Abstract
Staphylococci are increasingly aggressive human pathogens suggesting that active evolution is spreading novel virulence and resistance phenotypes. Large staphylococcal plasmids commonly carry antibiotic resistances and virulence loci, but relatively few have been completely sequenced. We determined the plasmid content of 280 staphylococci isolated in diverse geographical regions from the 1940s to the 2000s and found that 79% of strains carried at least one large plasmid >20 kb and that 75% of these large plasmids were 20–30 kb. Using restriction fragment length polymorphism (RFLP) analysis, we grouped 43% of all large plasmids into three major families, showing remarkably conserved intercontinental spread of multiresistant staphylococcal plasmids over seven decades. In total, we sequenced 93 complete and 57 partial staphylococcal plasmids ranging in size from 1.3 kb to 64.9 kb, tripling the number of complete sequences for staphylococcal plasmids >20 kb in the NCBI RefSeq database. These plasmids typically carried multiple antimicrobial and metal resistances and virulence genes, transposases and recombinases. Remarkably, plasmids within each of the three main families were >98% identical, apart from insertions and deletions, despite being isolated from strains decades apart and on different continents. This suggests enormous selective pressure has optimized the content of certain plasmids despite their large size and complex organization.
Keywords: plasmid, resistance, mobile element genomics, MRSA, horizontal gene transfer
Staphylococci are common commensals and opportunistic pathogens mainly found on the skin and in the nose of humans and also in domestic and companion animals (Epstein et al. 2009; Lindsay 2010; Middleton et al. 2005; Rubin et al. 2011; Rutland et al. 2009; Spohr et al. 2011; Sung et al. 2008; Van Duijkeren et al. 2011; Vanderhaeghen et al. 2011; Walther et al. 2009; Wassenberg et al. 2011). Staphylococcus aureus, carried by ∼30% of humans in developed countries (Lindsay and Holden 2004; Plata et al. 2009), is a leading cause of healthcare-associated (HA) infections and is increasingly responsible for life-threatening community-acquired (CA) infections in otherwise healthy persons (Diekema et al. 2001; Lindsay and Holden 2004, 2006; Navarro et al. 2008; Plata et al. 2009). S. aureus is the number one cause of bloodstream, skin, and lower respiratory infections (Diekema et al. 2001; Goetghebeur et al. 2007; Plata et al. 2009), and multiantibiotic resistances common to HA staphylococci are now increasingly present in the community strains (Lindsay and Holden 2006; McDougal et al. 2010; Navarro et al. 2008). Coagulase-negative staphylococci (CNS; e.g. Staphylococcus epidermidis) are also a major cause of HA bloodstream infections, and oxacillin resistance and methicillin resistance are found in over 70% of strains (Diekema et al. 2001).
Analyses of S. aureus complete genomes revealed that most virulence factors and antibiotic resistance genes are carried on mobile genetic elements (MGE) (Baba et al. 2002; Feng et al. 2008; Highlander et al. 2007; Holden et al. 2004; Omoe et al. 2003) such as pathogenicity islands, chromosomal cassettes, transposable elements, bacteriophages, and plasmids (Lindsay and Holden 2004, 2006; Lindsay 2010; Malachowa and Deleo 2010; Novick 2003). Thus, understanding the MGEs in staphylococci is critical to controlling dissemination of these virulence factors that markedly increase the hazard of these pathogens.
Multilocus sequence typing (MLST) of methicillin-resistant Staphylococcus aureus (MRSA) strains has shown that outbreaks are caused by relatively few clonal complexes or by strains with closely related genotypes (Diekema et al. 2001; Enright et al. 2002; Feil et al. 2003; Feil and Enright 2004; Feng et al. 2008; Highlander et al. 2007; Lindsay and Holden 2004, 2006). The success of these clonal complexes as pathogens may partially be explained by their enhanced ability to receive MGEs by horizontal gene transfer (HGT). Type I (Sung and Lindsay 2007; Waldron and Lindsay 2006) and type III–like (Corvaglia et al. 2010) restriction modification systems in S. aureus provide natural barriers to HGT. Strains with defective restriction modification systems acquire MGEs at higher frequencies and have greater potential to become “superbugs” by accumulating virulence factors and resistance genes (Corvaglia et al. 2010; Sung and Lindsay 2007; Waldron and Lindsay 2006).
Staphylococcal plasmids carry resistances to antibiotics, metals, antiseptics, and disinfectants, as well as virulence genes, such as enterotoxins (Bayles and Iandolo 1989; Omoe et al. 2003) and exfoliative toxins (Jackson and Iandolo 1986; Yamaguchi et al. 2001). Plasmids in staphylococci may be horizontally transferred through conjugation, mobilization, and/or transduction (Apisiridej et al. 1997; Berg et al. 1998; Francia et al. 2004; Lindsay and Holden 2004; Malachowa and Deleo 2010; Smith and Thomas 2004; Udo and Grubb 2001; Varella Coelho et al. 2009). Staphylococci also carry virulence plasmids originating from Bacillus (Gill et al. 2005) and Enterococcus (Clewell et al. 1985; Noble et al. 1992; Périchon and Courvalin 2009; Sung and Lindsay 2007; Weigel et al. 2003).
Staphylococcal plasmids are taxonomically grouped by replication mechanism and conjugation ability: the small, usually <5 kb, rolling-circle replicating (RCR) plasmids; the larger theta-replicating plasmids, which are subdivided into the pSK41-like conjugative plasmids; and the nonconjugative antimicrobial and metal resistance plasmids (Berg et al. 1998; Firth et al. 2000; Firth and Skurray 2006; Khan 1997; Malachowa and Deleo 2010; Novick 1989). The small RCR plasmids often carry a single antibiotic resistance gene (Khan 1997) that is transferred by transducing phages, mobilized by conjugative plasmids, or can form unresolved cointegrates with conjugative or mobilizable plasmids, arising from replicative transposition by IS257 or homologous recombination between IS257 elements (Berg et al. 1998; Leelaporn et al. 1996; Smith and Thomas 2004; Varella Coelho et al. 2009). The theta-replicating nonconjugative plasmids can also be transferred by mobilization or transduction (Apisiridej et al. 1997; Francia et al. 2004; Lindsay and Holden 2006; Malachowa and Deleo 2010; Smillie et al. 2010). As of 2010, the NCBI RefSeq database had 102 complete staphylococcal plasmid sequences, but only 29 (28%) of them were >20 kb and only 15% were >30 kb and, thus, large enough to encode conjugation machinery. Of the latter group, only 6% had annotated conjugative transfer loci (McDougal et al. 2010). Conjugative plasmids spread readily among Staphylococcus strains and to and from other genera such as Enterococcus (Lindsay 2010; Malachowa and Deleo 2010; Périchon and Courvalin 2009; Zhu et al. 2008), and they are implicated in spreading virulence loci and vancomycin resistance among clinical strains (Périchon and Courvalin 2009; Zhu et al. 2008). Multiresistance plasmids between 20 and 30 kb are common in staphylococci from several continents (Baba et al. 2002; Bayles and Iandolo 1989; Highlander et al. 2007; Holden et al. 2004; Shalita et al. 1980; Toh et al. 2007; Zuccarelli et al. 1990). Although unlikely to be conjugative, these 20–30 kb multiresistance virulence plasmids can be transferred by mobilization or by the generalized transducing phages prevalent among S. aureus strains (Lindsay and Holden 2004, 2006; Lindsay 2010).
We undertook the work reported here to identify the plasmid composition of naturally occurring isolates of staphylococci and found considerably more large plasmids than previously recorded. We used this opportunity to examine archived strains from the mid-twentieth century known to contain large plasmids. We screened 280 strains of staphylococci collected from 1946 to 2007 in diverse geographical regions to determine the number, size, and restriction type of plasmids they carried (aka the plasmid profile of each strain). We chose 100 strains containing distinct plasmids >20 kb for sequencing to increase the representation of large staphylococcal plasmid sequences available. The 93 complete new plasmid sequences included 59 plasmids >20 kb, tripling the number of large staphylococcal plasmid sequences in RefSeq. We also acquired 57 partial plasmid and phage sequences.
Materials and Methods
Strain collections
We screened 280 strains of staphylococci from eight geographically and epidemiologically distinct collections (supporting information, Table S1), including 251 S. aureus, 14 S. epidermidis, 3 S. lentus, 2 S. pseudintermedius, 2 S. schleiferi, and 8 CNS of unspecified species. In addition, we sequenced plasmid DNA from strain CM05, a human clinical isolate from Columbia (Toh et al. 2007) and from vancomycin-resistant Enterococcus faecium strain 5753c (McDougal et al. 2010).
Isolation and characterization of plasmid DNA
Strains were grown in BHI broth without shaking at 37° for 18–24 hr. Total plasmid DNA was isolated from 1 to 5 ml of culture with the CosMC Prep Kit (Agencourt Biosciences Corp., Beverly, MA), as described (Williams et al. 2006). Cell pellets were suspended in 100 μl RE1 buffer containing 200 μg/ml lysostaphin and 6% PEG (MW 8000) and incubated for 5 min at room temperature before proceeding with the CosMC Prep Kit protocol. CosMC Kit preparations were done in duplicate, and pooled plasmid DNA was electrophoresed using 0.5% SeaKem Gold agarose (Cambrex BioScience, Walkersville, MD) 16 cm gels in 1X TAE (40 mM Tris-acetate, 2 mM Na2EDTA–2H2O) for 15–18 hr at 30–35 V and stained with Sybr Green I (Invitrogen, Carlsbad, CA). Plasmid band sizes were estimated by BacTracker supercoiled DNA ladder (Epicentre, Madison, WI). DNA preparations found initially to have high sheared DNA background were in subsequent preparations treated with lambda exonuclease and RecJf (New England Biolabs) (Balagurumoorthy et al. 2008) and/or Plasmid-safe DNase (Epicentre) prior to electrophoresis. Sixteen plasmids were further purified by electroelution (Williams et al. 2006) into 1X TAE followed by TE dialysis. Three plasmids were prepared by QIAprep Spin Miniprep Kit (Qiagen) or Qiagen Midi Kit (Enterococcus faecium 5753c), and one plasmid (pCM05) was prepared by CsCl gradient ultracentrifugation.
Plasmid bands were characterized by restriction fragment length polymorphism (RFLP) analysis, and plasmids were assigned a restriction type (RT) according to their AccI RFLP pattern; a unique RT number was given to each unique pattern. In-slice restriction digests were performed on gel slices containing individual plasmid bands excised with a razor blade. Gel slices were rocked at 4° for 30 min in 1 ml TE (10 mM Tris-HCl, 1 mM EDTA, pH 8), transferred to 100 μl 1X restriction buffer (New England Biolabs), rocked at 4° for 30 min to 1 hr, transferred to 100 μl fresh 1X restriction buffer with 15–20 units enzyme, and then incubated at 37°, 50–75 rpm for 16–24 hr. Digested gel slices were transferred to 1 ml 1X TBE and rocked at 4° for 30–60 min, sealed with molten agarose into a well of a 16 cm 1.5% agarose gel (medium EEO) (Sigma, Inc., St. Louis, MO), and electrophoresed in 1X TBE for 15–18 hr at 30–35 V. Gels were stained with Sybr Green I (Invitrogen). RFLP patterns with several enzymes guided the choice of those for sequencing and subsequently tested the accuracy of the sequence assemblies.
Plasmid DNA sequencing
Plasmid DNA was sequenced according to standard high-throughput Sanger protocols at the J. Craig Venter Institute, and data was assembled using the Celera Assembler (Myers et al. 2000). Quality control inspections included coverage analysis, BLAST (http://blast.ncbi.nlm.nih.gov) (Zhang et al. 2000) of contigs >2000 bases, and visual inspection of mate pairing and scaffolding using Hawkeye (Schatz et al. 2007). Chromosomal and/or other contamination were filtered based on perfect BLAST hits to S. aureus and S. epidermidis references. DNA samples for each strain were assigned a unique SAP (S. aureus plasmids) project number. To distinguish multiple plasmids in the same strain, each was assigned the strain SAP number plus A, B, C, or D, starting with complete (closed) plasmid sequences in descending size, followed by partial sequences (Table S2).
Sequence analysis
Gene calling and annotations were done by P-RAST (http://cgat.mcs.anl.gov/plasmid-rast-dev/FIG/prast.cgi) and/or RAST (http://rast.nmpdr.org) (Aziz et al. 2008), which uses mobile element gene names from the ACLAME database (http://aclame.ulb.ac.be) (Leplae et al. 2004). Lasergene (DNAstar, Madison, WI) and Gene Construction Kit (Textco Biosoftware, West Lebanon, NH) were used to display sequences and predict restriction sites. Sequence alignments were done with Megalign (DNAstar) using ClustalW and/or with BLAST (http://blast.ncbi.nlm.nih.gov) (Zhang et al. 2000), and plasmid genome alignments were done using Mauve (Darling et al. 2004). One hundred fifty complete and partial plasmid and phage sequences were submitted to GenBank (http://www.ncbi.nlm.nih.gov) (Table S2). Partial sequences are those with one or more gaps in the sequence, including plasmid and phage fragments (Table S2). Partial sequences with the same plasmid name are fragments of the same plasmid (Table S2).
Results
Plasmid profiling revealed that the majority of staphylococcal strains carry 20–30 kb plasmids with three major families
We screened 280 staphylococci, 247 of which had not previously been examined for plasmid content. Ninety percent of strains had plasmids, and 78.5% of these had one or more large plasmids >20 kb (Table S3). Of the 184 typable >20 kb plasmids in these newly screened strains, 75% were 20–30 kb (Table 1). Indeed, plasmids from 20 to 30 kb are extremely abundant among staphylococci, but they were only 13.7% of previously sequenced staphylococcal plasmids in RefSeq. Restriction types (RT) were assigned by RFLP analysis (see Materials and Methods), and three RTs (RT1, RT2, and RT3) encompassed 60.7% of typable 20–30 kb plasmids in strains not previously examined for plasmids (Table 2). These three RTs were also 42.5% of all large plasmids >20 kb typed and 49.0% of those from newly examined strains (Table 2). We assigned 106 distinct RTs, and 80% of those were unique to a single plasmid; thus, only those three RTs were widely common.
Table 1 . Sizes of large typable staphylococcal plasmids.
| Number of Plasmids (%) | |
|---|---|
| Total typable >20 kba | 184 |
| >30 kb | 46 (25%) |
| 20–30 kb | 138 (75%) |
Large plasmids from newly examined strains (n = 247) typable by RFLP.
Table 2 . RFLP patterns of staphylococcal plasmids reveal three prevalent families.
| Family | Restriction Type | Estimated Size (kb)a | Number of Plasmidsb | 20–30 kbd | >20 kbe |
|---|---|---|---|---|---|
| pMW2-like | RT1 | 18–21 | 27 | 19.6% | 14.7% |
| pIB485-like | RT2 | 25–27 | 36c | 26.1% | 19.6% |
| pUSA300HOUMR-like | RT3 | 25–27 | 27 | 19.6% | 14.7% |
| Total | 90c | 65.3% | 49.0% |
Sizes estimated by comparisons of electrophoresed undigested DNA to size ladder (see Materials and Methods).
Includes all typable plasmids >20 kb (n = 184) from newly examined strains.
There was one additional pIB485-like plasmid in a strain previously examined for plasmids (for a total 37 of 214 typable >20 kb plasmids from all strains).
The representation of that family among the typable 20–30 kb plasmids from newly examined strains (n = 138)
The representation of that family among all typable >20 kb plasmids from newly examined strains (n = 184).
The RT1 family has the smallest plasmids of the three common groups, estimated by gels at 18–21 kb (Table 2), resembling previously sequenced pMW2 (Baba et al. 2002) and pSAS (Holden et al. 2004) of a U.S. MRSA strain and a U.K. MSSA strain, respectively. The RFLPs of 25–27 kb RT2 family resembled many previously unsequenced or partially sequenced plasmids, including 54 plasmids in California MRSA hospital strains isolated in the 1980s (Zuccarelli et al. 1990); plasmid pIB485 carrying the sed enterotoxin gene (Bayles and Iandolo 1989); and 8 pIB485-like S. aureus plasmids carrying ser and sej enterotoxin genes (Omoe et al. 2003). The RT3 plasmids also ranged from 25 to 27 kb (Table 2), but the RT3 restriction pattern resembled that of previously sequenced plasmid pUSA300HOUMR (NC_010063) (Highlander et al. 2007). Six pMW2-like RT1, 5 pIB485-like RT2, and 3 pUSA300HOUMR-like RT3 plasmids from diverse epidemiological and geographical backgrounds were chosen for sequencing, along with 86 plasmids with less common or unique RTs.
Analysis of the sequenced staphylococcal plasmid genomes
Prior to submission of the sequences from this project, only 29 of the 102 complete staphylococcal plasmid sequences in RefSeq were larger than 20 kb (28%) (Table 3). The 93 new complete staphylococcal plasmid sequences (Table 3 and Table S2) increased the number of large >30 kb plasmids from 15 to 42, the number of 20–30 kb plasmids from 14 to 46, and the number of small <20 kb plasmids from 73 to 107. Thus, 45% of staphylococcal complete plasmid sequences are now >20 kb, which more closely represents the plasmid content observed in the 247 newly examined staphylococcal strains (Table S3). For the partial plasmid sequences that were deposited, 16 are plasmids with one small gap (∼20 bp), and the other 41 are large plasmid or phage contigs (1.4–56.7 kb) (Table S2). As expected (Malachowa and Deleo 2010; Novick 2003), the plasmids carry multiple antibiotic resistance genes and virulence genes, such as enterotoxins and exfoliative toxins (Table S2), including three new enterotoxins. Plasmid maintenance and transmissibility genes and transposons were also identified (Table S2).
Table 3 . Complete staphylococcal plasmid genome sequences.
| <20 kb | 20–30 kb | >30 kb | Total | |
|---|---|---|---|---|
| RefSeq | 73 | 14 | 15 | 102 |
| This project | 34 | 32 | 27 | 93 |
| Total | 107 | 46 | 42 | 195 |
A RepA_N-type replication initiation gene (Weaver et al. 2009) occurs in 90% (54) of the 60 completely sequenced staphylococcal plasmids >10 kb [excepting the phage GQ900400, p5753cA GQ900435 from E. faecium and potential integrative conjugative element (ICE) GQ900429, Table S2]. The remaining 6, corresponding to pMW2-like RT1 plasmids, have only remnants of a RepA_N-type gene but contain a member of the Rep_3 superfamily first identified in the S. epidermidis plasmid pSK639 (Apisiridej et al. 1997; Firth and Skurray 2006). As further evidence of replicon fusions, 11 of 54 plasmids carrying a RepA_N-type gene also have a pSK639-like rep gene or a remnant thereof (Table S2). All 60 plasmids >10 kb encode some partitioning function: 78% have a homolog of the pSK1 par locus (Simpson et al. 2003), 12% have a type Ib partitioning system, and 10% have a type II system (Table S2).
Conjugation and mobilization loci:
Among plasmids large enough to encode conjugation loci (>12 kb) (Berg et al. 1998; Caryl and O'Neill 2009), only 6 of the 27 closed >30 kb plasmids have a clearly annotated conjugative transfer region, doubling the conjugation loci sequences available in RefSeq. An additional complete tra region was found in partial sequence SAP015B [GQ900500] (Table S2). Only 17 plasmids from this project (Table 4) have predicted mob loci (13 closed and 4 partial sequences), with only 3 from S. aureus (Table S2). An additional 31 plasmids (Table 4 and Table S2) encode a potential relaxase, tra or pre, which could function in mobilization (Francia et al. 2004; Garcillan-Barcia et al. 2009; Smith and Thomas 2004; Varella Coelho et al. 2009), for a total of 48 plasmids with putative mobilization genes.
Table 4 . Predicted transfer loci in new staphylococcal plasmid sequences.
| Total Sequences | tra Region | Mob | Relaxase (pre or tra) | |
|---|---|---|---|---|
| Complete | 93 | 6 | 13 | 22 |
| Partial | 57 | 1 | 4 | 9 |
| Total | 150 | 7 | 17 | 31 |
Transfer loci as predicted by RAST and/or P-RAST (http://rast.nmpdr.org) (Aziz et al. 2008).
sin and res plasmid maintenance recombinases:
Palindromic sequences in the recognition (res) sites, used by resolvases to separate plasmid multimers and thereby promote segregational stability, are also hotspots for insertion of Tn552 and other transposons (Derbise et al. 1995; Lebard et al. 2008; Paulsen et al. 1994; Rowland and Dyke 1989, 1990). Such res sites are adjacent to plasmid resolvase sin genes in nonconjugative multiresistance plasmids and to res in the pSK41-like conjugative plasmids (Berg et al. 1998; Lebard et al. 2008; Rowland et al. 2002). Insertion of Tn552-, Tn4002-, or Tn5404-like transposons at these recombination hotspots produces a region of DNA flanked by inverted repeats (resL and resR) that can be inverted by the Bin recombinase (Derbise et al. 1995; Rowland and Dyke 1989, 1990; Rowland et al. 2002). This continual flipping process results in a region of sequence heterogeneity, complicating plasmid sequence assembly. The common occurrence of transposable elements such as Tn552 and IS257 and recombinases Bin, Sin, and Res (Table 5 and Table S2) likely explains several of the 57 partial and gapped plasmid sequences we obtained.
Table 5 . Recombinases and Tn552 in staphylococcal plasmids.
Forty-four complete staphylococcal plasmid sequences and 32 partial sequences carry the sin gene (Table 5). The sin gene occurs in 47% (44 of 93, Table 5) of staphylococcal plasmids completely sequenced in this project. sin, almost exclusively found on plasmids (where it is sometimes annotated as bin3), is not associated with a transposable element (Paulsen et al. 1994; Rowland and Dyke 1989; Rowland et al. 2002). A resolvase/invertase site-specific recombinase family member, sin most resembles Gram-positive resolvases like Res from Enterococcus faecalis plasmid pAMβ1 (Paulsen et al. 1994; Swinfield et al. 1991), which reduce multimers to ensure plasmid inheritance. sin may play this role for staphylococcal plasmids (Paulsen et al. 1994; Rowland et al. 2002). As expected (Berg et al. 1998; Lebard et al. 2008; Rowland et al. 2002), sin occurred on plasmids lacking tra genes, and res was found on the pSK41-like plasmids encoding conjugation genes, with only one exception: res also occurred on the mobilizable plasmid SAP016A from a S. epidermidis strain (Table S2). The same 75 bp internal deletion is in sin in 5 of the pMW2-like plasmids, and 8 newly sequenced S. aureus plasmids (4 complete sequences and 4 partial sequences) have truncated sin genes. Of the 60 complete staphylococcal plasmids >10 kb (Table S2) likely to require a resolvase, 51 or 85% have sin or res. Only 16 RefSeq plasmid sequences carry sin and only 7 have res (Table 5); in total, 74 of 195 (37.9%) complete staphylococcal plasmid sequences have a sin or res recombinase gene, presumably to enable stable plasmid inheritance.
Prevalence of bin recombinase and Tn552:
Besides sin, the single most common gene in the staphylococcal plasmids is bin, mostly found with the β-lactamase genes blaZ, blaR1, and blaI of the replicative transposon Tn552 as the transposon’s resolvase. Of the plasmids >20 kb in size that we sequenced, 68 (68.7%) carry at least one bin (Table 5 and Table S2). Tn552, including bin, is also found on staphylococcal chromosomes (Rowland and Dyke 1989; Rowland and Dyke 1990). Full or truncated Tn552 occurs on 53 complete staphylococcal plasmids (18 in RefSeq and 35 from this project, Table 5), isolated from strains found in Australia, the United Kingdom, the United States, and Columbia from the 1940s to the 2000s (Table S2), demonstrating intercontinental plasmid-mediated spread of antibiotic resistances and their persistence over six decades. We found the full Tn552 located on 10 complete and 7 partial staphylococcal plasmids, and we found that 25 complete and 15 partial plasmids encode one of two truncated Tn552 variants (Table 5).
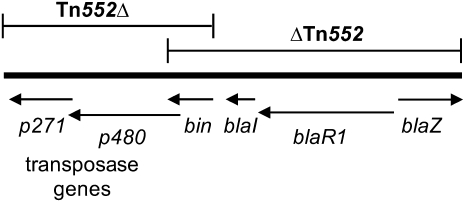
Tn552 encodes two transposition genes, p271 and p480, the resolvase bin, and the β-lactamase genes blaI, blaR1, and blaZ, and it generates 6 bp direct repeats of target DNA flanking its ∼120 bp inverted repeats (Rowland and Dyke 1989, 1990). The full or truncated variants of Tn552 (Figure 1) occur in 38% (57 of 150) of the new complete and partial staphylococcal plasmid sequences but in only 18% of those in RefSeq (Table 5). Tn552 insertion adjacent to transposons, within other transposons, or near res sites can create DNA segments that can then be inverted or deleted by Bin (Rowland and Dyke 1989, 1990). The prototypical ΔTn552 in pI258 likely arose by Bin-mediated deletion (Rowland and Dyke 1990). In pI258, ΔTn552 lacks transposition genes but has bin and all three β-lactamase–associated genes (Figure 1). Twenty-three complete and 12 partial newly sequenced staphylococcal plasmids, including pI258, have ΔTn552; only four examples of ΔTn552 are in RefSeq. We first observed here the converse truncated version of Tn552, Tn552Δ, which has the transposase genes and bin but lacks the bla genes (Figure 1). Tn552Δ was only observed in 2 complete and 3 partial staphylococcal plasmids (Table 5).
Figure 1 .
Dominant variants of Tn552 found on Staphylococcus plasmids. The full-length Tn552 is characterized by two transposase genes, p271 and p480, a bin recombinase, and three β-lactamase–associated genes, blaI, blaR1, and blaZ. Tn552Δ is missing the bla genes, and ΔTn552 is missing the transposase genes.
Three major Staphylococcus plasmid families revealed by plasmid profiling were confirmed and extended by sequencing
pIB485-like enterotoxin plasmids are found worldwide:
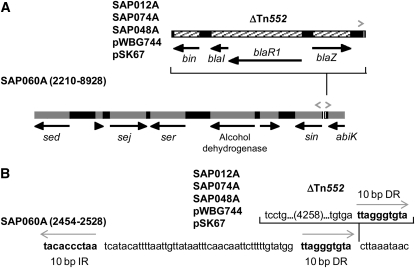
At 26% of 20–30 kb newly profiled plasmids, the pIB485-like RT2 plasmids are the most prevalent of the three major RTs identified by RFLP profiling (Table 2). No pIB485-like plasmid had been completely sequenced before this project, but 63 plasmids with a similar RFLP had been reported (see above) (Bayles and Iandolo 1989; Omoe et al. 2003; Zuccarelli et al. 1990), including the SED enterotoxin–containing plasmid pIB485 (Bayles and Iandolo 1989). The five strains whose plasmids we sequenced were isolated between 1949 and 2001 (Table 6) in the United States (2), the United Kingdom (1), and Australia (2), and they have ΔTn552 (Figure 1) similar to that of pI258. Over 99% identical (Table 6), these five plasmids clearly demonstrate geographic spread and stability in terms of gene sequence and organization over five decades. A sixth otherwise identical plasmid, SAP060A, from a pre-1960 US isolate, lacks ΔTn552 (Figure 2A) but is otherwise 99.97% identical to SAP012A (Table 6), a pIB485-like plasmid isolated from a 1995 US MRSA strain. In the other five pIB485-like plasmids, a 10 bp direct repeat of plasmid DNA flanks the ΔTn552 segment (Figure 2B), suggesting it was inserted by transposition; this same 10 bp also occurs as an inverted repeat in the predicted sin res site (Rowland et al. 2002). Thus, SAP060A has two copies of the 10 bp sequence in an inverted repeat, and the 5 other pIB485-like plasmids have three copies of the 10 bp sequence, two in direct repeat flanking ΔTn552 and the third on the opposite strand (Figure 2B).
Table 6 . Completely sequenced pIB485-like S. aureus plasmids.
| Plasmid | Percentage Identity to SAP012Aa | Strain Source | Year Isolated | Location |
|---|---|---|---|---|
| SAP012A | — | Human clinical | 1995 | Georgia, United States |
| SAP048A | 99.97 | Human clinical | 2006 | Nebraska, United States |
| SAP074A | 99.90 | CA infection | 1999 | Oxford, United Kingdom |
| pWBG744 | 99.96 | Screening | 2001 | Western Australia |
| pSK67 | 99.30 | Human clinical | 1949 | Melbourne, Australia |
| SAP060A | 84.29 (99.97)b | Not from infection | Pre-1960 | United States |
CA, community-acquired.
Mismatches and gaps were determined by ClustalW alignment. Gaps were considered mismatches in calculating percentage identity.
Percentage identity to SAP012A without ΔTn552.
Figure 2 .
pIB485-like enterotoxin plasmids with or without ΔTn552. (A) Diagram of pIB485-like plasmid SAP060A (open reading frames in gray), positions 2210–8928, showing the insert position of the 4278 bp ΔTn552 (hatched) in the sin res site (Rowland et al. 2002) in the other pIB485-like plasmids (Table 6). The ΔTn552 (hatched) insert has flanking 10 bp direct repeats (right-facing gray arrowheads) of res site DNA, likely resulting from transposition. An inverted repeat of that same 10 bp insertion site is also present upstream from sin, as part of the predicted res site (left-facing gray arrowhead). The black arrows show predicted open reading frames. (B) The sequence details of the ΔTn552 inserted in the sin res site. The sequence without the insertion is SAP060A positions 2454–2528. The other pIB485-like plasmid sequences contain ΔTn552, and the nucleotides shown are identical in all. The gray arrows show the 10 bp direct and inverted repeats.
In addition to ΔTn552, all pIB485-like plasmids, including SAP060A, carry the cluster of Staphylococcus enterotoxin genes sed, sej, and ser previously reported in this plasmid family (Bayles and Iandolo 1989; Omoe et al. 2003) that have contributed to several outbreaks of S. aureus food-borne illness (Omoe et al. 2003; Ono et al. 2008; Plata et al. 2009). The SAP012A and SAP060A sequences have a frameshift in sed due to a missing T in a run of eight Ts. This may be an occurrence of a common sequencing error (Li and California 2006), as only those two sequences have the sed frameshift; the question can be resolved by testing for the presence of the relevant gene transcript or product. The pIB485-like plasmids also carry cadmium resistance genes cadX and cadD. Energy-dependent Cd(II) efflux was first described by Tynecka (Tynecka et al. 1981a, 1981b) with pII147 and later in several staphylococcal plasmids, including pI258 (Crupper et al. 1999; Massidda et al. 2006; Nies 1992; Nucifora et al. 1989).
pMW2-like plasmids occur in human and animal S. aureus strains:
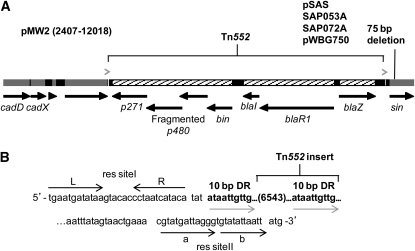
Like the pIB485-like plasmids, the pMW2-like RT1 plasmids are common (Table 2) with a wide geographical distribution, composing ∼20% (27 of 138) of the 20–30 kb plasmids and coming from US, UK, and Australian S. aureus strains between 1995 and 2004 (Table 7). Only three pMW2-like plasmids (pMW2, pSAS, and p21) were available in RefSeq when we sequenced the six described here. All pMW2-like plasmids are 99% identical (Table 7), differing mainly by a 75 bp deletion in sin only in pMW2, pSAS, SAP053A, SAP072A, and pWBG750 (Figure 3A). SAP072A, from the most recently isolated S. aureus strain (2004 UK), was from an animal isolate (Table 7); thus, these plasmids are not limited to human strains. SAP072A is 99.99% identical to pMW2, a plasmid from a 1998 US human CA-MRSA clinical isolate (Baba et al. 2002).
Table 7 . Completely sequenced pMW2-like S. aureus plasmids.
| Plasmid | Percentage Identity to pMW2a | Strain Source | Year | Location | Referenceb |
|---|---|---|---|---|---|
| pMW2 | — | Human clinical | 1998 | North Dakota, United States | Baba et al. 2002 |
| pSAS | 99.93 | Human clinical | 1998 | United Kingdom | Holden et al. 2004 |
| p21 | 98.95 | Clinical | unknown | unknown | Unpublished |
| SAP053A | 99.87 | Human clinical | 2007 | Nebraska, United States | This work |
| SAP072A | 99.99 | Animal clinical | 2004 | United Kingdom | This work |
| SAP073A | 99.58 | CA infection | 1999 | Oxford, United Kingdom | This work |
| pWBG750 | 99.96 | CA infection | 1995 | Western Australia | This work |
| pWBG757 | 99.54 | Screening | 1995 | Western Australia | This work |
| pWBG763 | 99.58 | Screening | 1995 | Western Australia | This work |
CA, community-acquired.
Mismatches and gaps were determined by ClustalW alignment. Gaps were considered mismatches in calculating percentage identity.
Figure 3 .
pMW2-like plasmids with full-length Tn552. (A) Diagram of the pMW2 sequence positions 2407–12018, including the complete Tn552 with flanking 10 bp direct repeats (gray arrowheads). All pMW2-like plasmids (Table 7) have a frameshift that truncates the p480 transposase and generates a second putative ORF. pMW2 and the four plasmids listed have a 75 bp deletion in sin; the four remaining pMW2-like plasmids have the full sin gene. (B) The pMW2 sequence that flanks Tn552, positions 4831–11469. Tn552 (6553 bp, including the 10 bp DR) is inserted into the putative sin res site; res siteI and res siteII predicted for p21 (Rowland et al. 2002) are labeled (black arrows). This sequence is identical in all pMW2-like plasmids. The gray arrows show the 10 bp direct repeats.
The pMW2-like RT1 plasmids are the smallest (20.7 kb) of the prevalent RT groups (Table 7). They carry putative bacteriocin and bacteriocin-immunity genes and several short ORFs annotated as pre, indicating possible mobilizability (Francia et al. 2004; Garcillan-Barcia et al. 2009; Smith and Thomas 2004; Varella Coelho et al. 2009). As with the pIB485-like plasmids, the pMW2-like plasmids carry cadmium resistance (cadD, cadX) and have a full Tn552 in the sin res site predicted for p21 (Rowland et al. 2002) (Figure 3, A and B). All pMW2-like plasmids contain a frameshift mutation in the Tn552 transposase p480, fragmenting it into two separate, overlapping predicted open reading frames (Figure 3A), both of which encode putative transposases, unlike the prototypical Tn552 of pI9789 where p480 is a single gene (Rowland and Dyke 1989, 1990). Like the ΔTn552 in the pIB485-like plasmids (Figure 2, A and B), the full Tn552 in the RT1 plasmids is flanked by 10 bp direct repeats of plasmid DNA (Figure 3B). Although no pMW2-like plasmids lacking Tn552 have been reported, such a plasmid (14.1 kb) would be much smaller than the >20 kb plasmids we focused on because the Tn552 insert is 6553 bp.
pUSA300HOUMR-like multiresistance plasmids are common in the United States:
The pUSA300HOUMR-like RT3 plasmids are the third common plasmid group identified by RFLP analysis and, like the pMW2-like plasmids, comprised ∼20% of the 20–30 kb plasmids we typed (Table 2). Unlike the geographically diverse pIB485-like and pMW2-like plasmids, they were found only in US S. aureus isolates (Table 8). However, like the pMW2-like plasmids, the pUSA300HOUMR-like RT3 plasmids occurred in both animal and human strains. The three plasmids identified as RT3 by RFLP analysis are 99.8–99.9% identical at the sequence level (Table 8) to plasmid pUSA300HOUMR from a human clinical MRSA isolated in Houston, TX (Highlander et al. 2007). Four other pUSA300HOUMR-like plasmids were identified by sequencing to have >98% identity with pUSA300HOUMR, apart from insertions and deletions (Table 8).
Table 8 . Completely sequenced pUSA300HOUMR-like S. aureus plasmids.
| Plasmid | Percentage Identity to pUSA300HOUMRa | Strain Source | Year | Location | Referencec |
|---|---|---|---|---|---|
| pUSA300HOUMR | — | Human clinical | 2002–4 | Texas, United States | Highlander et al. 2007 |
| SAP015A | 99.83 | Human clinical | 2002 | California, United States | This work |
| SAP046A | 99.89 | Canine abscess | 2005 | Georgia, United States | This work |
| SAP050A | 99.89 | Human clinical | 2007 | Nebraska, United States | This work |
| SAP027A | 89.58 (98.04)b | Human clinical | 2006 | Nebraska, United States | This work |
| SAP049A | 92.31 (99.88)b | Human clinical | 2007 | Nebraska, United States | This work |
| SAP051A | 85.06 (99.89)b | Human clinical | 2007 | Nebraska, United States | This work |
| SAP052A | 83.28 (99.85)b | Human clinical | 2007 | Nebraska, United States | This work |
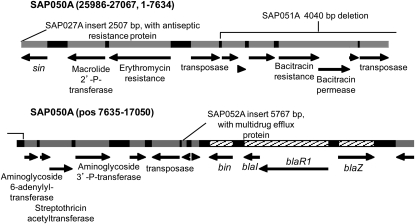
All pUSA300HOUMR-like plasmids carry a ΔTn552 (bin, blaI, blaR1, blaZ) (Figure 4) similar to pI258 and the pIB485-like RT2 plasmids, but it is not flanked by 10 bp repeats or located within the truncated sin res site. Lacking the Tn552 transposases, the pUSA300HOUMR-like plasmids have a transposase similar to that carried by IS257 and IS431mec, three copies of which are found in these plasmids. A deletion event like that described for pI258 (Rowland and Dyke 1990) may have removed the Tn552 transposases adjacent to bin.
Figure 4 .
Multiresistant pUSA300HOUMR-like plasmids with a ΔTn552 that lacks flanking direct repeats. Diagram of the pUSA300HOUMR-like plasmid SAP050A, positions 25986 through the end of the sequence (27067) and positions 1 to 17050, showing insertions and deletions for the other pUSA300HOUMR-like plasmids (Table 8). The ΔTn552 genes are hatched; all other predicted genes are gray. SAP027A has a 923 bp region of high mismatch (to positions 25058–25980) and a 2507 bp insertion just prior to the illustrated sequence that includes a predicted antiseptic resistance protein. SAP052A has a 5767 bp insertion after position 11570 that includes a predicted multidrug efflux protein. SAP051A does not encode bacitracin resistance genes (deleted positions 3654–7693). The SAP049A 2047 bp deletion is not shown (positions 19771–21817) and includes three hypothetical genes upstream of cadX.
Besides the β-lactamase and cadmium resistances carried by the pIB485-like and pMW2-like plasmids, the pUSA300HOUMR-like plasmids also encode macrolide and aminoglycoside resistances, and all but SAP015A carry bacitracin resistance (Figure 4). SAP027A has an insertion encoding antiseptic resistance, and SAP053A has an additional predicted multidrug efflux protein (Figure 4). The four non-RT3 pUSA300HOUMR-like plasmids identified by sequencing, because insertions and/or deletions gave them different RFLP profiles, show how easily these plasmids may gain even more resistance genes. Eight additional plasmids belonging to this family were recently sequenced from clinical S. aureus USA300 isolates from multiple locations in the United States (Kennedy et al. 2010).
Discussion
The prevalence of 20–30 kb plasmids, almost half of which belong to only three restriction types by RFLP analysis, in staphylococci isolated from sources very distant in time and space suggests that these nonconjugative plasmids are surprisingly widespread for non–self-mobile plasmids. Plasmids in this size range can potentially be transferred by transducing phages (Lindsay and Holden 2006; Malachowa and Deleo 2010; Smillie et al. 2010); most phage genomes identified in staphylococci are >40 kb, and transduction is thought to be restricted by phage genome size (Smillie et al. 2010). More of these 20–30 kb plasmids may be mobilizable than is apparent with current genome data if they contain mob genes not yet identified as such. However, the scarcity of conjugative plasmids (only 12 in total) implies that mobilization is rare and that staphylococcal plasmid transfer occurs mainly by transduction (Lindsay and Holden 2004; Lindsay 2010). The now larger dataset makes the mechanism of intercellular movement of these strongly peripatetic 20–30 kb plasmids ripe for examination.
Serine recombinases important for many mobile elements are prevalent in staphylococci (Rowland et al. 2002), playing roles in stable plasmid inheritance and transposon movement. Sin recombinase may have a role in plasmid multimer resolution (Rowland et al. 2002) in 47% of staphylococcal plasmids completely sequenced in this project and in 31% of all complete sequences, including those in RefSeq. Other serine recombinases are associated with the movement of antibiotic resistances, such as transposition of vancomycin resistance genes on Tn1546 (Katayama et al. 2000) and the β-lactamase genes carried on Tn552 whose movement is associated with bin. In addition to being the Tn552 resolvase, bin is commonly found on large staphylococcal plasmids and may play a role in deletions and rearrangements (Murphy and Novick 1980; Rowland and Dyke 1989, 1990).
The β-lactamase–encoding transposon Tn552 in plasmids of staphylococci isolated between the 1940s and the 2000s from distant locations shows the persistence of antibiotic resistance genes over time and geography. All three major families we observed carried β-lactamase genes associated with partial or full Tn552 and with cadmium resistance genes. Virtually identical pIB485-like plasmids with (or without) a ΔTn552 and carrying three enterotoxin genes (Figure 2A) occurred in strains from distant locations isolated 50 years apart. Our work markedly expands and enriches the evidence that plasmids have retained and gained gene content as they spread across the globe among epidemiologically and geographically diverse S. aureus strains.
Sequence alignments revealed four pUSA300HOUMR-like plasmids not identified as such by RFLP analysis, emphasizing the spread of this family among US S. aureus strains and showing they are even more abundant than detectable by plasmid profiling (Table 2). The pUSA300HOUMR-like plasmids all carry multiple antimicrobial resistances, and two of them, SAP027A and SAP052A, have insertions with additional resistance genes (Figure 4). These plasmids are a snapshot of how easily resistance genes are gained and spread among pathogenic staphylococci. The pUSA300HOUMR-like group of plasmids was found only in S. aureus strains isolated in the United States, unlike the other two major families, but it is likely that these plasmids will spread worldwide as have the pMW2-like and pIB485-like plasmids.
The three major families identified show that these plasmids are persistent and widespread on a global scale. The sequenced pMW2-like (Table 7) and pIB485-like (Table 6) plasmids were from strains isolated on three continents, and the latter group's strains were isolated decades apart (Table 6). The pMW2-like and pUSA300HOUMR-like plasmids came from both human and animal isolates, reemphasizing (Lindsay 2010) that properly assessing the spread of S. aureus strains and their mobile elements requires study of both animal and human strains to determine whether human strains are infecting animals or whether strains are simply sharing mobile elements.
In summary, we aimed to increase the number of large staphylococcal plasmid sequences to better assess the global and temporal diversity and spread of these mobile elements. We have tripled the number of large plasmid sequences available and identified three major plasmid families and the most common genes found on large plasmids, opening several key areas for future investigation. The phylogeny of the plasmids should be examined, but their varying sizes and the large number and variety of their transposable elements challenges classical cladistics approaches. These plasmids also carry several classes of genes providing clues to their ecology within the worldwide population of staphylococci. The abundance of arsenic, mercury, and cadmium resistances suggests that nonantibiotic environmental toxicants foster persistence of these multiresistant mobile elements. Arsenic is widely used in animal agriculture (Jackson and Bertsch 2001; Rutherford et al. 2003), approximately 30% of the US population is directly exposed to mercury via dental amalgam restorations (Richardson et al. 2011), and cadmium exposure can be occupational (Cespon-Romero and Yebra-Biurrun 2007; Wang et al. 2008) or from tobacco use (Butler Walker et al. 2006). There are several enterotoxins, exfoliative toxins, and pls (antiadhesion to nasal epithelial cells) (Savolainen et al. 2001), as well as predicted bacteriocins/lantibiotic genes and a very common predicted abiK gene (Table S2) that may have antibacterial or antiviral effects, any or all of which might be involved in the equilibrium between being a benign commensal or life-threatening pathogen. The mechanism of spread of these plasmids, including the potential for mobilization, especially of those shared among animal and human strains, is particularly important to elucidate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Naveen Aitha (Computer Sciences Department, University of Georgia) for Python scripting to aid analysis of BLAST outputs and Ross Overbeek, Victoria Vonstein, and Gordon Pusch of Argonne National Laboratory for gracious and valuable assistance with RAST and P-RAST. We also thank Jean Patel and Brandi Limbago of the CDC for strains and advice. Plasmid laboratory work and bioinformatics at the University of Georgia were partially supported by a Microbial Sequencing Center subcontract to A.O.S. from JCVI under its NIH NIAID contract and by a small grant to A.O.S. from Alliance for the Prudent Use of Antibiotics (APUA) through NIH Grant U24 AI 50139. Work on strain CM05 was supported by NIH (NIAID) grant RO1AI072445 to A.M. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Literature Cited
- Apisiridej S., Leelaporn A., Scaramuzzi C. D., Skurray R. A., Firth N., 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38: 13–24 [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., et al. , 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K., et al. , 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359: 1819–1827 [DOI] [PubMed] [Google Scholar]
- Balagurumoorthy P., Adelstein S. J., Kassis A. I., 2008. Method to eliminate linear DNA from mixture containing nicked circular, supercoiled, and linear plasmid DNA. Anal. Biochem. 381: 172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles K. W., Iandolo J. J., 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171: 4799–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T., Firth N., Apisiridej S., Hettiaratchi A., Leelaporn A., et al. , 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180: 4350–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler Walker J., Houseman J., Seddon L., Mcmullen E., Tofflemire K., et al. , 2006. Maternal and umbilical cord blood levels of mercury, lead, cadmium, and essential trace elements in Arctic Canada. Environ. Res. 100: 295–318 [DOI] [PubMed] [Google Scholar]
- Caryl J. A., O'Neill A. J., 2009. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the staphylococci. Plasmid 62: 35–38 [DOI] [PubMed] [Google Scholar]
- Cespon-Romero R. M., Yebra-Biurrun M. C., 2007. Flow injection determination of lead and cadmium in hair samples from workers exposed to welding fumes. Anal. Chim. Acta 600: 221–225 [DOI] [PubMed] [Google Scholar]
- Clewell D. B., An F. Y., White B. A., Gawron-Burke C., 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J. Bacteriol. 162: 1212–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvaglia A. R., François P., Hernandez D., Perron K., Linder P., et al. , 2010. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. USA 107: 11954–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crupper S. S., Worrell V., Stewart G. C., Iandolo J. J., 1999. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181: 4071–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R., Perna N. T., 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14: 1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbise A., Dyke K. G., El Solh N., 1995. Rearrangements in the staphylococcal beta-lactamase-encoding plasmid, pIP1066, including a DNA inversion that generates two alternative transposons. Mol. Microbiol. 17: 769–779 [DOI] [PubMed] [Google Scholar]
- Diekema D. J., Pfaller M. A., Schmitz F. J., Smayevsky J., Bell J., et al. , 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific Region for the SENTRY antimicrobial surveillance program, 1997–1999. Clin. Infect. Dis. 32: S114–S132 [DOI] [PubMed] [Google Scholar]
- Enright M. C., Robinson D. A., Randle G., Feil E. J., Grundmann H., et al. , 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99: 7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C. R., Yam W. C., Peiris J. S., Epstein R. J., 2009. Methicillin-resistant commensal staphylococci in healthy dogs as a potential zoonotic reservoir for community-acquired antibiotic resistance. Infect. Genet. Evol. 9: 283–285 [DOI] [PubMed] [Google Scholar]
- Feil E. J., Cooper J. E., Grundmann H., Robinson D. A., Enright M. C., et al. , 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185: 3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil E. J., Enright M. C., 2004. Analyses of clonality and the evolution of bacterial pathogens. Curr. Opin. Microbiol. 7: 308–313 [DOI] [PubMed] [Google Scholar]
- Feng Y., Chen C. J., Su L. H., Hu S., Yu J., et al. , 2008. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 32: 23–37 [DOI] [PubMed] [Google Scholar]
- Firth N., Apisiridej S., Berg T., O'rourke B. A., Curnock S., et al. , 2000. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 182: 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth N., Skurray R. A., 2006. Genetics: accessory elements and genetic exchange, pp. 413–426 Gram-Positive Pathogens, edited by Fischetti R. P., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I., 2nd Edition American Society for Microbiology, Washington, DC [Google Scholar]
- Francia M. V., Varsaki A., Garcillan-Barcia M. P., Latorre A., Drainas C., et al. , 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28: 79–100 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia M. P., Francia M. V., De La Cruz F., 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33: 657–687 [DOI] [PubMed] [Google Scholar]
- Gill S. R., Fouts D. E., Archer G. L., Mongodin E. F., Deboy R. T., et al. , 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187: 2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetghebeur M., Landry P. A., Han D., Vicente C., 2007. Methicillin-resistant Staphylococcus aureus: a public health issue with economic consequences. Can. J. Infect. Dis. Med. Microbiol. 18: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander S. K., Hulten K. G., Qin X., Jiang H., Yerrapragada S., et al. , 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T. G., Feil E. J., Lindsay J. A., Peacock S. J., Day N. P. J., et al. , 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101: 9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B. P., Bertsch P. M., 2001. Determination of arsenic speciation in poultry wastes by IC–ICP-MS. Environ. Sci. Technol. 35: 4868–4873 [DOI] [PubMed] [Google Scholar]
- Jackson M. P., Iandolo J. J., 1986. Sequence of the exfoliative toxin B gene of Staphylococcus aureus. J. Bacteriol. 167: 726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Ito T., Hiramatsu K., 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44: 1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. D., Porcella S. F., Martens C., Whitney A. R., Braughton K. R., et al. , 2010. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J. Clin. Microbiol. 48: 4504–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 61: 442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebard R. J., Jensen S. O., Arnaiz I. A., Skurray R. A., Firth N., 2008. A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol. Lett. 284: 58–67 [DOI] [PubMed] [Google Scholar]
- Leelaporn A., Firth N., Paulsen I. T., Skurray R. A., 1996. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J. Bacteriol. 178: 6070–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R., Hebrant A., Wodak S. J., Toussaint A., 2004. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 32: D45–D49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., 2006. Statistical models of sequencing error and algorithms of polymorphism detection. Ph.D. Thesis, University of Southern California, Los Angeles [Google Scholar]
- Lindsay J., Holden M., 2006. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genomics 6: 186–201 [DOI] [PubMed] [Google Scholar]
- Lindsay J. A., Holden M. T. G., 2004. Staphylococcus aureus: Superbug, super genome? Trends Microbiol. 12: 378–385 [DOI] [PubMed] [Google Scholar]
- Lindsay J. A., 2010. Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300: 98–103 [DOI] [PubMed] [Google Scholar]
- Malachowa N., Deleo F. R., 2010. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O., Mingoia M., Fadda D., Whalen M. B., Montanari M. P., et al. , 2006. Analysis of the beta-lactamase plasmid of borderline methicillin-susceptible Staphylococcus aureus: focus on bla complex genes and cadmium resistance determinants cadD and cadX. Plasmid 55: 114–127 [DOI] [PubMed] [Google Scholar]
- McDougal L. K., Fosheim G. E., Nicholson A., Bulens S. N., Limbago B. M., et al. , 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob. Agents Chemother. 54: 3804–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton J. R., Fales W. H., Luby C. D., Oaks J. L., Sanchez S., et al. , 2005. Surveillance of Staphylococcus aureus in veterinary teaching hospitals. J. Clin. Microbiol. 43: 2916–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Novick R. P., 1980. Site-specific recombination between plasmids of Staphylococcus aureus. J. Bacteriol. 141: 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E. W., Sutton G. G., Delcher A. L., Dew I. M., Fasulo D. P., et al. , 2000. A whole-genome assembly of Drosophila. Science 287: 2196–2204 [DOI] [PubMed] [Google Scholar]
- Navarro M. B., Huttner B., Harbarth S., 2008. Methicillin-resistant Staphylococcus aureus control in the 21st century: beyond the acute care hospital. Curr. Opin. Infect. Dis. 21: 372–379 [DOI] [PubMed] [Google Scholar]
- Nies D. H., 1992. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid 27: 17–28 [DOI] [PubMed] [Google Scholar]
- Noble W. C., Virani Z., Cree R. G., 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72: 195–198 [DOI] [PubMed] [Google Scholar]
- Novick R. P., 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43: 537–565 [DOI] [PubMed] [Google Scholar]
- Novick R. P., 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49: 93–105 [DOI] [PubMed] [Google Scholar]
- Nucifora G., Chu L., Misra T. K., Silver S., 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 86: 3544–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoe K., Hu D.-L., Takahashi-Omoe H., Nakane A., Shinagawa K., 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 71: 6088–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H. K., Omoe K., Imanishi K. I., Iwakabe Y., Hu D.-L., et al. , 2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 76: 4999–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen I. T., Gillespie M. T., Littlejohn T. G., Hanvivatvong O., Rowland S.-J., et al. , 1994. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene 141: 109–114 [DOI] [PubMed] [Google Scholar]
- Périchon B., Courvalin P., 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53: 4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plata K., Rosato A. E., Wegrzyn G., 2009. Staphylococcus aureus as an infectious agent: overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim. Pol. 56: 597–612 [PubMed] [Google Scholar]
- Richardson G. M., Wilson R., Allard D., Purtill C., Douma S., et al. , 2011. Mercury exposure and risks from dental amalgam in the US population, post-2000. Sci. Total Environ. 409: 4257–4268 [DOI] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G., 1989. Characterization of the staphylococcal β-lactamase transposon Tn552. EMBO J. 8: 2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G., 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4: 961–975 [DOI] [PubMed] [Google Scholar]
- Rowland S. J., Stark W. M., Boocock M. R., 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol. Microbiol. 44: 607–619 [DOI] [PubMed] [Google Scholar]
- Rubin J. E., Ball K. R., Chirino-Trejo M., 2011. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from various animals. Can. Vet. J. 52: 153–157 [PMC free article] [PubMed] [Google Scholar]
- Rutherford D. W., Bednar A. J., Garbarino J. R., Needham R., Staver K. W., et al. , 2003. Environmental fate of roxarsone in poultry litter. Part II. Mobility of arsenic in soils amended with poultry litter. Environ. Sci. Technol. 37: 1515–1520 [DOI] [PubMed] [Google Scholar]
- Rutland B. E., Weese J. S., Bolin C., Au J., Malani A. N., 2009. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 15: 1328–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen K., Paulin L., Westerlund-Wikstrom B., Foster T. J., Korhonen T. K., et al. , 2001. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 69: 3013–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M. C., Phillippy A. M., Shneiderman B., Salzberg S. L., 2007. Hawkeye: an interactive visual analytics tool for genome assemblies. Genome Biol. 8: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalita Z., Murphy E., Novick R. P., 1980. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid 3: 291–311 [DOI] [PubMed] [Google Scholar]
- Simpson A. E., Skurray R. A., Firth N., 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 185: 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P. C., De La Cruz F., 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74: 434–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. C., Thomas C. D., 2004. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J. Bacteriol. 186: 3363–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohr M., Rau J., Friedrich A., Klittich G., Fetsch A., et al. , 2011. Methicillin-resistant Staphylococcus aureus (MRSA) in three dairy herds in southwest Germany. Zoonoses Public Health 58: 252–261 [DOI] [PubMed] [Google Scholar]
- Sung J. M., Lindsay J. A., 2007. Staphylococcus aureus strains that are hypersusceptible to resistance gene transfer from enterococci. Antimicrob. Agents Chemother. 51: 2189–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J. M., Lloyd D. H., Lindsay J. A., 2008. Staphylococcus aureus host specificity: comparative genomics of human vs. animal isolates by multi-strain microarray. Microbiology 154: 1949–1959 [DOI] [PubMed] [Google Scholar]
- Swinfield T. J., Janniere L., Ehrlich S. D., Minton N. P., 1991. Characterization of a region of the Enterococcus faecalis plasmid pAM beta 1 which enhances the segregational stability of pAM beta 1-derived cloning vectors in Bacillus subtilis. Plasmid 26: 209–221 [DOI] [PubMed] [Google Scholar]
- Toh S. M., Xiong L., Arias C. A., Villegas M. V., Lolans K., et al. , 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64: 1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynecka Z., Gos Z., Zajac J., 1981a Energy-dependent efflux of cadmium coded by a plasmid resistance determinant in Staphylococcus aureus. J. Bacteriol. 147: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynecka Z., Gos Z., Zajac J., 1981b Reduced cadmium transport determined by a resistance plasmid in Staphylococcus aureus. J. Bacteriol. 147: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo E. E., Grubb W. B., 2001. New Staphylococcus aureus incompatibility group 1 plasmids encoding penicillinase production and resistance to different antibacterial agents. J. Chemother. 13: 34–42 [DOI] [PubMed] [Google Scholar]
- Van Duijkeren E., Ten Horn L., Wagenaar J. A., De Bruijn M., Laarhoven L., et al. , 2011. Suspected horse-to-human transmission of MRSA ST398. Emerg. Infect. Dis. 17: 1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen W., Van De Velde E., Crombe F., Polis I., Hermans K., et al. , 2011. Screening for methicillin-resistant staphylococci in dogs admitted to a veterinary teaching hospital. Res. Vet. Sci. 2: 2. [DOI] [PubMed] [Google Scholar]
- Varella Coelho M. L., Ceotto H., Madureira D. J., Nes I. F., Bastos Mdo C., 2009. Mobilization functions of the bacteriocinogenic plasmid pRJ6 of Staphylococcus aureus. J. Microbiol. 47: 327–336 [DOI] [PubMed] [Google Scholar]
- Waldron D. E., Lindsay J. A., 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188: 5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther B., Wieler L. H., Friedrich A. W., Kohn B., Brunnberg L., et al. , 2009. Staphylococcus aureus and MRSA colonization rates among personnel and dogs in a small animal hospital: association with nosocomial infections. Berl. Munch. Tierarztl. Wochenschr. 122: 178–185 [PubMed] [Google Scholar]
- Wang D., Du X., Zheng W., 2008. Alteration of saliva and serum concentrations of manganese, copper, zinc, cadmium and lead among career welders. Toxicol. Lett. 176: 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenberg M. W., Bootsma M. C., Troelstra A., Kluytmans J. A., Bonten M. J., 2011. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin. Microbiol. Infect. 17: 316–319 [DOI] [PubMed] [Google Scholar]
- Weaver K. E., Kwong S. M., Firth N., Francia M. V., 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61: 94–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel L. M., Clewell D. B., Gill S. R., Clark N. C., McDougal L. K., et al. , 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302: 1569–1571 [DOI] [PubMed] [Google Scholar]
- Williams L. E., Detter C., Barry K., Lapidus A., Summers A. O., 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72: 4899–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Hayashi T., Takami H., Ohnishi M., Murata T., et al. , 2001. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 69: 7760–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203–214 [DOI] [PubMed] [Google Scholar]
- Zhu W., Clark N. C., McDougal L. K., Hageman J., Mcdonald L. C., et al. , 2008. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 52: 452–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarelli A. J., Roy I., Harding G. P., Couperus J. J., 1990. Diversity and stability of restriction enzyme profiles of plasmid DNA from methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 28: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.