Abstract
All cloned sialyltransferases from vertebrates are classified into four subfamilies and are characterized as having type II transmembrane topology. The catalytic domain has highly conserved motifs known as sialylmotifs. Besides sialylmotifs, each family has several unique conserved cysteine (Cys) residues mainly in the catalytic domain. The number and loci of conserved amino acids, however, differ with each subfamily, suggesting that the conserved Cys-residues and/or disulphide linkages they make may contribute to linkage specificity. Using Matrix Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF)-mass spectrometry, the present study performed disulphide linkage analysis on soluble mouse ST6Gal-I, which has six Cys-residues. Results confirmed that there were no free Cys-residues, and all six residues contributed to disulphide linkage formation, C139–C403, C181–C332 and C350–C361. Study of single amino acid-substituted mutants revealed that the disulphide linkage C181–C332 was necessary for molecular expression of the enzyme, and that the disulphide linkage C350–C361 was necessary for enzyme activity. The remaining disulphide linkage C139–C403 was not necessary for enzyme expression or for activity, including substrate specificity. Crystallographic study of pig ST3Gal I has recently been reported. Interestingly, the loci of disulphide linkages in ST6Gal-I differ from those in ST3Gal I, suggesting that the linkage specificity of sialyltransferase may results from significant structural differences, including the loci of disulphide linkages.
Sialic acids are widely distributed in organisms from bacteria to mammals and have various biological functions (1). The synthesis of sialylglycoconjugates involves a family of glycosyltransferases called sialyltransferases that catalyse the transfer of a sialic acid from Cytidine monophosphate (CMP)-Sia to an acceptor carbohydrate. The enzymatic properties of sialyltransferases have been extensively studied in terms of linkage- and substrate specificities with respect to synthetic acceptors as well as their glycoproteins and glycolipids. Thus far, the cDNA cloning of 20 members of the mammalian sialyltransferase family has been performed and these members have been grouped into four subfamilies according to the carbohydrate linkages they synthesize: α-2,3-sialyltransferases (ST3Gal I–VI), α-2,6-sialyltransferases (ST6Gal I–II), GalNAc α-2,6-sialyltransferases (ST6GalNAc I–VI) and α-2,8-sialyltransferases (ST8Sia I–VI) (2). All of the cloned sialyltransferases from vertebrates are localized in the Golgi apparatus and are characterized as having type II transmembrane topology consisting of a short N-terminal cytoplasmic tail, a transmembrane domain followed by a stem region, and a large C-terminal catalytic domain facing the luminal side. The catalytic domain has highly conserved motifs called sialyl motifs L (long), S (short), III and VS (very short) (3–5). Besides these motifs, each subfamily has its own unique conserved amino acids mainly in the catalytic domain. For example, several cysteine (Cys) residues are well conserved in each subfamily (Fig. 1). The number and loci of conserved Cys-residues; however, differ with each subfamily suggesting that the conserved Cys-residues and/or disulphide linkages they make may contribute to linkage specificity. Despite extensive study of ST6Gal I; however, information on the disulphide linkages within the regions from stem to the catalytic domain, which has six Cys-residues is lacking. Using Matrix Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF)-mass spectrometry, the present study performed disulphide linkage analysis and functionally analysed each disulphide linkage with single amino acid-substituted mutants. Crystallographic study of pig ST3Gal I has recently been reported (6). Interestingly, the loci of disulphide linkages in ST6Gal I differ from those in ST3Gal I suggesting substantial structural differences in the two.
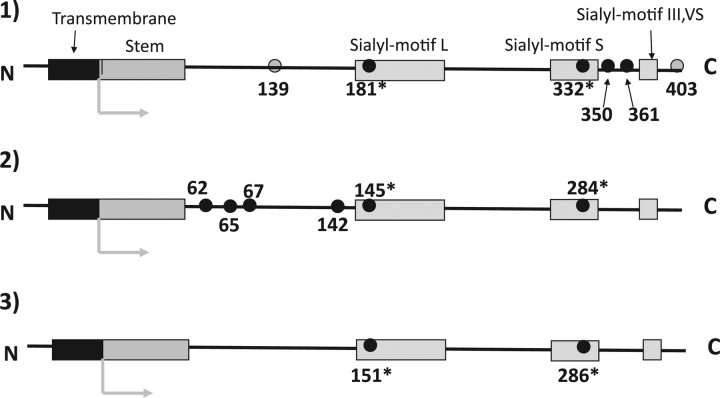
Fig. 1.
Illustrated comparison of conserved Cys-residue locations among mammalian ST6Gal, ST6GalNAc, ST3Gal and ST8Sia family. The conserved Cys-residues of enzymes located from the N-terminal stem region to the C-terminal are depicted with small black or grey circles. The number indicates the location of conserved Cys-residues in mouse ST6Gal-I (7), porcine ST3Gal-I (6) and mouse ST8Sia-I (8), respectively. Two Cys-residues with an asterisk in sialylmotifs were conserved in all of the cloned eukaryotic enzymes. (1) ST6Gal and ST6GalNAc (III, IV, V and VI) families: the four black and two grey-circled Cys-residues were conserved in the ST6Gal family. The four black-circled Cys-residues were also conserved in ST6GalNAc III, IV, V and VI, but two grey-circled Cys-residues were not conserved in the mammalian enzymes. (2) ST3Gal family: the six black-circled Cys-residues were conserved. Note: Except in sialylmotifs, all of the conserved Cys-residues were located in different positions than those in members of the ST6Gal family. (3) ST8Sia family and ST6GalNAc-I and -II: only two Cys-residues in sialylmotifs (marked with an asterisk) were conserved. Note: Conserved Cys-residues in ST6GalNAc I and -II differed substantially from those in other members of the ST6GalNAc family but resembled those in the ST8Sia family.
Materials and Methods
Materials
Gal β-1,3GalNAc; Gal β-1,3GlcNAc; Gal β-1,4GlcNAc and Triton CF-54 were purchased from Sigma (St Louis, MO, USA). Lactose was from Wako (Tokyo). Lacto-N-tetraose and lacto-N-neotetraose were from Glyko (Novato, CA, USA). IgG Sepharose 6 fast flow was from GE Healthcare Bio-Science AB (Uppsala, Sweden). CMP-[3H]NeuAc (3.7 MBq/mmol) was from Amersham Biosciences (Piscataway, NJ, USA).
Preparation of soluble and released mouse ST6Gal-I
The mouse ST6Gal-I cDNA clone used in this study was from the mouse cDNA sequence in GenBank (Accession No. D16106) (7). The expression vector of soluble mouse ST6Gal-I lacking the cytosolic and transmembrane domain (85–403; from the stem region to the C-terminal) as a secretable protein fused with the IgG-binding domain of Staphylococcus aureus protein A (designated pcDSA-mST6Gal-I) was constructed as described previously (9, 10). Soluble forms of sialyltransferases were produced by transfecting COS-7 cells with the above pcDSA vectors using LipofectAMINE™ Reagent (Invitrogen, Carlsbad, CA, USA) and culturing them as described previously (9, 10). The protein A-fused sialyltransferases expressed in the medium were adsorbed to IgG-Sepharose gel (Amersham Biosciences) and then used as the enzyme sources (9, 10).
In some cases, the enzyme was released from IgG Sepharose by incubation with 10 mM glycine buffer pH 3.0 containing 0.15 M NaCl for a few minutes. After centrifugation, the pH of the supernatant was immediately adjusted to pH 7.0 with 0.5 M Tris–HCl buffer containing 0.15 M NaCl. Here, this enzyme is denoted as released enzyme or released ST6Gal-I. The substrate specificity and specific activity of released enzyme was indistinguishable from those of IgG Sepharose-bound enzyme within a few days (data not shown). The released enzyme (20 µg) produced a single band in a SDS–PAGE with Coomassie Brilliant Blue R-250 staining.
Preparation of Cys-residue mutants of ST6Gal-I replaced with Ser or Ala
Five pcDSA-based constructs with a single amino acid mutation of ST6Gal-I (Table I) were created as briefly described below. Each construct was transfected in COS cells as described above. After correction of the culture medium, the expressed mutant enzyme was collected with IgG Sepharose. IgG Sepharose-bound enzyme was used as an enzyme source.
Table I.
PCR primer sequence for preparation of ST6gal-I Cys-residue mutants.
| Mutant | Mutation | Primer for the first PCR | ||
|---|---|---|---|---|
| name | Forward | Reverse | Plasmid names | |
| 139C/S | 195C→S | 5′-GCGCTGCGCTCCCACCTTCGAGAC | 5′-GTCTCGAAGGTGGCAGCGCAGCGC | pcDSA-ST6Gal-I-195C/S |
| 139C/A | 195C→A | 5′-GCGCTGCGCGCCCACCTTCGAGAC | 5′-GTCTCGAAGGTGGGCGCGCAGCGC | pcDSA-ST6Gal-I-195C/A |
| 181C/S | 237C→S | 5′-CCTTGGCATAAGTCTGCCGTCGTG | 5′-CACGACGGCAGACTTATGCCAAGG | pcDSA-ST6Gal-I-237C/S |
| 350C/S | 406C→S | 5′-CAGATGTGTCCTACTATCACCAG | 5′-CTGGTGATAGTAGGACACATCTG | pcDSA-ST6Gal-I-406C/S |
| 350C/A | 406C→A | 5′-CAGATGTGGCCTACTATCACCAG | 5′-CTGGTGATAGTAGGCCACATCTG | pcDSA-ST6Gal-I-406C/A |
Underline shows the sequence corresponding to the mutation.
A single amino acid mutation of ST6Gal-I was produced by first performing Polymerase Chain Reaction (PCR) using the primers in Table I and PUC119 encoding ST6Gal-I as a template. The first PCR product was then used as a template and the second round of PCR was done with 5′-GTGTGGGAATTCGGGAGCGACTATGAGGCTC and 5′-GGATGCTCGAGCCTGGCTCAACAGCGATTGTTCCGGAAGC. Then, the 1.1 kb EcoRI–XhoI fragment encoding a truncated form of ST6Gal mutant (from stem region to the C-terminal) was prepared and subcloned into the EcoRI–XhoI site of pcDSA. After the nucleotide sequence was confirmed, the resulting plasmid was designated as shown in Table I.
Enzyme assay
Sialyltransferase assays were performed as described previously (9, 10). In brief, enzyme activity was measured in 50 mM 2-(N-morpholino)ethanesulfonic acid buffer (pH 6.0), 1 mM MgCl2, 0.5% Triton CF-54, 100 µM CMP-[3H]-NeuAc with 10 µg of oligosaccharide as an acceptor substrate and enzyme preparation in a total volume of 10 µl. The enzyme reaction was performed at 37°C for 10 h. Afterwards, the reaction mixtures were directly subjected to High performance thin layer chromatography with a solvent system of 1-propanol, aqueous ammonia and water (6 : 1 : 2.5). The radioactive materials were visualized and quantified with a AR-2000 radio-TLC Imaging Scanner. The intensity of the radioactivity was converted into moles with the radioactivity of various amounts of CMP-[3H]NeuAc (3.7 MBq/mmol) serving as a reference. Quantification was performed within the linear range of standard radioactivity.
Amino acids analysis of carboxymethylated soluble mouse ST6Gal-I
The measurement of the total amounts of carboxymethylcysteine
After released ST6Gal-I (20 µg) was dissolved in 50 µl of 100 mM Tris pH 8.5 with 8 M urea, 1 µl of 1 M dithiothreitol (DTT) was added and the reduction reaction proceeded for 4 h at 37°C. Then, 2.5 µl of 1 M iodoacetic acid (IAA) was added and the mixture was allowed to stand for 30 min in the dark after which 2 µl of 1 M DTT was added to quench the IAA. The solvent was then exchanged for 5 mM ammonium acetate using spin columns. The amounts of free Cys-residues were determined following the procedure above excluding the reducing step with DTT.
Carboxymethylcysteine analysis
Carboxymethylated protein was hydrolysed with 6 N HCl in vacuo for 24, 48 or 72 h. The starting concentration of amino acids was extrapolated by time-course analysis of carboxymethylcysteine (CMC) at different hydrolysis times. The amounts of CMC were determined using a slightly modified AccQTag system (Waters) and Shimadzu LC10A with CapcellPack ACR (4.6 × 150 mm; Shiseido) column (11).
Electrophoresis and electroblotting of API digests
The released enzyme was digested with Achromobacter protease I (API) in 0.1% SDS/100 mM Tris–HCl (pH 9.0)/1 mM EDTA, and the resulting peptides were separated by Tricine–SDS–PAGE under non-reducing conditions as described previously (12). The gel was stained with 0.2% Coomassie brilliant blue R-250 in 50% methanol and 7% acetic acid, or the gel-separated peptides were electroblotted onto a polyvinylidene difluoride membrane (Millipore, MA, USA) as described previously (13).
In-gel digestion and mass spectroscopic analysis
For mass spectroscopic analysis, the peptide bands after electrophoresis were excised out of the gel and de-stained with 50% acetonitrile. The gel pieces were dried and treated with 40 ng endoproteinase Asp-N in 20 µl of 20 mM Tris–HCl (pH 8.0) for 16 h at 37°C under non-reducing conditions, and then subjected to mass spectrometry. Reduction of disulphide bonds was done by incubating, 10 µl of digestion mixture in 10 mM DTT/1.5 M Tris–HCl (pH 8.5) for 2 h at 37°C and then cysteines were carboxymethylated with 20 mM iodoacetate for 30 min at 25°C. The resulting samples were desalted with a ZipTip C18 column (Millipore), and then analysed with an Ultraflex mass spectrometer (Bruker Daltonics, Bremen, Germany) in positive reflector mode using α-cyano-4-hydroxycinnamic acid as a matrix.
Peptide sequencing
The electroblotted peptides were subjected to Edman degradation using a Procise HT protein sequencing system (Applied Biosystems, Foster City, CA, USA).
Results
Quantitative analysis of free Cys-residues within released mouse ST6Gal-I
COS-7 cells were first transfected with pcDSA vectors encoding secretable soluble forms of sialyltransferases [fusing with the IgG-binding domain of S. aureus protein A in place of the cytosolic and transmembrane domain (85–403; from the stem region to the C-terminal)] and cultured as described previously. The protein A-fused secretable sialyltransferases expressed in the medium were adsorbed to IgG-Sepharose gel (Amersham Biosciences) (9, 10). Next, the enzyme was released from IgG-Sepharose by incubation with 10 mM glycine buffer pH 3.0 containing 0.15 M NaCl. After centrifugation, the pH of the supernatant was immediately adjusted to pH 7.0 with 0.5 M Tris–HCl buffer containing 0.15 M NaCl. This enzyme was designated released mouse enzyme or ST6Gal-I.
First the number of free Cys-residues in the enzymes released from IgG-Sepharose was estimated. After acid hydration of 20 µg of enzyme alkylated with IAA and 8 M urea (with no DTT), amino acid analysis of hydrolysates revealed no detectable amounts of CMC. However, the hydrolysates of 20 µg of alkylated enzyme with IAA, 8 M urea and DTT revealed 5.4 ± 0.5 pmoles of CMC/pmole of enzyme protein, suggesting that this enzyme does not have a free Cys-residue but has six Cys-residues contributing to the formation of disulphide linkages, i.e. this enzyme has three disulphide bonds.
Estimation of the molecular weight of ST6Gal-I according to SDS–PAGE with or without reducing agents
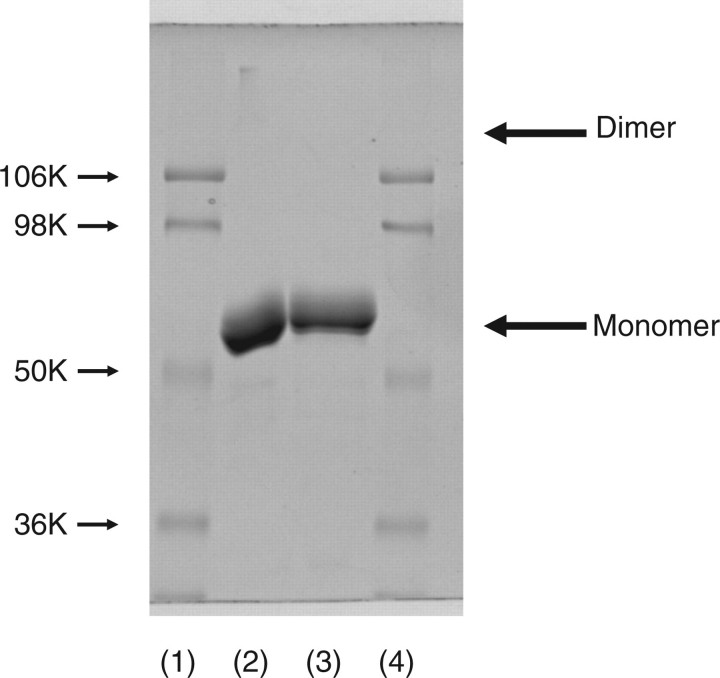
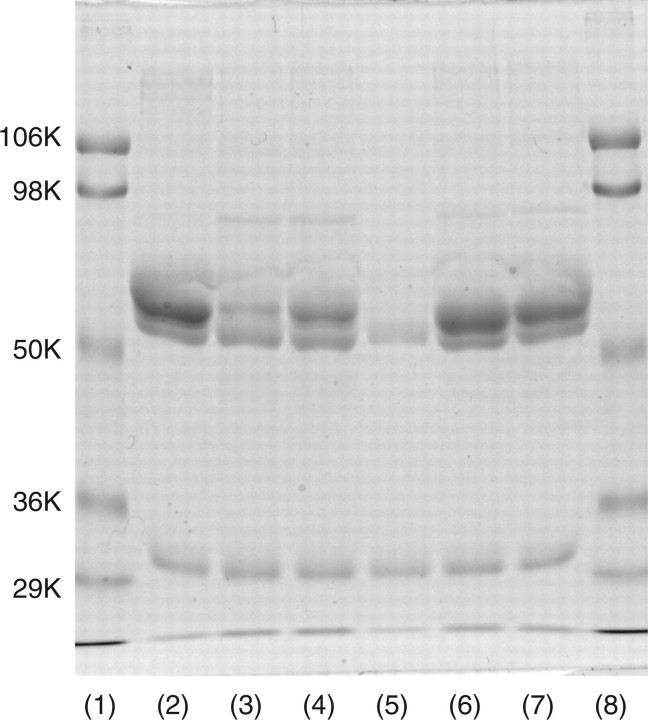
If some of the disulphide linkages were formed with inter- but not intra-molecular Cys-residues, this enzyme should assume dimer or oligomeric forms. As far as seen in SDS–PAGE with or without reducing treatment (Fig. 2) revealed only the monomer form of enzyme. Thus, this enzyme assumes a monomer form rather than an oligomeric form and there are three disulphide bonds in an enzyme molecule.
Fig. 2.
SDS–PAGE analysis of released mouse ST6Gal-I with or without β-mercaptoethanol (β-ME) treatment. Twenty microgram of released mouse ST6Gal-I was treated with reagent for 3 min for SDS–PAGE with or without β-ME. After SDS–PAGE, the gel was stained with Coomassie brilliant blue R-250. Lanes 1 and 4, molecular markers (106 K, phosphorylase B; 98 K, bovine serum albumin; 50 K, ovalbumin; 36 K carbonic anhydrase); lane 2, without β-ME treatment; and lane 3, with β-ME treatment.
Amino acid sequence analysis of API fragments of soluble mouse ST6Gal-I
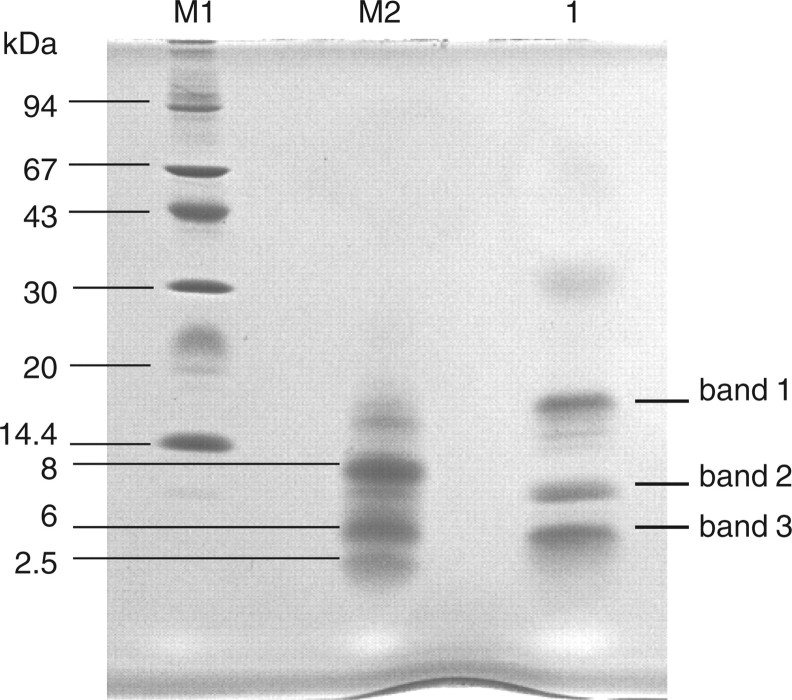
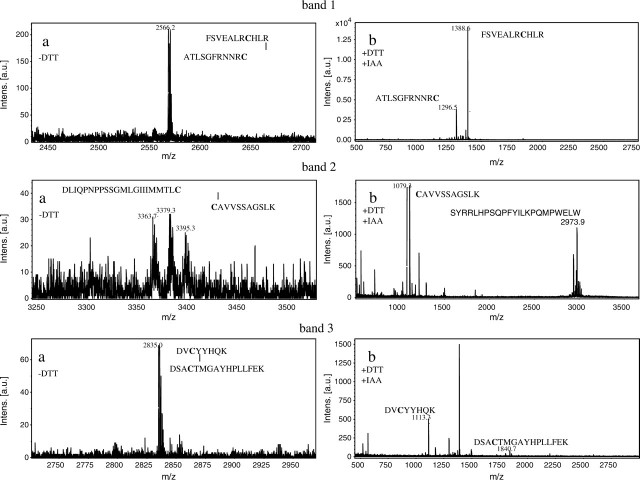
When ST6Gal-I (29–403; from the stem region to the C-terminal) was digested with API under non-reducing conditions and subjected to Tricine–SDS–PAGE, three polypeptide bands (bands 1, 2 and 3) appeared as shown in Fig. 3. Table II shows N-terminal sequences of these three bands. Band 1 yielded two sequences corresponding to F132–N146 and A393–C403 of ST6Gal I. This indicates that band 1 contains two peptide fragments cleaved between K131 and F132, and between K392 and A393 by API digestion; the two peptide fragments are linked by disulphide bond between C139 in the former fragment and C403 in the latter fragment. Bands 2 and 3 yielded two sequences corresponding to C181–K191 and S280–S309, and R345–K355 and F356–L370, respectively. These suggest that disulphide bonds are formed between C181 and C332 in band 2 and between C350 and C361 in band 3. To confirm the location of these disulphide bonds, MALDI-TOF-mass spectrometry analysis was carried out. Peptide bands 1, 2 and 3 (Fig. 3) were treated with endoproteinase Asp-N using in-gel digestion method under non-reducing conditions, and subjected to MALDI-TOF-mass spectrometry. As shown in Fig. 4a, ion peaks were observed at m/z 2,566.2, 3,363.7 and 2,835.0 in bands 1, 2 and 3, respectively. The assignments of these mass spectroscopic peaks are shown in Table III. The peak at m/z 2,566.2 obtained in band 1 is attributed to F132–R142 and A393–C403 in which a disulphide bond is formed between C139 and C403. The peaks at m/z 3,363.7 in band 2 and at m/z 2,835.0 in band 3 are attributed to C181–K191 and D311–C332, and to D348–K355 and D358–L370, in which disulphide bonds are formed between C181 and C332 in band 2, and between C350 and C361 in band 3. (Table III). Two additional ion peaks appeared in band 2 at 3,379.3 and 3,395.3. These may be peptides containing oxidized methionine in the fragments. To verify this result, the disulphide bonds were cleaved with DTT and then carboxymethylated with iodoacetate before MALDI-TOF-mass spectrometry analysis. As shown in Fig. 4b, new peaks appeared at m/z 1,388.6 and 1,296.5 in band 1, m/z 1,079.3 in band 2 and m/z 1,113.3 and 1,849.8 in band 3. These ion peaks corresponded to F132–R142 and A393–C403 in band 1, C181–K191 in band 2, and D348–K355 and D358–L370 in band 3 (Table III). However, the carboxymethylated peptide corresponding to D311–C332 was not observed in band 2 (Fig. 4b). Thus, these results clearly indicate that ST6Gal-I contains three disulphide bonds at C139–C403, C181–C332 and C350–C361.
Fig. 3.
Tricine–SDS–PAGE profile of API-digested ST6Gal-I. Lane M1, molecular marker; lane M2, CNBr-treated horse myoglobin and lane3, API-digested ST6Gal-I.
Table II.
N-terminal amino acid sequences of the bands 1, 2 and 3 obtained in Tricine–SDS–PAGE of API-digested ST6Gal-I.
| Band | N-terminal amino acid sequences |
|---|---|
| 1 | 132 FSVEALRcHLRDHVN |
| 393 ATLSGFRNNRc | |
| 2 | 181 cAVVSSAGSLK |
| 280 SYRRLHPSQPFYILKPQMPWELWDIIQEIS | |
| 3 | 345 RKTDVcYYHQK |
| 356 FFDSAcTMGAYHPLL |
Fig. 4.
MALDI-TOF-mass spectra of endoproteinase Asp-N-treated bands 1, 2 and 3. Peptide bands 1, 2 and 3 were digested with Asp-N under non-reducing conditions (a). After digestion with Asp-N; the peptide fragments were reduced with DTT and then carboxymethylated (b).
Table III.
Assignments for peptide fragments obtained from Asp-N digestion of bands 1, 2 and 3 before and after reduction and carboxymethylation.
| Band | Peptides | Observed mass | Theoretical mass |
|---|---|---|---|
| Non-reduced peptides | |||
| 1 | FSVEALRCHLR/ATLSGFRNNRC | 2,566.2 | 2,566.3 |
| 2 | CAVVSSAGSLK/DLIQPNPPSSGMLGIIIMMTLC | 3,363.7 | 3,362.7 |
| 3 | DVCYYHQK/DSACTMGAYHPLL | 2,835.0 | 2,835.3 |
| Reduced and carboxymethylated peptides | |||
| 1 | FSVEALRCHLR | 1,388.6 | 1,388.7 |
| ATLSGFRNNRC | 1,296.5 | 1,296.6 | |
| 2 | CAVVSSAGSLK | 1,079.3 | 1,079.5 |
| DLIQPNPPSSGMLGIIIMMTLC | ND | 2,402.2 | |
| 3 | DVCYYHQK | 1,113.3 | 1,113.5 |
| DSACTMGAYHPLL | 1,840.8 | 1,840.8 | |
ND, not detected.
Functional analysis of disulphide linkages using single amino acid-substituted mutants
Five single amino acid-substituted mutants 139C/S, 139C/A, 181C/S, 350C/S and 350C/A (Table I) were created using PCR. All of the mutants except 181C/S expressed significant amounts of dual bands (Fig. 5). Mouse ST6Gal-I has two N-glycosylation sites, and thus the two bands of each mutant may correspond to the heterogeneity of N-glycosylation. The enzyme activity and substrate specificity of each mutant revealed that 181C and 350C were necessary for the enzyme activity. (Table IV) Mutants 139C/S and C/A; however, both had almost the same activity and specificity as those of native enzyme.
Fig. 5.
SDS–PAGE analysis of mutants. Fifteen microlitre of native or mutant enzyme-bound IgG-Sepharose was treated with reagent for 3 min for SDS–PAGE with β-ME. The gel was stained with Coomassie brilliant blue R-250. Fifteen microlitre of enzyme-bound IgG-Sepharose corresponded to 15 ml of culture medium under the same culture conditions. Thus, the staining intensity of each band is comparable to the level of expression in each mutant. Lanes 1 and 8, molecular markers (106 K, phosphorylase B; 98 K, bovine serum albumin; 50 K, ovalbumin; 36 K carbonic anhydrase); lane 2, native ST6Gal I; lane 3, 139C/S; lane 4, 139C/A; lane 5, 181C/S; lane 6, 350C/S; and lane 7, 350C/A.
Table IV.
Enzyme activity and substrate specificity of ST6Gal-I mutants.
| Oligosaccharide acceptors | Structure | Native | 139C/S | 139C/A | 181C/S | 350C/S | 350C/A |
|---|---|---|---|---|---|---|---|
| Type II | Gal β-1,4GlcNAc | 100a | 94 | 98 | ND | ND | ND |
| Type I | Gal β-1,3GlcNAc | 2 | 1 | 1 | ND | ND | ND |
| Type III | Gal β-1,3GalNAc | 10 | 11 | 9 | ND | ND | ND |
| Lactose | Gal β-1,4Glc | 14 | 11 | 16 | ND | ND | ND |
| Lacto-N-tetraose | Gal β-1,3GlcNAc β-1,3 Gal β-1,4Glc | 35 | 42 | 25 | ND | ND | ND |
| Lacto-N-neotetraose | Gal β-1,4GlcNAc β-1,3 Gal β-1,4Glc | 108 | 102 | 95 | ND | ND | ND |
Each mutant's level of activity in relation to oligosaccharide is shown in proportion to that of the native enzyme for Type II oligosaccharide (Gal β-1,4GlcNAc), which represents 100.
a45.2 pmol/h/ml medium.
ND, not detected.
Discussion
This study used expressed enzyme as a secretable protein lacking the N-terminal and transmembrane domain (85–403; from the stem region to the C-terminal) and fused it with the IgG-binding domain of S. aureus protein A. In some experiments, enzyme released from IgG- Sepharose was used as a released enzyme. Although the data are not shown, these enzymes had almost the same enzymatic parameters as those of full length, i.e. native enzymes (7). Here, discussion will focus on the relationship between enzymatic function and the structure from the stem region to the C-terminal, including the catalytic domain. Examination of the nucleotide sequence revealed that the enzyme had six Cys-residues, but the full-length enzyme had two more residues, one in the cytosolic domain and another in the transmembrane domain. Analysis of the extra two Cys-residues was not possible, and the possible existence of a dimer form of ST6Gal-I via Cys24 located in the transmembrane region cannot be ruled out (14, 15). The present data shows that mouse ST6Gal-I (85–403; from the stem region to the C-terminal) has three disulphide linkages that are intra-molecular but not inter-molecular, and this ST6Gal-I assumes a monomer form. The data revealed three disulphide linkages between the Cys sets C139–C403, C181–C332 and C350–C361. All six Cys-residues are conserved in members of the mammalian ST6Gal family, suggesting that the three disulphide linkages may also be conserved.
All eukaryotic sialyltransferases share several unique conserved peptide regions, referred to as sialylmotifs L, S, III and VS, in their catalytic domain (3, 16, 17). Sequence comparison of all eukaryotic sialyltransferase clones thus far revealed that each sialylmotif L and S has a single conserved Cys-residue. Mutational analyses of other groups suggested that disulphide linkages occur between sialylmotifs L and S (3, 18, 19). A recent crystallographic study confirmed such linkages in the case of porcine ST3Gal-I (6). The present data also clearly show that a C181–C332 linkage is formed between sialylmotifs L and S. As reported previously, sialylmotifs L and S have binding affinity for acceptor substrate and CMP-Sia (18, 19). The single disulphide linkage between sialylmotifs L and S may help form a domain for the binding of acceptor substrate and CMP-Sia. Furthermore, the 181C/S mutant seldom expressed the enzyme molecule suggested that this Cys-residue, may be a C181–C332 linkage and may also be essential for proper folding of the enzyme structure.
The 350C/S and C/A mutants were both expressed but had no activity at all, suggesting that the disulphide linkage C350–C361 is not directly correlated with protein folding but may play an important role in forming a domain related to enzyme activity. A crystallographic study of porcine ST3Gal I showed that sialylmotif VS has a catalytic centre (6). The disulphide linkage C350–C361 is located between the sialylmotif L/S domain and sialylmotif VS domain, suggesting that the disulphide linkage may contribute to the proper orientation of both domains. The loop between the disulphide bond C350–C361 consists of 10 amino acids in ST6Gal family, while it consists of ∼20 amino acids in the members of ST6GalNAc family (III, IV, V and VI). Although further experiments are necessary, the number of amino acids in this linkage may contribute to the formation of a Gal- or GalNAc-recognizing domain.
The 139C/S and C/A mutants were both expressed and had almost the same activity and the substrate specificity as those of native enzyme. Thus, the disulphide linkage C139–C403 was not necessary for protein folding nor for enzymatic activity. There are numerous amino acids i.e. 264, between this disulphide linkage, suggesting that the enzyme molecule must be folded into its proper structure without this disulphide linkage. Further details will become apparent after crystallographic analysis.
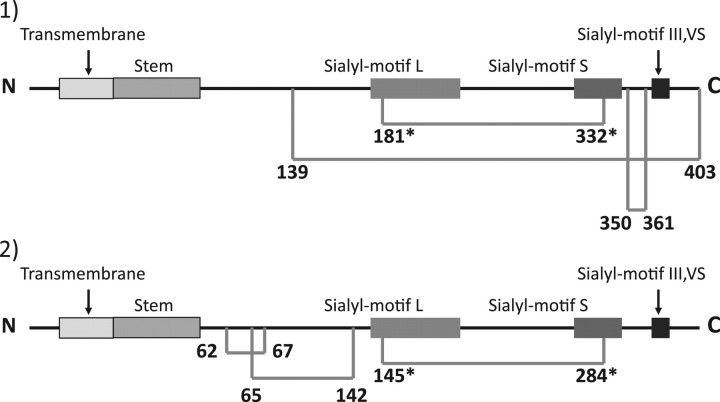
Results of the crystallographic study of porcine ST3Gal-I and the current results for mouse ST6Gal-I revealed striking differences in the location of disulphide linkages except for the linkage between sialylmotifs L and S (Fig. 6). These differences must be due to differences in enzymes but not in animals, because in all mammalian enzymes thus far, the six Cys-residues are mostly conserved in ST6Gal-I and in ST3Gal-I, That said their conserved loci differ vastly except in sialylmotifs L and S. In addition, ST8Sia family members also have completely different conserved Cys-residues from those of the ST3Gal and ST6Gal families except for the two Cys- residues in sialylmotifs L and S (Fig. 1). These data suggest that the linkage specificity of sialyltransferases may be a result of significant structural differences, including the loci of disulphide linkages.
Fig. 6.
Illustrated comparison of the location of disulphied linkages in mouse ST6Gal-I (1) and porcine ST3Gal-I (2). The number indicates the Cys location. Cys-residues conserved in all of the cloned eucaryotic siaiyltransferases are indicated with an asterisk. The horizontal bar indicates a disulphide linkage. The data on porcine ST3Gal-I are from reference (6).
Looking to the future, the three-dimensional structure of each of the member of the ST6Gal, ST6GalNAc and ST8Sia families should be determined. Three-dimensional analysis is crucial to clarifying the mechanisms of linkage specificity and of donor and acceptor substrate binding to mammalian sialyltransferases.
Acknowledgements
The authors wish to thank to Dr Otsuka, Department of molecular science, Tokai University, Ishehara Teaching and Research Support Center for his analysis of amino acids including CMC.
Conflict of interest
None declared.
Glossary
Abbreviations†
- API
Achromobacter protease I
- CMC
carboxymethylcysteine
- DTT
dithiothreitol
- IAA
iodoacetic acid
- β-ME
β-mercaptoethanol
Footnotes
†The abbreviated nomenclature for cloned sialyltransferases follows the system of Tsuji, S., Datta, A.K., and Paulson, J.C. (1996) Systematic nomenclature for sialyltransferases. Glycobiology 6, v–vii.
References
- 1.Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 2.Takashima S, Tsuji S, Tsujimoto M. Characterization of the second type of human β-galactoside α2,6-sialyltransferase (ST6Gal II) that sialylates Galβl,4GlcNAc structures on oligosaccharides preferentially. J. Biol. Chem. 2002;277:45719–45728. doi: 10.1074/jbc.M206808200. [DOI] [PubMed] [Google Scholar]
- 3.Drickamer K. A conserved disulphide bond in sialyltransferases. Glycobiology. 1993;3:2–3. doi: 10.1093/glycob/3.1.2. [DOI] [PubMed] [Google Scholar]
- 4.Livingston BD, Paulson JC. Polymerase chain reaction cloning of a developmentally regulated member of the sialyltransferase gene family. J. Biol. Chem. 1993;268:11504–11507. [PubMed] [Google Scholar]
- 5.Geremia RA, Harduin-Lepers A, Delannoy P. Identification of two novel conserved amino acid residues in eukaryotic sialyltransferases: implications for their mechanism of action. Glycobiology. 1997;7:v–vii. doi: 10.1093/glycob/7.2.161. [DOI] [PubMed] [Google Scholar]
- 6.Rao FV, Rich JR, Rakic B, Buddai S, Schwartz MF, Johnson K, Bowe C, Wakarchuk WW, DeFrees S, Withers SG, Strynadka NCJ. Structural insight into mammalian sialyltransferases. Nat. Struct. Mol. Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- 7.Hamamoto T, Kawasaki M, Kurosawa N, Nakaoka T, Young-Choon Lee Y-C, Tsuji S. Two step single primer mediated polymerase chain reaction. Application to cloning of putative mouse β-galactoside α2,6-sialyltrans- ferase cDNA. Bioorg. Med. Chem. 1993;1:141–145. doi: 10.1016/s0968-0896(00)82111-2. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki K, Kurata K, Kojima N, Kurosawa N, Ohta S, Hanai N, Tsuji S, Nishi T. Expression cloning of a GM3-specific α2,8-sialyltransferase (GD3 synthase) J. Biol. Chem. 1994;269:15950–15956. [PubMed] [Google Scholar]
- 9.Kono M, Yoshida Y, Kojima N, Tsuji S. Molecular cloning and expression of a fifth type of α2, 8-sialyltrans-ferase (STSSia V) J. Biol. Chem. 1996;271:29366–29371. doi: 10.1074/jbc.271.46.29366. [DOI] [PubMed] [Google Scholar]
- 10.Takashima S, Tsuji S, Tsujimoto M. Comparison of the enzymatic properties of mouse β-galactoside α2,6-sialyltransferases, ST6Gal I and II. J. Biochem. 2003;134:287–296. doi: 10.1093/jb/mvg142. [DOI] [PubMed] [Google Scholar]
- 11.Cohen SA, Michaud DP. Synthesis of a fluorescent derivatizing reagent, 6-Aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate aminoacids via high-performance liquid chromatography. Anal Biochem. 1993;211:279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 12.Schaegger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 13.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 14.Qian R, Chen C, Colley KJ. Location and mechanism of a2,6-sialyltransferase dimer formation. J. Biol. Cem. 2001;276:28641–28649. doi: 10.1074/jbc.M103664200. [DOI] [PubMed] [Google Scholar]
- 15.Fenteany FH, Colley KJ. Multiple signals are required for a2,6-sialyltransferase (ST6Gal_I) oligomerization and Golgi location. J. Biol. Chem. 2005;280:5423–5429. doi: 10.1074/jbc.M412396200. [DOI] [PubMed] [Google Scholar]
- 16.Livingston BD, Paulson JC. Polymerase chain reaction cloning of a developmentally regulated member of the sialyltransferase gene family. J. Biol. Chem. 1993;268:11504–11507. [PubMed] [Google Scholar]
- 17.Geremia RA, Harduin-Lepers A, Delannoy P. Identification of two novel conserved aminoacid residues in eukaryotic sialyltransferase: implications for their mechanism of action. Glycobiology. 1997;7:v–vii. doi: 10.1093/glycob/7.2.161. [DOI] [PubMed] [Google Scholar]
- 18.Datta AK, Paulson JC. The sialyltransferase sialylmotif participates in binding the donor substrate CMP-NeuAc. J. Biol. Chem. 1995;270:1497–1500. doi: 10.1074/jbc.270.4.1497. [DOI] [PubMed] [Google Scholar]
- 19.Datta AK, Sinha A, Paulson JC. Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J. Biol. Chem. 1998;273:9608–9614. doi: 10.1074/jbc.273.16.9608. [DOI] [PubMed] [Google Scholar]