Abstract
Genetic linkage maps play fundamental roles in understanding genome structure, explaining genome formation events during evolution, and discovering the genetic bases of important traits. A high-density cotton (Gossypium spp.) genetic map was developed using representative sets of simple sequence repeat (SSR) and the first public set of single nucleotide polymorphism (SNP) markers to genotype 186 recombinant inbred lines (RILs) derived from an interspecific cross between Gossypium hirsutum L. (TM-1) and G. barbadense L. (3-79). The genetic map comprised 2072 loci (1825 SSRs and 247 SNPs) and covered 3380 centiMorgan (cM) of the cotton genome (AD) with an average marker interval of 1.63 cM. The allotetraploid cotton genome produced equivalent recombination frequencies in its two subgenomes (At and Dt). Of the 2072 loci, 1138 (54.9%) were mapped to 13 At-subgenome chromosomes, covering 1726.8 cM (51.1%), and 934 (45.1%) mapped to 13 Dt-subgenome chromosomes, covering 1653.1 cM (48.9%). The genetically smallest homeologous chromosome pair was Chr. 04 (A04) and 22 (D04), and the largest was Chr. 05 (A05) and 19 (D05). Duplicate loci between and within homeologous chromosomes were identified that facilitate investigations of chromosome translocations. The map augments evidence of reciprocal rearrangement between ancestral forms of Chr. 02 and 03 versus segmental homeologs 14 and 17 as centromeric regions show homeologous between Chr. 02 (A02) and 17 (D02), as well as between Chr. 03 (A03) and 14 (D03). This research represents an important foundation for studies on polyploid cottons, including germplasm characterization, gene discovery, and genome sequence assembly.
Keywords: cotton (Gossypium spp.) genomes, genetic linkage map, simple sequence repeat (SSR), single nucleotide polymorphism (SNP), recombinant inbred line (RIL) population
Cotton belongs to the Gossypium genus, which consists of approximately 45 diploid and 5 allotetraploid species of global distribution (Beasley 1942; Endrizzi et al. 1985; Kohel et al. 2001; Stewart 1994; Wendel and Cronn 2003). The gametic chromosome number of all diploid species is 13, but significant differences among the genomes in meiotic affinity and relative size led to the recognition of eight genome groups: A through G and K (Beasley 1942; Endrizzi et al. 1985; Stewart 1994). Of the approximately 50 Gossypium species, four have been domesticated independently: two diploid species, G. arboreum L. and G. herbaceum L. (n = x = 13) with A1 and A2 genomes, and two allotetraploid species, G. hirsutum L. and G. barbadense L. (n = 2x = 26) with (AD)1 and (AD)2 genomes (Bowers et al. 2003; Lee 1984; Percival and Kohel 1990). The allotetraploid cotton species are the products of a presumed single polyploidization event between ancient A-genome and D-genome diploids that occurred approximately 1-2 million years ago (Stelly et al. 2005; Wendel and Cronn 2003). Chromosome numbers assigned in allotetraploid cottons are based on pairing relationships in diploid x tetraploid crosses, with chromosomes 1−13 corresponding to the At subgenome and chromosomes 14−26 to the Dt subgenome (Brown 1980).
Cotton species serve as a model system for polyploid plants and plant cell elongation, cell wall and cellulose biosynthesis because they are the only known plants that produce single-celled fibers (Jiang et al. 1998; Kim and Triplett 2001). The genes that make cotton valuable function in unique ways, requiring long-term research into the development of molecular tools such as DNA markers and genome maps to translate genomic information into agronomic benefits and to other biological systems.
Cotton researchers have explored genetic mapping with multiple types of DNA markers, including restriction fragment-length polymorphism (RFLP) (Reinisch et al. 1994; Rong et al. 2004; Shappley et al. 1998), amplified fragment-length polymorphism (Lacape et al. 2003), random-amplified polymorphic DNA (Kohel et al. 2001), and simple sequence repeats (SSRs) (Guo et al. 2007; Lacape et al. 2009; Park et al. 2005; Xiao et al. 2009; Yu et al. 2011). Although early genetic mapping with hybridization-based markers such as RFLP opened the door to important genomic studies (Jiang et al. 1998; Shappley et al. 1998), recent genetic mapping with polymerase chain reaction (PCR)-based markers such as SSR have facilitated portable applications among different mapping populations and research programs (Abdurakhmonov et al. 2008; Zhang et al. 2003). As such, the cotton research community has made efforts to develop many portable markers to overcome the problem of low DNA polymorphism rates among various cultivated cotton breeding programs (http://www.cottonmarker.org/; Blenda et al. 2006). To date, approximately 17,000 pairs of SSR primers have been developed from four cotton species (G. arboreum, G. barbadense, G. hirsutum, and G. raimondii Ulbrich) and a portion of this number have been surveyed for polymorphism against a 12-genotype panel of six Gossypium species (Blenda et al. 2006; Yu 2004). As single nucleotide polymorphism (SNP) markers are explored in other plant species (Ganal et al. 2009), new research has been initiated to examine nucleotide sequence diversity in Gossypium genomes (An et al. 2008; Van Deynze et al. 2009). These findings are laying the groundwork for developing large numbers of SNP markers in cotton. The growing collection of portable markers in cotton provides a cost-effective tool for genome mapping and gene discovery to understand and improve the cotton plant.
High-resolution mapping in cotton has been conducted with segregating populations that were derived from interspecific crosses between Gossypium species because of limited DNA polymorphism within a cotton species. The resulting segregating populations used in major mapping projects often were either F2 or BC1 progeny (Guo et al. 2007; Lacape et al. 2003; Rong et al. 2004; Yu et al. 2011). In addition, these maps relied heavily on a single marker type such as RFLP or SSR markers derived from limited sources. Rong et al. (2004) reported the first high-density map in cotton using 57 F2 plants derived from an interspecific cross between G. hirsutum race “palmeri” and G. barbadense acc. “K101.” The majority of markers used in this map were RFLP markers. This map provided one of the first insights into the allotetraploid cotton genome structure and evolution, although the RFLP markers have proven to have limited portability and utility for marker assisted breeding (Ulloa et al. 2005).
Guo et al. (2007) reported the first comprehensive SSR map by using 138 BC1 plants derived from an interspecific cross of G. hirsutum TM-1/G. barbadense Hai 7124//G. hirsutum TM-1. The majority of SSR markers in this map were derived from cotton expressed sequence tag (EST) sequences. Lacape et al. (2009) reported a genetic linkage map that consisted of a total of approximately 800 (amplified fragment-length polymorphism, RFLP, and SSR) marker loci via the use of 140 recombinant inbred lines (RILs); derived from an interspecific cross between G. hirsutum Guazuncho 2 and G. barbadense VH8-4602. Recently, Yu et al. (2011) used 141 BC1 plants derived from an interspecific cross of G. hirsutum Emian 22/G. barbadense 3-79//G. hirsutum Emian 22. As with Guo et al. (2007), this map also contained SSR markers, the majority of which were derived from ESTs. In addition, a whole-genome radiation hybrid population of 93 plants derived from an interspecific cross of G. barbadense 3-79/G. hirsutum TM-1 was also explored for mapping the cotton genome (Gao et al. 2004, 2006).
Here we report the development of a high-density cotton (Gossypium spp.) genetic map by using representative sets of SSR markers and the first public set of SNP markers to genotype 186 RILs derived from an interspecific cross between G. hirsutum TM-1 and G. barbadense 3-79. Both TM-1 and 3-79 are considered genetic standards for their respective species because of breeding and history of genetic/genomic research conducted by the cotton community. These two lines are highly homozygous, and extensive genetic and cytogenetic materials have been developed using them as reference parents, including mutants and hypoaneuploids (Kohel et al. 1970; Stelly 1993; Stelly et al. 2005). RILs possess several advantages over F2 or BC1 populations for mapping genes and quantitative trait loci (QTL), and high levels of homozygosity and recombination in the RILs enable replicate studies across different environments by different research groups. This immortal TM-1 × 3-79 RIL population is maintained at USDA-ARS, College Station, Texas, USA, and it is used by the cotton research community for genetic investigations, including QTL mapping studies. In addition, we selected SSR markers derived from different sequence sources (EST, genomic, and BAC clones). These markers were developed by 16 research groups (all 16 sources available to the public at Cotton Marker Database (http://www.cottonmarker.org/). This combined high-density genetic map will facilitate the advancement of many basic and applied genomic studies in cotton.
Materials and Methods
Plant materials and DNA extraction
The mapping population was an immortalized set of 186 RILs. At the time of genomic DNA extraction for this study, the average generation was F7. These lines were derived from selfing via single-seed descent original individual F2 plants from a cross between G. hirsutum TM-1 and G. barbadense 3-79, two highly homozygous parents (Kohel et al. 1970; Niles and Feaster 1984). Factors in selecting TM-1 and 3-79 as parents in creation of a segregating population for genetic mapping are the unique high-quality fiber characteristics of extra long staple cotton 3-79 and the high productivity and modest environmental sensitivity of Upland cotton TM-1 (Kohel et al. 2001). The parents (TM-1 and 3-79) and their 186 RIL progeny are maintained as living specimens to produce seed, fiber, and leaf tissue for this mapping effort and other genetic studies.
Interspecific F1 hypoaneuploid hybrids for specific chromosomes were used for deficiency mapping by means of loss of heterozygosity. All but one were derived previously by pollinating monosomic and monotelodisomic aneuploids quasi-isogenic to TM-1 with pollen from euploid 3-79, and recovering the respective deficiency among F1 progeny. The F1 aneuploid monosomic for chromosome 26 was unusual in that the deficiency arose de novo in 3-79 pollen, i.e. not via transmission from the maternal TM-1−like stock. The general procedures for mapping with cotton monosomic (2n = 51) and monotelodisomic stocks have been described previously (Beasley 1942; Stelly 1993; Stelly et al. 2005).
Genomic DNA was extracted from fresh young leaf tissue of individual cotton plants grown in the greenhouse in accordance with the modified CTAB DNA extraction procedure as described by Kohel et al. (2001).
PCR primers and assays
The primer pairs used for PCR were developed by collaborators of the cotton research community (Table 1). Approximately 10,000 pairs of SSR primers from 16 different research projects (http://www.cottonmarker.org/) were first analyzed to identify polymorphic markers between TM-1 and 3-79. Nine genomic DNA sources for SSR primer pairs included BNL, CIR, CM, DOW, DPL, GH, JESPR, MUSB, and TMB, and seven EST sources of SSR primer pairs included HAU, MGHES, MUCS, MUSS, NAU, STV, and UCD. While EST SSR primer pairs were developed from Gossypium cDNA clones that contain SSR, genomic primer pairs were developed from Gossypium random enriched small insert libraries except MUSB and TMB. MUSB was developed from the end sequences of the bacterial artificial chromosome (BAC) clones of G. hirsutum acc. Acala Maxxa (Frelichowski Jr. et al. 2006). TMB was developed from the BAC clones and/or physical contigs of TM-1 (Guo et al. 2008). MUSB and TMB markers facilitate an integration of genetic and physical maps of the allotetraploid cotton genome (Xu et al. 2008). The first public SNP set (UC) also was included in this mapping project (Van Deynze et al. 2009). SNP primer pairs were largely derived from G. arboreum EST unigenes. The actual sequence of the individual primer pairs and source clone for each SSR or SNP marker set can be found at http://www.cottonmarker.org/.
Table 1 . Primer sources of cotton molecular markers (http://www.cottonmarker.org/).
| Marker set | No. Mapped Marker Loci | No. Mapped Primer Pairs |
|---|---|---|
| Genomic SSRs | ||
| BNL | 304 | 239 |
| CIR | 123 | 104 |
| CM | 32 | 24 |
| DOW | 60 | 60 |
| DPL | 213 | 200 |
| GH | 149 | 144 |
| JESPR | 122 | 89 |
| MUSB | 155 | 123 |
| TMB | 310 | 266 |
| Subtotal | 1468 | 1249 |
| EST SSRs | ||
| HAU | 12 | 8 |
| MGHES | 20 | 14 |
| MUCS | 63 | 54 |
| MUSS | 112 | 93 |
| NAU | 113 | 90 |
| STV | 9 | 7 |
| UCD | 28 | 17 |
| Subtotal | 357 | 283 |
| SNPs | ||
| UC | 247 | 247 |
| Total | 2072 | 1779 |
EST, expressed sequence tag; SNP, single nucleotide polymorphism; SSR, simple sequence repeat.
PCR assays for amplifying SSR markers were performed in a cocktail of 10 μL containing 20 ng of DNA, 0.25 μM forward primer, 0.25 μM reverse primer, 0.25 mM dNTPs, 2.5 mM MgCl2, and 0.65 unit DNA Taq polymerase. Thirty-five PCR cycles were used to amplify SSR products, using a primer annealing temperature of 55° or 60°. For nonlabeled SSR primers, amplified DNA products were electrophoresed in a 20-cm-long horizontal agarose gel system (Owl Separation Systems, Portsmouth, NH) with 1X TBE (45 mM tris-borate, 1 mM EDTA, pH 8) running buffer and 3.5% Hi-Resolution agarose (e.g. Metaphor agarose, Cambrex, East Rutherford, NJ; or SFR agarose, Amresco, Solon, OH). PCR product sizes were estimated by comparison with DNA size standard ladders (E and K Scientific, Santa Clara, CA). For fluorescently labeled primers (forward primer only with 6-FAM, HEX, or NED), amplified DNA products were separated using 36-cm or 50-cm capillary electrophoresis of automated ABI PRISM 3130xl or ABI PRISM 3730 Genetic Analyzer (Applied Biosystems/Life Technology, Foster City, CA). In a separate project, an array for Ilumina (San Diego, CA) Golden Gate assay was designed to analyze 384 SNP markers between TM-1 and 3-79 (Van Deynze et al. 2009). Polymorphic SNP markers based on the parental survey were used to genotype the 186 RILs.
Marker data acquisition and linkage map construction
SSR data collection was performed either manually for gel-based assays or with the GeneMapper 3.7. Among nearly 10,000 pairs of primers that were surveyed, more than 2000 primer pairs that detected the best resolution of polymorphisms between TM-1 and 3-79 were selected to genotype the 186 RILs. These primer pairs included subsets (54 MUCS, 123 MUSB, and 93 MUSS) that were previously used to genotype the same population (Park et al. 2005; Frelichowski Jr. et al. 2006), and the genotyping data were incorporated into this mapping project.
Genotyping of the RIL population for SSR and SNPs was performed as previously described (Park et al. 2005; Frelichowski Jr. et al. 2006; Van Deynze et al. 2009). SSR markers were generally codominant, but the calling or scoring of the tetraploid cotton alleles at a specific locus required careful examination of gel images or electrographs. Allotetraploid cottons likely had multiple copies of DNA fragments or alleles amplified with a single primer-pair. To distinguish dominant markers from codominant markers, any RIL missing one pair of the parental polymorphic fragments/alleles indicated that alleles were nonallelic or simply an existence of two dominant marker loci after all pairing attempts had failed. A missing data point of a RIL was determined if there was a lack of any signal attributable to failed PCR amplification. Duplicate marker loci were designated by adding a lower-case letter in alphabetical order after the primer name. The raw scores were first inspected for any coding error and segregation distortion before using the data as input for the JoinMap 4.0 program (Van Ooijen 2006) for mapping analysis. Using the JoinMap’s function “identify identical loci,” we identified 47 identical or cosegregating loci (supporting information, Table S2) and removed them in subsequent mapping. The Kosambi mapping function (Kosambi 1944) was selected to convert a recombination frequency to a genetic distance (centiMorgan, or cM), and 40 cM was the threshold to determine linkage between two markers. Linkage groups and marker orders were determined on the basis of likelihood ratio statistic (or LOD) 10 or greater (up to LOD 15). Chromosome assignment was determined by the common markers that were located by authors in previous publications (Frelichowski Jr. et al. 2006; Guo et al. 2007, 2008; Lacape et al. 2003; Liu et al. 2000; Park et al. 2005; Yu et al. 2011) and by use of the subsets of new SSR markers (GH, Table 3) with the cotton hypoaneuploid stocks described previously. SSR loci localized to one of the chromosomes (Chr.) 1 to 13 were assigned to the A-subgenome (At), whereas loci localized to Chr. 14 to 26 were assigned to the D-subgenome (Dt).
Table 3 . Assignment of 37 GH SSR markers to specific allotetraploid cotton chromosomes.
| Marker Name | Fragment Size, bp | Hypoaneuploid | Mapped Chromosome | |
|---|---|---|---|---|
| TM-1 allele | 3-79 allele | |||
| GH002 | 75 | 65 | H16 | Chr.16(D07) |
| GH027 | 70 | 80 | H09 | Chr.09(A09) |
| GH034 | 130 | 120 | H07 | Chr.13(A13) |
| GH039 | 125 | 120 | H06 | Chr.06(A06) |
| GH048 | 90 | 98 | H20 | Chr.20(D10) |
| GH055 | 175 | 170 | Te18sh | Chr.18(D13) |
| GH082 | 175 | 155 | H06 | Chr.06(A06) |
| GH098 | 130 | 145 | H09 | Chr.09(A09) |
| GH110 | 105 | 80 | H20 | Chr.20(D10) |
| GH119 | 150 | 165 | H20 | Chr.20(D10) |
| GH295 | 95 | 75 | H16 | Chr.16(D07) |
| GH312 | 110 | 102 | H12 | Chr.12(A12) |
| GH330 | 105 | 115 | Te22Lo | Chr.22(D04) |
| GH336 | 98 | 86 | H01 | Chr01(A01) |
| GH345 | 115 | 103 | H16 | Chr.16(D07) |
| GH422 | 116 | 126 | Te5Lo | Chr.05(A05) |
| GH428 | 195 | 170 | H20 | Chr.20(D10) |
| GH433 | 168 | 150 | H06 | Chr.06(A06) |
| GH441 | 175 | 150 | H06 | Chr.06(A06) |
| GH443 | 150 | 120 | H18 | Chr.18(D13) |
| GH462 | 170 | 152 | Te14Lo | Chr.14(D03) |
| GH463 | 150 | 165 | H12 | Chr.12(A12) |
| GH478 | 90 | 100 | H25 | Chr.25(D06) |
| GH484 | 140 | 145 | H09 | Chr.09(A09) |
| GH486 | 155 | 130 | H09 | Chr.09(A09) |
| GH495 | 80 | 72 | H09 | Chr.09(A09) |
| GH499 | 148 | 144 | H09 | Chr.09(A09) |
| GH501 | 200 | 202 | H18 | Chr.18(D13) |
| GH506 | 134 | 160 | H07 | Chr.07(A07) |
| GH511 | 135 | 130 | H20 | Chr.20(D10) |
| GH526 | 100 | 200 | Te22Lo | Chr19(D05) |
| GH537 | 175 | 170 | H25 | Chr.25(D06) |
| GH548 | 120 | 140 | H07 | Chr.07(A07) |
| GH584 | 140 | 120 | H09 | Chr.09(A09) |
| GH603 | 154 | 158 | H26 | Chr.26(D12) |
| GH629 | 128 | 132 | H26 | Chr.26(D12) |
| GH684 | 102 | 90 | H16 | Chr.16(D07) |
SSR, simple sequence repeat.
Results
Parental polymorphisms and genotype frequencies of the mapping population
Approximately 25% of the genomic SSR markers and approximately 15% of the cDNA SSR markers were polymorphic between TM-1 and 3-79. A total of 1601 pairs of polymorphic SSR primers were selected and analyzed for genotyping 186 RILs. Of the 1601 SSR primer pairs that revealed 1895 marker loci, 1344 primer pairs revealed one locus, 234 revealed two loci, and the remaining 23 revealed more than two loci. Among the 1895 SSR marker loci, 1785 were codominant; 43 were dominant loci that received alleles from TM-1, and 67 were dominant loci that received alleles from 3-79. Of these 1895 marker loci, 1825 were mapped (Table 1). The remaining 70 loci were not mapped because of highly skewed segregation (χ2 > 8.5) and high levels of missing data. Fifty-five of the unmapped loci were dominant loci. In addition, 247 of the 384 SNP primer pairs were polymorphic between parents and used to genotype the 186 RILs. All 247 SNP markers were codominant and revealed 247 loci (Table S1). Of these, 207 SNP loci were mapped in unique positions, and the remaining 40 SNP loci were identical to other mapped loci (Table S2). In summary, a total of 1848 pairs of SSR and SNP primers were used to genotype the 186 RILs, and 2142 marker loci were scored, of which 2072 marker loci revealed by 1532 pairs of SSR and 247 pairs of SNP primers were mapped (Table 1). Approximately 98% of 2072 total marker loci were mapped in unique positions, with only 47 identical or cosegregating markers including 40 SNP markers (Table S2).
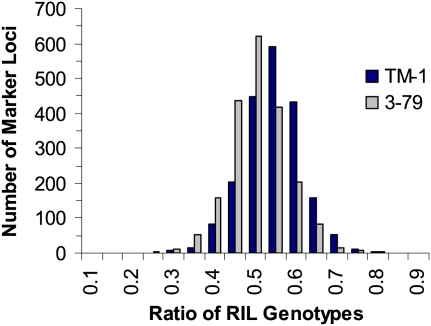
This RIL population displayed a greater-than-expected level of residual heterozygosity, i.e. 4.2% instead of the expected 1.6% for an F7 population derived by single-seed descent. Residual heterozygosity in individual lines ranged from 0.8% to 19.9%. Among the 2032 codominant SSR and SNP loci, the average residual heterozygosity for individual markers was 4.2%, ranging from 0% to 66.7% with SSR marker STV129 demonstrating the greatest heterozygosity. Markers that detected more than 20% residual heterozygosity of the RIL population were usually difficult to map because determining linkage of these markers conflicted with more than one marker. Analysis of the genotyping data revealed a statistically significant preference of TM-1 alleles to 3-79 alleles (χ2 = 768; Figure 1). Overall, the allele frequencies of TM-1 and 3-79 were 52.3% and 47.7%, respectively.
Figure 1 .
Distribution of the TM-1 and 3-79 allele frequencies in the RIL mapping population (χ2 = 768 and P < 0.0001).
Genetic linkage maps of the allotetraploid cotton
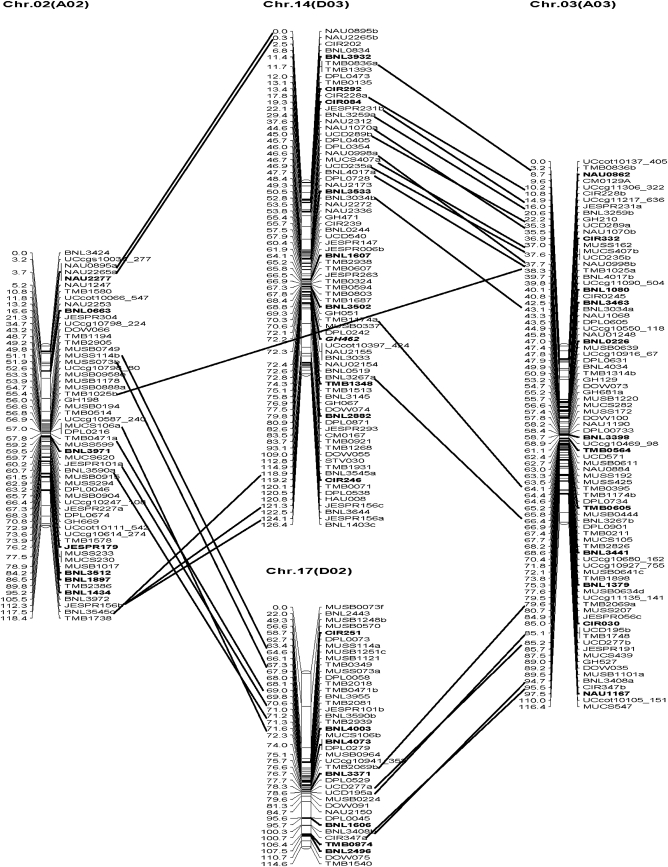
The genetic linkage map comprises 2072 SSR and SNP loci mapped to the 26 linkage groups, corresponding to 26 chromosomes of allotetraploid cotton, for a total map distance of 3380 cM (Table 2 and Figure 2). The average marker interval in this map is 1.63 cM. Forty-seven pairs of marker loci were found to be either identical or cosegregated (Table S2), and therefore only one locus from each pair is shown on the map. For example, BNL3545b is identical to or cosegregated with BNL3545a, so only BNL3545a is shown on Chr. 14 (D03).
Table 2 . Distribution of 2072 SSR and SNP marker loci among the 26 allotetraploid cotton chromosomes.
| Chromosome | No. Marker Loci | Recombinational Size, cM | Average Marker Interval, cM | No. Gaps >10 cM (Largest) |
|---|---|---|---|---|
| A-subgenome | ||||
| Chr.01(A01) | 66 | 144.4 | 2.19 | 2 (14.46) |
| Chr.02(A02) | 60 | 118.4 | 1.97 | 1 (13.46) |
| Chr.03(A03) | 87 | 116.4 | 1.34 | 2 (13.09) |
| Chr.04(A04) | 56 | 101.6 | 1.81 | 2 (15.88) |
| Chr.05(A05) | 139 | 199.2 | 1.43 | 1 (10.03) |
| Chr.06(A06) | 89 | 131.0 | 1.47 | 0 (8.35) |
| Chr.07(A07) | 87 | 128.9 | 1.48 | 1 (10.23) |
| Chr.08(A08) | 92 | 140.0 | 1.52 | 2 (16.52) |
| Chr.09(A09) | 99 | 139.4 | 1.41 | 0 (9.00) |
| Chr.10(A10) | 75 | 109.0 | 1.45 | 0 (6.55) |
| Chr.11(A11) | 140 | 166.5 | 1.19 | 0 (9.98) |
| Chr.12(A12) | 84 | 122.8 | 1.46 | 0 (8.79) |
| Chr.13(A13) | 64 | 109.3 | 1.71 | 0 (7.17) |
| Subtotal-At | 1138 | 1726.8 | 1.52 | 11 (16.52) |
| D-subgenome | ||||
| Chr.15(D01) | 93 | 118.0 | 1.27 | 1 (10.05) |
| Chr.17(D02) | 42 | 114.6 | 2.73 | 2 (22.01) |
| Chr.14(D03) | 79 | 126.4 | 1.60 | 1 (15.92) |
| Chr.22(D04) | 45 | 77.9 | 1.73 | 1 (14.09) |
| Chr.19(D05) | 132 | 227.2 | 1.72 | 1 (15.78) |
| Chr.25(D06) | 70 | 126.9 | 1.81 | 0 (9.249) |
| Chr.16(D07) | 58 | 124.4 | 2.15 | 2 (17.02) |
| Chr.24(D08) | 62 | 118.8 | 1.92 | 1 (10.47) |
| Chr.23(D09) | 83 | 146.0 | 1.76 | 1 (11.07) |
| Chr.20(D10) | 76 | 119.0 | 1.57 | 0 (5.03) |
| Chr.21(D11) | 80 | 136.8 | 1.71 | 0 (9.27) |
| Chr.26(D12) | 53 | 112.3 | 2.12 | 0 (9.23) |
| Chr.18(D13) | 61 | 104.8 | 1.72 | 0 (7.23) |
| Subtotal-Dt | 934 | 1653.1 | 1.77 | 10 (22.01) |
| Total | 2072 | 3380 | 1.63 | 21 (22.01) |
SNP, single nucleotide polymorphism; SSR, simple sequence repeat.
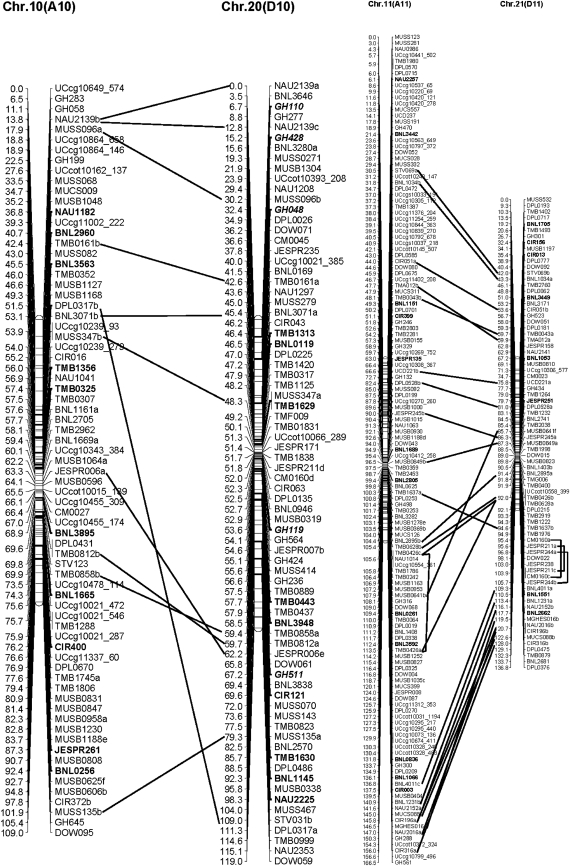
Figure 2 .
Genetic linkage maps of 26 allotetraploid cotton chromosomes that are presented in 13 At and Dt subgenome homeologous pairs (in parentheses). The names of DNA markers are shown on the right, and the positions of the markers are shown in Kosambi centiMorgan (cM) on the left. A line bar connects duplicate marker loci between a pair of homeologous chromosomes. Marker loci in bold are assigned to cotton chromosomes by previously published studies (Frelichowski Jr. et al. 2006; Guo et al. 2007, 2008; Lacape et al. 2003; Liu et al. 2000; Park et al. 2005; Yu et al. 2011) and marker loci in italic bold are assigned to cotton chromosomes in this study (Table 3). Homeologous marker linkage relationships indicate of reciprocal rearrangement between ancestral forms of Chr. 02 and 03 and/or 14 and 17 relative to each other; they also indicate that centromeric regions are homeologous between Chr. 02 (A02) and Chr. 17 (D02), as well as between Chr. 03 (A03) and Chr. 14 (D03). Intrachromosomal duplications were noted in Chr. 5, 11, and 21, the latter two in homeologous segments.
The At subgenome consisted of 1138 marker loci (927 SSR and 211 SNP), and the total genetic distance was 1726.8 cM with an average marker interval of 1.52 cM. The largest chromosome in terms of recombination frequency was Chr. 05 (A05), which spans 199.2 cM with 139 marker loci. The second largest was Chr. 11 (A11), which spans 166.5 cM with 140 loci. The shortest was Chr. 04 (A04), which spans 101.6 cM with 56 loci (Table 2 and Figure 2). In the At subgenome, there were 11 gaps greater than 10 cM, and the largest gap between two loci was 16.52 cM on Chr. 08 (A08).
The Dt subgenome consisted of 934 marker loci (898 SSR and 36 SNP), and the total genetic distance was 1653.1 cM, with an average marker interval of 1.77 cM. The largest chromosome with respect to recombination frequency was Chr. 19 (D05), which spans 227.2 cM with 132 loci, and the shortest chromosome was Chr. 22 (D04), which spans 77.9 cM with 45 loci (Table 2 and Figure 2). There were 10 gaps greater than 10 cM, and the largest gap between two loci was 22.01 cM on Chr. 17 (D02). Although SNP marker loci were largely mapped in the At subgenome because of the A-genome origin of SNP primers (Van Deynze et al. 2009), the At subgenome and Dt subgenome had virtually similar numbers of SSR marker loci and total genetic distances. Furthermore, there were similar amounts of recombination between each of 13 pairs of cotton homeologous chromosomes.
Complete assignment of linkage groups to cotton chromosomes
A complete set of 26 cotton chromosomes (13 At subgenome and 13 Dt subgenome) were identified that correspond to 26 respective linkage groups (Figure 2). Assignment of SSR markers and linkage groups to the cotton chromosomes was achieved in part by comparison of the common markers (bold font in Figure 2) with the previous SSR mapping reports (Frelichowski Jr. et al. 2006; Guo et al. 2007; Lacape et al. 2003; Park et al. 2005; Yu et al. 2011) and with the three aneuploid studies for TMB markers (Guo et al. 2008) and BNL markers (Gutiérrez et al. 2009; Liu et al. 2000), respectively. In addition, hypoaneuploid cottons were also analyzed to identify TM-1 deficiency with 37 newly developed GH markers (bold italic in Figure 2) from G. hirsutum and other SSR markers of interest in the mapping study (Table 3 and Figure 3) (Hoffman et al. 2007). Although most SSR markers generally agreed with published reports, a few incongruities, such as GH034 and GH526, between various data types were encountered when cotton hypoaneuploid stocks were used along with individual mapping populations. Additional mapping analyses in the present research confirmed or reassigned such SSR markers to the corresponding cotton chromosomes (Table 3).
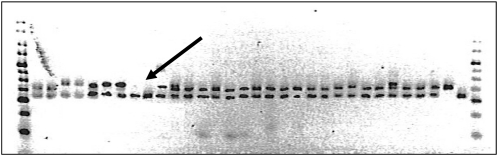
Figure 3 .
Deletion analysis of cotton SSR markers. GH584 amplified (from L to R) cotton hemizygous F1 hypoaneuploids as well as homozygous TM-1 and 3-79. TM-1 allele (140 bp) was missing in both lanes (see arrow) with the H09 template, suggesting the location of GH584 locus on chromosome 09 (A09).
Genomic duplication and chromosomal translocation of allotetraploid cottons
Among 1601 SSR primer pairs that amplified 1895 loci in TM-1 and 3-79, 257 SSR primer pairs amplified two or more loci, resulting in a total of 551 duplicate loci. Excluding dominant loci amplified by these SSRs, there were 494 codominant loci that were duplicated, resulting in 247 pairs (Table S3). Most of the duplicate loci were mapped on the homeologous chromosome pairs (Table 4 and Figure 2). The relative orders of most duplicate loci on the homeologous chromosomes were similar (Figure 2). The duplicate loci identified by these SSR markers demonstrated the complex but linear features of the allotetraploid cotton genomes. A few duplicate loci also were present between nonhomeologous chromosomes and/or within the same subgenome, which indicated likely genome rearrangements (Table 4 and Table S3). For example, an intrasubgenome duplication was revealed by the marker BNL1044 between Chr. 04 (A04) and Chr. 05 (A05). Distinct intrachromosome duplications were indicated by one SSR duplication in Chr. 11 (A11) and three SSRs in Chr. 21 (D11) (Figure 2). In chromosome 11 (A11), TMB0426 revealed two loci that were mapped 8.1 cM apart. In chromosome 21 (D11), three markers (i.e. CM0160, JESPR211, and JESPR244) each revealed two loci. In the latter, the recombination rates remained similar (~8-9 cM) but the relative orders among duplicated loci were altered.
Table 4 . Pairs of duplicate marker loci between homeologous and nonhomeologous chromosomes in cotton.
| Homeologous Chromosomes | No. Pairs of Duplicate Loci | Nonhomeologous Chromosomes | No. Pairs of Duplicate Loci |
|---|---|---|---|
| Chr.01(A01)-Chr.15(D01) | 19 | Chr.02(A02)-Chr.14(D03) | 5 |
| Chr.02(A02)-Chr.17(D02) | 6 | Chr.03(A03)-Chr.17(D02) | 5 |
| Chr.03(A03)-Chr.14(D03) | 13 | Chr.02(A02)-Chr.03(A03) | 1 |
| Chr.04(A04)-Chr.22(D04) | 15 | Chr.04(A04)-Chr.05(A05) | 1 |
| Chr.05(A05)-Chr.19(D05) | 27 | Chr.05(A05)-Chr.22(D04) | 1 |
| Chr.06(A06)-Chr.25(D06) | 12 | ||
| Chr.07(A07)-Chr.16(D07) | 18 | ||
| Chr.08(A08)-Chr.24(D08) | 9 | ||
| Chr.09(A09)-Chr.23(D09) | 27 | ||
| Chr.10(A10)-Chr.20(D10) | 10 | ||
| Chr.11(A11)-Chr.21(D11) | 24 | ||
| Chr.12(A12)-Chr.26(D12) | 13 | ||
| Chr.13(A13)-Chr.18(D13) | 11 | ||
| Totals | 204 | 13 |
A postpolyploidization reciprocal translocation of chromosomes 02 (A02) and 03 (A03) was suggested by 10 pairs of duplicate loci (Figure 2 and Table 4). Five pairs of duplicate loci were identified between chromosomes 02 (A02) and 14 (D03) and 5 pairs between chromosomes 03 (A03) and 17 (D02). The marker TMB1025 revealed duplicate loci between chromosomes 02 (A02) and 03 (A03), which inferred a possible breakpoint for the reciprocal translocation in these two At subgenome chromosomes. Additional mapping data in the vicinity of TMB1025 will be necessary to confirm this conclusion. Another translocation between At subgenome chromosomes 04 (A04) and 05 (A05), as previously suggested by Guo et al. (2007), was observed by the marker BNL1044 loci (BNL1044a at 33.6 cM of A05) and (BNL1044c at 48.1 cM of A04) (Figure 2 and Table S3). Furthermore, the marker GH252 loci showed a translocation between non-homeologous chromosomes 05 (A05) with GH252a at 136.4 cM and 22 (D04) with GH252b at 18.3 cM.
Discussion
The high-density genetic linkage map created in this research is composed of 2072 SSR and SNP loci representing many individual groups of the cotton research community, and it provides a transferable platform that is essential for a broad spectrum of basic and applied studies aimed at understanding and manipulating complex cotton genomes. Among the 17 sets of SSR and SNP marker loci, BAC-derived SSRs (310 TMB Table S1 and 155 MUSB) facilitate an integration of genetic and physical maps of the cotton chromosomes (Frelichowski Jr. et al. 2006; Xu et al. 2008). The markers linked to the novel genes can be used to screen cotton BAC clones or physical contigs from which the SSR markers were developed (Yin et al. 2006). The 357 EST-derived SSR markers mapped herein offer an opportunity to study functional genes and gene islands for fiber development and other important traits of interest. In addition, the genetic mapping of the 247 SNP markers is the first major public effort to use nucleotide sequence diversity in cotton species by mapping SNP loci (Table S1). Localization of these SNP markers to the 26 individual cotton chromosomes and their integration with large numbers of SSR markers will facilitate other studies in cotton genomics. We believe that the high-density genetic map reported herein is a saturated one for the allotetraploid cotton, as evidenced by a separate mapping analysis (data not shown). Further increase in the map density may not significantly change the total genetic length of this map but will facilitate whole-genome physical alignment, sequencing, and mapping of genes for cotton improvement.
Deviation from a Mendelian segregation ratio is common in intra- and interspecific crosses (Causse et al. 1994; Lacape et al. 2009; Rong et al. 2004; Ulloa et al. 2002; Yu et al. 2011). An extremely severe distortion (99%) toward G. hirsutum was observed by Lacape et al. (2009) when 140 RILs were used to produce a low-density map of approximately 800 loci. Only 15 of the 140 RILs exhibited 50% or more G. barbadense parental alleles. In this research, TM-1 was less environmentally sensitive than 3-79, as reflected by the allele transmission preference in the advancement of generations of the RIL population (Figure 1). Of the 2072 mapped marker loci, 1391 (67.1%) fit an expected 1:1 segregation ratio, and 681 (32.9%) deviated significantly (χ2 > 3.8) from expectations among 186 RILs. The 681 segregation-distorted loci (SDL) were mapped in all 26 groups with 349 mapped in At subgenome, and 332 in Dt subgenome chromosomes. Four chromosomes, i.e. Chr. 15 (D01), Chr. 05 (A05), Chr. 07 (A07), and Chr. 08 (A08), had the most SDL, with 68, 60, 57, and 42 loci, respectively. However, Chr. 26 (D12) has the greatest percentage of SDL, 75%, followed by Chr. 15 (D01) with 73.9%, Chr. 07 (A07) with 68.7% and Chr. 05 (A05) with 45.1%. In most cases, the SDL were mapped at centromeric regions.
Our mapping studies indicate that the two subgenomes of allotetraploid cottons are equivalent in recombination frequencies despite the extra repetitive DNA in the At subgenome (Zhao et al. 1998). This result is consistent with other independent mapping studies in which the authors used different allotetraploid cotton populations (F2 or BC1) where variation between At and Dt map sizes supports the ratio of our genetic distances between the two subgenomes. Rong et al. (2004) mapped a total of 2584 STS loci that span 4447 cM, with the A subgenome being 9.5% larger genetically than the D subgenome. To the contrary, Guo et al. (2007) mapped a total of 1790 SSR loci that span 3426 cM, with the D subgenome being 4.5% larger genetically than the A subgenome. Yu et al. (2011) mapped a total of 2316 SSR loci that span 4419 cM with the A subgenome being 3.9% larger genetically than the D subgenome. In this study, the tetraploid cotton were mapped with 1106 loci (54.5%) on 13 At chromosomes at 1726.8 cM (51.1%) and 922 loci (45.5%) on 13 Dt chromosomes at 1653.1 cM (48.9%). Variation in the ratio of subgenome map distances is likely the result of differences in mapping population sizes, as well as in the numbers and sources of DNA markers.
As evidenced in our mapping data, two reciprocal translocations (between Chr. 02 and 03 and between Chr. 04 and 05) are inferred during or after the polyploidization process of two ancestral diploid genomes (A and D). The translocation breakpoint between Chr. 02 and Chr. 03 may be at or near homeologous SSR marker TMB1025. Further investigation is needed to identify additional markers in the vicinity of TMB1025. On the basis of homeologous markers of the two chromosome pairs (A02-D02 and A03-D03), the majority of duplicate loci were mapped to individual pairs of Chr. 02 vs. Chr. 17 and Chr. 03 vs. Chr. 14. The centromeric cores of these chromosomes seem to show the homeologous relationship, either reciprocal insertional translocations or two temporally separate traditional reciprocal translocations. Thus, we propose to name Chr. 17 as D02 and Chr. 14 as D03, whereas Chr. 02 and Chr. 03 remain as A02 and A03, respectively, which is a revision to Wang et al. (2006b) and Guo et al. (2007). Duplication of marker loci revealed genome rearrangements within the same individual chromosomes and/or between nonhomeologous chromosomes (Table S3). We recognize that nomenclature revision of cotton chromosomes and linkage groups would be needed in the future, but this could be accomplished by an international committee of experts in the subject matter.
Genetic mapping coupled with physical alignment of genomic regions into chromosomal maps will expedite the discovery of resistance (R) or pathogen-induced R genes underlying QTL involved in resistance to nematode and Fusarium wilt (Ulloa et al. 2011). Chromosomes 11 (A11) and 21 (D11) are homologs that harbor important genes for cotton improvement because these chromosomes contain genes for resistance to reniform (Dighe et al. 2009) and root-knot nematodes (Wang et al. 2006a), race 1 of Fusarium (Ulloa et al. 2010) and other traits affecting fiber yield and quality. The high-density genetic map will facilitate and expedite the analysis of plant defense genes against nematodes and other biotrophic pathogens.
This high-density cotton map was constructed with an immortal RIL mapping population. A high level of homozygosity in this RIL population (currently in F8-F9) was achieved with less than 5% genome-wide residual heterozygosity. The RILs are maintained as living stocks to produce seed sources for multilocation research on fiber among other traits and to extract fresh DNA samples for a broad spectrum of genomic studies. Our mapping population of 186 RILs is the largest population ever used in high-density cotton genetic mapping. The accuracy of mapping results can be improved substantially as the proportion of recombination between the two linked markers in an inbred population is about twice that of a single meiotic event F2 or BC1 population when linkage distances are small (<12.5 cM) and increase nonlinearly to 50% for unlinked markers (Burr et al. 1988; Haldane and Waddington 1931). This population provides the greatest mapping power currently known in cotton to detect additional loci between closely linked markers by members of the cotton research community who are interested in SSR and SNP augmentation. The advantages of this immortal RIL population and its parental lines make it practical for high-resolution consensus mapping with additional sequence-based portable markers, enabling better understanding and exploitation of complex Gossypium genomes (Mace et al. 2009). This information will complement other work because of the use of the same parents in developing genetic resources, such as hypoaneuploid cytogenetic stocks, chromosome substitution lines, chromosome specific RILs, and QTL mapping populations in other research programs (Jenkins et al. 2006, 2007; Saha et al. 2006, 2010, 2011; Stelly et al. 2005).
The International Cotton Genome Initiative (http://icgi.tamu.edu/) has proposed to map and sequence the Gossypium genomes (Brubaker et al. 2000; Chen et al. 2007; Paterson 2008; Wilkins 2008; Yu et al. 2008), but large amounts of dispersed repetitive elements and duplicate loci between and within the allotetraploid cotton chromosomes present great challenges to properly assemble a complex Gossypium genome. Development of additional numbers of SSR and SNP markers from the fingerprinted and sequenced BAC clones or physical contigs, such as the 310 TMB and 155 MUSB markers on the present map, would provide a unique opportunity to facilitate the mapping the gaps (5−15 cM) of genomic regions (Lin et al. 2010; Xu et al. 2008; Yin et al. 2006). A high-density genetic map is essential in the reconciliation with a whole-genome physical map to facilitate genome sequencing, sequence assembly, gene mapping, and the design of targeted genetic markers for better understanding and improvement of the cotton plant.
Supplementary Material
Acknowledgments
We offer our appreciation to many technical staff residing in the various laboratories who contributed in the development and mapping of SSRs and SNPs on the RIL population. Special thanks go to Ping Cui, Jianmin Dong, Nicole Steele, and Jewel Stroupe for their technical assistance in surveying parental DNA polymorphism and in genotyping the RIL mapping population, and special thanks to Jared Harris for his assistance in maintaining the immortal RIL population. We also would like to thank Cotton Incorporated, Cary, NC, for facilitating the dye-labeled SSR primers, and Dr. Roy Cantrell for his contribution and valuable discussion on this research project. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The U.S. Department of Agriculture is an equal opportunity provider and employer.
Literature Cited
- Abdurakhmonov I. Y., Kohel R. J., Yu J. Z., Pepper A. E., Abdullaev A. A., et al. , 2008. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 92: 478–487 [DOI] [PubMed] [Google Scholar]
- An C., Saha S., Jenkins J. N., Ma D. P., Scheffler B. E., et al. , 2008. Cotton (Gossypium spp.) R2R3-MYB transcription factors SNP identification, phylogenomic characterization, chromosome localization, and linkage mapping. Theor. Appl. Genet. 116: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Beasley J. O., 1942. Meiotic chromosome behavior in species, species hybrids, haploids and induced polyploids of Gossypium. Genetics 27: 25–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenda A., Scheffler J., Scheffler B., Palmer M., Lacape J. M., et al. , 2006. CMD: a Cotton Microsatellite Database resource for Gossypium genomics. BMC Genomics 7: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J. K., Paterson A. H., 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Brown M. S., 1980. Identification of the chromosomes of Gossypium hirsutum L. by means of translocations. J. Hered. 71: 266–274 [Google Scholar]
- Brubaker C., Cantrell R., Giband M., Lyon B., Wilkins T., 2000. Letter to Journal of Cotton Science Community from the Steering Committee of the International Cotton Genome Initiative (ICGI). J. Cotton Sci. 4: 149–151 [Google Scholar]
- Burr B., Burr F. A., Thompson K. H., Albertson M. C., Stuber C. W., 1988. Gene-mapping with recombinant inbreds in maize. Genetics 118: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse M. A., Fulton T. M., Cho Y. G., Ahn S. N., Chunwongse J., et al. , 1994. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Scheffler B. E., Dennis E., Triplett B., Zhang T., et al. , 2007. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145: 1303–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dighe N. D., Robinson A. F., Bell A. A., Menz M. A., Cantrell R. G., et al. , 2009. Linkage mapping of Gossypium longicalyx resistance to reniform nematode during introgression into cotton Gossypium hirsutum. Crop Sci. 49: 1151–1164 [Google Scholar]
- Endrizzi J. E., Turcotte E. L., Kohel R. J., 1985. Genetics, cytology, and evolution of Gossypium, pp. 271–375 Advances in Genetics, edited by Caspari E. W., John G. S. Academic Press, San Diego [Google Scholar]
- Frelichowski J. E., Jr, Palmer M. B., Main D., Tomkins J. P., Cantrell R. G., et al. , 2006. Cotton genome mapping with new microsatellites from Acala 'Maxxa' BAC-ends. Mol. Genet. Genomics 275: 479–491 [DOI] [PubMed] [Google Scholar]
- Ganal M. W., Altmann T., Röder M. S., 2009. SNP identification in crop plants. Curr. Opin. Plant Biol. 12: 211–217 [DOI] [PubMed] [Google Scholar]
- Gao W., Chen Z. J., Yu J. Z., Raska D., Kohel R. J., et al. , 2004. Wide-cross whole-genome radiation hybrid mapping of cotton (Gossypium hirsutum L.). Genetics 167: 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Chen Z. J., Yu J. Z., Kohel R. J., Womack J. E., et al. , 2006. Wide-cross whole-genome radiation hybrid mapping of the cotton (Gossypium barbadense L.) genome. Mol. Genet. Genomics 275: 105–113 [DOI] [PubMed] [Google Scholar]
- Guo W. Z., Cai C. P., Wang C. B., Han Z. G., Song X. L., et al. , 2007. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics 176: 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. F., Saha S., Yu J. Z., Jenkins J. N., Kohel R. J., et al. , 2008. BAC-derived SSR markers chromosome locations in cotton. Euphytica 161: 361–370 [Google Scholar]
- Gutiérrez O., Stelly D., Saha S., Jenkins J., McCarty J., et al. , 2009. Integrative placement and orientation of non-redundant SSR loci in cotton linkage groups by deficiency analysis. Mol. Breed. 23: 693–707 [Google Scholar]
- Haldane J. B., Waddington C. H., 1931. Inbreeding and linkage. Genetics 16: 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman S. M., Yu J. Z., Grum D. S., Xiao J., Kohel R. J., et al. , 2007. Identification of 700 new microsatellite loci from cotton (G. hirsutum L.). J. Cotton Sci. 11: 208–241 [Google Scholar]
- Jenkins J. N., Wu J., McCarty J. C., Saha S., Gutierrez O. A., et al. , 2006. Genetic effects of thirteen Gossypium barbadense L. chromosome substitution lines in topcrosses with Upland cotton cultivars: I. Yield and yield components. Crop Sci. 46: 1169–1178 [Google Scholar]
- Jenkins J. N., Wu J., McCarty J. C., Saha S., Gutierrez O. A., et al. , 2007. Genetic effects of thirteen Gossypium barbadense L. chromosome substitution lines in topcrosses with Upland cotton cultivars: II. Fiber quality traits. Crop Sci. 47: 561–570 [Google Scholar]
- Jiang C. X., Wright R. J., El-Zik K. M., Paterson A. H., 1998. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. U S A. 95: 4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Triplett B. A., 2001. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Kohel R. J., Richmond T. R., Lewis C. F., 1970. Texas Marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Sci. 10: 670–671 [Google Scholar]
- Kohel R. J., Yu J. Z., Park Y.-H., Lazo G., 2001. Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121: 163–172 [Google Scholar]
- Kosambi D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Lacape J. M., Nguyen T. B., Thibivilliers S., Bojinov B., Courtois B., et al. , 2003. A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum x Gossypium barbadense backcross population. Genome 46: 612–626 [DOI] [PubMed] [Google Scholar]
- Lacape J. M., Jacobs J., Arioli T., Derijcker R., Forestier-Chiron N., et al. , 2009. A new interspecific, Gossypium hirsutum × G. barbadense, RIL population: towards a unified consensus linkage map of tetraploid cotton. Theor. Appl. Genet. 119: 281–292 [DOI] [PubMed] [Google Scholar]
- Lee J. A., 1984. Cotton as a world crop, pp. 1–25 Cotton, Agronomy Monograph 24, edited by Kohel R. J., Lewis C. F. American Society of Agronomy, Madison, WI [Google Scholar]
- Lin L., Pierce G., Bowers J., Estill J., Compton R., et al. , 2010. A draft physical map of a D-genome cotton species (Gossypium raimondii). BMC Genomics 11: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Saha S., Stelly D., Burr B., Cantrell R. G., 2000. Chromosomal assignment of microsatellite loci in cotton. J. Hered. 91: 326–332 [DOI] [PubMed] [Google Scholar]
- Mace E., Rami J.-F., Bouchet S., Klein P., Klein R., et al. , 2009. A consensus genetic map of sorghum that integrates multiple component maps and high-throughput Diversity Array Technology (DArT) markers. BMC Plant Biol. 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles G. A., Feaster C. V., 1984. Breeding, pp. 201–231 Cotton. Monograph series Agronomy No. 24, edited by Kohel R. J., Lewis C. F. American Society of Agronomy, Madison, WI [Google Scholar]
- Park Y. H., Alabady M. S., Ulloa M., Sickler B., Wilkins T. A., et al. , 2005. Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol. Genet. Genomics 274: 428–441 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., 2008. Sequencing the cotton genomes—Gossypium spp. Cotton Sci. 20: 3 [Google Scholar]
- Percival A. E., Kohel R. J., 1990. Distribution, collection, and evaluation of Gossypium. Adv. Agron. 44: 225–256 [Google Scholar]
- Reinisch A. J., Dong J., Brubaker C. L., Stelly D. M., Wendel J. F., et al. , 1994. A detailed RFLP map of cotton, Gossypium hirsutum X Gossypium barbadense—chromosome organization and evolution in a disomic polyploid genome. Genetics 138: 829–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J. K., Abbey C., Bowers J. E., Brubaker C. L., Chang C., et al. , 2004. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166: 389–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Jenkins J. N., Wu J., McCarty J. C., Gutiérrez O. A., et al. , 2006. Effects of chromosome-specific introgression in Upland cotton on fiber and agronomic traits. Genetics 172: 1927–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Wu J., Jenkins J. N., McCarty J. C., Hayes R., et al. , 2010. Genetic dissection of chromosome substitution lines of cotton to discover novel Gossypium barbadense L. alleles for improvement of agronomic traits. Theor. Appl. Genet. 120: 1193–1205 [DOI] [PubMed] [Google Scholar]
- Saha S., Wu J., Jenkins J., McCarty J., Hayes R., et al. , 2011. Delineation of interspecific epistasis on fiber quality traits in Gossypium hirsutum by ADAA analysis of intermated G. barbadense chromosome substitution lines. Theor. Appl. Genet. 122: 1351–1361 [DOI] [PubMed] [Google Scholar]
- Shappley Z. W., Jenkins J. N., Meredith W. R., McCarty J. C., 1998. An RFLP linkage map of Upland cotton, Gossypium hirsutum L. Theor. Appl. Genet. 97: 756–761 [Google Scholar]
- Stelly D. M., 1993. Interfacing cytogenetics with the cotton genome mapping effort. Proc. Beltwide Cotton Improvement Conference: 1545–1550 [Google Scholar]
- Stelly D. M., Saha S., Raska D. A., Jenkins J. N., McCarty J. C., et al. , 2005. Registration of 17 Upland (Gossypium hirsutum) cotton germplasm lines disomic for different G. barbadense chromosome or arm substitutions. Crop Sci. 45: 2663–2665 [Google Scholar]
- Stewart J. M., 1994. Potential for crop improvement with exotic germplasm and genetic engineering, pp. 13–327 Challenging the Future: Proceedings of the World Cotton Research Conference-1, edited by Constable G. A., Forrester N. W. CSIRO, Melbourne, Brisbane, Australia [Google Scholar]
- Ulloa M., Meredith W. R., Jr, Shappley Z. W., Kahler A. L., 2002. RFLP genetic linkage maps from four F2.3 populations and a joinmap of Gossypium hirsutum L. Theor. Appl. Genet. 104: 200–208 [DOI] [PubMed] [Google Scholar]
- Ulloa M., Saha S., Jenkins J. N., Meredith W. R., McCarty J. C., et al. , 2005. Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) joinmap. J. Hered. 96: 132–144 [DOI] [PubMed] [Google Scholar]
- Ulloa M., Wang C., Roberts P. A., 2010. Gene action analysis by inheritance and quantitative trait loci mapping of resistance to root-knot nematodes in cotton. Plant Breed. 129: 541–550 [Google Scholar]
- Ulloa M., Wang C., Hutmacher R., Wright S., Davis R., et al. , 2011. Mapping Fusarium wilt race 1 resistance genes in cotton by inheritance, QTL and sequencing composition. Mol. Genet. Genomics 286: 21–36 [DOI] [PubMed] [Google Scholar]
- Van Deynze A., Stoffel K., Lee M., Wilkins T. A., Kozik A., et al. , 2009. Sampling nucleotide diversity in cotton. BMC Plant Biol. 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen J. W., 2006. JoinMap 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma B.V., Wageningen, The Netherlands [Google Scholar]
- Wang C., Ulloa M., Roberts P. A., 2006a Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 112: 770–777 [DOI] [PubMed] [Google Scholar]
- Wang K., Song X. L., Han Z. G., Guo W. Z., Yu J. Z., et al. , 2006b Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor. Appl. Genet. 113: 73–80 [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Cronn R. C., 2003. Polyploidy and the evolutionary history of cotton, pp. 139–186 Advances in Agronomy. Academic Press, San Diego [Google Scholar]
- Wilkins T. A., 2008. Sequencing of a cultivated diploid cotton genome - Gossypium arboreum. Cotton Sci. 20: 5 [Google Scholar]
- Xiao J., Wu K., Fang D. D., Stelly D. M., Yu J., et al. , 2009. New SSR markers for use in cotton (Gossypium spp.) improvement. J. Cotton Sci. 13: 75–157 [Google Scholar]
- Xu Z. Y., Kohel R. J., Song G. L., Cho J. M., Yu J., et al. , 2008. An integrated genetic and physical map of homoeologous chromosomes 12 and 26 in Upland cotton (G. hirsutum L.). BMC Genomics 9: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Guo W., Yang L., Liu L., Zhang T., 2006. Physical mapping of the Rf 1 fertility-restoring gene to a 100 kb region in cotton. Theor. Appl. Genet. 112: 1318–1325 [DOI] [PubMed] [Google Scholar]
- Yu J. Z., 2004. A standard panel of Gossypium genotypes established for systematic characterization of cotton microsatellite markers. Plant Breeding News Edition 148 (1.07), an electronic newsletter of applied plant breeding sponsored Food and Agriculture Organization of the United Nations. [Google Scholar]
- Yu S. X., Wang K. B., Li F. G., Kohel R. J., Percy R. G., et al. , 2008. Sequencing of the cultivated tetraploid cotton genome - Gossypium hirsutum. Cotton Sci. 20: 4 [Google Scholar]
- Yu Y., Yuan D. J., Liang S. G., Li X. M., Wang X. Q., et al. , 2011. Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 12: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T. Z., Yuan Y. L., Yu J., Guo W. Z., Kohel R. J., 2003. Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theor. Appl. Genet. 106: 262–268 [DOI] [PubMed] [Google Scholar]
- Zhao X. P., Si Y., Hanson R. E., Crane C. F., Price H. J., et al. , 1998. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 8: 479–492 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.