Abstract
Large-scale DNA sequence data may enable development of genetic resources in endangered species, thereby facilitating conservation efforts. Ziziphus celata, a federally endangered, self-incompatible plant species occurring in Florida, USA, is one species for which genetic resources are necessary to facilitate new introductions and augmentations essential for recovery of the species. We used 454 pyrosequencing of a Z. celata normalized floral cDNA library to create a genomic resource for gene and marker discovery. A half-plate GS-FLX Titanium run yielded 655 337 reads averaging 250 bp. A total of 474 025 reads were assembled de novo into 84 645 contigs averaging 408 bp, while 181 312 reads remained unassembled. Forty-seven and 43% of contig consensus sequences had BLAST matches to known proteins in the Uniref50 and TAIR9 annotated protein databases, respectively; many contigs fully represented orthologous proteins in TAIR9. A total of 22 707 unique genes were sequenced, indicating substantial coverage of the Z. celata transcriptome. We detected single-nucleotide polymorphisms and simple sequence repeats (SSRs) and developed thousands of SSR primers for use in future genetic studies. As a first step towards understanding self-incompatibility in Z. celata, we identified sequences belonging to the gene family encoding self-incompatibility. This study demonstrates the efficacy of 454 transcriptome sequencing for rapid gene and marker discovery in an endangered plant.
Keywords: transcriptome, conservation genetics, microsatellites, S-locus, Ziziphus
1. Introduction
While genetic information is just one of many tools that may be used to conserve endangered species, the use of such information is often valuable to facilitate recovery efforts and the management of surviving populations.1 Genetic data from endangered species have been used for multiple conservation applications, e.g. to carry out parentage analysis in ex situ breeding and translocation programmes,2,3 and to quantify levels of genetic diversity and identify factors such as founder effects, genetic drift, genetic bottlenecks, and inbreeding that may threaten endangered species.4–6 However, because of the difficulty of isolating genetic markers and the considerable cost and time associated with extensive genotyping of individuals in populations, many conservation-genetic studies have employed relatively small numbers of genetic loci, such as RFLPs, AFLPs, and SSRs (simple sequence repeats, or microsatellites). Increasing the number of sampled loci may increase the precision and accuracy of estimates of genetic structure and population genetic parameters such as levels of genetic diversity and inbreeding coefficients.7 Furthermore, large numbers of genetic loci are necessary to unequivocally assign parentage, which may be very useful for establishing genetically diverse populations of endangered species through the augmentation of existing genetically depauperate populations or the creation of new populations.
Recently, however, the number of genomic-scale sequence collections has increased, enabling rapid gene and marker discovery for an increased number of taxa.8 Previously, the development of such genomic resources was largely limited to model organisms; however, recent advances in DNA sequencing technology have reduced the cost and time required for the development of genomic resources, resulting in a rapid growth in the availability of such data, particularly in non-model organisms.9–11 Transcriptome sequencing, which is DNA sequencing of the mRNA pool of a given tissue, has allowed sequencing efforts to focus on the protein-coding portion of the genome, and has become a valuable approach for developing genomic-level resources of the transcribed portion of the genome.8 Next-generation sequencing technologies, such as 454 pyrosequencing, remove many time-consuming steps involved in Sanger sequencing and now facilitate transcriptome sequencing at a minute fraction of the previously required time and cost.10–15 A single-plate run on the 454 GS-FLX titanium pyrosequencing platform typically produces around a million reads averaging 400 bp in length, often resulting in near complete transcriptome coverage.11,16,17 The generation of such large-scale sequence data is enabling gene discovery, molecular marker development, and comparative analyses in ecologically important, non-model plant taxa,11,18–22 including those of conservation concern.23
One species for which conservation efforts would be greatly assisted by the establishment of a genome-level resource is Ziziphus celata (Rhamnaceae), a federally endangered, diploid shrub endemic to central Florida, which is currently known from only 14 populations along the Lake Wales Ridge in Polk and Highlands Counties. The species is highly clonal and previous genetic analyses using allozymes,24 RAPDs,25 AFLPs, and SSRs26 found that 9 of the 14 extant wild populations are uniclonal (i.e. each population comprises a single genetic individual). Depending on the method of determining the number of genotypes, the remaining five populations comprise between 22 and 32 genotypes;26 however, unambiguous quantification of the number of genotypes in these populations will require the employment of a larger number of genetic markers. Very little genetic data are presently available for Z. celata, complicating the development of genetic markers, e.g. as of April 2011, only six DNA sequences had been deposited at NCBI for Z. celata, and none of these represented expressed genes. Furthermore, the entire Rhamnaceae family, which contains ∼900 species,27 was represented by fewer than 2000 sequences at NCBI. The development of a genome-level resource for Z. celata stands to improve conservation-genetic approaches in Z. celata and to provide a future basis for comparative genomic studies in the Rhamnaceae, which contains economically important crop and ornamental tree and shrub species.
Given the small numbers of populations and genotypes of Z. celata, its recovery relies on the augmentation of uniclonal populations and the introduction of multiple genotypes to publicly protected sites containing appropriate habitat;28,29 however, the ability to establish self-sustaining, sexually reproducing, translocated populations has been complicated by the reproductive failure of most experimental crosses. Such reproductive failure has been attributed to self-incompatibility in Z. celata. Self-incompatibility has been documented or proposed in at least five genera in the Rhamnaceae, including Colletia,30 Discaria,31,32 Frangula,33 Trevoa,34 and Ziziphus.25,35,36
Gametophytic self-incompatibility (GSI) has been proposed as the system of self-incompatibility in Z. celata based on results from hundreds of experimental hand pollinations, analysis of RAPD data, and studies of pollen tube inhibition.25,35,37 GSI is a system whereby pollen is rejected when its S-haplotype is the same as either of the S-haplotypes present in the pistil.38,39 In four of the five plant families with well-characterized GSI systems, GSI is mediated by the S-RNase gene.38,39 Previous research suggests that a S-RNase-based system of GSI likely evolved only once in Angiosperms,38,39 and the presence of S-RNase-based GSI in the Rosaceae, a plant family which is closely related to the Rhamnaceae, suggests that this is the most likely mechanism causing GSI in Rhamnaceae.
An understanding of the genetic basis of self-incompatibility and the ability to assess mating types in Z. celata would greatly facilitate recovery efforts. Previous research has shown that as few as two mating types may be present in Z. celata wild populations,25,35 the minimum necessary to maintain sexual reproduction in a GSI species; however, more mating types may exist, as the compatibility of most genotypes has not yet been tested. The most efficient approach to determine cross-compatibility is to directly sequence the genes that encode the GSI reaction. Another efficient approach is parentage analysis of progeny to provide indirect evidence of compatible mating types; however, more genetic data and molecular markers are required to carry out either of these approaches in Z. celata.
In this study, we conducted 454 pyrosequencing of a normalized cDNA library isolated from flowers of three genotypes of Z. celata. Our goals were to (i) characterize the floral transcriptome of Z. celata, (ii) identify and characterize a large number of gene-based markers, including single-nucleotide polymorphisms (SNPs) and SSRs, for future genetic analyses, and (iii) determine whether analysis of the floral transciptome includes genes belonging to gene family that encodes the GSI reaction. The increased genomic information produced in this study will aid in creating self-sustaining, sexually reproducing populations of this endangered species and bolster comparative genomic studies in a plant family for which very little genetic data are currently available.
2. Materials and methods
2.1. Tissue sampling, cDNA library creation, and 454 sequencing
During January–February 2010, we collected several hundred newly opened flowers from each of three Z. celata genets, including the two S-locus mating types confirmed in the wild and one genet of unknown mating type. We collected flowers in sterile RNase-free tubes, which were placed immediately into liquid nitrogen. Flowers from each of the three mating genotypes were then shipped on dry ice to GATC Inc., in Konstanz, Germany. Flowers from the three genotypes were pooled in equal amounts before RNA extraction. RNA extraction, cDNA synthesis, cDNA library normalization, and 454 sequencing were performed by GATC staff following previously outlined methods.17 The Z. celata floral cDNA library was sequenced in a half-plate run on a 454 GS XLR70 Titanium genomic sequencer (Roche, Inc.).
2.2. Assembly and annotation
We trimmed 454 primer sequences from all reads prior to assembly and removed reads with average quality scores lower than 18. We used Seqman Ngen v2.0 (DNAstar, Inc.) to assemble reads into contigs, as this program has been successful in assembling 454 sequences from transcriptomes.13,17 Because no reference genome exists for Ziziphus, reads were assembled de novo. The assembly was run with a minimum match size of 19 nucleotides, match percentage of 95%, mismatch penalty of 18, and gap penalty of 30 (further information on assembly is available from the authors by request). The resulting contig consensus sequences and remaining singletons were then combined into a single set for the following analyses, except where noted.
We annotated the 454 sequences by using local BLASTx40 to align the consensus sequences from the assembled contigs and the singleton sequences to the Uniref50 15.441 and the TAIR9 Arabidopsis thaliana42 annotated protein databases using an E-value threshold of 10−11. BLAST results were passed through a custom Perl pipeline that produced tab-delimited tables containing accession numbers, gene name, taxonomic ID, query length, orthologue sequence length, sequence alignment, E-value, and bit score for each protein matching to the Z. celata 454 sequences. To determine the number of unique genes represented, we filtered these files for redundancy in protein accessions. To assess the taxonomic distribution of BLAST hits, we used a custom perl script that employs a BioPerl package to retrieve hierarchical taxonomic identities for each protein accession ID. Assignment of gene ontology (GO) terms to sequences with BLAST matches to known proteins was then performed by importing the accession numbers for the BLAST hits to unique proteins into Blast2go (version 2.3.6; www.blast2go.org). We based these analyses on the contig consensus sequences with positive BLAST matches to 13 401 unique accessions in the TAIR annotated protein database.
2.3. Search for the S-locus
In most other taxa, the female component of GSI is encoded by an S-RNase, a member of the T2 RNase family.38,39 Thus, as a first step towards characterizing the genes involved in GSI in Z. celata, we searched annotation results and carried out BLAST searches to find Z. celata 454 sequences belonging to the T2 RNase gene family. We searched the Z. celata annotation results for the terms ‘T2 RNase’, ‘RNase’, ‘ribonuclease T2’, and ‘S-RNase’. We also created a blastable database of characterized S-RNase sequences deposited at NCBI's dbEST, including Nicotiana alata S2, Antirrhinum hispanicum S2, Prunus avium S1, and Pyrus pyrifolia S4.38 We used BLASTx to align all Z. celata contig consensus sequences and singletons to these sequences with an E-value threshold of 10−10. Once we identified sequences that putatively belonged to the T2 RNase gene family, we translated them to proteins, manually aligned them with amino acid sequences from verified members of the T2 RNase gene family38 using Se-Al,43 and inspected their protein-coding motifs for homology with verified members of the gene family.
2.4. Molecular marker characterization
We used custom perl scripts to locate di-, tri-, and tetra-nucleotide SSRs in the 454 sequences with a minimum length of 12 bp and six contiguous repeating units for di-nucleotide motifs, four contiguous repeats for tri-nucleotide motifs, and three repeating units for tetra-nucleotide motifs. We determined which SSRs occurred in coding regions of genes by extracting the aligned portions of contig consensus sequences that had BLAST matches to annotated protein-coding orthologues in Uniref50, and then used the same algorithm as above to detect SSRs in both the aligned and remaining portions of these contigs. To construct polymerase chain reaction (PCR) primers in the flanking regions of SSRs, we used the program BatchPrimer3.44 We created primers with a minimum GC content of 30%, a GC clamp (the last two nucleotides were G or C), a melting temperature between 52 and 55, and we positioned primers to obtain PCR products between 100 and 450 bp.
We isolated SNPs in contigs with high coverage depths using the SNP reporter feature in Seqman Pro (DNAstar, Inc.). We identified SNPs with coverage depth of at least 10 and with an alternate allele in a minimum of 20% in all contigs containing >25 reads, in order to obtain an estimate of the transcriptome-wide occurrence of high-quality SNPs. We then enumerated SNPs at sites where coverage depth was at least 8 and where alternate alleles were present at a minimum frequency of 20%.
3. Results
3.1. Assembly and annotation
A half-plate run on the 454 GS-FLX Titanium platform produced 655 337 sequences that, after adaptor and vector trimming, averaged 254 bases in length (Supplementary Fig. S1). Files containing the 454 DNA sequences and quality scores have been deposited at NCBIs Short Read Archive (accession SRA045662). A total of 474 025 reads (72% of total) were assembled into 84 645 contigs, with 181 312 reads remaining as singletons. The average contig length was 408 bases (min = 19, max = 2438), with an average of 5.6 (min = 2, max = 1337) reads assembled per contig (Supplementary Fig. S2) and a GC content of 0.39. The average read length of the singleton sequences was 255 bp, similar to the average length of the reads assembled into contigs (253 bp), and these sequences had an average GC content of 0.37. The mean coverage depth per nucleotide position in the assembled contigs was 3.1 (min = 1, max = 868), indicating reasonably good coverage depth for a half-plate 454 GS-FLX Titanium run. As expected, contig length increased as a function of coverage depth and the number of reads assembled into each contig (Supplementary Fig. S2).
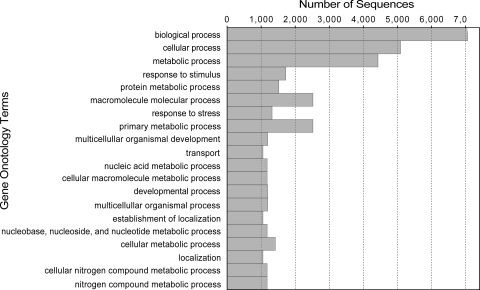
BLAST annotation of contig consensus and singleton sequences to a large number of unique genes indicated extensive coverage of the Z. celata floral transcriptome. Results of BLAST searches against the Uniref50 and TAIR9 databases, including the species name, accession number, identity percentage, and E-value, are presented in Supplementary Tables S1 and S2, respectively. Forty-seven per cent of the contig consensus sequences had BLAST hits to annotated proteins in Uniref50, and 43% had BLAST hits to annotated proteins in TAIR9 (Table 1). Smaller percentages of the singleton sequences also had BLAST matches to the annotated protein databases (25% UniRef; 23% TAIR). Nonetheless, the large number of singleton sequences with BLAST matches (Table 1) indicates that unassembled sequences still provide an abundance of valuable sequence information and improve overall transcriptome coverage breadth. In many cases, multiple sequences representing both contigs and singletons had BLAST matches to the same protein. After correcting for redundancy, the combined set of contig consensus sequences and singleton sequences had matches to 22 707 unique proteins in Uniref50, and 16 089 unique proteins in TAIR, indicating that many Z. celata genes are represented (Table 1). Of the 13 514 unique BLAST matches of contig sequences to protein accessions in the TAIR database, 10 808 were annotated to GO terms. Of the assigned GO terms, 6083 were to biological processes, 8036 to molecular function, and 6565 to cellular components, indicating a large functional diversity of genes in the trancriptomic data. The even distribution of assignments of proteins to more specialized GO terms further indicates that the Z. celata 454 sequences represent proteins from a diverse range of functional classes (Fig. 1).
Table 1.
The number and percentage of the total Ziziphus celata 454 contigs and singletons that matched to annotated protein databases
| Contigs |
Singletons |
Combined set |
||||
|---|---|---|---|---|---|---|
| Database | Total hits | Unique | Total hits | Unique | Total hits | Unique hits |
| Uniref | 39 815 (47%) | 17 420 | 46 124 (25%) | 16 332 | 85 939 (32%) | 22 707 |
| TAIR | 36 801 (43%) | 13 514 | 42 707 (23%) | 12 479 | 79 508 (30%) | 16 089 |
Figure 1.
Functional gene diversity in the Z. celata transcriptome data. Bars represent the number of assignments of Z. celata proteins with BLAST matches in the TAIR9 database to each GO term.
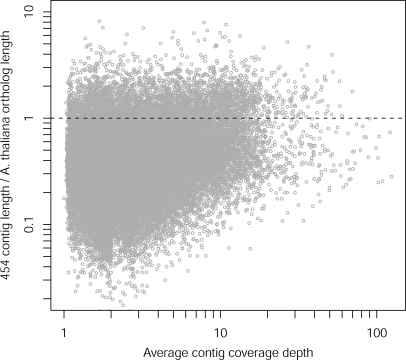
Many contig consensus sequences were sufficiently long to cover full or nearly full transcripts (Fig. 2). Examination of the 36 801 contigs with BLAST matches to protein-coding sequences in the TAIR9 protein database indicates that many contigs were as long as their corresponding full-length orthologous Arabidopsis transcripts (Fig. 2). As coverage depth increased, the ratio of Z. celata contig length/Arabidopsis orthologue length increased, with a large number of Z. celata contig sequences likely covering full-length transcripts (Fig. 2). This indicates that the assembly was of high quality and that we obtained extensive coverage breadth and depth for much of the floral transcriptome.
Figure 2.
The ratio of Z. celata contig length to A. thaliana orthologue length as a function of contig coverage depth. The dotted line corresponds to a ratio of 1, where 454 contigs are as long or longer than the BLAST matched A. thaliana orthologues.
As expected, the vast majority of BLAST hits matched to known plant proteins (Table 2). The abundance of BLAST matches to plant-specific proteins was further indicated by sequences matching nearly as many unique proteins in searches of TAIR as those with Uniref50. There were a small number of BLAST hits to non-plant proteins (Table 2), which may be due to representation of these proteins in Uniref50 by non-plant taxa, short or poor quality consensus sequences in 454 Z. celata contigs, or because the sampled tissues contained other organisms such as pathogens.
Table 2.
Number of unique BLAST matches to annotated proteins in different taxonomic groups
| Taxonomic group | Unique BLAST hits |
|---|---|
| Plants | 19 790 |
| Algae | 100 |
| Other eukaryote | 496 |
| Fungi | 138 |
| Bacteria | 131 |
| Virus | 553 |
| Other | 1499 |
Annotation and BLAST searches identified six Z. celata sequences that putatively belonged to the T2 RNase gene family, five of which were isolated from contig consensus sequences and one of which was isolated from a singleton sequence. We translated these Z. celata sequences into amino acid sequences and aligned them with previously characterized, complete T2-RNase amino acid sequences from 12 plant families. Although the 5′ and 3′ ends of the amino acid sequences were highly variable and difficult to align across taxa, we were able to easily align ∼110 highly conserved amino acids, suggesting that the six Z. celata sequences identified in this study are likely members of the T2 RNase gene family. The Z. celata T2 RNases were 115–206 residues in length (347–647 bp) and were shorter than all 12 of the complete T2 RNase sequences (which ranged from 222 to 275 residues). Alignments revealed that the Z. celata T2 RNase sequences did not span the full T2 RNase protein-coding region, as all were truncated on either the 5′ or 3′ end. We detected significant diversity among the six Z. celata T2 RNases, but additional analyses of these alignments will be necessary to determine whether any of these T2 RNase sequences are orthologues of S-RNase genes that encode the female component of GSI in other taxa.
3.2. Molecular marker characterization
SSRs were highly abundant in the 454 DNA sequences, occurring in 17% of sequences from the combined set of contig consensus sequences and singletons. We identified 10 954 di-, tri-, and tetra-nucleotide repeats in the 84 465 contig consensus sequences, and an additional 33 587 SSRs in the 181 312 singleton sequences (Table 3). Although influenced by the criteria used to identify these SSRs, tri-nucleotide repeats were the most common, followed by di- and tetra- nucleotide repeats (Table 3). A total of 11 926 SSRs occurred in contigs with BLAST matches to Uniref50 annotated proteins, of which 4246 occurred in protein-coding regions of these sequences. The density of SSRs was much higher in non-coding regions that followed coding regions than in coding regions (0.0016 SSRs per basepair in coding regions vs. 0.0075 in non-coding regions). Of the three repeat motifs, di-nucleotide repeats were the most abundant in coding regions (2551), followed by tri- (1684) and tetra-nucleotide (11) repeats. Primer design was successful for a much larger percentage of contig consensus sequences (76%) than singleton sequences (23%), presumably because the shorter length of the singleton sequences limited the availability of suitable priming sites. Supplementary Tables S3 and S4 provide primer sequences, GC content, melting temperatures, expected product length, SSR motif, SSR sequence, SSR length, and sequence identification for the SSRs developed from the Z. celata 454 sequences. SNPs were also highly abundant in the Z. celata 454 contigs. Across 1.4 million bases represented by a minimum of 8× coverage, we identified 11 056 SNPs that were present at a minimum frequency of 25%, resulting in a SNP occurrence rate of 0.008 per base, similar to that reported in other 454 transcriptome studies.13,16,17
Table 3.
Number of each type of SSRs detected in the Z. celata transcriptome. Values in parentheses indicate the number of sequences for which PCR primers were successfully designed.
| Repeat motif | Contigs | Singletons | Total |
|---|---|---|---|
| Di | 3493 (2145) | 6620 (2075) | 10 113 (4220) |
| Tri | 4195 (3836) | 11 638 (3188) | 15 833 (7024) |
| Tetra | 3266 (2313) | 15 329 (2379) | 18 595 (4692) |
| Total | 10 954 (8294) | 33 587 (7643) | 44 541 (15 936) |
4. Discussion
4.1. Transcriptome assembly, coverage breadth and depth, and gene annotation
454 pyrosequencing has arisen as a powerful tool for the sequencing of transcriptomes, and many studies have used de novo assembly of such data to produce and characterize genome-level resources for non-model organisms.11,13,16,23,45 Similarly, a half-plate run on the 454 GS-FLX Titanium platform provided substantial coverage of the Z. celata floral transcriptome, and de novo assembly placed a large fraction (72%) of the 454 sequences into contigs, a large number of which were of considerable length and coverage (Fig 2). BLAST hits to more than 22 000 unique proteins indicate that a large portion of the floral transcriptome was likely sequenced. The large number of diverse GO assignments of these transcripts also highlights the diversity of genes likely represented by these data. Furthermore, many contigs were as long as, or longer than, the corresponding Arabidopsis orthologues, indicating thorough transcript coverage for many genes. These sequences provide a significant genomic-level resource for an endangered plant species and plant family for which very little DNA sequence data previously existed.
Nearly all of the unique BLAST matches were to proteins characterized in green plants (Table 3). A small subset, however, were to proteins characterized in other organisms, and could represent contaminant RNA in the floral tissues we sampled. In particular, a reasonably large number of matches were to plant viral genes (Table 3). However, low frequencies of BLAST hits to distantly related taxonomic groups are common in such transcriptome sequencing studies and are often attributable to factors other than contaminant RNA,13,17,45 e.g. these sequences could be short contigs containing little information, genes that have not been well characterized in plants, or novel Z. celata genes.
Transcriptome sequencing studies have proven highly valuable for gene discovery, characterization, and variant analysis in both model and non-model organisms.11,16,17,46,47 Here, we detected genes expressed in the floral tissues of Z. celata that are putative members of the T2 RNase gene family, demonstrating that 454 transcriptome sequencing is a useful approach for identifying sequences belonging to specific gene families in non-model organisms. We detected significant variation among amino acid sequences within the gene family, indicating that we may have isolated multiple paralogues. Further phylogenetic analyses and analyses of protein-coding motifs for these sequences will be necessary to determine whether these sequences are orthologues or paralogues of S-RNases. If any of the sequences identified in this study encode the S-RNase gene, future research will focus on genotyping this gene in all individuals of Z. celata, which will allow us to identify rare mating types and determine the compatibility of extant genotypes in this self-incompatible species. An understanding of S-locus diversity in Z. celata will help to establish genetically diverse populations capable of reproducing sexually, thereby increasing levels of genetic diversity in this highly endangered species, with the ultimate goal of forming self-sustaining and demographically viable populations.
4.2. Molecular marker characterization
The 454 pyrosequencing and other next-generation sequencing technologies have increased the opportunity for molecular marker development in non-model study organisms at an unprecedented scale.10,16,17 In addition, the gene-based markers developed from transcriptome sequencing projects have the advantages of higher cross-species transferability.8,48 We detected and characterized an enormous number of SSR and SNP loci that will likely facilitate future evolutionary, ecological, and conservation-genetic-oriented studies in Z. celata. The primer sets developed for candidate SSRs will allow higher density genotyping that will facilitate accurate assessment of the number of extant genotypes in this species. These markers will also be used for parentage analysis of seedlings to identify the parents with the highest fitness and to ensure that translocated populations contain the highest possible levels of genetic diversity. Furthermore, if we are unable to identify the genes encoding GSI, we will employ these SSR markers to carry out parentage analysis to provide indirect estimates of compatible mating types. The thousands of high-quality SNPs contained in deeply covered contigs offer an enormous number of informative sites that will facilitate genome-wide analyses of molecular variation in future studies.
4.3. Conclusions
In this study, we characterized the floral transcriptome, isolated thousands of molecular markers, and identified transcripts that may encode the S-locus in Z. celata, a highly endangered shrub species. This work has taken an important initial step in characterizing the protein-coding portion of the genome of Z. celata, providing a substantial genomic-level resource for an endangered plant species and plant family for which little previous DNA sequence data existed. Indeed, given that very few endangered plant species have been the focus of genomic or next-generation DNA sequencing efforts, Z. celata may now have one of the best-characterized genomes of any endangered plant species. This work also highlights the utility of using next-generation sequencing for marker and gene discovery; we isolated thousands of genetic markers to be used for parentage analysis, and identified members of a specific gene family that may encode GSI in Z. celata. These resources will be employed to facilitate recovery efforts for Z. celata, highlighting the utility of the new technology in rapidly expanding the resources available to conservation biologists to aid in the recovery of highly endangered species.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the US Fish and Wildlife Service (Grant #40181AG003 to C.W.W.), the Florida Division of Forestry Plant Conservation Grants Program, and Archbold Biological Station.
Supplementary Material
Acknowledgements
We thank Alex Buerkle and Johan Grahnen for assistance with data analysis, Martin Heine and staff at GATC Inc. for technical work; Stacy Smith and Megan Larson for field assistance in collecting Florida Ziziphus flower samples under difficult field conditions; David Bender for acquisition and administration of the USFWS grant; Dennis Hardin and Mike Jenkins of the Florida Division of Forestry for DOF funding, and Eric Menges, Satoshi Tabata, and two anonymous reviewers for comments on previous versions of this manuscript.
References
- 1.DeSalle R., Amato G. The expansion of conservation genetics. Nat. Rev. Genet. 2004;5:702–12. doi: 10.1038/nrg1425. [DOI] [PubMed] [Google Scholar]
- 2.Krauss S.L., Dixon B., Dixon K.W. Rapid genetic decline in a translocated population of the endangered plant Grevillea scapigera. Conserv. Biol. 2002;16:986–94. [Google Scholar]
- 3.Mino C.I., Sawyer G.M., Benjamin R.C., Del Lama S.N. Parentage and relatedness in captive and natural populations of the roseate spoonbill (Aves: Ciconiiformes) based on microsatellite Data. J. Exp. Zool. Part A. 2009;311A:453–64. doi: 10.1002/jez.543. [DOI] [PubMed] [Google Scholar]
- 4.Riley L., McGlaughlin M.E., Helenurm K. Genetic diversity following demographic recovery in the insular endemic plant Galium catalinense subspecies acrispum. Conserv. Genet. 2010;11:2015–25. [Google Scholar]
- 5.Funk W.C., Forsman E.D., Johnson M., Mullins T.D., Haig S.M. Evidence for recent population bottlenecks in northern spotted owls (Strix occidentalis caurina) Conserv. Genet. 2010;11:1013–21. [Google Scholar]
- 6.McCraney W.T., Goldsmith G., Jacobs D.K., Kinziger A.P. Rampant drift in artificially fragmented populations of the endangered tidewater goby (Eucyclogobius newberryi) Mol. Ecol. 2010;19:3315–27. doi: 10.1111/j.1365-294X.2010.04755.x. [DOI] [PubMed] [Google Scholar]
- 7.Allendorf F.W., Hohenlohe P.A., Luikart G. Genomics and the future of conservation genetics. Nat. Rev. Genet. 2010;11:697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- 8.Bouck A., Vision T. The molecular ecologists guide to expressed sequence tags. Mol. Ecol. 2007;16:907–24. doi: 10.1111/j.1365-294X.2006.03195.x. [DOI] [PubMed] [Google Scholar]
- 9.Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Hudson M.E. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Resour. 2008;8:3–17. doi: 10.1111/j.1471-8286.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- 11.Wheat C.W. Rapidly developing functional genomics in ecological model systems via 454 transcriptome sequencing. Genetica. 2010;138:433–51. doi: 10.1007/s10709-008-9326-y. [DOI] [PubMed] [Google Scholar]
- 12.Margulies M., Egholm M., Altman W.E., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vera J.C., Wheat C.W., Fescemyer H.W., et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008;17:1636–47. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 14.Ellegren H. Sequencing goes 454 and takes large-scale genomics into the wild. Mol. Ecol. 2008;17:1629–31. doi: 10.1111/j.1365-294X.2008.03699.x. [DOI] [PubMed] [Google Scholar]
- 15.Holt R.A., Jones S.J.M. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–46. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
- 16.Meyer E., Aglyamova G.V., Wang S., et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GS-FLX. BMC Genomics. 2009;10:219. doi: 10.1186/1471-2164-10-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parchman T.L., Geist K.S., Grahnen J.A., Benkman C.W., Buerkle C.A. Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics. 2010;11:180. doi: 10.1186/1471-2164-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta S., Kumawat G., Singh B.P., et al. Development of genic SSR markers by deep transcriptome sequencing in pigeonpea Cajanus cajan (L.) Millspaugh. BMC Plant Biol. 2011;11:17. doi: 10.1186/1471-2229-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angeloni F., Wagemaker C.A.M., Jetten M.S.M., et al. De novo transcriptome characterization and development of genomic tools for Scabiosa columbaria L. using next-generation sequencing techniques. Mol. Ecol. Resour. 2011;11:662–74. doi: 10.1111/j.1755-0998.2011.02990.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaur S., Cogan N.O.I., Pembleton L.W., et al. Transcriptome sequencing of lentil based on second-generation technology permits large-scale unigene assembly and SSR marker discovery. BMC Genomics. 2011;12:265. doi: 10.1186/1471-2164-12-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg R., Patel R.K., Tyagi A.K., Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajgain P., Richardson B.A., Price J.C., Cronn R.C., Udall J.A. Transcriptome characterization and polymorphism detection between subspecies of big sagebrush (Artemisia tridentata) BMC Genomics. 2011;12:370. doi: 10.1186/1471-2164-12-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale M.C., McCormick C.R., Jackson J.R., DeWoody J.A. Next-generation pyrosequencing of gonad transcriptomes in the polyploid lake sturgeon (Acipenser fulvescens): the relative merits of normalization and rarefaction in gene discovery. BMC Genomics. 2009;10:203. doi: 10.1186/1471-2164-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godt M.J.W., Race T., Hamrick J.L. A population genetic analysis of Ziziphus celata, an endangered Florida shrub. J. Hered. 1997;88:531–3. [Google Scholar]
- 25.Weekley C.W., Kubisiak T.L., Race T.M. Genetic impoverishment and cross-incompatibility in remnant genotypes of Ziziphus celata (Rhamnaceae), a rare shrub endemic to the Lake Wales Ridge, Florida. Biodivers. Conserv. 2002;11:2027–46. [Google Scholar]
- 26.Gitzendanner M., Weekley C.W., Germain-Aubrey C., Soltis D.E., Soltis P.S. Microsatellite evidence for high clonality and limited genetic diversity in Ziziphus celata (Rhamnaceae), an endangered, self-incompatible Lake Wales Ridge, Florida, USA endemic. Conserv. Genet. in review. [Google Scholar]
- 27.Judd W.S., Campbell C.S., Kellogg E.A., Stevens P.F., Donoghue M. Plant Systematics: A Phylogenetic Approach. Sunderland: Sinauer Associates, Inc; 2008. [Google Scholar]
- 28.USFWS. Multi-species Recovery Plan for the Threatened and Endangered Species of South Florida. Atlanta, GA: U.S. Fish and Wildlife Service; 1999. Florida Ziziphus; pp. 1986–99. [Google Scholar]
- 29.USFWS. Florida Ziziphus (Ziziphus celata 5-year Status Review: Summary and Evaluation. Vero Beach, FL: U.S. Fish and Wildlife Service; 2009. [Google Scholar]
- 30.Medan D., Basilio A.M. Reproductive biology of Colletia spinosissima (Rhamnaceae) in Argentina. Plant Syst. Evol. 2001;229:79–89. [Google Scholar]
- 31.Webb C.J. Protandry, pollination, and self-incompatibility in Discaria toumatou. N. Z. J. Bot. 1985;23:331–5. [Google Scholar]
- 32.Medan D., Vasellati V. Nonrandom mating in Discaria americana (Rhamnaceae) Plant Syst. Evol. 1996;203:179–80. [Google Scholar]
- 33.Medan D. Reproductive biology of Frangula alnus (Rhamnaceae) in southern Spain. Plant Syst. Evol. 1994;193:173–86. [Google Scholar]
- 34.Medan D., D'Ambrogio A.C. Reproductive biology of the andromonoecious shrub Trevoa quinquenervia (Rhamnaceae) Bot. J. Linn. Soc. 1998;12:191–206. [Google Scholar]
- 35.Weekley C.W., Race T. The breeding system of Ziziphus celata Judd and DW Hall (Rhamnaceae), a rare endemic plant of the Lake Wales Ridge, Florida, USA: implications for recovery. Biol. Conserv. 2001;100:207–13. [Google Scholar]
- 36.Zietsman P.C., Botha F.C. Flowering of Ziziphus mucronata subsp. mucronata (Rhamnaceae)—anthesis, pollination and protein synthesis. Bot. Bull. Acad. Sinica. 1992;33:33–42. [Google Scholar]
- 37.Faivre A.E. Summary of 2005–2007 observations of pollen grains and pollen tube growth in Florida ziziphus and observations of open-pollinations and aborted fruits. In: Weekley C.W., Menges E.S., editors. Continuation of Research on the Federally-listed Lake Wales Ridge Endemic Florida Ziziphus (Ziziphus celata) Tallahassee, FL: Final Report to Plant Conservation Program, Florida Division of Forestry; 2007. [Google Scholar]
- 38.Vieira J., Fonseca N.A., Vieira C.P. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. J. Mol. Evol. 2008;67:179–90. doi: 10.1007/s00239-008-9137-x. [DOI] [PubMed] [Google Scholar]
- 39.Igic B., Kohn J.R. Evolutionary relationships among self-incompatibility RNases. Proc. Natl Acad. Sci. USA. 2001;98:13167–71. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzek B.E., Huang H.Z., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–8. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 42.Swarbreck D., Wilks C., Lamesch P., et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–14. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rambaut A. Se-Al: A Manual Sequence Alignment Editor. 2003. 2.0 a11. [Google Scholar]
- 44.You F.M., Huo N., Gu Y.Q., et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Der J.P., Barker M.S., Wickett N.J., dePamphilis C.W., Wolf P.G. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics. 2011;12:99. doi: 10.1186/1471-2164-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emrich S.J., Barbazuk W.B., Li L., Schnable P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novaes E., Drost D.R., Farmerie W.G., et al. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis J.R., Burke J.M. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99:125–32. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.