2005). In this study, PI 94749, which harbors only the dominant Vrn-B1 allele, headed up to two weeks later than Lebsock, which had only the dominant Vrn-A1 allele (Table S3). However, in the DH population, the lines that carried only the dominant Vrn-B1 allele had similar heading dates as the lines that only carried the dominant Vrn-A1 (Table S3). It seems that the effect of the retrotrans_VRN–containing Vrn-B1 allele on growth habit might be similar to that of Vrn-A1 in tetraploid wheat. No further experiments were conducted to explain variation in flowering within the spring or winter groups, as this research was focused on the identification of allelic variation between winter lines that had a vernalization requirement and spring lines that lacked this requirement for flowering.
Supplementary Material
Acknowledgments
We thank N. Jiang at Michigan State University for help with annotation of the retrotransposon sequences. This work was supported by USDA–ARS CRIS (Current Research Information System) Project 5442-22000-033-00D and by the Oklahoma Center of Advanced Science and Technology (OCAST). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Literature Cited
- Barrett B., Bayram M., Kidwell K., 2002. Identifying AFLP and microsatellite markers for vernalization response gene Vrn-B1 in hexaploid wheat using reciprocal mapping populations. Plant Breed. 121: 400–406 [Google Scholar]
- Chu C. G., Chao S., Friesen T. L., Faris J. D., Zhong S., et al. , 2010. Identification of novel tan spot resistance QTLs using an SSR-based linkage map of tetraploid wheat. Mol. Breed. 25: 327–338 [Google Scholar]
- Chu C. G., Friesen T. L., Faris J. D., Xu S. S., 2008a Evaluation of seedling resistance to tan spot and Stagonospora nodorum blotch in tetraploid wheat. Crop Sci. 48: 1107–1116 [Google Scholar]
- Chu C. G., Xu S. S., Friesen T. L., Faris J. D., 2008b Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol. Breed. 22: 251–266 [Google Scholar]
- Danyluk J., Kane N. A., Breton G., Limin A. E., Fowler D. B., et al. , 2003. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Li C., Dubcovsky J., 2009. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Lijavetzky D., Appendino L., Tranquilli G., 1998. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor. Appl. Genet. 97: 968–975 [Google Scholar]
- Dubcovsky J., Loukoianov A., Fu D., Valarik M., Sanchez A., et al. , 2006. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J. D., Haen K. M., Gill B. S., 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Szücs P., Yan L., Helguera M., Skinner J. S., et al. , 2005. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273: 54–65 [DOI] [PubMed] [Google Scholar]
- Golovnina K. A., Kondratenko E. Y., Blinov A. G., Goncharov N. P., 2010. Molecular characterization of vernalization loci VRN1 in wild and cultivated wheats. BMC Plant Biol. 10: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. H., Wang G. X., Liu Z., Liu J. H., Yue W., et al. , 2010. Divergence in centromere structure distinguishes related genomes in Coix lacryma-jobi and its wild relative. Chromosoma 119: 89–98 [DOI] [PubMed] [Google Scholar]
- Hanocq E., Niarquin M., Heumez E., Rousset M., Le Gouis J., 2004. Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor. Appl. Genet. 110: 106–115 [DOI] [PubMed] [Google Scholar]
- Hemming M. N., Fieg S., Peacock W. J., Dennis E. S., Trevaskis B., 2009. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomics 282: 107–117 [DOI] [PubMed] [Google Scholar]
- Hollister J. D., Smith L. M., Guo Y.-L., Ott F., Weigel D., et al. , 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 108: 2322–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane N. A., Agharbaoui Z., Diallo A. O., Adam H., Tominaga Y., et al. , 2007. TaVRT2 represses transcription of the wheat vernalization gene TaVRN1. Plant J. 51: 670–680 [DOI] [PubMed] [Google Scholar]
- Kato K., Miura H., Sawada S., 1999. QTL mapping of genes controlling ear emergence time and plant height on chromosome 5A of wheat. Theor. Appl. Genet. 98: 472–477 [Google Scholar]
- Kosambi D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Lander E. S., Green P., Abrahamson J., Barlow A., Daly M. J., et al. , 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Law C. N., Worland A. J., Giorgi B., 1976. The genetic control of ear-emergence time by chromosomes 5A and 5D of wheat. Heredity 36: 49–58 [Google Scholar]
- Leonova I., Pestsova E., Salina E., Efremova T., Röder M., et al. , 2003. Mapping of the Vrn-B1 gene in Triticum aestivum using microsatellite markers. Plant Breed. 122: 209–212 [Google Scholar]
- Li C., Dubcovsky J., 2008. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukoianov A., Yan L., Blechl A., Sanchez A., Dubcovsky J., 2005. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol. 138: 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B., Yanofsky M. F., 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Manly K. F., Cudmore R. H., Jr, Meer J. M., 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Murai K., Miyamae M., Kato H., Takumi S., Ogihara Y., 2003. WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 44: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Oliver R. E., Cai X., Friesen T. L., Halley S., Stack R. W., et al. , 2008. Evaluation of Fusarium head blight resistance in tetraploid wheat (Triticum turgidum L.). Crop Sci. 48: 213–222 [Google Scholar]
- Olivera Firpo P. D., Jin Y., Xu S., Klindworth D., 2010. Resistance to race TTKSK of Puccinia graminis f. sp. tritici in tetraploid wheat. APS 2010 Annual Meeting, August 7–11, 2010, Charlotte, NC. Phytopathology 100: S93 [Google Scholar]
- Pidal B., Yan L., Fu D., Zhang F., Tranquilli G., et al. , 2009. The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. J. Hered. 100: 355–364 [DOI] [PubMed] [Google Scholar]
- Pugsley A. T., 1971. A genetic analysis of the spring-winter habit of growth in wheat. Aust. J. Agric. Res. 22: 21–31 [Google Scholar]
- Pugsley A. T., 1972. Additional genes inhibiting winter habit in wheat. Euphytica 21: 547–552 [Google Scholar]
- Rousset M., Bonnin I., Remoué C., Falque M., Rhoné B., et al. , 2011. Deciphering the genetics of flowering time by an association study on candidate genes in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 123: 907–926 [DOI] [PubMed] [Google Scholar]
- Shimada S., Ogawa T., Kitagawa S., Suzuki T., Ikari C., et al. , 2009. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 58: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape J. W., Sarma R., Quarrie S. A., Fish L., Galiba G., et al. , 2001. Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica 120: 309–315 [Google Scholar]
- Stelmakh A. F., 1987. Growth habit in common wheat (Triticum aestivum L. EM. Thell.). Euphytica 36: 513–519 [Google Scholar]
- Stelmakh A. F., 1993. Genetic effects of Vrn genes on heading date and agronomic traits in bread wheat. Euphytica 65: 53–60 [Google Scholar]
- Takahashi R., Yasuda S., 1971. Genetics of earliness and growth habit in barley, pp. 388–408 Barley Genetics II, Proceedings of the Second International Barley Genetics Symposium, edited by Nilan R. A. Washington State University Press, Pullman, WA [Google Scholar]
- Tóth B., Galiba G., Fehér E., Sutka J., Snape J. W., 2003. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor. Appl. Genet. 107: 509–514 [DOI] [PubMed] [Google Scholar]
- Tranquilli G., Dubcovsky J., 2000. Epistatic interactions between vernalization genes Vrn-Am1 and Vrn-Am2 in diploid wheat. J. Hered. 91: 304–306 [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Bagnall D. J., Ellis M. H., Peacock W. J., Dennis E. S., 2003. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland A. J., 1996. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89: 49–57 [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., et al. , 2003. Positional cloning of wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Helguera M., Kato K., Fukuyama S., Sherman J., et al. , 2004a Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109: 1677–1686 [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., et al. , 2004b The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., et al. , 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. K., Xiao Y. G., Zhang Y., Xia X. C., Dubcovsky J., et al. , 2008. Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Sci. 48: 458–470 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.