Summary
Signal molecules of the Diffusible Signal Factor (DSF) family have been shown recently to be involved in regulation of pathogenesis and biofilm formation in diverse Gram-negative bacteria. DSF signals are reported to be active not only on their cognate bacteria, but also on unrelated bacteria and the pathogenic yeast, Candida albicans. DSFs are monounsaturated fatty acids of medium chain length containing an unusual cis-2 double bond. Although genetic analyses had identified genes involved in DSF synthesis, the pathway of DSF synthesis was unknown. The DSF of the important human pathogen Burkholderia cenocepacia (called BDSF) is cis-2-dodecenoic acid. We report that BDSF is synthesized from a fatty acid synthetic intermediate, the acyl carrier protein (ACP) thioester of 3-hydroxydodecanoic acid. This intermediate is intercepted by protein Bcam0581 and converted to cis-2-dodecenoyl-ACP. Bcam0581 is annotated as a homologue of crotonase, the first enzyme of the fatty acid degradation pathway. We demonstrated Bcam0581to be a bifunctional protein that not only catalyzed dehydration of 3-hydroxydodecanoyl-ACP to cis-2-dodecenoyl-ACP, but also cleaved the thioester bond to give the free acid. Both activities required the same set of active site residues. Although dehydratase and thioesterase activities are known activities of the crotonase superfamily, Bcam0581 is the first protein shown to have both activities.
Introduction
Many species of bacteria use quorum sensing mechanisms to coordinate gene expression by measuring local cell concentrations (Deng et al., 2011, Ng & Bassler, 2009, Winans, 2011). Quorum sensing generally involves secretion of small molecules called autoinducers or pheromones. In gram-negative bacteria the signaling molecules often contain a fatty acid moiety whereas gram-positive bacteria generally use peptides. The fatty acid based autoinducers are the widely distributed acyl-homoserine lactones (AHLs), 3-hydroxypalmitate methyl ester and the diffusible single factor (DSF) family of 2-enoic acids of medium chain lengths. AHLs vary in acyl group length (C4–C18), in substitutions at C3 (hydrogen, hydroxyl or oxo groups) and in the presence or absence of one or more carbon-carbon double bonds in the fatty acid chain. Indeed, the acyl chains of the known AHLs sample each of the intermediates of fatty acid synthesis (Churchill & Chen, 2011). The R configuration of the 3-hydroxy species (Cao & Meighen, 1993) as well as use of acyl chains derived from unsaturated fatty acid synthesis (Churchill & Chen, 2011) plus many in vitro and in vivo investigations (Parsek et al., 1999, Val & Cronan, 1998, More et al., 1996) indicate that the first step in AHL synthesis is acylation of S-adenosyl-L-methionine by transfer of an acyl group from an acyl thioester of acyl carrier protein (ACP) (Churchill & Chen, 2011). The 3-hydroxypalmitate methyl ester acyl chain is also thought to be derived from an acyl-ACP (Flavier et al., 1997). Similar diversions of fatty acid synthetic intermediates also occur in housekeeping synthetic pathways such as that of the lipid A component of the outer membrane (Raetz et al., 2007) and the enzyme cofactor, lipoic acid (Cronan et al., 2005).
In contrast to the other acyl chain-based autoinducers, the acyl chains of the DSF family of 2-enoic acids despite their structural simplicity cannot be directly diverted from the fatty acid synthetic pathway because the 2-enoic double bond has the cis configuration (Deng et al., 2011) rather than the usual trans configuration (often called enoyl when in thioester linkage) that is the last intermediate of the fatty acid elongation cycle. To our knowledge no cis-2 enoic acids other than the DSF family members have been reported. Acyl chains having a cis-3 bond are intermediates in the anaerobic pathway of long chain unsaturated fatty acid synthesis, but since the cis-3 bond results from isomerization of a trans-2 intermediate (Cronan & Thomas, 2009), a variation of this pathway was not a plausible route to DSFs. Thus far, production the DSF family of quorum sensing molecules seems restricted to fewer bacterial species than produce AHLs (Deng et al., 2011, Ryan & Dow, 2011). However, unique among bacterial signaling molecules, DSF and BDSF (the DSF of Burkholderia cenocepacia) are reported to be active on diverse bacteria and on fungi. Nanomolar concentrations of BDSF (or the two-carbon shorter homologue synthesized by Pseudomonas aeruginosa) are reported to induce the dispersion of biofilms, formed by Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Streptococcus pyogenes, Bacillus subtilis, Staphylococcus aureus and the yeast Candida albicans (Boon et al., 2008, Davies & Marques, 2009, Ryan et al., 2008). Moreover, BDSF (Zhang et al., 2011) and DSF (Wang et al., 2004) are reported to inhibit the dimorphic transition of C. albicans at physiologically relevant concentrations and thereby block biofilm formation by this fungus.
Clues to the DSF synthetic route were the sequences of two proteins, RpfF of the plant pathogen Xanthomonas campestris pv. campestris and Bcam0581 of the highly virulent opportunistic human pathogen Burkholderia cenocepacia, thought to be responsible for synthesis of DSF (11-methyl- cis-2 dodecenoic acid) and BDSF (cis-2 dodecenoic acid), respectively (Fig. 1A). The two proteins are functionally interchangeable and show good sequence conservation (Deng et al., 2011). However, more important is that both protein sequences align well with a thoroughly studied enzyme called crotonase, the archetype of a large protein superfamily (PF00378). Moreover, an x-ray crystal structure of RpfF demonstrates that the protein can also be structurally aligned with crotonase (Cheng et al., 2010). Crotonase catalyzes the hydration of a trans-2-acyl-CoA (enoyl-CoA) to the 3-hydroxy species, the last step in the b-oxidative cycle of fatty acid degradation (Fig 1C). (Note that a more proper name for crotonase is enoyl-CoA hydratase, but since enoyl-CoA hydratases are known that are not of the crotonase superfamily, we shall retain the classic nomenclature). Although the crotonase reaction is readily reversed to produce trans-2-acyl-CoAs from 3-hydroxyacyl-CoAs, the stereochemistry of the 3-hydroxyacyl-thioester substrate (3(S)-hydroxyacyl-CoA) is the opposite that of the 3-hydroxy intermediate of fatty acid synthesis (3(R)-hydroxyacyl-ACP) (Hamed et al., 2008). Therefore, invoking a crotonase-like mechanism for DSF (BDSF) synthesis encounters three potential problems, the opposite stereochemistry, the use of ACP thioesters in place of CoA thioesters and the fact that DSF and BDSF are produced as free acid indicating that thioester hydrolysis is required. We report that Bcam0581 has properties that overcome all three potential problems. The protein is active with ACP thioesters and is a bifunctional enzyme that both dehydrates 3-hydroxydodecanoyl-ACP to cis-2-dodecenoyl-ACP and cleaves the acyl thioester bond to yield the final product, BDSF. Although, dehydratase/hydratase activity is characteristic of the crotonase superfamily and two members of this large protein family have demonstrated thioesterase activity, Bcam0581 is the first example of a superfamily member that has both enzymatic activities.
Fig. 1.
Structures, alignments, the crotonase and proposed Bcam 0581 dehydratase reactions. A. Structures of BDSF (cis-2-dodecenoic acid) and DSF (11-methyl-cis-2-dodecenoic acid). B. Relevant partial alignments of rat liver mitochondrial crotonase, Bcam0581 and RpfF. The solid circles denote the catalytic glutamate residues whereas the arrows denote the oxyanion hole residues. C. The crotonase reaction. The physiological crotonase reaction is to the right. D. The proposed Bcam0581 dehydratase reaction. The formal names of BDSF and DSF are 2-(Z)-dodecenoic acid and 11-methyl-2-(Z)-dodecenoic acid, respectively.
Results
Sequence alignments suggest that B. cenocepacia Bcam0581 catalyzes a reaction similar to crotonase
The classical crotonase, that of rat mitochondria, catalyzes the second step of the β-oxidative pathway of fatty acid degradation, hydration of the trans-2 double bonds introduced by the prior enzyme of the pathway (the enzyme name comes from the four carbon substrate, the CoA ester of crotonic acid). However, the crotonase reaction is freely reversible and at equilibrium the hydrated product is only slightly favored (Stern & Del Campillo, 1956). The product of the reverse crotonase reaction, hydration of a trans-2-enoyl-CoA, is a 3(S)-hydroxyacyl-CoA as dictated by the stereochemistry of the reaction (Feng et al., 2002, Hamed et al., 2008). Crotonase also produces 3(R)-hydroxyacyl-CoAs (although this is an exceedingly rare event) (Feng et al., 2002, Hamed et al., 2008). Moreover, mutation of the crotonase active site can markedly increase the 3(R) to 3(S) product ratio (albeit at the expense of enzyme activity) (Feng et al., 2002, Hamed et al., 2008), and thus it seemed likely that B. cenocepacia Bcam0581 could catalyze the reaction depicted in Fig. 1D. The most straightforward precursor to the cis-2 double bond of BDSF would be 3(R)-hydroxydodecanoyl-ACP, an intermediate of saturated fatty acid synthesis. Substitution of ACP for CoA should be possible since both contain a 4’-phosphopantotheine moiety used to form the thioester that polarizes C2 of the acyl chain. We chose to study B. cenocepacia Bcam0581 rather than RpfF because BDSF lacks the terminal methyl group of DSF which simplified acquisition of substrates and standards.
Expression and purification of Bcam0581
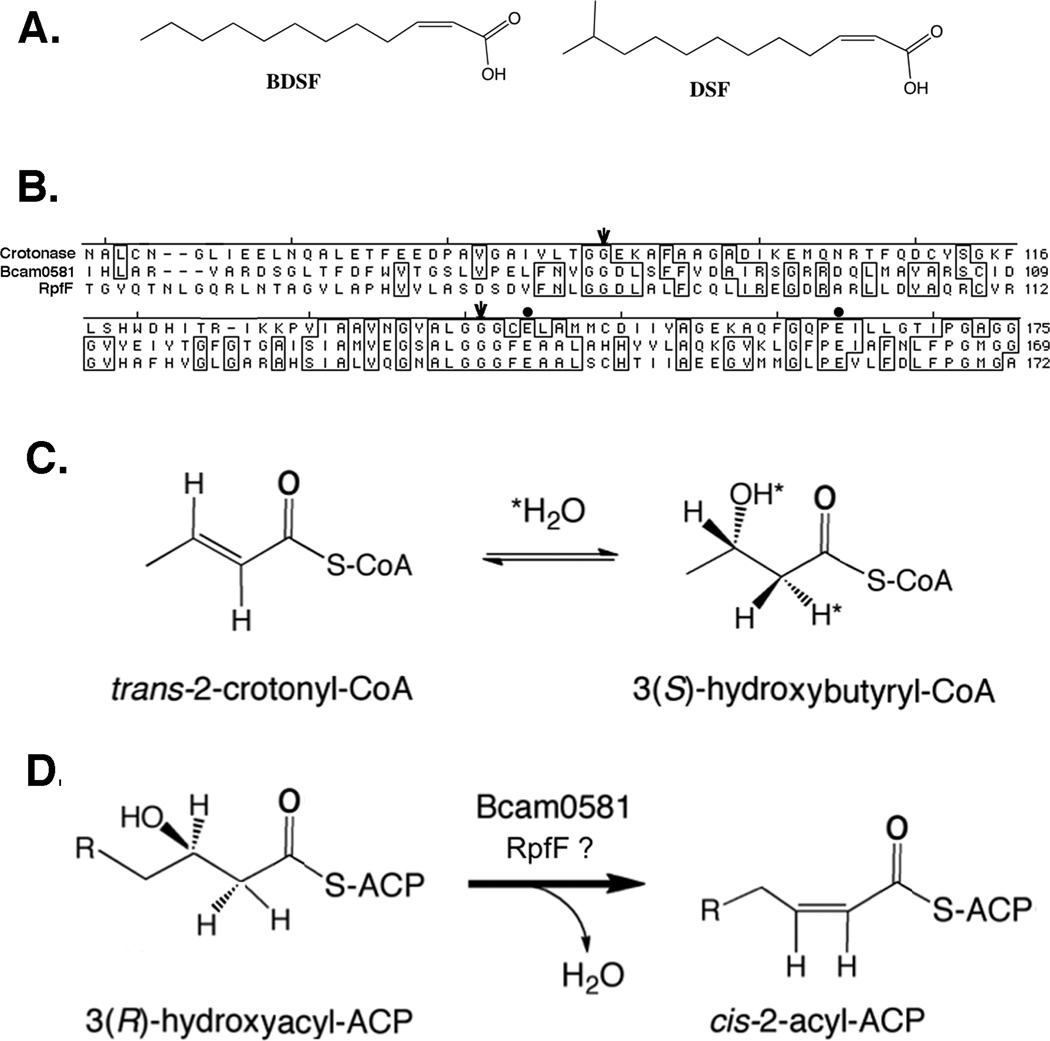
We expressed the protein encoded by Bcam0581 in E. coli. The native Bcam0581 gene contains an abundance of codons rarely used in E. coli and thus a synthetic gene optimized for E. coli expression was used to obtain high-level expression of the protein using the inducible phage T5 promoter vector pQE-2. The hexahistidine-tagged fusion protein was expressed and purified by affinity chromatography followed by size exclusion chromatography (Fig. 2A). Most crotonase superfamily proteins function as trimers or hexamers (Hamed et al., 2008) and RpfF crystallized as a trimer (Cheng et al., 2010). The Bcam0581 elution profile showed that Bcam0581 exists in two states (trimer and monomer with perhaps some dimer) in solution (Fig. 2A). The purified proteins collected from both have an apparent monomeric molecular mass of 36 kDa. Chemical cross-linking indicated that the polymer is probably a trimer, although the dimer may also be present (Fig. 2B).
Fig. 2. Purification and solution structure of recombinant Bcam0581 protein.
A. Size exclusion chromatography of the His-tagged Bcam0581 protein expressed in E. coli. The inset is a 10% SDS-PAGE analysis of the proteins of the peaks 1 and 2 where M, is a protein standard marker (BioRad). The elution peaks of the molecular weight standards are given at the top of the panel. B. Chemical cross-linking analyses of the purified Bcam0581 protein where EGS denotes ethylene glycol bis succinimidylsuccinate (Experimental procedures). The samples were separated by 10% SDS-PAGE where M is the Precision Plus Protein Standard (BioRad).
Bcam0581 has acyl-ACP thioesterase activity
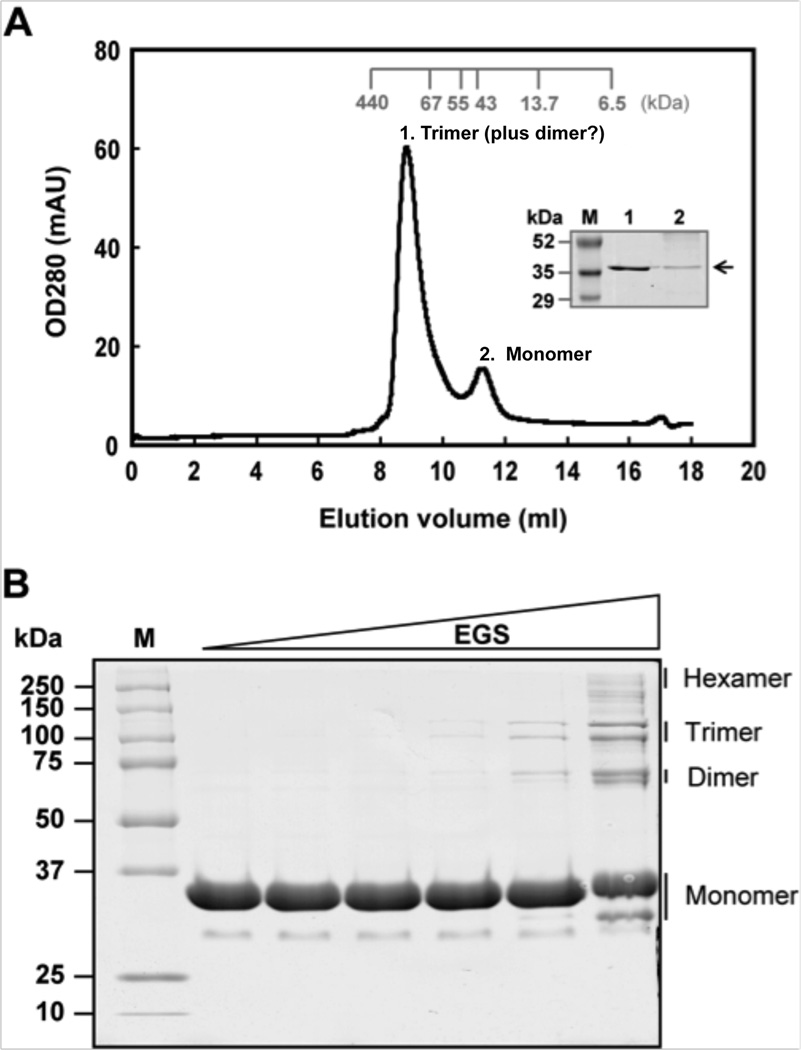
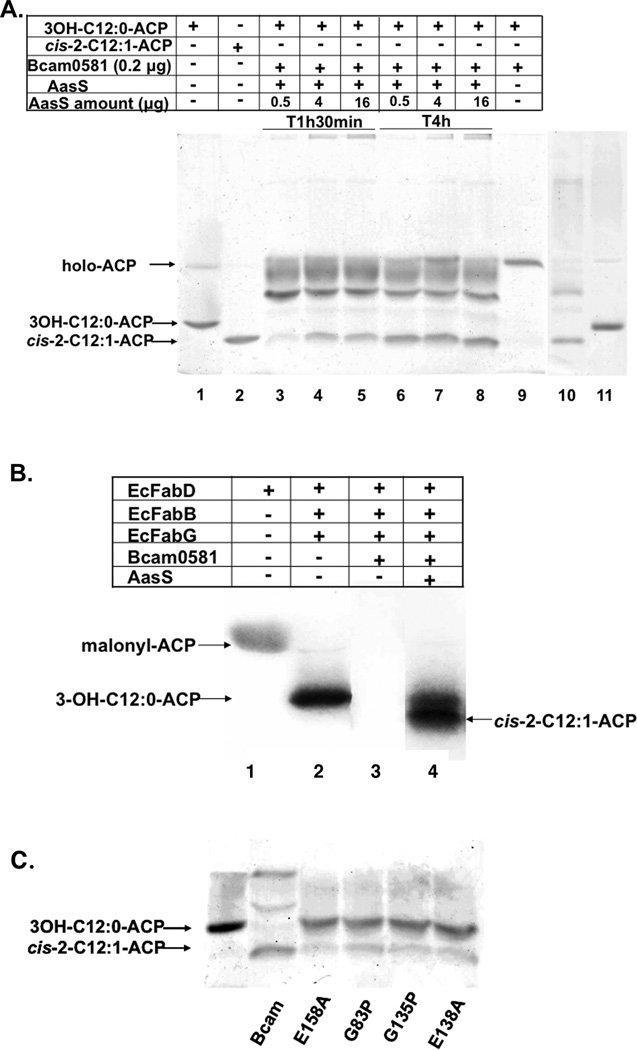
We first assayed purified Bcam0581 for enoyl-CoA hydratase activity in vitro (Experimental procedures) and detected no activity using crotonyl-CoA or the CoA thioesters of the C12 trans-2 and cis-2 isomers as substrates (data not shown). Acyl-ACP substrates made using the Vibrio harveyi acyl-ACP synthetase were then tested. The key substrate, 3-hydroxydodecanoyl-ACP, was shown to be a substrate for E. coli 3-hydroxyacyl-ACP dehydrase (EcFabZ) which produced trans-2-dodecenoyl-ACP (Fig. 3A, lane 3). Following incubation of apparently homogenous Bcam0581with 3-hydroxydodecanoyl-ACP we found that all of the substrate had been converted to holo-ACP, the unacylated species, indicating the presence of thioesterase activity (Fig. 3A, lane 4). This conversion was verified by electrospray mass spectrometry (data not shown). When dodecanoyl-ACP, trans-2- or cis-2-dodecenoyl-ACPs were used as substrates, holo-ACP was also released (Fig. 3A, lanes 6 and 8), indicating that Bcam0581 cleaves acyl-ACP thioester bonds to give free fatty acids plus holo-ACP (Fig. 4). We first supposed that our Bcam0581 preparations were contaminated with an E. coli thioesterase active on acyl-ACPs. However, this seemed improbable because we have used similar purification protocols to purify many enzymes acting on acyl-ACP substrates without interference by thioester hydrolysis. The possibility of contaminating thioesterase activity was eliminated by the finding that mutagenesis of Bcam0581 could eliminate thioesterase activity.
Fig. 3. Bcam0581 catalyzes the hydrolysis of acyl-ACP thioesters.
A. Acyl-ACPs were first prepared as described in Experimental procedures. The reaction mixture for assays of Bcam0581 thioesterase activity contained 0.1 M Tris-HCl (pH7.5), 2 mM β-mercaptoethanol, 0.2 µg Bcam0581 and 20 µM acyl-ACP (3-hydroxydodecanoyl-ACP, trans-2-dodecenoyl-ACP or cis-2-dodecenoyl-ACP). The assay mixtures were incubated at 37°C for 30 min and the reaction products were resolved by conformationally sensitive gel electrophoresis on 18% polyacrylamide gels containing a concentration of urea optimized for the separation (Post-Beittenmiller et al., 1991).
B. Bcam0581thioesterase activity.
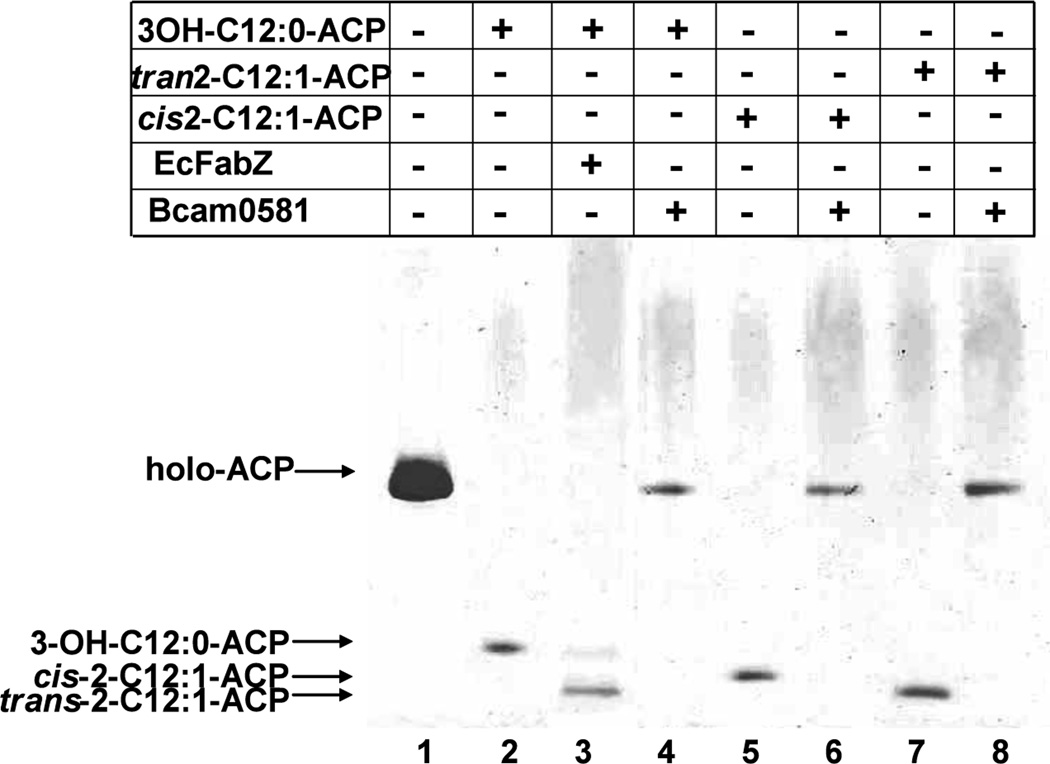
Fig. 4. Mutagenesis indicates that thioesterase activity is intrinsic to Bcam0581 and involves essential residues conserved in crotonase.
Acyl-ACPs were prepared and Bcam0581 variants were purified as described in Experimental procedures. Bcam0581 thioesterase activity assays were performed using (Panel A) 3-hydroxydodecanoyl-ACP (3-OH-C12:0-ACP), (Panel B) cis-2-dodecenoyl-ACP (cis-2-C12:1-ACP), (Panel C) trans-2-dodecenoyl-ACP (trans-2-C12:1-ACP) and (panel D) dodecanoyl-ACP (C12:0-ACP) as substrates. The reactions were initiated by the addition of 0.2 µg of wild type Bcam0581 or one of the the mutant proteins carrying amino acid substitutions E158A, E158Q, G83P, G135P, E138A or E138Q. In panels A and B the reaction mixtures were incubated at 37°C for 10 min whereas panels C and D are 30 min incubations. The reaction products were resolved by conformationally sensitive gel electrophoresis as in Fig. 3.
Active site residues required for Bcam0581 thioesterase activity
In rat crotonase two conserved glutamate residues (E144 and E164) are required for catalysis (Hofstein et al., 1999). In the crotonase hydration reaction (Fig. 1C) E144 activates a water molecule for nucleophilic attack at C3 whereas E164 protonates C2 of the unsaturated fatty acid (Hamed et al., 2008). Sequence alignments of Bcam0581 with crotonase indicated that glutamate residues E138 and E158 of Bcam0581 that correspond to E144 and E164 of the crotonase (Fig. 1B). To test if Bcam0581 thioesterase activity required these residues, we substituted either alanine or glutamine for residues E138 and E158. The four mutant proteins were purified and assayed for thioesterase activity. All failed to cleave either trans-2-dodecenoyl-ACP or dodecanoyl-ACP although the mutant enzymes retained traces of activity toward 3-hydroxydodecanoyl-ACP and cis-2-dodecenoyl-ACP (Fig. 4).
The common theme of reactions catalyzed by crotonase family enzymes is the stabilization of enolate anion intermediates of acyl-CoA substrates by two structurally conserved peptidic NH groups that form an “oxyanion hole” (Hamed et al., 2008). Detection of such conserved groups is difficult because all residues except proline have peptidic nitrogens. The crotonase oxyanion hole is formed by the peptidic nitrogens of residues A98 and G141 both of which are essential for activity (Feng et al., 2002). The Bcam0581 analogue, G135, of crotonase residue G141 was readily identified because it is located three residues before the E138 catalytic residue within a highly conserved sequence. Alignments (Fig. 1B) suggested that Bcam0581 residue A90 is the analogue of crotonase residue A98. However, sequence conservation between crotonase and the bacterial proteins is poor within the crotonase A98 region and the analogous residue in RpfF (L94) is located far from the active site (Cheng et al., 2010). Indeed, the RpfF structure argued that Bcam0581 residue G83 would provide the second oxyanion hole peptidic NH group. The hypothesis that G83 and G135 formed the oxyanion hole was tested by substitution of proline for each of the glycine residues. Both substitutions resulted in a complete loss of thioesterase activity in vitro (Fig. 4).
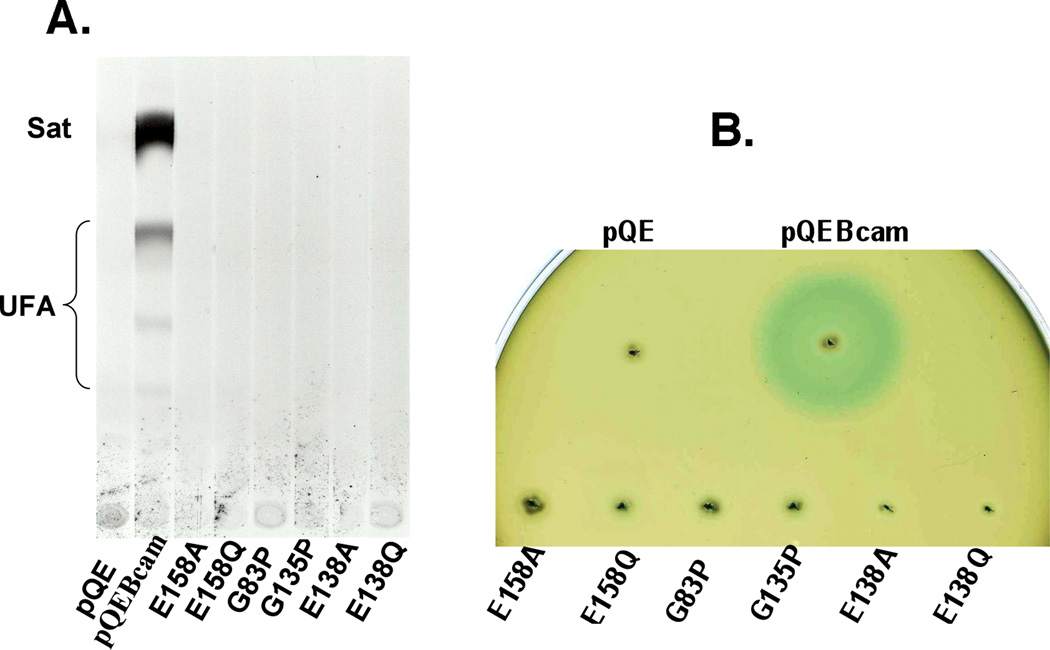
Many bacterial and plant thioesterases had been identified by their expression in fatty acid β-oxidation deficient E. coli strains (Voelker & Davies, 1994, Cho & Cronan, 1995, Feng & Cronan, 2009). The liberated fatty acids are secreted to the medium and the composition of the secreted acids accurately reflects the substrate specificity of the thioesterase. To test if Bcam0581 had thioesterase activity in vivo, individual colonies containing plasmids encoding the wild type or mutant Bcam0581proteins were grown in the presence of [14C]acetate and induced. The medium was collected followed by extraction of the fatty acids. After conversion of the fatty acids their methyl esters they were analyzed by argentation thin layer chromatography (Fig. 5A). We found that the level of de novo synthesized fatty acids including saturated and unsaturated fatty acids secreted to the culture medium was much higher in the wild type Bcam0581-expressing strain than in the strain carrying the empty vector plasmid. In contrast expression of any of the mutant Bcam0581 proteins resulted in no detectable accumulation of free fatty acids released to the medium (Fig. 5A). These data indicate that the thioesterase activity of Bcam0581 is nonspecific and is not an artifact of our in vitro system.
Fig. 5. Functional characterization of Bcam0581 and its mutant derivatives in E. coli.
Panel A. Argentation thin-layer chromatographic analysis of [1-14C] acetate-labeled esters present in the medium from E. coli strain K19 carrying either pBHK06 (Bcam0581m) or its mutant derivatives as given. Cultures of plasmid-containing strains were induced with IPTG. The fatty acid methyl esters were obtained from the medium as described in Experimental procedures. The methyl esters were then separated by argentation thin layer chromatography followed by autoradiography. The migration positions of the fatty acid species are shown. Sat, saturated fatty acid esters; UFA, unsaturated fatty acid esters.
Panel B. BDSF bioassay of E. coli DH5α expressing Bcam0581 or the mutant derivatives. The formation of a blue halo due to hydrolysis of 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid around the site of inoculation indicates the presence of DSF-like activity. pQE is the vector (pQE-2) control whereas pQEBcam expressed wild type Bcam0581. The mutant proteins were also expressed in pQE-2.
BDSF is produced upon expression of Bcam0581 in E. coli
The BDSF bioassay takes advantage of the fact that BDSF can substitute for DSF in the X. campestris signaling pathway (the two fatty acids differ only by the 11-methyl group of DSF). The bioassay is done with a X. campestris strain (rpfF::Tn5lac) that lacks the ability to produce DSF and carries a plasmid-borne fusion of the promoter and ribosome binding sites of the DSF-inducible gene encoding the major X. campestris endoglucanase (engXCA) to the coding sequence for E. coli b-glucuronidase (Slater et al., 2000). Addition of either DSF or BDSF results in greatly increased b-glucuronidase activity that is readily assayed colorimetrically by cleavage of 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid.
E. coli strain DH5α carrying pBHK06 which encodes wild type Bcam0581 gave a very large blue halo in the bioassay due to cleavage of 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (Fig. 5B) indicating that as recently reported by others (Deng et al., 2011, Deng et al., 2010), Bcam0581 confers the ability to produce BDSF on E. coli. In contrast derivatives of E. coli strain DH5α expressing the mutant Bcam0581 proteins failed to give detectable halos (Fig. 5B) indicating that the two oxyanion hole glycine residues and both catalytic glutamate residues of Bcam0581 all play essential roles in BDSF biosynthesis. Similar mutagenesis of the analogous glutamate residues of RpfF were reported to block DSF production in vivo (Cheng et al., 2010).
The observed production of BDSF in E. coli raised the possibility that this bacterium could somehow produce cis-2-dodecenoyl-ACP and that Bcam0581 functioned only as a broad substrate specificity thioesterase allowing release of cis-2-dodecenoic acid (BDSF) to the culture medium. If so, then other thioesterases of broad substrate specificity should have the same effect. This was not the case. Derivatives of E. coli strain DH5α carrying plasmids that expressed E. coli thioesterase I lacking its export sequence (Cho & Cronan, 1995), Cinnamonum camphorum thioesterase (Feng & Cronan, 2009) or a mutant C. camphorum thioesterase of altered specificity (Feng & Cronan, 2009) were all negative for BDSF production in the bioassay (data not shown). These results lead us to expect that Bcam0581 must have another enzymatic activity required for BDSF synthesis that was hidden by its thioesterase activity.
Bcam0581 has dehydratase activity
As discussed above based on the mechanism of the crotonase reaction, we thought it likely that 3-hydroxydodecanoyl-ACP was the fatty acid synthetic intermediate used to make cis-2-dodecenoic acid (BDSF). The crotonase reaction mechanism (Hamed et al., 2008) also argued that the thioester bond would be required for dehydratase activity and thus thioester cleavage would abort the dehydratase reaction. Therefore in order to counteract the Bcam0581 thioesterase activity we added Vibrio harveyi acyl-ACP synthetase (AasS), ATP and Mg2+ to our in vitro reactions. The AasS reaction was expected to recycle the 3-hydroxydodecanoic acid released by the Bcam0581 thioesterase activity back to the ACP thioester in order to allow it another chance to be used as a dehydratase substrate.
Indeed, upon addition of AasS to the dehydratase assays a new product was formed, the level of which increased as the AasS concentration was increased (Fig. 6A). The migration rate of the new band in gel electrophoresis was essentially identical to that of the cis-2-dodecenoyl-ACP standard. Note that addition of AasS at the end of the reaction (after inactivation of Bcam0581) failed to give detectable cis-2-dodecenoyl-ACP indicating that the role of AasS is to facilitate the dehydratase activity rather than to convert free cis-2-dodecenic acid to its ACP thioester and thereby allows its detection of the gels (Fig. 6A lanes 10 & 11).
Fig. 6.
AasS allows production of cis-2-C12:1-ACP by Bcam0581by reversal of thioesterase action. A. in vitro Bcam0581 reaction with the addition of AasS using 3-hydroxydodecanoyl-ACP (3-OH-C12:0-ACP) as substrate. Lanes 3–8 are reactions in which Bcam0581 and AasS were added together. The reaction mixture contained 0.1 M Tris-HCl (pH7.5), 2 mM β-mercaptoethanol, 20 µM 3-hydroxydodecanoyl-ACP, 10 mM ATP, 10 mM MgCl2 and 0.2 µg purified Bcam0581 and various amounts of AasS in a final volume of 30 µl. The assay mixtures were incubated at 37°C for either 90 min or 4 h. Lane 9 is a reaction as described above except with Bcam0581 alone (AasS was not added) and incubation at 37°C for 10 min. Lanes 10 and 11 are boiled samples of the reactions of Lanes 8 and 9, respectively, in which additional AasS was added to convert any free fatty acid to acyl-ACPs. These assay mixtures were incubated at 37°C for additional 2 h after addition of 4 µg AasS, 10 mM ATP and 10 mM MgCl2. Boiling precipitated the high molecular weight proteins whereas the ACP species remained soluble.
B. Reconstruction of fatty acid synthesis in vitro showing the new band is derived from 3-hydroxydodecanoyl-ACP. Lanes 2–4, 14C-labeled 3-hydroxydodecanoyl-ACP was synthesized in 20 min reactions as described in Experimental procedures. Then either Bcam0581 (0.2 µg) alone or Bcam0581 (0.2 µg) plus AasS (4 µg) were added to the reaction mixtures of lanes 3 and 4, respectively, followed by incubation at 37°C for additional 30 min. Note that the reaction mixture in lane 4 also contains 10 mM ATP and 10 mM MgCl2. The reaction products were analyzed as in Figs. 3 and 4. Note that two irrelevant lanes were excised from the middle of this radiogram to simplify the figure.
C. Mutagenesis of the Bcam0581 active site residues impairs dehydratase activity in vitro.
In vitro Bcam0581 or its variants (E158A, G83P, G135P and E138A) reactions with 3-hydroxydodecanoyl-ACP (3-OH-C12:0-ACP) as substrate. All reactions contained 0.2 µg of a Bcam0581 protein and 4 µg of AasS and were performed as in A.
To confirm that this band was derived from 3-hydroxydodecanoyl-ACP, we used a system in which in vitro E. coli fatty acid synthesis was reconstructed from purified proteins. The assay employed FabD to generate [2-14C]malonyl-ACP from [2-14C]malonyl-CoA and holo-ACP (Fig. 6B, lane 1). Incubation of decanoyl-ACP, NADPH, NADH, [2-14C]malonyl-CoA, holo-ACP, FabB, FabG and FabD resulted in the formation of [14C]-labeled 3-hydroxydodecanoyl-ACP (Fig. 6B, lane 2). The addition of Bcam0581 resulted in the loss of acyl-ACPs (Fig. 6B, lane 3) due to the Bcam0581 thioesterase activity. However, simultaneous addition of Bcam0581 and AasS resulted in the restoration of the [14C]-labeled 3-hydroxydodecanoyl-ACP plus the putative cis-2-dodecenoyl-ACP product labeled with [14C] (Fig. 6B, lane 4,). Note that E. coli 3-ketoacyl-ACP reductase (FabG) produces the R-enantiomer of the 3-hydroxyacyl moiety (Majerus et al., 1965, Toomey & Wakil, 1966) and thus the activity observed with the reconstructed E. coli system indicates that the Bcam0581 substrate is 3(R)-hydroxydodecanoyl-ACP (as depicted in Fig. 1D). The mutant Bcam0581 protein previously shown to have impaired thioesterase activities also had impaired dehydratase activities indicating that the same active site catalyzed both reactions (Fig. 6C) as expected from prior studies of crotonase superfamily members.
Identification of the Bcam0581 dehydration product
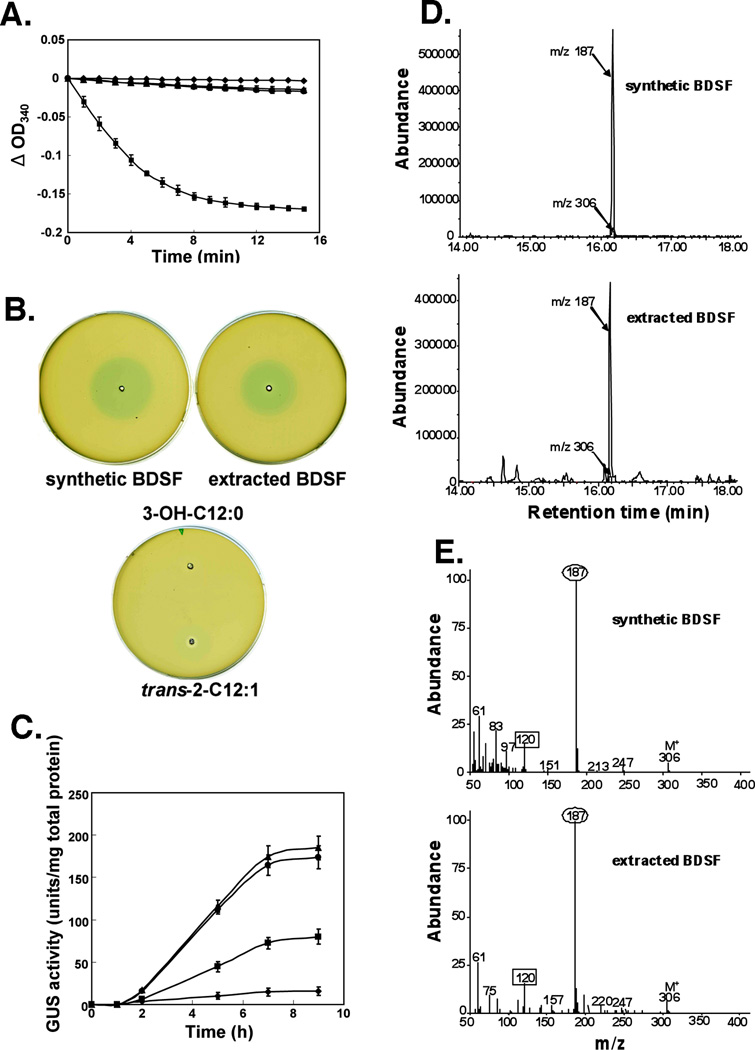
The product formed in the experiments of Fig. 6 was confirmed to be cis-2-dodecenoyl-ACP by several tests. First we showed that, like authentic cis-2-dodecenoyl-ACP, the product was not a substrate for the E. coli FabI enoyl-ACP reductase indicating that it was not trans-2-dodecenoyl-ACP (Fig.7A). We also analyzed the free acid released upon addition of hexokinase plus glucose (an “ATP-trap”) to consume ATP and thereby inactivate AasS such that the Bcam0581 thioesterase activity was no longer counteracted and any-dodecenoyl-ACP would be hydrolyzed to the free acid. The resulting product was bioassayed and showed a response identical to that of authentic cis-2-dodecenoic acid whereas trans-2-dodecenoic and 3-hydroxydodecanoic acids had no activity (Fig. 7B & C). Another portion of the free acid was converted to the methyl ester and then to the dimethyldisulfide (DMDS) adduct to allow determination of the double bond position by gas chromatography-mass spectroscopy. The extracted product displayed a major peak with a chromatographic retention time of 16.18 min, a value identical to that of synthetic BDSF (Fig. 7D). The mass spectrum of the DMDS adduct showed weak ions [M]+ at m/z 306, corresponding to the theoretical mass of molecular ions of the DMDS adduct of a C12 monounsaturated fatty acid (Fig. 7E). Upon cleavage in the mass spectrometer two major ions were formed by bond scission between the methylthio-substituted (CH3S) carbons located at the site of the double bond. The strong ions at m/z 187 and m/z 120 indicate that the position of the double bond was at Δ2 of a dodecenoic acid (Fig. 7E). These data, the bioassay results and the lack of reactivity of the ACP thioester with FabI indicate that double bond configuration must be cis. Hence, the Bcam0581 product is cis-2-dodecenoic acid (BDSF).
Fig. 7. Identification of the reaction products formed in vitro.
A. NADH oxidation activity of EcFabI using different substrates. NADH oxidation was monitored at 340 nm for the EcFabI reaction using trans-2-dodecenoyl-ACP(■), synthetic cis-2-dodecenoyl-ACP (●) or purified cis-2-dodecenoyl-ACP (○) from in vitro reactions run with simultaneous addition of Bcam0581 and AasS as described in Experimental procedures. Note that the ● and ○ symbols largely overlap. The □ symbol denotes background without addition of substrates. The controls without substrate showed no significant change in absorbance. The curves have been adjusted to the same zero time absorbance in the figure. The data are the means± standard error of the mean of three independent assays.
B. BDSF bioassay. Each well contains 10 µl of BDSF (either extracted from reaction mixtures or synthetic), trans-2-dodecenoic acid or 3-hydroxydodecanoic acid at a concentration of 60 µM. Synthetic BDSF was the positive control.
C. BDSF induction of endoglucanase expression in the DSF biosensor strain, Xcc8523 (pL6engGUS). The biosensor strain in which E. coli gusA encoding β-D-glucuronidase is fused to the engXCA promoter was cultured and exposed to BDSF samples. Samples of the cultures were collected at different time points after the addition (final concentration 60 µM) of trans-2-dodecenoic acid (■), synthetic BDSF (●) or BDSF extracted (○) from in vitro reactions. The in vitro reactions received simultaneous additions of Bcam0581 and AasS. □ denotes background without any additions. The β-D-glucuronidase activities were determined as described in Experimental procedures. The data are the means± standard error of the mean of three independent assays.
D. GC-MS chromatogram from analyses of the fatty acid extracted from a reaction mixture and synthetic BDSF. The fatty acids were derivatized first to their methyl esters and then to their dimethyl disulfide adducts which were analyzed by gas chromatography-mass spectroscopy. The elution profiles of DMDS adducts of the methyl esters show mass chromatogram peaks of m/z 187 and m/z 306.
E. Mass spectroscopy of the cleavage products of the dimethyl disulfide adducts of fatty acid methyl esters either extracted from a reaction mixture or synthetic BDSF. The unsaturated esters gave a cleavage fragment of m/z 187 (numbers in ovals) corresponding to the methyl end of the molecule plus a second fragment m/z 120 (numbers in squares) corresponding to the ester end of the molecule (expected m/z 119).
Bioinformatic Analysis of Bcam0581
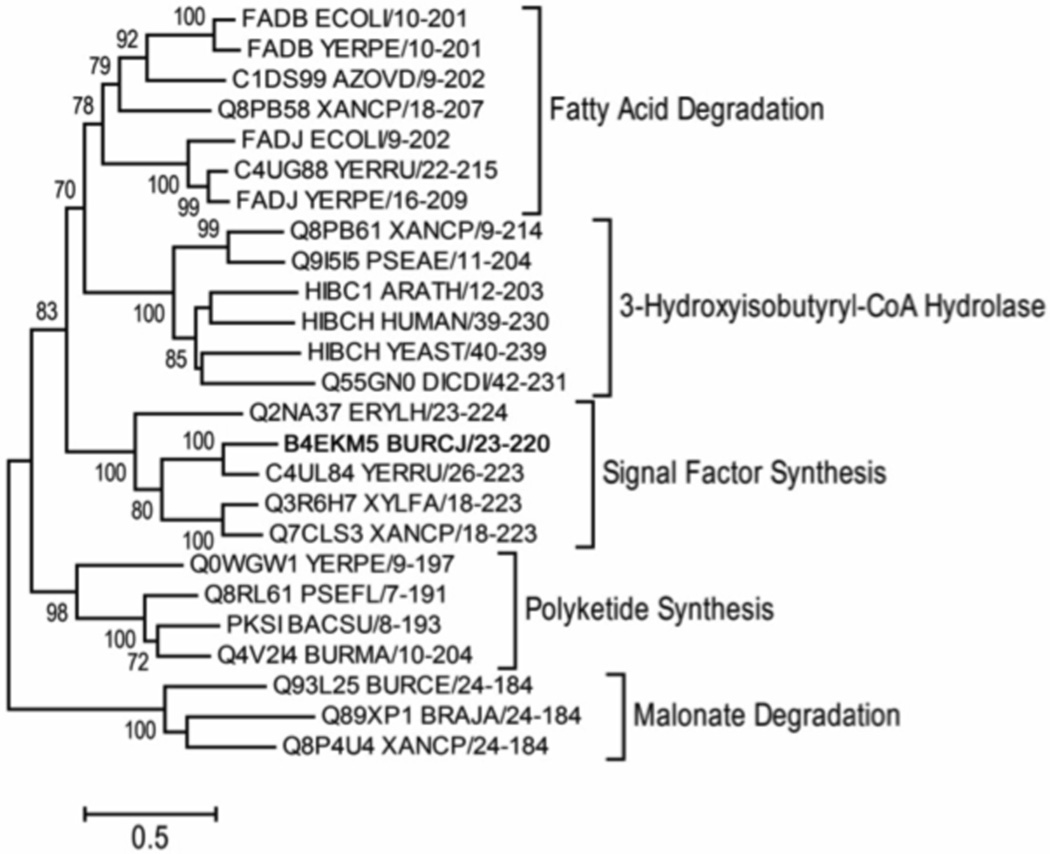
The phylogeny of Bcam0581 was determined in relation to other members of the crotonase family (PF00378), of which it is a member (Fig. 8). Members from the related malonyl-CoA decarboxylase family (PF06833) were also included as an out group in order to infer the ancestry of Bcam0581. Phylogenetic analysis revealed that Bcam0581 homologues form a clade distinct from other groups of the crotonase family (Fig. 8). A deeply branching clade of the crotonase superfamily contains proteins involved in biosynthesis of various polyketides. These systems use acyl carrier protein domains (thiolation domains), which are highly similar to the acyl carrier proteins of fatty acid biosynthesis. Interestingly, Bcam0581 homologues are found in many bacterial plant pathogens such as Xylella fastidiosa and Xanthomonas oryzae pv. oryzae, other opportunistic human pathogens such as Stenotrophomonas maltophilia and other members of the Burkholderia cepacia complex, although not in B. pseudomallei.
Fig. 8. Phylogeny of Bcam0581.
The minimum evolution tree of selected Pfam00378 protein sequences with bootstrap percentage confidence values shown for each branch is given. Phylogenetic analyses were conducted as described in Experimental procedures. The scale bar corresponds to a 50% difference in compared residues, on average, per branch length. Protein sequences from the malonyl-CoA decarboxylase superfamily, Pfam06833, were used as a related out-group. Each branch is labeled with the Uniprot sequence identifier followed by the species identifier. “B4EKM5 BURCJ” corresponds to Bcam0581 and is in bold.
Discussion
We report the first in vitro synthesis of an autoinducer of the DSF family and show that the reaction is catalyzed by a single protein having both dehydratase and thioesterase activities. This is the first example of bifunctional crotonase homologue and hence a protein annotated as an enzyme of fatty acid degradation plays a role in fatty acid synthesis. Moreover, our results indicate that Bcam0581 uses 3-hydroxydodecanoyl-ACP as a precursor for BDSF biosynthesis rather than the CoA thioester suggested as the RpfF substrate (Cheng et al., 2010). Thus, the BDSF biosynthesis pathway branches from the classic fatty acid biosynthesis pathway by “tapping off” the 3-hydroxydodecanoyl-ACP intermediate (Fig. 9) in a process analogous to that which provides the 3-hydroxyacyl chains of lipid A (Raetz et al., 2007). It seems clear that the BDSF 3-hydroxydodecanoyl precursor comes from the fatty acid synthetic pathway rather than from fatty acid degradation. This is due to the stereochemical considerations and data discussed above as well as recent reports that various Burkholderia cepacia complex and Xanthomonas species produce not only BDSF (cis-2-dodecenoic acid) but also a fatty acid containing two cis double bonds, the one expected at Δ2 plus a second at Δ5 (Deng et al., 2011). Cis-5-dodecenoyl-ACP is an intermediate in the synthesis of the C16 and C18 cis acids that are the major unsaturated fatty acid species (palmitoleic and cis-vaccenic acid) found in the membrane phospholipids of these bacteria (Krejci & Kroppenstedt, 2006) and hence both the saturated and unsaturated fatty acid synthetic pathways can provide Bcam0581 with twelve carbon 3-hydroxyacyl-ACP intermediates. Surprisingly, the relative use of the unsaturated pathway intermediate is reported not to be a property of the Bcam0581 homologue but rather of the genetic background of the Burkholderia strain (Deng et al., 2010).
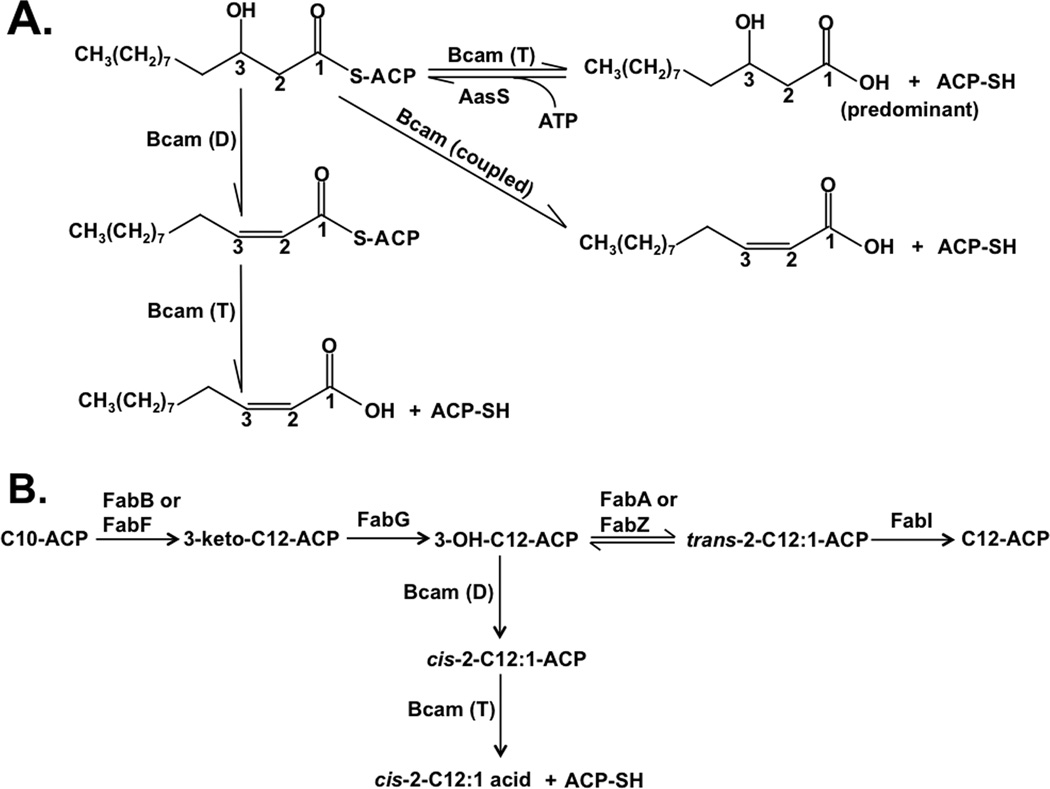
Fig. 9. Current models of BDSF biosynthesis in B. cenocepacia.
BDSF biosynthesis is catalyzed by Bcam0581. A. In the presence of acyl-ACP synthetase (AasS) Bcam0581 functions as a dehydratase, Bcam (D), to convert 3-hydroxydodecanoyl-ACP to cis-2-dodecenoyl-ACP and then as a thioesterase, Bcam(T) to release free BDSF. Although we did not detect cis-2-C12:1 in the absence of AasS, a coupled reaction occurred in vivo in E. coli.
B. The intersection of BDSF synthesis and fatty acid synthesis. Bcam0581 taps off a portion of the 3-hydroxydodecanoyl-ACP intermediate of long chain fatty acid synthesis
Our finding that Bcam0581 has thioesterase activity was unexpected, but not without precedent. The first example of a crotonase superfamily thioesterase was 3-hydroxyisobutyryl-CoA hydrolase, an enzyme of amino acid degradation (Wong & Gerlt, 2003). More recently a second example was reported, CarB, a protein involved in the synthesis of an unusual amino acid carboxymethylproline, (Batchelar et al., 2008). Although the mechanisms of thioester hydrolysis proposed for the two enzymes differ in that an anhydride intermediate was proposed for the hydrolase (Wong & Gerlt, 2003) whereas direct attack of an activated water molecule on the thioester was proposed for CarB (Batchelar et al., 2008) the differing results may reflect differing states of the art of mass spectroscopy. CarB like Bcam0581 has activities other than thioester hydrolysis in that it also catalyzes decarboxylation and C-C bond formation (Batchelar et al., 2008).
Bcam0581 shares about 37% identity with DSF synthase RpfF and can replace RpfF in X. campestris pv. campestris (Boon et al., 2008). Our mutagenesis studies identified the active site residues of Bcam0581 that are strictly conserved in RpfF. Given this conservation we expressed hexahistidine-tagged RpfF in E. coli, purified the protein and assayed its enzymatic activities. RpfF behaved much like Bcam0581 in these assays. RpfF had thioesterase activity of broad substrate specificity and converted 3-hydroxydodecanoyl-ACP to cis-2-dodecenoyl-ACP in the presence of AasS (data not shown). Although RpfF functions with 3-hydroxydodecanoyl-ACP in vitro, the substrate in its cognate bacteria must be 11-methyl-3-hydroxydodecanoyl-ACP. The 11-methyl substitution is presumably derived from leucine via the classical branched chain fatty acid synthetic pathway.
Although we have observed thioesterase activity uncoupled from dehydratase activity, it seems likely that our observations are artifacts perhaps due to use of an in vitro system and use of E. coli as a surrogate in vivo system. We believe it probable that dehydration and thioester hydrolysis are coupled when Bcam0581 functions in B. cenocepacia. Coupling would make the dehydration reaction irreversible (due to loss of the thioester which imparts acidity to the substrate a-protons) and avoid the wasteful cleavage of acyl-ACPs destined for membrane lipid synthesis. The most parsimonious coupling mechanism would use the activated water molecule liberated by the dehydration reaction to cleave the thioester bond. If so, uncoupled thioesterase activity would be avoided by preventing water molecules from entering the active site from solution. In this scenario B. cenocepacia would contain a protein that interacts with Bcam0581 to prevent water from entering the active site from solution. In the RpfF system a protein called RpfC binds RpfF and thereby inhibits DSF synthesis (Deng et al., 2011). This inhibition is thought to be reversed by autophosphorylation of RpfC (Deng et al., 2011). Although a similar mechanism may exist in B. cenocepacia, the sequenced Burkholderia genomes contain no recognizable RpfC homologues. Another more remote possibility is that the “plug” protein might be the B. cenocepacia ACP. Although, the sequence of B. cenocepacia ACP is 73% identical to that of E. coli ACP, there may be subtle structural features of the native ACP that wall the Bcam0581 active site off from solution water molecules. In this scenario binding of the 3-hydroxydodecanoyl thioester of B. cenocepacia ACP would displace any water molecules from the Bcam0581 active site and (unlike E. coli ACP) the native ACP would block solution water from entering the active site such that the coupled dehydration and thioesterase reactions could proceed. Upon cleavage of the thioester bond with the dehydratase-generated activated water molecule, the ACP moiety would dissociate from Bcam0581 allowing release of BDSF as the free acid for release to the culture medium. However, this mechanism would not prevent hydrolysis of acyl-ACPs other than 3-hydroxydodecanoyl-ACP. A third possibility is a protein that would specifically block access of acyl-ACPs other than the dehydration substrate to the Bcam0581 active site. Finally, we propose the gene name dfsA (diffusible factor synthase A) for the Bcam0581 locus. It should be noted that members of the Burkholderia cepacia complex are opportunistic human pathogens that produce infections that are difficult to treat due to the multiple antibiotic resistances of the bacterium whereas Xanthomonas species infect many plants of commercial value. Therefore, compounds that inhibit DSF (BDSF) synthesis or utilization could be effective antimicrobials.
Experimental procedures
Materials
Sodium [1-14C]acetate and [2-14C]malonyl-CoA were purchased from Moravek (Brea, CA). The cis-2-dodecenoic and trans-2-dodecenoic acids were synthesized by Advanced Synthesis Technologies (San Ysidro, CA). Racemic 3-hydroxydodecanoic acid was purchased from Matreya, LLC. Other fatty acids, malonyl-CoA, NADH, NADPH, antibiotics and most other chemicals were purchased from Sigma. Qiagen provided plasmid isolation and PCR product purification kits plus Fast T4 DNA ligase and vector pQE-2. Oligonucleotide primers were synthesized by Integrated DNA Technologies and DNA sequencing was done by AGCT, Inc. Invitrogen provided Ni2+-agarose columns. The Bcam0581m gene of Burkholderia cenocepacia J2315 optimized for expression in E. coli was synthesized by Epoch Biolabs (Missouri City, TX).
Bacterial strains, plasmids and growth conditions
E. coli strains and plasmids used in this study are listed in Table 1. E. coli was grown at 37°C in Luria–Bertani (LB) medium (tryptone, 10 g l−1; yeast extract, 5 g l−1; NaCl, 10 g l−1; pH 7.0). X. campestris was grown at 30°C in NYG or YEB medium (Deng et al., 2010). When required, antibiotics were added as follows (in µg ml−1): sodium ampicillin, 100; kanamycin sulfate, 50; tetracycline HCl, 15 and chloramphenicol, 20 µg. Arabinose was added to a final concentration of 0.02%. Bacterial growth was determined by measuring optical density at 600 nm.
Table 1.
Bacterial strains, plasmid and primers used in this study
| Strain, plasmid or primer |
Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DH5α | Δ(argF–lac)169 φ80dlacZ58(M15) ΔphoA8, glnV44deoR481 gyrA96 recA1 endA1 hsdR17 | Lab stock |
| E. coli K19 | fadE62 | Coli Genetic Stock Center |
| Xanthomonas campestris pv. campestris 8523 | rpfF::Tn5lac | (Tang et al., 1991) |
| Plasmids | ||
| pQE-2 | Ampr, T5 promoter-based expression vector | Qiagen |
| pET-28b | Kmr, T7 promoter-based expression vector | Novagen |
| pBSKCronan871 | Ampr, pBluescript II SK(+) carrying B. cenocepacia Bcam0581m | Epoch Biolabs |
| pBHK06 | PCR-amplified Bcam0581m of pBSKCronan871 inserted between the NdeI and HindIII sites of pQE-2 | This work |
| pBHK21 | Ampr, Bcam0581 E158A of pBHK06 | This work |
| pBHK22 | Ampr, Bcam0581 E158Q of pBHK06 | This work |
| pBHK23 | Ampr, Bcam0581 G83P of pBHK06 | This work |
| pBHK24 | Ampr, Bcam0581 G135P of pBHK06 | This work |
| pBHK126 | Ampr, Bcam0581 E138A of pBHK06 | This work |
| pBHK127 | Ampr, Bcam0581 E138Q of pBHK06 | This work |
| pL6engGUS | Tetr, engXCA::gusA fusion in pLAFR6 | (Slater et al., 2000) |
| pCY792 | Ampr, C. camphorum thioesterase in pQE-2 | (Feng & Cronan, 2009) |
| pCY793 | Ampr, Cc-short thioesterase in pQE-2 | (Feng & Cronan, 2009) |
| pHC122 | Ampr, pBAD22 carrying E. coli ‘tesA | (Cho & Cronan, 1995) |
| Primers (5’-3’) | ||
| Bcam0581-f | AGTCATATGCAGCTGCAGAGTCATCCT | |
| Bcam0581-r | CTGAAGCTTAAACAGTACGCAGCTTGC | |
| G83P-f | CTGGTCCCAGAATTATTTAATGTTGGACCCGATTTAAGTTTCTTCGTG | |
| G83P-r | CACGAAGAAACTTAAATCGGGTCCAACATTAAATAATTCTGGGACCAG | |
| G135P-f | GGATCAGCTCTGGGGCCTGGTTTTGAGGCAGC | |
| G135P-r | GCTGCCTCAAAACCAGGCCCCAGAGCTGATCC | |
| E158A-f | GTGTAAAACTGGGGTTTCCGGCAATTGCGTTCAATTTATTTCC | |
| E158A-r | GGAAATAAATTGAACGCAATTGCCGGAAACCCCAGTTTTACAC | |
| E158Q-f | GGTGTAAAACTGGGGTTTCCGCAGATTGCGTTCAATTTATTTCCG | |
| E158Q-r | CGGAAATAAATTGAACGCAATCTGCGGAAACCCCAGTTTTACACC | |
| E138A-f | CTGGGGGGTGGTTTTGCGGCAGCATTAGC | |
| E138A-r | GCTAATGCTGCCGCAAAACCACCCCCCAG | |
| E138Q-f | AGCTCTGGGGGGTGGTTTTCAGGCAGCATT | |
| E138Q-r | AATGCTGCCTGAAAACCACCCCCCAGAGCT | |
Protein expression and purification
The gene Bcam0581m was amplified from pBSKCronan871 (Epoch Biolabs) with primers containing designed restriction sites (an NdeI site overlapping the initiation codon and a HindIII site downstream of the termination codon) was ligated into the vector pQE-2 cut with NdeI and HindIII to produce plasmid pBHK06. Bcam0581 with a vector-encoded N-terminal hexahistidine (His)-tag was expressed in E. coli DH5α grown at 30°C in LB medium containing 500 mM glycylglycine. At an OD600 of 1.0, the cultures were induced with 0.15 mM isopropyl-β-D-thio-D-galactoside (IPTG) and grown at 25°C for an additional 12 h prior to harvest. The cells were collected, resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 15 mM imidazole, 1 mM dithiothreitol, pH8.0), lysed by French pressure cell treatment and centrifuged. The clarified bacterial supernatant was loaded onto a nickel-ion affinity column (Qiagen). The column was washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 40 mM imidazole, 1 mM dithiothreitol, pH 8.0) to remove contaminant proteins and the His-tagged Bcam0581 protein was eluted in elution buffer containing 200 mM imidazole. The protein was concentrated by ultrafiltration (10-kDa cutoff) and exchanged into sodium phosphate buffer (50 mM NaH2PO4, 150 mM NaCl, 1 mM dithiothreitol, pH 8.0). The purity of the samples was monitored by SDS-PAGE. The E. coli FabD, FabB, FabG, FabZ, FabI, holo-ACP and Vibro harveyi AasS were purified by their hexahistidine tags described previously (Cronan & Thomas, 2009, Heath & Rock, 1995, Zhu et al., 2010).
Solution structure of Bcam0581
The solution structure of Bcam0581 was analyzed by size exclusion chromatography on a Superdex 75 10/300 GL column (GE Healthcare) using an AKTA purifier10 at 0.45 ml/min in phosphate running buffer (135 mM NaCl, 2.7 mM KCl, 1.5 mM Na2HPO4, and 8 mM K2HPO4, 10% glycerol, pH 7.4) (Feng & Cronan, 2011). To further assess the solution structure of Bcam0581, chemical cross-linking with ethylene glycol bis succinimidylsuccinate (Pierce) was performed as described previously (Feng & Cronan, 2010). In each chemical cross-linking reaction (20 µl in total), the purified Bcam0581 protein (~10 mg ml−1) in sodium phosphate buffer (50 mM NaH2PO4, 150 mM NaCl, 1 mM dithiothreitol, pH 8.0) was mixed with cross-linker at different concentrations (0, 0.005, 0.01, 0.05, 0.2 or 2 mM), and kept 30 min at room temperature before analysis of the reaction products by SDS-PAGE.
Site-Directed Mutagenesis of Bcam0581
Plasmids pBHK21, pBHK22, pBHK23 and pBHK24 each carrying a single mutation within the Bcam0581m coding sequence were obtained using the QuickChange site-directed mutagenesis kit (Stratagene) with pBHK06 as the PCR template following the manufacturer’s instructions (Stratagene). The mutations were verified by DNA sequencing. The primers used in PCR and mutagenesis are listed in Table 1. The constructed plasmids were transformed into E. coli DH5α by CaCl2 treatment. All of the mutant Bcam0581 proteins were expressed and purified as described above with yields comparable to that of the wild type protein.. All constructs were verified by DNA sequencing.
Labeling and analysis of fatty acids
For fatty acid analysis, each of the plasmids encoding a mutated Bcam0581m locus together with the empty vector (pQE-2) and pBHK06 which encodes the wild type Bcam0581m was transformed into E. coli strain K19 which carries the fadE62 mutation that blocks fatty acid degradation. When a thioesterase active on acyl-ACPs is expressed in E. coli, the free fatty acids produced are exported to the culture (Cho & Cronan, 1994, Cho & Cronan, 1995, Voelker & Davies, 1994, Feng & Cronan, 2009).
The transformed strains were grown at 37°C in RB medium to log phase. IPTG (0.2 mM) was added to the medium to induce thioesterase expression together with [1-14C]acetate to label the newly synthesized fatty acids. The cultures were incubated at 37°C with shaking for further 18 h. Then the cells were pelleted at 4750 × g for 10 min and each 3 ml supernatant was decanted into a second tube. The free fatty acids present the supernatant were extracted and then methyl esters were prepared as described previously (Feng & Cronan, 2009). The resulting methyl esters were separated by argentation thin-layer chromatography and analyzed by autoradiography (Feng & Cronan, 2009).
Acyl-ACP preparations
3-Hydroxydodecanoyl-ACP, decanoyl-ACP, dodecanoyl-ACP, trans-2 and cis-2-dodecenoyl-ACPs were synthesized as described previously (Cronan & Thomas, 2009). Briefly, a typical reaction mixture consists of 20 µM holo-ACP, 200 µM fatty acid and 170 nM V. harveyi AasS in a buffer containing 100 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 1 mM tris [2-carboxyethyl] phosphine and 10 mM ATP in a reaction volume of 1 ml. The reaction mixtures were incubated at 37°C for 4 h and stopped by addition of two volumes of acetone and the proteins allowed to precipitate at −20°C overnight. The precipitates were pelleted at 20,000 ×g for 30 min at 4°C and then washed twice with 3 volumes of acetone. The pellets were air dried and resuspended in 20 mM Tris-HCl (pH 7.4) containing 1 mM tris [2-carboxyethyl] phosphine ACP-SH was removed from the acyl-ACP preparations by chromatography on Octyl-Sepharose as described previously (Rock et al., 1981). The final samples were concentrated with an Amicon ultracentrifugation filter device from Millipore (5,000 MWCO). Acyl-ACP synthesis was verified by electrophoresis on 18% gels containing 2.5 M urea (Cronan & Thomas, 2009) and by electrospray mass spectrometry.
Assay of Bcam0581 thioesterase activity and its mutants in vitro
The ability of Bcam0581 to function as a thioesterase was assessed in a reaction mixture containing 0.1 M Tris-HCl (pH 7.5), 2 mM β-mercaptoethanol, 20 µM acyl-ACP (3-hydroxydodecanoyl-ACP, trans-2-dodecenoyl-ACP, or cis-2-dodecenoyl-ACP) and 0.2 µg of a purified His-tagged enzyme (Bcam0581 or a mutant derivative) in a final volume of 30 µl. The assay mixtures were incubated at 37°C for 10 min or 30 min and the reaction products were resolved by conformationally sensitive gel electrophoresis on 18% polyacrylamide gels containing a concentration of urea optimized for the separation (Post-Beittenmiller et al., 1991). The gels were stained with Coomassie brilliant blue R250.
Reconstruction of Fatty Acid Synthesis in Vitro
A single cycle of fatty acid elongation was reconstructed in vitro by sequentially adding the individual enzymes that catalyze each reaction of the cycle (Zhu et al., 2009). The fatty acid synthesis assay mixtures contained, in a volume of 30 µl, 0.1 M sodium phosphate (pH 7.0) 2 mM β-mercaptoethanol, 100 µM decanoyl-ACP, 100 µM NADH, 100 µM NADPH, 100 µM holo-ACP and 50 µM [2-14C]malonyl-CoA (specific activity, 55 mCi/mmol). Purified His-tagged E. coli enzymes FabB and FabG enzymes were added at 0.3 µg/assay with FabD being added last to initiate the reaction to produce 14C-labeled 3-hydroxydodecanoyl-ACP. After incubation at 37 °C for 20 min, the reactions were stopped by placing the tubes in an ice slush. Samples of the mixtures were mixed with gel loading buffer and analyzed by conformationally sensitive gel electrophoresis on 18% polyacrylamide gels containing a urea concentration optimized for the separation. The gels were fixed, soaked in Enlightning (DuPont), dried and exposed to x-ray film.
Extraction and gas chromatography-mass spectroscopy analysis of BDSF from in vitro Bcam0581 reactions
BDSF was extracted into ethyl acetate from in vitro reaction systems as described previously (Barber et al., 1997). The ethyl acetate was evaporated to dryness and the residue was suspended in methanol. Methyl esters were prepared and analyzed on an Agilent system consisting of a 5975C Mass Selective Detector, a 7683B autosampler, and a 7890A gas chromatograph equipped with a ZB-WAX (Phenomenex Inc) capillary column (30 m × 0.25 mm inner diameter and 250 µm film thickness). Extracted BDSF concentration was quantitated using a standard curve prepared with defined concentrations of chemically synthesized BDSF. The position of the BDSF double bond in was determined by GC-MS analysis of the dimethyldisulfide (DMDS) adduct prepared from ethyl acetate samples. Analysis was done on the same instrument as above except that an Agilent 60-m HP-5MS column with 0.25-mm inner diameter and 0.25-mm film thickness was used. The procedure for GC-MS was described previously (Feng & Cronan, 2009).
BDSF Bioassay
The assay was performed as described previously using the biosensor strain X. campestris 8523/pL6engGUS (Slater et al., 2000). Briefly, 4 mm diameter wells were cut into the agar of prepared bioassay plates and 10 µl of a diluted fatty acid solution was added to each well. Alternatively, single colonies were spotted on bioassay plates. The plates were incubated at 30 °C overnight. BDSF activity is indicated by the presence of a blue halo around the well or colony.
Quantitative analysis of the β-glucuronidase (GUS) activities after BDSF induction
The BDSF-dependent expression pattern of the engXCA gene, which encodes endoglucanase, was determined by analyzing the activity of GUS encoded by gusA under the control of the engXCA promoter. Biosensor strain Xcc8523 (pL6engGUS) was grown at 30°C in YEB medium to an OD600 of 0.6. The culture was split and either BDSF or trans-2-C12:1 was added to a final concentration of 60 µM. Aliquots of the bacterial cell culture were taken and total soluble protein samples were prepared using a cell lysis reagent (Sigma) according to the recommendations of the manufacturer. Protein concentrations were determined using the Bradford assay kit (Bio-Rad). Quantitative determination of the GUS activity was performed with phenolphthalein glucuronide (Sigma) as substrate according to the procedure of Enzymatic Assay of β-glucuronidase (EC 3.2.1.31) from E. coli (Sigma). A standard curve was prepared using phenolphthalein (Sigma). One unit is defined as 1.0 µg of phenolphthalein produced per hour at pH 6.8 at 37°C.
Phylogenetic Analysis and Bioinformatics
Bcam0581 was aligned with selected sequences using T-Coffee http://www.ebi.ac.uk/Tools/msa/tcoffee/. The evolutionary history of Bcam0581 was inferred using the Minimum Evolution method. The bootstrap consensus tree inferred from 10000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1. The Neighbor-joining algorithm was used to generate the initial tree. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 120 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
Acknowledgements
This work was supported by National Institutes of Health (NIH) Grant AI15650 from National Institute of Allergy and Infectious Diseases (NIAID). We thank Drs. Steven Lindow and Max Dow for strains.
REFERENCES
- Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol. 1997;24:555–566. doi: 10.1046/j.1365-2958.1997.3721736.x. [DOI] [PubMed] [Google Scholar]
- Batchelar ET, Hamed RB, Ducho C, Claridge TD, Edelmann MJ, Kessler B, Schofield CJ. Thioester hydrolysis and C-C bond formation by carboxymethylproline synthase from the crotonase superfamily. Angew Chem Int Ed Engl. 2008;47:9322–9325. doi: 10.1002/anie.200803906. [DOI] [PubMed] [Google Scholar]
- Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Biosynthesis and stereochemistry of the autoinducer controlling luminescence in Vibrio harveyi. J Bacteriol. 1993;175:3856–3862. doi: 10.1128/jb.175.12.3856-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, He YW, Lim SC, Qamra R, Walsh MA, Zhang LH, Song H. Structural basis of the sensor-synthase interaction in autoinduction of the quorum sensing signal DSF biosynthesis. Structure. 2010;18:1199–1209. doi: 10.1016/j.str.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Cho H, Cronan JE., Jr "Protease I" of Escherichia coli functions as a thioesterase in vivo. J Bacteriol. 1994;176:1793–1795. doi: 10.1128/jb.176.6.1793-1795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Cronan JE., Jr Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J Biol Chem. 1995;270:4216–4219. doi: 10.1074/jbc.270.9.4216. [DOI] [PubMed] [Google Scholar]
- Churchill ME, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111:68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Zhao X, Jiang Y. Function, attachment and synthesis of lipoic acid in Escherichia coli. Adv Microb Physiol. 2005;50:103–146. doi: 10.1016/S0065-2911(05)50003-1. [DOI] [PubMed] [Google Scholar]
- Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu J, Eberl L, Zhang LH. Structural and functional characterization of diffusible signal factor family quorum-sensing signals produced by members of the Burkholderia cepacia complex. Appl Environ Microbiol. 2010;76:4675–4683. doi: 10.1128/AEM.00480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu J, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies - an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Overlapping repressor binding sites result in additive regulation of Escherichia coli FadH by FadR and ArcA. J Bacteriol. 2010;192:4289–4299. doi: 10.1128/JB.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol. 2011;80:195–218. doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Hofstein HA, Zwahlen J, Tonge PJ. Effect of mutagenesis on the stereochemistry of enoyl-CoA hydratase. Biochemistry. 2002;41:12883–12890. doi: 10.1021/bi020382g. [DOI] [PubMed] [Google Scholar]
- Flavier AB, Clough SJ, Schell MA, Denny TP. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol. 1997;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- Hamed RB, Batchelar ET, Clifton IJ, Schofield CJ. Mechanisms and structures of crotonase superfamily enzymes-how nature controls enolate and oxyanion reactivity. Cell Mol Life Sci. 2008;65:2507–2527. doi: 10.1007/s00018-008-8082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. Enoyl-acyl carrier protein reductase (FabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J Biol Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- Hofstein HA, Feng Y, Anderson VE, Tonge PJ. Role of glutamate 144 and glutamate 164 in the catalytic mechanism of enoyl-CoA hydratase. Biochemistry. 1999;38:9508–9516. doi: 10.1021/bi990506y. [DOI] [PubMed] [Google Scholar]
- Krejci E, Kroppenstedt RM. Differentiation of species combined into the Burkholderia cepacia complex and related taxa on the basis of their fatty acid patterns. J Clin Microbiol. 2006;44:1159–1164. doi: 10.1128/JCM.44.3.1159-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, Alberts AW, Vagelos PR. Acyl Carrier Protein. 3. An Enoyl Hydrase Specific for Acyl Carrier Protein Thioesters. J Biol Chem. 1965;240:618–621. [PubMed] [Google Scholar]
- More MI, Finger LD, Stryker JL, Fuqua C, Eberhard A, Winans SC. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci U S A. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CO, Garwin JL, Cronan JE., Jr Preparative enzymatic synthesis of acyl-acyl carrier protein. Methods Enzymol. 1981;72:397–403. doi: 10.1016/s0076-6879(81)72029-9. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011;19:145–152. doi: 10.1016/j.tim.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol Microbiol. 2000;38:986–1003. doi: 10.1046/j.1365-2958.2000.02196.x. [DOI] [PubMed] [Google Scholar]
- Stern JR, Del Campillo A. Enzymes of fatty acid metabolism. II. Properties of crystalline crotonase. J Biol Chem. 1956;218:985–1002. [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, Daniels MJ. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris var campestris. Mol Gen Genet. 1991;226:409–417. doi: 10.1007/BF00260653. [DOI] [PubMed] [Google Scholar]
- Toomey RE, Wakil SJ. Studies on the mechanism of fatty acid synthesis. XV. Preparation and general properties of β-ketoacyl acyl carrier protein reductase from Escherichia coli. Biochim Biophys Acta. 1966;116:189–197. doi: 10.1016/0005-2760(66)90001-4. [DOI] [PubMed] [Google Scholar]
- Val DL, Cronan JE., Jr In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker TA, Davies HM. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J Bacteriol. 1994;176:7320–7327. doi: 10.1128/jb.176.23.7320-7327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, Fang RX, Zhang LH. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol. 2004;51:903–912. doi: 10.1046/j.1365-2958.2003.03883.x. [DOI] [PubMed] [Google Scholar]
- Winans SC. A new family of quorum sensing pheromones synthesized using S-adenosylmethionine and acyl-CoAs. Mol Microbiol. 2011;79:1403–1406. doi: 10.1111/j.1365-2958.2011.07551.x. [DOI] [PubMed] [Google Scholar]
- Wong BJ, Gerlt JA. Divergent function in the crotonase superfamily: an anhydride intermediate in the reaction catalyzed by 3-hydroxyisobutyryl-CoA hydrolase. J Am Chem Soc. 2003;125:12076–12077. doi: 10.1021/ja037652i. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cai C, Yang Y, Weng L, Wang L. Blocking of Candida albicans biofilm formation by cis-2-dodecenoic acid and trans-2-dodecenoic acid. J Med Microbiol. 2011:1643–1650. doi: 10.1099/jmm.0.029058-0. [DOI] [PubMed] [Google Scholar]
- Zhu L, Cheng J, Luo B, Feng S, Lin J, Wang S, Cronan JE, Wang H. Functions of the Clostridium acetobutylicium FabF and FabZ proteins in unsaturated fatty acid biosynthesis. BMC Microbiol. 2009;9:119. doi: 10.1186/1471-2180-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Lin J, Ma J, Cronan JE, Wang H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother. 2010;54:689–698. doi: 10.1128/AAC.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]