Abstract
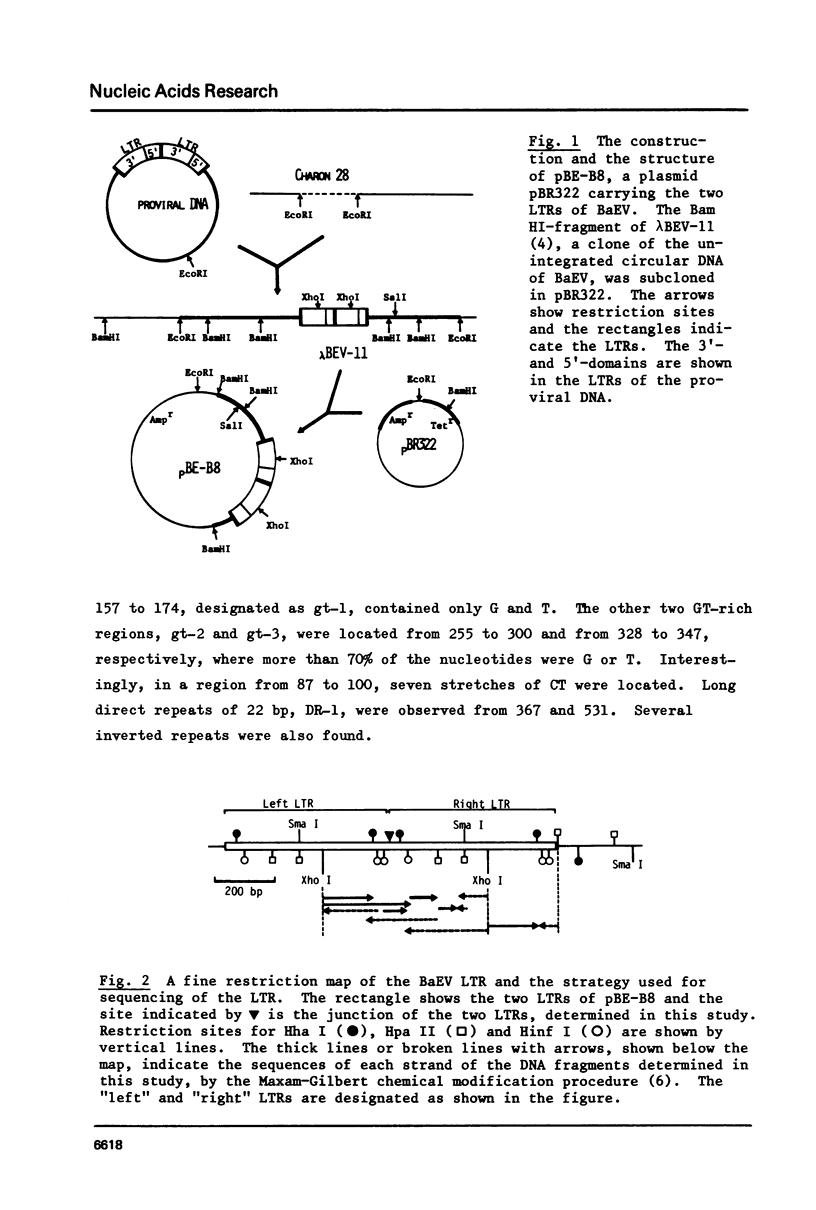
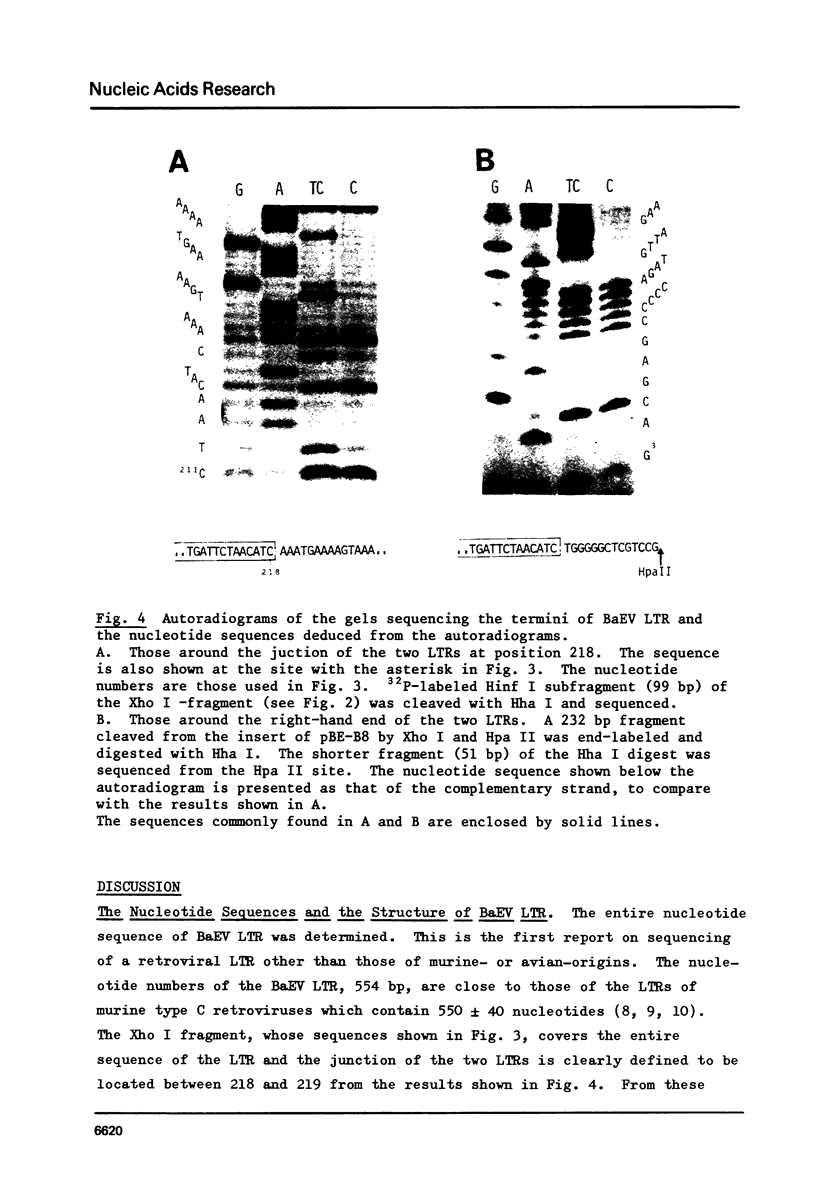
The entire nucleotide sequence of the long terminal repeat (LTR) of baboon endogenous virus (BaEV) M7 was determined, which consisted of 554 base pairs (bp). At both ends of the LTR, 13 bp sequences, AAATGAAAAGTAA and TGATTCTAACATC, were detected to be inverted repeats. The structure with these inverted repeats resembles those of other retroviruses and transposable elements. a Hogness box, TATAAAA, and a putative poly(A)-addition signal, AGTAAA, were present within the right-hand half of the LTR, where the initiation and termination of the viral RNA synthesis seems to occur in the integrated BaEV genome. The primer-binding site of at least 14 bp long was found just outside of the LTR where the strong stop DNA started, and the primer for reverse transcription in BaEV seemed to be tRNAPro. Several structural features are commonly detected in the LTRs of BaEV and other retroviruses. Our studies suggest that BaEV has evolved from a common ancestor with other mammalian type C viruses. Close relationships between BaEV and a feline endogenous virus, RD114, are demonstrated.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baralle F. E., Brownlee G. G. AUG is the only recognisable signal sequence in the 5' non-coding regions of eukaryotic mRNA. Nature. 1978 Jul 6;274(5666):84–87. doi: 10.1038/274084a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste R. E., Lieber M. M., Livingston D. M., Sherr C. J., Todaro G. J., Kalter S. S. Infectious C-type virus isolated from a baboon placenta. Nature. 1974 Mar 1;248(5443):17–20. doi: 10.1038/248017a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Smith R., Peters G. In vitro synthesis of polypeptides encoded by the long terminal repeat region of mouse mammary tumour virus DNA. Nature. 1981 Jun 11;291(5815):511–513. doi: 10.1038/291511a0. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Haseltine W. A., Kleid D. G. A method for classification of 5' termini of retroviruses. Nature. 1978 Jun 1;273(5661):358–364. doi: 10.1038/273358a0. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., DeBona P. J., Astrin S., Skalka A. M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981 Jan;23(1):155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Ju G., Skalka A. M. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980 Nov;22(2 Pt 2):379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Lovinger G. G., Schochetman G. 5' terminal nucleotide sequences of type C retroviruses: features common to noncoding sequences of eucaryotic messenger RNAs. Cell. 1980 Jun;20(2):441–449. doi: 10.1016/0092-8674(80)90630-3. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Matsubara K. Genetic structure and regulation of a replicon of plasmid lambdadv. J Mol Biol. 1976 Apr 15;102(3):427–439. doi: 10.1016/0022-2836(76)90325-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Wagatsuma M., Tamura T., Takano T., Matsubara K. Structure of the baboon endogenous virus genome: cloning of circular virus DNA in bacteriophage lambda. Nucleic Acids Res. 1981 May 11;9(9):2173–2185. doi: 10.1093/nar/9.9.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Isolation of inverted repeat sequences, including IS1, IS2, and IS3, in Escherichia coli plasmids. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2316–2320. doi: 10.1073/pnas.73.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Nomura N., Sugimoto, Sugisaki H., Takanami M. Nucleotide sequence at the insertion sites of a kanamycin transposon. Nature. 1978 Dec 21;276(5690):845–847. doi: 10.1038/276845a0. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Gagnon J., Vogt V. M., Ripley S., Eisenman R. N. The NH2-terminal sequence of the avian oncovirus gag precursor polyprotein (Pr76gag). Virology. 1978 Dec;91(2):423–433. doi: 10.1016/0042-6822(78)90388-4. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Farabaugh P. J., Chaleff D. T., Fink G. R. The origins of gene instability in yeast. Science. 1980 Sep 19;209(4463):1375–1380. doi: 10.1126/science.6251544. [DOI] [PubMed] [Google Scholar]
- Sabran J. L., Hsu T. W., Yeater C., Kaji A., Mason W. S., Taylor J. M. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol. 1979 Jan;29(1):170–178. doi: 10.1128/jvi.29.1.170-178.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Sugimoto K., Takanami M., Shiroki K., Saito I., Shimojo H., Sawada Y., Uemizu Y., Uesugi S., Fujinaga K. Structure and gene organization in the transformed Hind III-G fragment of Ad12. Cell. 1980 Jul;20(3):777–786. doi: 10.1016/0092-8674(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G., Shinnick T. M., Verma I. M., Lerner R. A. Nucleotide sequence of Moloney leukemia virus: 3' end reveals details of replications, analogy to bacterial transposons, and an unexpected gene. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3302–3306. doi: 10.1073/pnas.77.6.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Goddard J. G., Berns A., Verma I. M. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. The reverse transcriptase. Biochim Biophys Acta. 1977 Mar 21;473(1):1–38. doi: 10.1016/0304-419x(77)90005-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., de Crombrugghe B., Pastan I. Identification of a functional promoter in the long terminal repeat of Rous sarcoma virus. Cell. 1980 Dec;22(3):787–797. doi: 10.1016/0092-8674(80)90555-3. [DOI] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]



