Abstract
Tauopathies, characterized by neurofibrillary tangles (NFTs) of phosphorylated tau proteins, are a group of neurodegenerative diseases, including frontotemporal dementia and both sporadic and familial Alzheimer's disease. Forebrain-specific over-expression of human tauP301L, a mutation associated with frontotemporal dementia with parkinsonism linked to chromosome 17, in rTg4510 mice results in the formation of NFTs, learning and memory impairment and massive neuronal death. Here, we show that the mRNA and protein levels of NMNAT2 (nicotinamide mononucleotide adenylyltransferase 2), a recently identified survival factor for maintaining neuronal health in peripheral nerves, are reduced in rTg4510 mice prior to the onset of neurodegeneration or cognitive deficits. Two functional cAMP-response elements (CREs) were identified in the nmnat2 promoter region. Both the total amount of phospho-CRE binding protein (CREB) and the pCREB bound to nmnat2 CRE sites in the cortex and the hippocampus of rTg4510 mice are significantly reduced, suggesting that NMNAT2 is a direct target of CREB under physiological conditions and that tauP301L overexpression down-regulates CREB-mediated transcription. We found that over-expressing NMNAT2 or its homolog NMNAT1, but not NMNAT3, in rTg4510 hippocampi from 6 weeks of age using recombinant adeno-associated viral vectors significantly reduced neurodegeneration caused by tauP301L over-expression at 5 months of age. In summary, our studies strongly support a protective role of NMNAT2 in the mammalian central nervous system. Decreased endogenous NMNAT2 function caused by reduced CREB signaling during pathological insults may be one of underlying mechanisms for neuronal death in tauopathies.
INTRODUCTION
Hyperphosphorylated microtubule-associated protein tau is the major component of the intracellular neurofibrillary tangles (NFTs) that are tauopathy hallmarks. Tauopathies include several neurodegenerative diseases such as Pick's disease and frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17; reviewed in 1). Several tau mutations linked to FTDP-17 have been identified (2–7) and transgenic mice expressing human mutated tau showed NFT formation and neuronal loss, implying a causal relationship between tau over-expression and neurodegeneration (8–11). However, neuronal loss appears to be independent of tangle formation in some transgenic mouse models (12–14). Although there is evidence for the presence of activated caspase and morphological features of necrosis, autophagy and abnormal cell cycle events (10,13,15–18), the precise mechanism of cell death in tauopathies remains unknown.
Nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NMNAT) proteins are the central enzymes for nicotinamide mononucleotide (NAD) synthesis (19–22). Loss of nmnat in the Drosophila eye causes rapid neuronal degeneration (23). Three mammalian homologs, NMNAT1–3, have been identified (24–26), with NMNAT2 (nicotinamide mononucleotide adenylyltransferase 2) being the most labile of the three isoforms with a half-life of <4h (27). Reducing NMNAT2 levels in cultured mouse superior cervical ganglia neurons induced Wallerian-like degeneration, suggesting that NMNAT2 is required to maintain axonal health in the peripheral nervous system (27). Interestingly, several gene-array studies found that NMNAT2 levels were reduced in brain specimens from patients with Alzheimer's disease (AD; https://www.nextbio.com). Over-expression of NMNAT, on the other hand, provides neuroprotection against several degenerative conditions in Drosophila (21,23). In mammals, over-expressing NMNATs significantly delays Wallerian degeneration of peripheral nerves (28–33).
To explore whether impaired NMNAT function can contribute to the neurodegeneration found in tauopathies, we examined the expression levels of NMNAT1–2, two homologs present in the brain, in rTg4510 mice at various ages. In rTg4510 mice (34,35), the age-dependent loss of neurons and progressive cognitive dysfunction closely mimic the clinical features of tauopathy. We found that the levels of phospho-cAMP-response element binding protein (CREB) and NMNAT2 were decreased by tauP301L over-expression prior to the onset of neurodegeneration in rTg4510 mice. The reduction in nmnat2 transcription in rTg4510 mice likely results from reduced CREB activity. Conversely, over-expressing NMNAT2 or NMNAT1 in the hippocampus provided remarkable neuroprotection.
RESULTS
NMNAT2 is down-regulated in the rTg4510 mouse brain prior to neurodegeneration
In rTg4510 mice (34,35), tau with the mis-sense mutation P301L, found in some FTDP-17 cases (36), is over-expressed in the forebrain driven by the calcium-calmodulin kinase II (CaMKII) promoter. Mutant tau expression starts around postnatal day 7 and peaks at ∼1 month of age (34,35). From 2.5 months onward, phosphorylated tau protein and neuronal loss can be detected in the forebrain of these mice (14,34,35). Cognitive impairment in spatial reference memory and synaptic dysfunction in hippocampal CA1 pyramidal neurons begin to manifest at 4.5 months of age (37). Substantial neurodegeneration is observed after 5 months of age, being most apparent in the hippocampal CA1 area (14,34,35). Noticeable cortical thinning in rTg4510 mice occurs between 4 and 9 months of age and NFT formation and gliosis are often observed around 9 months.
To examine the level of NMNAT2 in brain tissue over-expressing tauP301L, western blot analysis was conducted to quantitatively compare NMNAT2 protein levels between rTg4510 mice and their littermate controls (see Supplementary Material, Fig. S1 demonstrating NMNAT2 antibody specificity). We prepared lysates of cortex, hippocampus and cerebellum from rTg4510 mice and their age-matched littermate controls at 2 weeks, 1, 2 and 7 months of age (Fig. 1; n = 3–10 for each age group and each genotype; Supplementary Material, Table S1). Cerebella, where tauP301L was not over-expressed, were used as an internal control for NMNAT2 expression. Indeed, no change in the expression of cerebellar NMNAT2 was found at any of the ages examined (Fig. 1A and D). Strikingly, we found that the level of NMNAT2 in the cortex and hippocampi of rTg4510 mice was often reduced to <60% of the level found in their age-matched controls at 1, 2 and 7 months of age (Fig. 1A–C; Supplementary Material, Table S1). The reduced NMNAT2 levels in the rTg4510 cortex and hippocampus at 1 month of age coincide with the time when tauP301L over-expression begins to reach its plateau. At this age, neuronal loss was not yet observed. Thus, the decrease in cortical NMNAT2 coincides with tauP301L over-expression and precedes significant neuronal loss. The expression level of NMNAT1, another NMNAT isoform expressed in the brain (http://mouse.brain-map.org/brain/Nmnat1.html), was not altered in the cortex, hippocampus or cerebellum of 2-month-old rTg4510 mice (Fig. 1A; Supplementary Material, Table S1), indicating specific expressional regulation for NMNAT2 in rTg4510 forebrain.
Figure 1.
NMNAT2 protein and mRNA levels were down-regulated in the forebrain of rTg4510 mice. (A) Western blots show NMNAT2, NMNAT1, NF-M, GFAP, actin and hTau immunoreactivity in the cortex, hippocampus and cerebellum of 1-month-old control and rTg4510 mice (abbreviated as Tau mice). (B–D) Normalized ratios of NMNAT2/actin in the cortex (B), hippocampus (C) and cerebellum (D) of control and Tau mice at 2 weeks, 1, 2 and 7 months of age. Western data are presented as percentages of their age-matched controls and are plotted as mean ± SEM (for 1-month-old, n= 10 for each genotype; for other age groups, n = 3 for each genotype group). (E) Real-time qPCR analyses for nmnat2 mRNA levels in the cortex, hippocampus and cerebellum of Tau mice relative to their littermate controls at 2 weeks, 1, 2 and 7 months of age. nmnat2 transcript levels were normalized to the level of the house keeping gene gapdh. Fold differences (Tau/control) were calculated with the standard 2-ΔΔCt method (see Materials and Methods) as 2(power, -ΔΔCt) and plotted as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001).
To further investigate a potential reduction in neuronal population or increased gliosis in rTg4510 mice, we determined the levels of neurofilament-medium (NF-M) and glial fibrillary acidic protein (GFAP) in rTg4510 and control mice at 2 weeks, 1 month and 2 months of age. No alterations in the levels of actin, NF-M or GFAP were observed in rTg4510 mice in the cerebellum, cortex or hippocampus (Fig. 1A; Supplementary Material, Table S1). These data are consistent with earlier immunohistochemical studies (14,34,35) that found no gross abnormalities in the glial and neuronal populations in these brain areas of rTg4510 mice up to 2 months of age. Despite the reduction in NMNAT2 protein levels, there was no significant change in NAD+ and NADH levels in the cortex or hippocampus of rTg4510 mice (Supplementary Material, Fig. S2).
Using real-time quantitative PCR (real-time qPCR) analysis, we examined whether tauP301L over-expression affected nmnat2 mRNA expression. Specific brain areas, including the cortex, hippocampus and cerebellum from rTg4510 mice and their littermate controls at 2 weeks, 1, 2 and 7 months of age were analyzed (n= 3 for each age group and each genotype). We found that nmnat2 transcription in rTg4510 mice was significantly down-regulated at 1, 2 and 7 months of age in the cortex and at 2 and 7 months of age in the hippocampus (Fig. 1E; Supplementary Material, Table S2). No down-regulation of nmnat2 transcription was seen in the cerebellum. Thus, the decrease in the NMNAT2 protein in rTg4510 mice was accompanied by a down-regulation of nmnat2 transcripts in the cortex at 1 month of age and older, and in the hippocampus at 2 months of age and older. The specific reduction in nmnat2 expression in tauP301L over-expressing regions and the temporal correlation between nmnat2 down-regulation and tauP301L peak expression suggest that the pathological down-regulation of nmnat2 transcription is a direct consequence of tauP301L expression.
CREB activity is reduced in rTg4510 forebrains before the onset of cognitive dysfunction
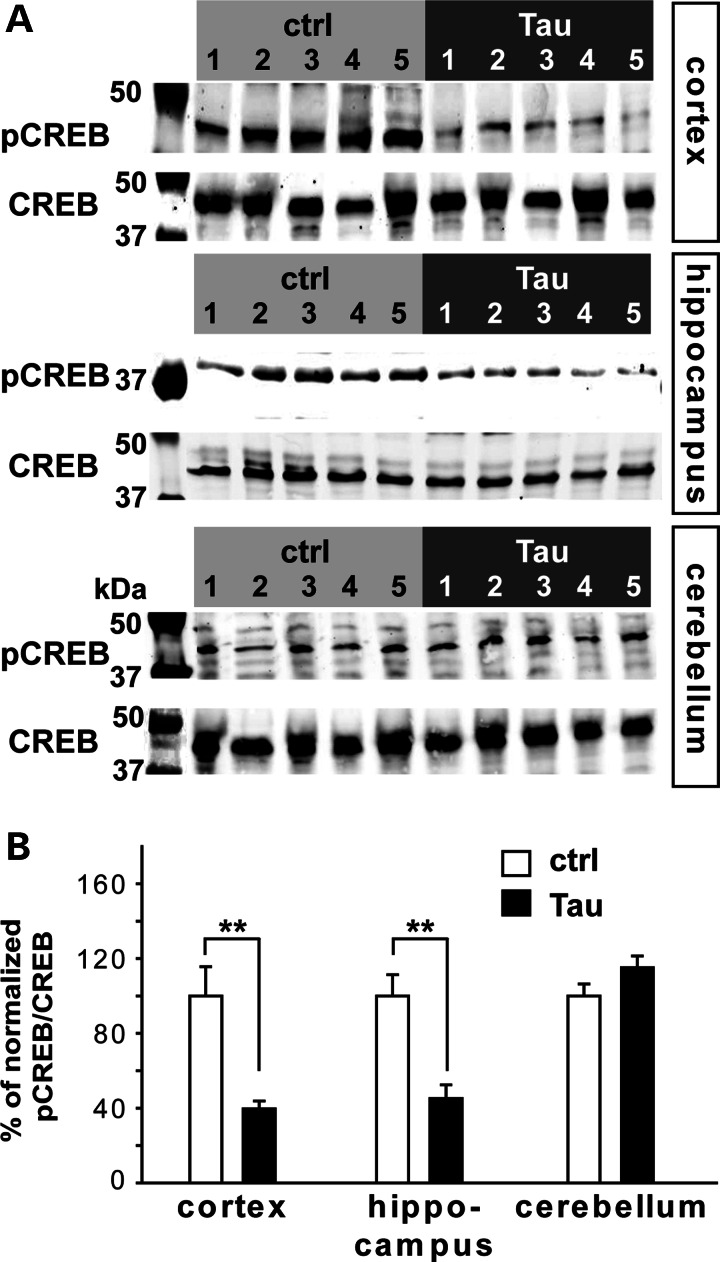
The CREB regulates gene transcription relevant to specific cognitive tasks and is critical for synaptic plasticity and learning/memory (reviewed in 38). When neurons are appropriately activated, CREB is phosphorylated at Ser133 (pCREB). pCREB then activates the transcription of several immediate early response genes (39,40). The expression of CREB-binding protein and many CREB downstream targets, for example brain-derived neurotrophic factor and cFOS are altered in models of AD (41–47). Interestingly, the abundance of pCREB (Ser133), but not the total level of CREB, is significantly reduced in 3xTg-AD mice (48), the transgenic mice harboring three mutated genes found in some familial AD: β-amyloid precursor protein (βAPPswe), presenilin-1 (PS1M146V) and tauP301L (49–51). Memory deficits and synaptic dysfunction in rTg4510 mice are first detected at 4.5 months of age (34,37). To determine whether CREB activity was reduced in the forebrain of rTg4510 mice when NMNAT2 level was reduced, we examined the levels of pCREB (Ser133) and total CREB in the brains of 2-month-old rTg4510 (n = 5) mice and their littermate controls (n = 5). Western blot analyses were conducted with tissue homogenates prepared from the cerebellum, cortex and hippocampus of these two groups of mice (Fig. 2). pCREB (Ser133) but not total CREB was significantly reduced in both the cortex and hippocampus of rTg4510 mice compared with control mice (Fig. 2A; Supplementary Material, Table S1). The ratios of pCREB (Ser133) to CREB in the cortex or hippocampus of rTg4510 mice were <50% of control values, while no difference was found in the cerebellum (Fig. 2B). The reduced abundance of two well-characterized CREB targets TrkB and cFos (52,53) provide further confirmation that CREB-mediated transcription is reduced in rTg4510 mice (Supplementary Material, Fig. S3). These data suggest that cAMP/CREB signaling is impaired in rTg4510 brains prior to the onset of cognitive deficits.
Figure 2.
pCREB levels are reduced in rTg4510 forebrains. (A) Western blots show a reduction in the level of pCREB (Ser133), but not total CREB, in 2-month-old rTg4510 cortex and hippocampus compared with littermate controls. pCREB (Ser133) levels were unaffected in the cerebellum of rTg4510 mice. (B) Summary of the ratios of pCREB (Ser133) to total CREB in the cortex, hippocampus and cerebellum (n = 5 for each genotype). Data are plotted as mean ± SEM (**P < 0.01).
nmnat2 transcription is regulated in a CREB-dependent manner
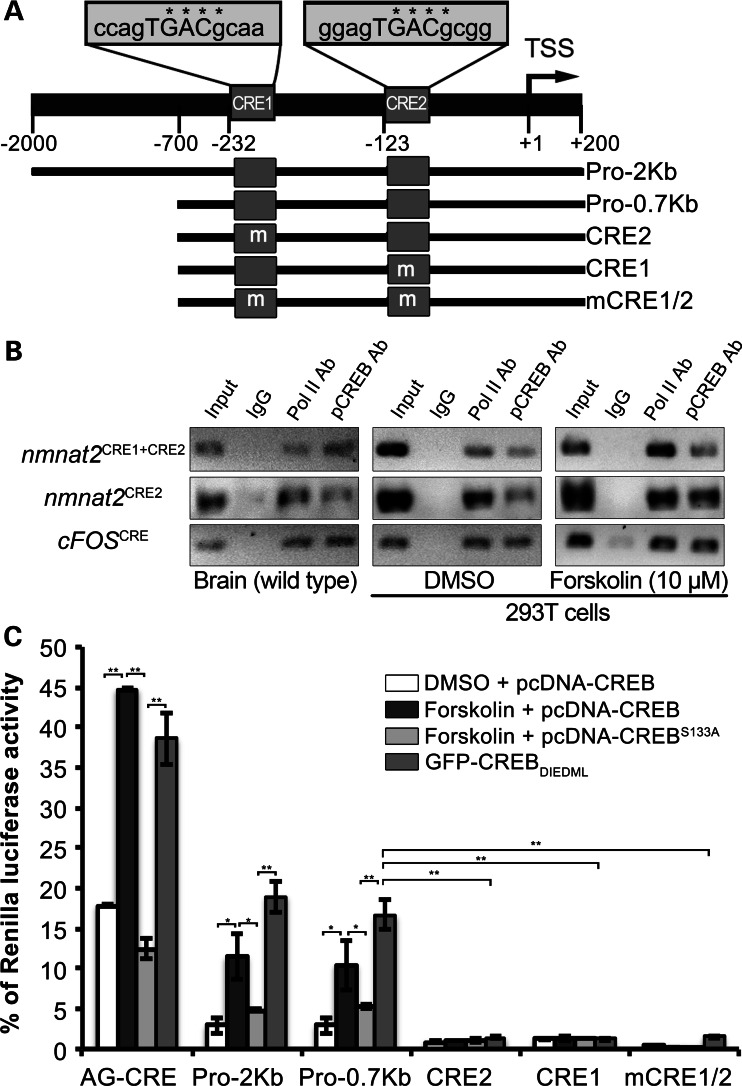
To examine whether reduced CREB activity can account for nmnat2 down-regulation in rTg4510 forebrains, we performed an in silico analysis of the mouse nmnat2 promoter region to search for possible CREB response elements (CRE) using Genomatix. This analysis identified two putative CREs close to the transcriptional start site (TSS) of the mouse nmnat2 gene (Fig. 3A). To test for potential pCREB binding to the putative CREs, we performed chromatin immunoprecipitation (ChIP) experiments with a pCREB (Ser133) antibody, using both mouse whole-brain extract and extracts from 293T cells treated with either dimethyl sulfoxide (DMSO) or 10 µm forskolin, an activator of adenylyl cyclases (54). Since previous reports have indicated a lack of NMNAT2 expression in 293T cells (25,26,55,56), we expressed the luciferase vector with the full nmnat2 promoter (Pro-2Kb, Fig. 3A) in these cells to detect binding of pCREB to either CREs. ChIP analysis in the wild-type brain demonstrated direct binding of pCREB to genomic regions containing both nmnat2 CRE sites as well as the single CRE2 region (Fig. 3B). Analysis in 293T cells also found pCREB binding to these regions and an increase in the amount of pCREB bound to the specific CRE-containing regions upon forskolin treatment (Fig. 3B). These results suggest the direct binding and occupation of pCREB at the CRE sites in the nmnat2 promoter region.
Figure 3.
nmnat2 transcription is directly regulated by CREB. (A) Diagram of the mouse nmnat2 promoter showing the sequence of the two predicted CREs proximal to the TSS. Schematic representation displays the different promoter fragments inserted in firefly luciferase vector. (B) Representative data from the ChIP assay with pCREB antibody using mouse whole-brain lysate or 293T cells transfected with Pro-2Kb luciferase vector and treated with either DMSO or forskolin. pCREB binds to both CRE1 + CRE2 and CRE2 containing regions in wild-type brain and 293T cells. Binding was enriched in 293T cells upon forskolin treatment. (C) Luciferase activity measured from 293T cells transfected with either pcDNA-CREB or pcDNA-CREBS133A or GFP-CREBDIEDML and either empty, AG CRE, nmnat2 promoter plasmid (Pro-2Kb or Pro-0.7Kb) as well as CRE mutants mCRE1:CRE2 (CRE2), CRE1:mCRE2 (CRE1) or mCRE1:mCRE2 (mCRE1/2) after 12h of treatment with or without 10 μm forskolin. Data are plotted as mean ± SEM (n = 3 for each group; *P < 0.05, **P < 0.001).
To determine the transcriptional functionality of the nmnat2 CRE sites, we then performed a Dual Luciferase assay (see Materials and Methods). The nmnat2 promoter region including either 2kb (Pro-2Kb) or 700bp (Pro-0.7Kb) upstream and 200 bp downstream of the TSS was cloned upstream of a luciferase expression cassette. The Pro-0.7Kb luciferase vector was then used as a template for site-directed mutagenesis of the specific CREs in various combinations (Fig. 3A). To render the CREs completely non-functional, the core 5′-TGACGC-3′ sequence was mutated to 5′-TGGGTA-3′. A luciferase vector containing three tandem CREs derived from the achorionic gonadotropin promoter (AG CRE) was used as a positive control (57). Furthermore, to ensure that any enhanced luciferase induction upon forskolin treatment was due to pCREB binding to either CREs, we transfected cells with either pcDNA-CREB or pcDNA-CREBS133A (a dominant-negative pCREB mutant), in addition to the specific firefly and Renilla luciferase vectors (Fig. 3C). Luciferase activity of cells transfected with vectors containing either AG CRE or nmnat2 promoter (Pro-2Kb or Pro-0.7Kb) was significantly increased upon forskolin treatment (Fig. 3C). Forskolin treatment increased the luciferase activity driven by the nmnat2 promoter (Pro-2Kb) by almost 4-fold from its basal level (2.9 ± 0.57% of normalized Renilla luciferase activity to 11.44 ± 1.62% of normalized Renilla luciferase activity; P = 0.001 between control and forskolin conditions). The luciferase activity driven by AG CRE was increased almost 3-fold from its basal level (17.79 ± 0.08% of normalized Renilla luciferase activity) upon forskolin treatment (44.69 ± 0.11% of normalized Renilla luciferase activity; P= 0.011 between control and forskolin conditions). Almost, no increase in luciferase activity was detected upon forskolin treatment if either or both of the CRE sites were mutated. Furthermore, the induction of luciferase activity seen with AG-CRE, Pro-2Kb or Pro-0.7Kb upon forskolin treatment was abolished in cells transfected with the dominant-negative pcDNA-CREBS133A, strongly suggesting the increase in luciferase activity was due to CREB phosphorylation.
To further confirm that CREB can regulate nmnat2 transcription, we examined the luciferase activity of all our constructs in the presence of a constitutively active form of CREB (CREBDIEDML). In cells transfected with CREBDIEDML, high levels of luciferase activity were found in the cells transfected with AG CRE (38.63 ± 3.2% of normalized Renilla luciferase activity; P < 0.001), Pro-2Kb (18.99 ± 1.9% of normalized Renilla luciferase activity; P < 0.001) or Pro-0.7Kb (16.61 ± 1.8% of normalized Renilla luciferase activity; P< 0.001) but not in cells transfected with a vector containing one or two mutated CRE sites (Fig. 3C). These data suggest that the induction of luciferase activity results from the direct binding of pCREB to the nmnat2 CRE sites. Furthermore, our luciferase studies indicated that both the CREs are required for nmnat2 transcription as mutating either one abolishes transcription of nmnat2, suggesting transcriptional regulation in a cooperative manner. Taken together with the data from ChIP assay with wild-type mouse brains, we conclude that nmnat2 is a direct target of CREB signaling via two CREs proximal to the transcription start site in its promoter.
To examine whether the reduced CREB-mediated transcription in rTg4510 brains could account for the decreased nmnat2 expression, we performed ChIP assays with the cortex, hippocampus and cerebellum from 2-month-old rTg4510 mice (n = 3) and their littermate controls (n = 3) to compare the amount of pCREB bound to the nmnat2 promoter area containing CRE sites (Fig. 4). ChIP analysis was repeated at least three times with all the appropriate controls (see Materials and Methods) and pCREB occupancy in the promoter region containing both CRE1 and CRE2 as well as the one containing only CRE2 were examined (Supplementary Material, Table S3). We found that pCREB occupancy was significantly reduced at both the promoter with CRE1 + CRE2 and the one with CRE2 alone in the rTg4510 cortex (P < 0.05 for CRE1 + CRE2; P < 0.001 for CRE2) and hippocampus (P < 0.001 for CRE1 + CRE2 and P < 0.05 for CRE2) (Fig. 4; Supplementary Material, Table S3). There was no difference in pCREB occupancy in rTg4510 cerebellum for either the promoter with CRE1 + CRE2 or CRE2 alone when compared with littermate controls (Fig. 4; Supplementary Material, Table S3). These data suggest that less pCREB binds to nmnat2 promoter in the brain regions of the rTg4510 mice that over-express tauP301L. In summary, our data suggest that cAMP/CREB signaling regulates nmnat2 transcription under physiological conditions. The significant nmnat2 down-regulation in brain regions with tauP301L over-expression is likely to be a direct consequence of reduced CREB activity.
Figure 4.
pCREB occupancy at the nmnat2 promoter is reduced in the cortex and hippocampus but not cerebellum of rTg4510 mice. (A and B) The ChIP assay was performed on the cortex, hippocampus and cerebellum from 2-month-old rTg4510 and littermate control mice. RNA pol II was used as a positive control and rabbit IgG as a negative control. pCREB occupancy at the nmnat2 promoter was studied in a region containing both CRE1 and CRE2 (A) as well as the one containing only CRE2 (B). (C and D) Summaries for the pCREB occupancies (normalized to input) onto nmnat2 promoter containing both CRE1 and CRE2 (C) or the promoter containing only CRE2 (D). Data are plotted as mean ± SEM (n = 3 for each group; *P < 0.05, **P < 0.001).
Exogenous expression of NMNAT1 or NMNAT2, but not NMNAT3, provides neuroprotection in rTg4510 hippocampi
The neurodegeneration found in Drosophila NMNAT loss-of-function mutants (23) and cultured mouse ganglia neurons with reduced NMNAT2 expression (27) suggests that NMNATs are required to maintain neuronal integrity. If reduced NMNAT2 levels in rTg4510 mice contribute to neuronal loss, over-expressing NMNAT2 or its homolog (NMNAT1 or NMNAT3) should reduce the neurodegenerative phenotype in rTg4510 mice. Recombinant adeno-associated viral (rAAV) vectors offer high-efficiency gene transfer and have gained popularity as gene therapy vectors (58–62). To examine the therapeutic potential of NMNAT1-3 over-expression in rTg4510 mice, serotype 6 rAAV vectors carrying human NMNAT1, -2 or -3 full-length cDNAs with IRES-enhanced green fluorescent protein (EGFP) (NMNAT1-, NMNAT2- and NMNAT3-rAAVs) were generated to over-express NMNATs in a tissue- and time-specific manner. rAAV with IRES-EGFP only (EGFP-rAAV) was used as a control for virus injection and ectopic protein expression. EGFP expression allows us to identify rAAV-transduced cells. Full-length cDNAs of NMNAT1–3 were individually cloned into the pAV4-EF-1-EGFP shuttle plasmid (Fig. 5A), permitting the transcription of cDNA-IRES-EGFP under direction of the elongation factor-1 (EF-1) promoter. A pseudotyped strategy was used to produce rAAV vectors packed with serotype 6 capsid proteins (see Materials and Methods).
Figure 5.
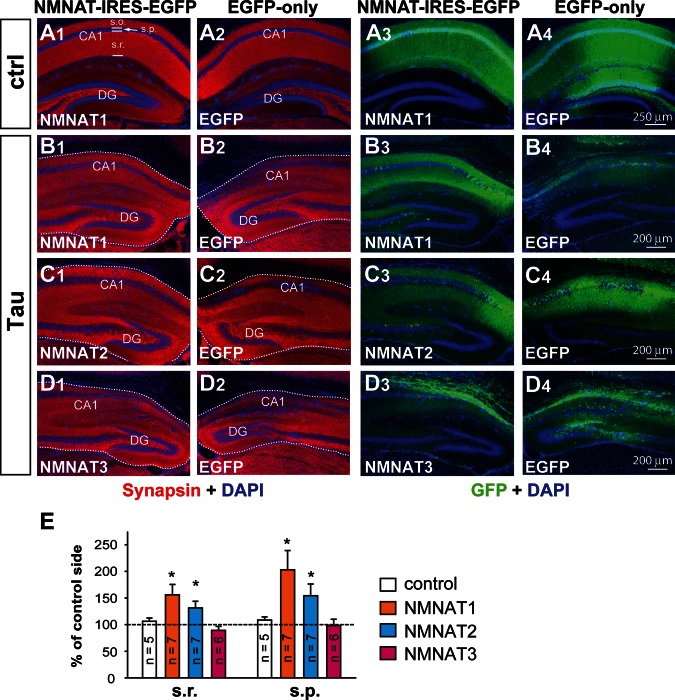
rAAV-mediated expression of NMNAT1–3 and EGFP in neurons. (A) Schematic representation of the AAV shuttle vector pAV4-EF-1-EGFP used for cloning NMNAT-AAV vectors. (B–D) High-titer rAAVs carrying NMNAT1-IRES-EGFP (N1-EGFP) or EGFP only were unilaterally injected into the hippocampal CA1 area of the right- or left-hemisphere of a 6-week-old Tau mouse, respectively. (B) Double EGFP (green) and NeuN (red) immunostaining showed that rAAV vectors primarily transduce neurons. B1 and B2 inserts show enlarged views (dashed box in B) for EGFP and NeuN immunostaining. (C and D) Dual EGFP (green) and NMNAT1 (N1, red) immunostaining shows strong ectopic expression of EGFP or NMNAT1 at 5 months of age (C). No NMNAT1 is detected in CA1 in neurons infected with EGFP alone (D). (E–G) The distinct subcellular distribution of ectopic NMNAT1-3 expression in cultured cortical neurons. Cortical neurons were infected by rAAVs at 2 DIV and stained with EGFP and NMNAT1, NMNAT2 or NMNAT3 antibodies at 8 DIV.
To assess the transduction efficiency of the rAAV vectors generated in this study and whether these rAAVs could drive strong and long-lasting expression in hippocampal neurons, 1 μl of 109–1010 particles of NMNAT1-EGFP-rAAV was stereotactically injected into the hippocampal CA1 area of the right hemisphere of a rTg4510 mouse at 6 weeks of age. The same amount of EGFP-rAAV was injected into the CA1 area within the left hemisphere of the same animal to serve as an internal control. NeuN (a neuronal marker), NMNAT1 and EGFP multiple fluorescence staining was conducted 3.5 months post-injection to examine the expression of NMNAT1 and EGFP in neurons. Almost all EGFP-positive cells were co-labeled with NeuN (Fig. 5B). EGFP was expressed at high levels in dendrites, and transgene expression had no detectable impact on the hippocampal structure (Fig. 6A). Strong NMNAT1 immunoreactivity was detected in the nuclei of EGFP-positive neurons within the NMNAT1-rAAV-injected hippocampus (Fig. 5C). No NMNAT1 immunoreactivity was detected in EGFP-positive cells within the EGFP-rAAV-injected side (Fig. 5D). Clearly, the rAAVs generated here support efficient and long-lasting expression of NMNAT and EGFP in neurons located near the injection site.
Figure 6.
NMNAT1 and NMNAT2 over-expression reduces neurodegeneration in Tau mice. NMNAT-IRES-EGFP or EGFP-only rAAVs were injected into the hippocampal CA1 area of 6-week-old control (A) and rTg4510 (B–D) mice. The neuroprotective effects of NMNAT over-expression on hippocampal morphology were assessed at 5 months of age. Double-staining images of EGFP (green) and synapsin (red) in sections prepared from the same rTg4510 mice at a similar level of the hippocampus (outlined with white dashed lines) show that the hippocampal CA1 and DG area appears larger on the NMNAT1 (B) or NMNAT2 (C) rAAV-injected side (B1, C1) compared with the contralateral side injected with EGFP-rAAV (B2, C2). (D) NMNAT3 over-expression did not have an impact on hippocampal size. Panels 3 and 4 show EGFP and DAPI staining of same slices shown in panels 1 and 2. The extensive EGFP-positive area in the CA1 region indicates successful rAAV injections. (E) Normalized thickness of the stratum radiatum and pyramidale layers (marked by white lines in A). Thicknesses were analyzed by taking six measurements per section and three sections per animal. The three 50 μm sections were separated by 200 μm. The value from each NMNAT rAAV-injected side was normalized to the value on the EGFP-rAAV-injected side in the same animal. Data are presented as percentages to the average value of control side and plotted as mean ± SEM (*P < 0.05). Abbreviations: s.p., stratum pyramidale; s.r., striatum radiatum; s.o., stratum oriens.
To examine the subcellular distribution of NMNAT1-3 driven by rAAVs generated here, cultured cortical neurons derived from ICR mice were transduced with rAAV at 2 days in vitro (DIV) and NMNAT expression was examined at six DIV. Only EGFP-positive neurons were detected by anti-NMNAT1, NMNAT2 or NMNAT3 antibody. As expected, NMNAT1 immunoreactivity was found in the nucleus (Fig. 5E), while NMNAT2 immunoreactivity was found in neurites (Fig. 5F), but not in the nucleus. NMNAT3 was mainly found in the cytoplasm (Fig. 5G). This differential subcellular distribution is consistent with previous observations that found NMNAT1 in the nucleus, NMNAT2 in the Golgi complex and neurites and NMNAT3 in the mitochondria (25,26,55,56).
To evaluate the effect of NMNATs over-expression on neurodegeneration, we injected 1 μl of 109–1010 particles of rAAVs into the hippocampi of 6-week-old rTg4510 mice. This age was chosen so NMNATs would be over-expressed after the onset of tauP301L expression (postnatal day 7) (35), but before the levels of endogenous NMNAT2 substantially declined (2 months in the hippocampus). NMNAT1-, NMNAT2- or NMNAT3-rAAV was injected into the CA1/dentate gyrus area of the hippocampus in the right hemisphere, while EGFP-rAAV was injected into the left hemisphere as an internal control. A total of 41 rTg4510 and 15 control mice were bilaterally injected with rAAVs (see Material and Methods). The neuroprotective potential of NMNAT-AAV was evaluated 3.5 months post-injection (at 5 months of age) with EGFP and synapsin (an abundant presynaptic protein) double staining (Fig. 6). Neurons transduced by rAAV were identified by the green fluorescence from the IRES-EGFP expression cassette. Only animals with clear and equal EGFP in the CA1 area of both hemispheres were selected for further analyses. No detectable abnormality was caused by rAAV injections into control mice (Fig. 6A; n= 5). The size of the hippocampal CA1 areas in rTg4510 mice receiving NMNAT1- (Fig. 6B; n = 7) or NMNAT2-AAV (Fig. 6C; n = 7) injections was qualitatively larger than the ones with control EGFP-AAV injections. In contrast, the hippocampi from NMNAT3- and EGFP-AAV injected hemispheres of rTg4510 mice were of similar size (Fig. 6D; n = 6).
To quantify the impact of NMNAT over-expression on CA1 structures, we measured the thickness of the stratum radiatum and the stratum pyramidale guided by synapsin immunoreactivity and DAPI staining (see Materials and Methods). To account for variability in the extent of neuronal loss among individual rTg4510 mice, the thickness of the stratum radiatum and stratum pyramidale layers measured from the test side (NMNAT1–3 injected side) was normalized to the EGFP-rAAV-injected side at comparable anatomical locations. The widths of the stratum radiatum and stratum pyramidale of NMNAT1 over-expressing CA1 were significantly larger than the EGFP expressing side (Fig. 6E; stratum radiatum, 156 ± 19%, P = 0.027; stratum pyramidale, 203 ± 36%, P = 0.029). NMNAT2 expression also offered substantial protection of the CA1 anatomical structures (Fig. 6E; stratum radiatum, 131 ± 13%, P = 0.047; stratum pyramidale, 154 ± 22%, P = 0.049). However, in mice with NMNAT3 over-expression, the widths of the stratum radiatum and stratum pyramidale were similar to the EGFP expressing side (stratum radiatum, 89 ± 8%, P = 0.657; stratum pyramidale, 98 ± 12%, P = 0.883). Our data suggest that over-expression of NMNAT1 or NMNAT2, but not NMNAT3, is protective against hippocampal neuronal loss in rTg4510 mice.
To further quantitatively evaluate the impact of NMNAT2 overexpression, western blot analysis was conducted with tissue homogenates prepared from EGFP-rAAV and NMNAT2-rAAV-transduced hippocampi from rTg4510 mice 3.5 months after injection (Fig. 7). These mice were injected at 2 months of age with NMNAT2-rAAV into the right hippocampus, while EGFP-rAAV was injected into the left. NMNAT2 abundance in the NMNAT2-rAAV-injected side was about twice the level found in the EGFP-rAAV side (Fig. 7A and B; ctrl, 100 ± 30%; NMNAT2, 202 ± 17%; P = 0.02). The significant increase in NF-M in NMNAT2 over-expressing side (Fig. 7A and C; ctrl, 100 ± 18%; NMNAT2, 207 ± 32%; P = 0.02) suggests that more neurons survived in rTg4510 hippocampi over-expressing NMNAT2. Interestingly, we also observed reduced levels of activated cleaved caspase 3 (Fig. 7A and D; ratios of activated cleaved caspase 3 to un-cleaved caspase 3: ctrl, 100 ± 26%; NMNAT2, 17 ± 12%; P = 0.01) and GFAP (Fig. 7A and E; ctrl, 100 ± 10%; NMNAT2, 65 ± 13%; P < 0.05) in the NMNAT2-injected rTg4510 hippocampi compared with the EGFP injected rTg4510 hippocampi. Thus, NMNAT2 overexpression reduces apoptosis and gliosis in rTg4510 hippocampi.
Figure 7.
Western analysis of rTg4510 hippocampi over-expressing EGFP- or NMNAT2-IRES-EGFP. (A) Western blots show immunoreactivity of NMNAT2, NF-M, GFAP, actin, caspase 3 and activated cleaved caspase 3 in the hippocampus of rTg4510 mice overexpressing either EGFP- or NMNAT2-IRES-EGFP (N2-EGFP). (B–E) Summary results for the normalized ratios of NMNAT2/actin (B), NF-M/actin (C), cleaved/total caspase 3 (D) and GFAP/actin (E). (F) Western blots show CP-13 (hyperphosphorylated tau), MC-1 (Tau with pathological conformation) and human Tau immunoreactivity in the hippocampus of rTg4510 mice over-expressing either EGFP- or N2-EGFP. (G and H) Summary results for the normalized ratios of CP-13/hTau (G) and MC-1/hTau (H). Data are presented as percentages of the average value of the control side and plotted as mean ± SEM (three animals; *P < 0.05, **P < 0.01).
We also investigated whether overexpression of NMNAT2 affects the levels of tau that are hyperphosphorylated or have abnormal conformations (Fig. 7F). CP-13 antibody recognizing tau phosphorylated on Ser202 is commonly used to detect tau pathology in both early and more advanced stages of NFT accumulation, while MC-1 antibody has been used to detect an abnormal tau conformation. We found abundant CP-13 and MC-1 reactivity in rTg4510 hippocampi as previously described (34). Expressing NMNAT2 in the rTg4510 hippocampi significantly reduced CP-13 (Fig. 7F and G; the ratios of CP-13 to human tau: ctrl, 100 ± 13%; NMNAT2, 39 ± 7%; P < 0.01) and MC-1 (Fig. 7F and H; the ratios of MC-1 to human tau: ctrl, 100 ± 13%; NMNAT2, 42 ± 10%; P = 0.01) immunoreactivity, compared with the control side expressing EGFP. These results suggest that overexpression of NMNAT2 protects against tau-induced degeneration by reducing the toxic hyperphosphorylated tau load and ameliorates tau-induced cytotoxicity.
DISCUSSION
In this study, we found that nmnat2 transcription was significantly reduced in brain areas over-expressing tauP301L in 1-month-old rTg4510 mice, an FTDP-17 tauopathy animal model. The initial reduction in nmnat2 expression coincided with peak tauP301L expression and remained low throughout the life. We demonstrated CREB-dependent regulation of nmnat2 transcription through two CREB-binding sites in the nmnat2 promoter using a combination of ChIP and luciferase activity assays. The early reduction in both the total pCREB (Ser133) levels and the pCREB bound to the nmnat2 promoter in rTg4510 forebrain suggests that defective CREB-mediated transcription in rTg4510 brains accounts at least in part for reduced NMNAT2 expression. Using rAAV to over-express NMNAT2 or its homolog NMNAT1 in the hippocampus of rTg4510 mice, we could reduce the extent of neurodegeneration. Taken together, our studies demonstrate down-regulation of NMNAT2 and CREB activity in the central nervous system (CNS) of a tauopathy animal model. Notably, these reductions precede the onset of neurodegeneration. This is the first demonstration that NMNAT1 or NMNAT2, but not NMNAT3, over-expression provides neuroprotection in the mammalian brain.
Reduced CREB signaling in rTg4510 mice
CREB has been called ‘the memory gene’ for several species and its activity is regulated through many activity-driven signaling cascades (38,63–65). Genetic manipulations that decrease CREB-mediated transcription lead to deficits in both synaptic plasticity and long-term memory, whereas over-expression of CREB results in memory enhancements (38). CREB is also important for cell survival (65,66). Removing the creb1 gene in adult mouse forebrain or chronic inhibition of CREB function leads to extensive neurodegeneration in the hippocampus (67,68). CREB binds to CREs in the promoter region of their target genes (69), but will only activate gene transcription upon phosphorylation at Ser133 (40). In human AD brains, pCREB immunoreactivity was significantly decreased (70). Defective CREB signaling has been postulated to mediate the effect of Aβ on hippocampal function (46,51,71,72) and pCREB levels are also reduced in 3xTg mice at 6 months of age (48). We found that pCREB(Ser133), but not total CREB levels, are reduced in rTg4510 mice prior to the onset of neurodegeneration. No difference was found in the levels of P300, cREL or NFATc (data not shown), transcription factors that have been implicated in different neurodegeneration models (73–76).
How might tauP301L overexpression reduce pCREB levels? NMDAR (N-methyl-D-aspartate receptors)-mediated calcium influx is known to increase pCREB levels through several signaling pathways (65). A recent study by Hoover et al. (37) found that rTg4510 neurons have fewer NMDARs on their dendritic spines due to the mislocalization of tau protein from axons to dendritic spines. NMDAR-dependent long-term potentiation was impaired in the Schaffer collateral pathway of rTg4510 mice at 4.5 months of age. Aβ oligomers also cause tau mislocalization (77,78). It remains to be determined whether reduced NMDAR function in rTg4510 mice accounts for lower levels of pCREB. NMDARs have been a popular target for enhanced cognition (reviewed in 38). Various transgenic and pharmacological approaches that augment NMDAR signaling have been found to increase learning and memory in mice, for example, NR2B overexpression and application of NMDAR partial agonist, D-cycloserine (79,80). It will be important to determine whether this strategy can reverse the age-dependent decrease in NMNAT2 and pCREB levels and the cognitive deficits in rTg4510 mice.
Such reduced CREB activity in rTg4510 mice could be the major cause for the down-regulation of nmnat2 transcription. We identified two functional CREs proximal to the TSS of the mouse nmnat2 gene. ChIP analysis demonstrated direct binding of pCREB to these CREs in mouse brains and less pCREB bound to the nmnat2 promoter in the cortex and hippocampus of 2-month-old rTg4510 mice. Mutating either one of the CRE sites located in nmnat2 promoter abolishes nmnat2 transcription. In addition to the reported regulation of NMNAT2 stability through the proteasome pathway (27), our results provide additional insight into the multi-faceted regulation of this neuronal maintenance factor at the transcriptional level. nmnat2 is likely to be a CREB target necessary for maintaining neuronal health at physiological conditions. It is likely that a small change in CREB activity results in a rapid decline of nmnat2 levels. Multiple signaling pathways, such as the AKT pathway, leading to regulation of pCREB activity would offer tight regulation of NMNAT2 levels (65). Our data suggest that tauP301L over-expression impairs CREB-mediated transcription and leads to a decrease in nmnat2 expression prior to the onset of neurodegeneration. The down-regulation of CREB activity in rTg4510 mice and the subsequent reduction in NMNAT2 could partially account for the cognitive deficits and neurodegeneration induced by tauP301L over-expression.
Over-expression of NMNAT2 and NMNAT1 reduce neurodegeneration in rTg4510 hippocampus
The role of NMNATs in neuroprotection was first implicated in studies using Wallerian Slow mice (WldS; reviewed in 81). In this spontaneous mutant, the Wallerian degeneration process is significantly delayed (82). The protective effects occurring in these mice are attributed to a chimeric gene Ube4b/NMNAT1, which encodes an in-frame fusion protein of the amino-terminal of ubiquitination factor E4B (UBE4B) and the complete coding region of NMNAT1 (83,84). The protective property of the WldS protein extends beyond injury-induced Wallerian degeneration. In the CNS, WldS has been shown to protect nigrostriatal axons from degeneration in a model of Parkinson's disease, to reduce the number of axonal spheroids in mice with gracile axonal dystrophy, and to protect axons from neuronal degeneration induced by toxic insults (85–87). Together with the broad protective effects of NMNAT found in Drosophila (21,23,88), it has been proposed that WldS or NMNATs protect neurons at a step common to many degenerative processes. In our study, we found that over-expression of NMNAT1 and NMNAT2, but not NMNAT3, is neuroprotective in a mouse FTDP-17 model. NMNAT3-transgenic mice show delayed Wallerian degeneration similar to WldS mice (33). This interesting difference in NMNAT3 neuroprotection efficacy for peripheral nerve damage versus central tauopathy suggests that the distinct NMNATs may have distinct modes of protection in different forms of neurodegeneration. Nevertheless, our study provides the first evidence for a role of NMNATs in protecting against neurodegeneration in the mammalian CNS.
Possible mechanisms of NMNAT neuroprotection
NMNATs are essential enzymes for NAD biosynthesis and several studies have found the enzymatic activity to be important for their neuroprotective effect (reviewed in 81,89). For example, the axon-protective function of NMNAT2 in cultured neurons derived from the superior cervical ganglia requires NAD synthesis activity (90). NAD+ is used to synthesize NADP+, whose reduced form NADPH is involved in defense against oxidative stress (91). Interestingly, NMNAT over-expression can prevent axonal damage resulting from exogenous application of oxidants (28). Mitochondria dysfunction has been demonstrated in tauP301L transgenic mice (92–94) and it is possible that increased levels of NMNAT1 and NMNAT2 protect tauP301L over-expressing neurons by reducing oxidative damage. However, normal NAD and NADP levels were found in the cortex and hippocampus of rTg4510 mice.
NMNATs can also act as chaperones (21). Over-expression of NMNAT substantially reduced the formation of protein aggregates such as hAtx-1[82Q] protein, the cause of spinocerebellar ataxia 1, in Drosophila. Thus, exogenous NMNAT1 or 2 may be neuroprotective by reducing the level of toxic tau species. Consistent with this idea, we found that NMNAT2 overexpression in rTg4510 hippocampus significantly reduced the levels of hyperphosphorylated tau and tau with pathological conformation(s). These findings support the hypothesis that NMNAT-mediated chaperone activity promotes the clearance of toxic species. In this study, we used the immunoreactivity of synapsin, a synaptic marker, to estimate neuronal morphology and anatomical integrity in rTg4510 mice. Using western blot analysis, we also observed reduced apoptosis and gliosis in the NMNAT2-injected hippocampus, compared with the control side. It remains to be determined whether rescue of neurodegeneration by NMNAT1 or -2 over-expression can alleviate the synaptic and cognitive deficits observed in rTg4510 mice.
In summary, we demonstrate that NMNAT2 is an important maintenance factor for mammalian brain health and its expression can be regulated through CREB-mediated transcription. Maintaining CREB activity and endogenous NMNAT2 levels may serve as novel avenues for therapies to protect against the neurodegeneration that accompany tauopathies.
MATERIALS AND METHODS
Mice and genotyping
In this study, we used mice that over-express the P301L mutation in 4R0N human tau associated with FTDP-17. The generation of rTg(tauP301L)4510 mice (abbreviated as rTg4510 mice) has been described previously (34,35). In brief, rTg4510 mice were generated by F1 crossing of responder and activator transgenic lines. Responder mice carried tauP301L cDNA with an upstream tetracycline-operon responsive element and were maintained on a FVB/N background. Activator mice contained a trans-activator gene consisting of the tetracycline-off open reading frame placed downstream of the CaMKII promoter and were maintained on a 129Sv6 background. Both transgenic lines were gifts from Michael Hutton (Mayo Foundation) and Karen Ashe (University of Minnesota Medical School). The resulting F1 progenies from the cross between activator and responder mice were on a mixed 129Sv6 x FVB/N genetic background. Littermates lacking both the activator and the responder transgenes were used as controls. Mouse genotypes were determined from tail biopsies using real-time qPCR with specific probes designed for each gene by a commercial service (Transnetyx, Cordova, TN, USA). Animal housing and use were in compliance with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the institutional animal care committee at Baylor College of Medicine.
Viral vector production and stereotaxic injection
Full-length human NMNAT1, NMNAT2 and NMNAT3 cDNAs were cloned into BamHI or NotI sites 5′ to the expression cassette-IRES-EGFP driven by the EF-1 promoter within the AV4-EF1-GFP shuttle plasmid (Fig. 5A). This shuttle plasmid contains the AAV serotype 2 5′- and 3′-inverted terminal repeats derived from pSub201 (95), and a p5 integration efficiency element (p5IEE) (96) 5′ to the expression cassette. Serotype 6 rAAVs were produced in 293T cells by transfecting both the shuttle plasmid carrying one NMNAT gene and a pDF6 helper plasmid (a gift from Mark A. Kay, Stanford University) using a calcium phosphate transfection protocol (97). Cells were harvested 48h after transfection and rAAV particles were purified by iodixanol density centrifugation (98), dialyzed against phosphate buffered saline (PBS) without Mg2+ and Ca2+ (Invitrogen, Carlsbad, CA, USA) and concentrated using an Amicon Ultra-15 centrifugal filter (100 000 Da cut off, 0.0062 µm, Millipore, MA). Titration of DNA-containing particles was carried out using SYBR Green (BioRad, Hercules, CA, USA) real-time qPCR with the following 5IEE specific primers, along with serial dilutions of AAV vector plasmid as standard. 5′ upstream primer: 5′-GTG GAG TCG TGA CGT GAA TTA C-3′; 3′ downstream primer: 5′-AAT GGA GAC CCT GCG TGC TC-3′. Agilent Mx3005P QPCR system and data analysis software (Santa Clara, CA, USA) were used for analysis. All viral stocks were adjusted to 5 × 109 virus particles/µl.
To assess the effect of NMNAT over-expression on the neurodegeneration found in rTg4510 mice, we injected rTg4510 and control mice with rAAVs (NMNAT1, -2 or -3, with EGFP alone as control) at 6 weeks of age and then processed the mice at 5 months of age. For viral injections, mice were deeply anesthetized with an intraperitoneal injection (3 ml/kg) of a rodent anesthetic cocktail containing ketamine 37.6 mg/ml, xylazine 1.92 mg/ml and acepromazine 0.38 mg/ml. Following establishment of anesthesia, mice were secured in a stereotactic frame (Narishige International, East Meadow, NY, USA). We injected 1 µl rAAV-NMNAT1, -2 or -3 into the right and 1 µl of rAAV-EGFP into the left hippocampus of 6-week-old rTg4510 (n = 16 for NMNAT1-rAAV, n = 14 for NMNAT2-rAAV, n = 11 for NMNAT3-rAAV) and control (n = 6 for EGFP-rAAV, n = 2 for NMNAT1-rAAV, n = 3 for NMNAT2-rAAV, n = 4 for NMNAT3-rAAV) mice at the following co-ordinates measured from bregma: A: 2 mm, L: 1–1.5 mm, V: 1.2 mm (tooth bar set at zero). The injections were done using a polished glass micropipette coupled to a Nanospritzer (Nanoject II; Drummond Scientific Company, Broomall, PA, USA) set to inject 50 ml per push, giving an injection speed of ∼0.75 µl/min. The micropipette was left in place for 1 min after injection in order to minimize back diffusion along the cannula track. A single injection of rAAV-EGFP particles usually resulted in selective EGFP expression in the CA1 of the injected side that spread as much as a millimeter in the rostrocaudal axis (data not shown).
Neuronal cultures
Cortical neurons were prepared essentially as described in Sasaki et al. (32). In brief, we isolated primary neurons from the forebrains of embryonic day 16.5 wild-type mice (ICR strain) using a papain dissociation kit (Worthington Biochemical Corp, Lakewood, NJ, USA) according to the manufacturer's directions. Cells were seeded onto poly-l-lysine (Sigma-Aldrich, St Louis, MO, USA) coated 12 mm diameter glass cover slips (VWR, West Chester, PA, USA) in 24-well tissue culture plates (BD, Franklin Lakes, NJ, USA). Neurons were cultured in Neurobasal media supplemented with B27, antibiotics and glutamine (all from Invitrogen). Cells were grown for 2 days before transduction with rAAV (∼7.5 × 109 particles/well), grown for another 6 days and then fixed in 4% paraformaldehyde (PFA) in PBS, pH 7.4 and processed for immunofluorescence.
Tissue processing
Mice (control and rTg4510) used for real-time qPCR and protein analysis were sacrificed at 2 weeks, 1 month and 2 months of age (n = 3 per genotype and age). Brains were dissected into the cortex, hippocampus and cerebellum and snap frozen. One half was used for qPCR, and the other for protein analysis. Tissue was kept at −80°C until use. Mice injected with rAAV at 6 weeks of age were processed at 5 months of age. Following establishment of anesthesia as described above, mice were transcardially perfused with ice-cold PBS, pH 7.4, followed by ice-cold 4% PFA in PBS, pH 7.4. The brains were then post-fixed with the same fixative overnight at 4°C, washed with PBS and stored in 0.1% sodium azide/PBS at 4°C until processing.
Brains were serially sectioned in the coronal plane through the hippocampus into 50 µm thick sections using a Leica VT-1000 vibrating microtome (Leica Microsystems, Bannockburn, IL, USA). These tissue sections stayed free-floating for all subsequent washes and incubations. Sectioned brains were scored for successful injection by evaluating the intensity and location of EGFP fluorescence. Only the brains with similar amounts of EGFP in the CA1 area of hippocampus in both hemispheres were chosen for further immunofluorescence analysis.
Immunofluorescence staining
Immunofluorescence staining was performed on both free-floating sections and cultured neurons. For the rAAV-injected animals, every third section through the hippocampus was selected for EGFP and synapsin double staining. Two sections from each mouse with rAAV-NMNAT1/rAAV-EGFP injections were labeled using anti-NMNAT1 antibodies. rAAV-transduced cultured neurons were labeled with NMNAT1, 2 or 3 antibodies, respectively, to verify that the rAAV expressed the appropriate NMNAT.
Free-floating sections or cultured neurons on cover slips were washed with PBS/0.01% Triton X-100 (PBST) and permeabilized with 0.2% Triton X-100 in PBS at room temperature for 20 (sections) or 5 (cells) min. Free-floating sections or cover slips were then washed with PBST and blocked for 1h with 3% normal goat serum in PBST at room temperature. Free-floating sections were then incubated in a mixture of the two following antibodies: GFP (chicken polyclonal, 1/1000, Aves Lab, Tigard, OR, USA), synapsin (rabbit polyclonal, 1/500, Synaptic Systems, Göttingen, Germany), NeuN (mouse monoclonal, 1/1000, Chemicon, Temecula, CA, USA), NMNAT1 (rabbit polyclonal, 1/500, Abcam, Cambridge, MA, USA), while cultured neurons were incubated in anti-GFP (as above) and one of the following three antibodies: anti-NMNAT1, anti-NMNAT2 (mouse monoclonal, 1/300, Abcam) or anti-NMNAT3 (mouse monoclonal, 1/200, Abcam). All primary antibodies were diluted in PBST/1% normal goat serum and incubations were at 4°C overnight. The next day, sections and cells were washed with PBST, and incubated with goat anti-chicken Alexa 488 and goat anti-rabbit or goat anti-mouse 594 secondary antibodies (all at 1/500, Invitrogen) in PBST at room temperature for 2h. Following this incubation, sections or cells were washed with PBST three times for 10min each. Sections and cells were mounted in Vectashield mounting media with DAPI (Vector Labs, Burlingame, CA, USA) and cover slipped for imaging. Fluorescence microscopy was performed using a Zeiss AxioImager M1 system equipped with epifluorescence filters, a Zeiss monochrome digital camera and AxioVision software. All images were processed in Adobe Photoshop for brightness/contrast, orientation and background correction to better illustrate staining patterns.
Morphometric analysis
Images of hippocampi in coronal sections of rAAV-injected brains stained with synapsin and GFP antibodies and with DAPI counterstain were taken using a 5× objective on a Zeiss AxioImager M1. Exposure time was adjusted to avoid saturation. Three comparable (in regard to rostrocaudal level and site of injection) sections separated by 200 μm in the rostrocaudal axis were imaged and analyzed for each animal. For morphometric analysis of the thickness of the pyramidal and stratum radiatum layers of CA1, images were scored using Zeiss Axiovision software. For each slice and each side (either NMNAT1, -2 or -3 on one side, EGFP control on the other) and layer, six measurements (spaced 100 μm apart) of the thickness of the layer were taken. The pyramidal layer of CA1 was identified by DAPI counterstaining, while the dendritic field of the stratum radiatum was defined using synapsin immunofluorescence. The NMNAT1, -2 or -3 side in each animal was compared with its own EGFP control side and data were expressed as percent change from control ± SEM.
RNA extraction and quantitative real-time PCR
Total RNA was extracted by TRIzol reagent (Invitrogen), according to the manufacturer's protocol. For each sample, RNA concentration was determined by spectrophotometry at 260 nm. Two micrograms of RNA were used for the reverse transcription reaction with the High Capacity RNA to cDNA kit (Applied Biosystems, Carlsbad, CA, USA). Quantification of nmnat2 mRNA was performed using an ABI PRISM 7000 sequence Detection System (Applied Biosystems) and a real-time PCR System (Epicentre Biotechnologies, Madison, WI, USA). A housekeeping gene, gapdh, was used as an internal control to normalize mRNA expression. Amplification mix (25 μl) consisted of 0.1 µg cDNA, 12.5 μl TaqMan Universal PCR Master Mix (Applied Biosystems) and 1 μl TaqMan Gene Expression Assay (nmnat2, Mm00615393_m1, GAPDH, Mm99999915_g1). Samples were amplified by a PCR program of 40 cycles of 10 s at 95°C, 15 s at 55°C and 1 min at 72°C. Raw Ct values were obtained for a set of three rTg4510 and age-matched control animals per group in triplicate for each of the repeated experiment. The Ct value was defined as the number of cycles required for the fluorescence to exceed the detection threshold and the data were analyzed using the 2−ΔΔCt method to quantitatively assess relative changes in gene expression (99). Briefly, the raw Ct values were averaged for animals per group (rTg4510 and the age-matched littermate control animals) for both nmnat2 and gapdh. ▵Ct values were calculated by subtracting the average gapdh Ct value from that of nmnat2. The relative expression of nmnat2 in rTg4510 animals compared with controls was obtained by calculating the ΔΔCt value for each group by subtracting the average ΔCt value of control nmnat2 from rTg4510 average ΔCt. The relative fold change of rTg4510 groups to control was expressed as 2−ΔΔCT.
Protein extraction and western blotting
For protein extraction, tissues were homogenized in lysis buffer consisting of 50 mm Tris (pH 7.5), 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, protease inhibitor cocktail (Sigma, St Louis, MO, USA) and phosphatase inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). Total protein concentration was measured using the Bradford Reagent (Thermo Scientific/Pierce Biotechnology, Rockford, IL, USA).
Ten micrograms of total protein lysate per sample were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane, and probed with antibodies against NMNAT1 (1/1000; Abcam), NMNAT2 (1/1000; Abcam), NMNAT2 Blocking Peptide (20 μg, Santa Cruz Biotechnology), NMNAT3 (1/1000; Abcam), GFAP (1/4000; DAKO, Carpinteria, CA, USA), neurofilament medium (NF-M, 1/4000; Encore, Gainesville, FA, USA), actin (1/4000; Sigma), CREB (1/1000; Abcam), phospho(Ser133)CREB (pCREB, 1/1000; Abcam), hTAU (1/1000; DAKO), MC-1 (1:1000, gift from Peter Davis), CP-13 (1:1000, gift from Peter Davis), anti-Caspase 3 (1:1000, Cell Signaling, Beverly, MA, USA), anti-cleaved Caspase 3 (Asp175) (1:1000, Cell Signaling), TrkB (1:500, Cell Signaling), cFos (1:500, Cell Signaling) or GFP (1:1000, Invitrogen). Western blot analysis was performed with infrared dye conjugated secondary antibodies, IR700 and IR800 (LI-COR Biosciences, Lincoln, NE, USA). Blots were imaged and processed on an Odyssey® Infrared Imaging System. Densitometry analysis was performed on all the blots using ImageJ software (NIH). Data are represented as means ± SEM. Statistical analysis was performed using Student's t-test unless specifically indicated otherwise. Values of P< 0.05 were considered statistically significant.
nmnat2 promoter cloning and mutagenesis
In order to study the functionality of the putative CREs identified in the nmnat2 promoter region, a 2 kb and 700 bp fragment upstream of the TSS of nmnat2 including the putative CREs was cloned into a minimal PGL4 luciferase vector (Promega, Madison, WI, USA). Mouse genomic DNA was used as an amplification template using a Failsafe PCR Kit (EpiCenter Technologies) with the following amplification conditions: 25 cycles of 50 s at 95°C, 50 s at 61°C and 1 min at 72°C. PCR amplification primers include: 2Kb-Pro (forward) 5′-GCT AGC ATA ATT ATG TTA ATT GCC CAG CTT CT, Pro-0.7Kb (forward) 5′-CTA GCT AGC ACC CAT AAT TCA ACC CAC ACA AAG GAC CAA C-3′, 2Kb/700bp-Pro (reverse) 5′-CCC AAG CTT GGA CTC ACC GAA CAT CTG AAT GTG CCC TTT A-3′. For mutagenesis of the CREs, the 700bp-Pro-PGL4 vector served as the template using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, CA, USA), according to the manufacturer's protocol. The 5′-TGACGC-3′ core sequence in both CREs was mutated to 5′-TGGGTA-3′ with the following PCR cycling conditions for an 18-cycle reaction: 30 s at 95°C, 60 s at 61°C and 5 min at 68°C. The following primers were used for mutagenesis: mCRE1 (forward) 5′-GAA CCA AGA CCA GTG GGT AAA AAG AAG CTA GAG AGG-3′, mCRE1 (reverse) 5′-CCT CTC TAG CTT CTT TTTA CCC ACT GGT CTT GGT TC-3′, mCRE2 (forward) 5′-GAG GCG GGG AGT GGG TAG GTT TGC GTC TAG AG-3′, mCRE2 (reverse) 5′-CTC TAG ACG CAA ACC TAC CCA CTC CCC GCC TC-3′.
Dual luciferase reporter assay
293T cells were double-transfected using Lipofectamine Reagent (Invitrogen) with a Renilla luciferase vector, -36prl-Renilla luciferase (a gift from Dr Michael Kapiloff, Univ. of Miami) and a firefly luciferase vector of either nmnat2 promoter-PGL4 luciferase (Pro-2Kb or Pro-0.7Kb containing both CREs), mCRE1:CRE2-PGL4 luciferase (CRE1 mutated only), CRE1:mCRE2-PGL4 luciferase (CRE2 mutated only), mCRE1:mCRE2-PGL4 luciferase (both CRE mutated) or -36prl-CRE- luciferase (gift of Dr Michael Kapiloff). -36prl-CRE- luciferase vector contains three copies of CRE in front of the −36 bp prolactin minimal promoter and was used as a positive control. In addition to these plasmids, cells were additionally transfected with either pcDNA-CREB or pcDNA-CREBS133A (a dominant-negative pCREB mutant) or GFP-CREBDIEDML (constitutively active CREB). Forty-eight hours after transfection, cells were treated with either DMSO or 10 μm forskolin in DMSO as described (100) for 12h. Cells were then lysed and assayed for luciferase activity using the Dual-Glo Luciferase System (Promega) by luminescence measurement using BMG Omega® plate reader (Imgen Technologies, Alexandria, VA, USA).
Chromatin immunoprecipitation (ChIP) analysis
For ChIP analysis, both wild-type and rTg4510 mouse brain and 293T cells were used to investigate pCREB binding to the nmnat2 promoter. For mouse brain samples, the crosslinking and shearing procedure according to Lee et al. (101) was followed. Briefly, whole brains from 1- or 2-month-old mice were finely minced, washed in 2 × volume of 1 × PBS, and fixed in 1/10 volume of fresh 11% formaldehyde for 10 min at RT. Formaldehyde was then quenched by adding 1/20 volume of 2.5 m glycine and cells were then homogenized by a mechanical homogenizer and passed through 10 μm nylon cell strainer. Cells were pooled and spun at 1100g for 5 min at 4°C and resuspended in 50 ml 1 × PBS by gentle inversion and spun again at 1100g for 5 min at 4°C to pellet the cells. The final cell pellet was resuspended in 10 ml of 1 × PBS, spun at 1350g for 5 min at 4°C. Samples were lysed in three different lysis buffers (lysis buffer 1: 50 mm HEPES pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100, protease inhibitor; lysis buffer 2: 10 mm Tris–HCl pH 8, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA; lysis buffer 3: 10 mm Tris–HCl pH 8, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.5% N-Lauroylsarcosine) and sonicated (Microson ultrasonic cell disruptor) to shear DNA to an average length of 500–700 bp. One percent Triton X-100 was added to the sonicated lysate which was then centrifuged at 20 000g for 10 min at 4°C to pellet debris. After the crosslinking and shearing, the ChIP assay was performed according to the EZCHIP Kit protocol (Millipore).
For 293T cells, in order to perform ChIP, cells were transfected with the 2Kb-nmnat2 promoter-PGL4 vector since prior work shows the lack of NMNAT2 expression in 293T cells (25,26,55,56). Forty-eight hours after transfection, cells were treated with either DMSO or 10 µm forskolin before crosslinking proteins with chromatin using formaldehyde. Following crosslinking, the chromatin was sheared using a Branson Sonifier 150, by seven 20 s bursts at high setting (6) with a 1min incubation on ice after each sonication.
Immunoprecipitation was performed with the following antibodies: anti-pCREB (Ser133) (Millipore), Rabbit IgG (negative control; Millipore) or anti-RNAPolII (positive control; Millipore). PCR of the ChIP DNA was performed using the Failsafe PCR Kit (EpiCenter Technologies) using the following primers: nmnat2 CRE1 + CRE2 (forward) 5′-GCT GTA AGG ATG CCA GGG-3′; nmnat2 CRE2 (forward) 5′-GCC AAA GGG AGA GCA ATA-3′; nmnat2 CRE1 + CRE2 (reverse) 5′-CAG CAG GAT AAC GTG GGT-3′. For a positive control, amplification primers included in the ChIP Ab+TM Phospho-CREB (Ser133) antibody kit (Millipore) were used to amplify cFos CRE (Millipore). ChIP experiments were replicated three times incorporating all the appropriate controls.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institute of Health (NS048884, DA029381 and HD065561 to H.-C.L.; NS64269 to R.G.Z.; HL73144 and DK79638 to K.O.), an American Heart Association Predoctoral Fellowship (09PRE2250608 to Y.O.A.) and the Pew Charitable Trust to R.G.Z.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Drs Joanna Jankowsky and Hui Zheng for their helpful comments on the manuscript. We also want to thank Drs Jada Lewis, Michael Hutton and Karen Ashe for providing rTg4510 mice, Drs Xuejun Zhang and Hong Zhang for human NMNAT cDNA clones and Steven Baker and Dr Huda Zoghbi for CREB and CREB(S133A) cDNA clones.
REFERENCES
- 1.Lee V.M., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 2.Hutton M. Molecular genetics of chromosome 17 tauopathies. Ann. N. Y. Acad. Sci. 2000;920:63–73. doi: 10.1111/j.1749-6632.2000.tb06906.x. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini M.G., Goedert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 4.Sperfeld A.D., Collatz M.B., Baier H., Palmbach M., Storch A., Schwarz J., Tatsch K., Reske S., Joosse M., Heutink P., et al. FTDP-17: an early-onset phenotype with parkinsonism and epileptic seizures caused by a novel mutation. Ann. Neurol. 1999;46:708–715. doi: 10.1002/1531-8249(199911)46:5<708::aid-ana5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 5.Bugiani O., Murrell J.R., Giaccone G., Hasegawa M., Ghigo G., Tabaton M., Morbin M., Primavera A., Carella F., Solaro C., et al. Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in tau. J. Neuropathol. Exp. Neurol. 1999;58:667–677. doi: 10.1097/00005072-199906000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Poorkaj P., Grossman M., Steinbart E., Payami H., Sadovnick A., Nochlin D., Tabira T., Trojanowski J.Q., Borson S., Galasko D., et al. Frequency of tau gene mutations in familial and sporadic cases of non-Alzheimer dementia. Arch. Neurol. 2001;58:383–387. doi: 10.1001/archneur.58.3.383. [DOI] [PubMed] [Google Scholar]
- 7.Goedert M., Jakes R. Mutations causing neurodegenerative tauopathies. Biochim. Biophys. Acta. 2005;1739:240–250. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Gotz J., Chen F., Barmettler R., Nitsch R.M. Tau filament formation in transgenic mice expressing P301L tau. J. Biol. Chem. 2001;276:529–534. doi: 10.1074/jbc.M006531200. [DOI] [PubMed] [Google Scholar]
- 9.Lewis J., McGowan E., Rockwood J., Melrose H., Nacharaju P., Van Slegtenhorst M., Gwinn-Hardy K., Paul Murphy M., Baker M., Yu X., et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 2000;25:402–405. doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- 10.Andorfer C., Kress Y., Espinoza M., de Silva R., Tucker K.L., Barde Y.A., Duff K., Davies P. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 2003;86:582–590. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- 11.Yoshiyama Y., Higuchi M., Zhang B., Huang S.M., Iwata N., Saido T.C., Maeda J., Suhara T., Trojanowski J.Q., Lee V.M. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Wittmann C.W., Wszolek M.F., Shulman J.M., Salvaterra P.M., Lewis J., Hutton M., Feany M.B. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293:711–714. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- 13.Andorfer C., Acker C.M., Kress Y., Hof P.R., Duff K., Davies P. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J. Neurosci. 2005;25:5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spires T.L., Orne J.D., SantaCruz K., Pitstick R., Carlson G.A., Ashe K.H., Hyman B.T. Region-specific dissociation of neuronal loss and neurofibrillary pathology in a mouse model of tauopathy. Am. J. Pathol. 2006;168:1598–1607. doi: 10.2353/ajpath.2006.050840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamblin T.C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A.L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., et al. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rissman R.A., Poon W.W., Blurton-Jones M., Oddo S., Torp R., Vitek M.P., LaFerla F.M., Rohn T.T., Cotman C.W. Caspase-cleavage of tau is an early event in Alzheimer disease tangle pathology. J. Clin. Invest. 2004;114:121–130. doi: 10.1172/JCI20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin W.L., Lewis J., Yen S.H., Hutton M., Dickson D.W. Ultrastructural neuronal pathology in transgenic mice expressing mutant (P301L) human tau. J. Neurocytol. 2003;32:1091–1105. doi: 10.1023/B:NEUR.0000021904.61387.95. [DOI] [PubMed] [Google Scholar]
- 18.de Calignon A., Fox L.M., Pitstick R., Carlson G.A., Bacskai B.J., Spires-Jones T.L., Hyman B.T. Caspase activation precedes and leads to tangles. Nature. 2010;464:1201–1204. doi: 10.1038/nature08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau C., Niere M., Ziegler M. The NMN/NaMN adenylyltransferase (NMNAT) protein family. Front. Biosci. 2009;14:410–431. doi: 10.2741/3252. [DOI] [PubMed] [Google Scholar]
- 20.Magni G., Amici A., Emanuelli M., Orsomando G., Raffaelli N., Ruggieri S. Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr. Med. Chem. 2004;11:873–885. doi: 10.2174/0929867043455666. [DOI] [PubMed] [Google Scholar]
- 21.Zhai R.G., Zhang F., Hiesinger P.R., Cao Y., Haueter C.M., Bellen H.J. NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration. Nature. 2008;452:887–891. doi: 10.1038/nature06721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belenky P., Bogan K.L., Brenner C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhai R.G., Cao Y., Hiesinger P.R., Zhou Y., Mehta S.Q., Schulze K.L., Verstreken P., Bellen H.J. Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol. 2006;4:e416. doi: 10.1371/journal.pbio.0040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emanuelli M., Carnevali F., Saccucci F., Pierella F., Amici A., Raffaelli N., Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J. Biol. Chem. 2001;276:406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 25.Raffaelli N., Sorci L., Amici A., Emanuelli M., Mazzola F., Magni G. Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem. Biophys. Res. Commun. 2002;297:835–840. doi: 10.1016/s0006-291x(02)02285-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Kurnasov O.V., Karthikeyan S., Grishin N.V., Osterman A.L., Zhang H. Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis. J. Biol. Chem. 2003;278:13503–13511. doi: 10.1074/jbc.M300073200. [DOI] [PubMed] [Google Scholar]
- 27.Gilley J., Coleman M.P. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 2010;8:e1000300. doi: 10.1371/journal.pbio.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Press C., Milbrandt J. Nmnat delays axonal degeneration caused by mitochondrial and oxidative stress. J. Neurosci. 2008;28:4861–4871. doi: 10.1523/JNEUROSCI.0525-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araki T., Sasaki Y., Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Zhai Q., Chen Y., Lin E., Gu W., McBurney M.W., He Z. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki Y., Milbrandt J. Axonal degeneration is blocked by nicotinamide mononucleotide adenylyltransferase (NMNAT) protein transduction into transected axons. J. Biol. Chem. 2010;285:41211–41215. doi: 10.1074/jbc.C110.193904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki Y., Vohra B.P., Baloh R.H., Milbrandt J. Transgenic mice expressing the Nmnat1 protein manifest robust delay in axonal degeneration in vivo. J. Neurosci. 2009;29:6526–6534. doi: 10.1523/JNEUROSCI.1429-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yahata N., Yuasa S., Araki T. Nicotinamide mononucleotide adenylyltransferase expression in mitochondrial matrix delays Wallerian degeneration. J. Neurosci. 2009;29:6276–6284. doi: 10.1523/JNEUROSCI.4304-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsden M., Kotilinek L., Forster C., Paulson J., McGowan E., SantaCruz K., Guimaraes A., Yue M., Lewis J., Carlson G., et al. Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L) J. Neurosci. 2005;25:10637–10647. doi: 10.1523/JNEUROSCI.3279-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santacruz K., Lewis J., Spires T., Paulson J., Kotilinek L., Ingelsson M., Guimaraes A., DeTure M., Ramsden M., McGowan E., et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 37.Hoover B.R., Reed M.N., Su J., Penrod R.D., Kotilinek L.A., Grant M.K., Pitstick R., Carlson G.A., Lanier L.M., Yuan L.L., et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y.S., Silva A.J. The molecular and cellular biology of enhanced cognition. Nat. Rev. Neurosci. 2009;10:126–140. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheng M., Thompson M.A., Greenberg M.E. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 40.Mayr B., Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 41.Dickey C.A., Loring J.F., Montgomery J., Gordon M.N., Eastman P.S., Morgan D. Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein + presenilin-1 transgenic mice. J. Neurosci. 2003;23:5219–5226. doi: 10.1523/JNEUROSCI.23-12-05219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Q.L., Harris-White M.E., Ubeda O.J., Simmons M., Beech W., Lim G.P., Teter B., Frautschy S.A., Cole G.M. Evidence of Abeta- and transgene-dependent defects in ERK-CREB signaling in Alzheimer's models. J. Neurochem. 2007;103:1594–1607. doi: 10.1111/j.1471-4159.2007.04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palop J.J., Chin J., Bien-Ly N., Massaro C., Yeung B.Z., Yu G.Q., Mucke L. Vulnerability of dentate granule cells to disruption of arc expression in human amyloid precursor protein transgenic mice. J. Neurosci. 2005;25:9686–9693. doi: 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palop J.J., Jones B., Kekonius L., Chin J., Yu G.Q., Raber J., Masliah E., Mucke L. Neuronal depletion of calcium-dependent proteins in the dentate gyrus is tightly linked to Alzheimer's disease-related cognitive deficits. Proc. Natl Acad. Sci. USA. 2003;100:9572–9577. doi: 10.1073/pnas.1133381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong L., Thornton P.L., Balazs R., Cotman C.W. Beta -amyloid-(1–42) impairs activity-dependent cAMP-response element-binding protein signaling in neurons at concentrations in which cell survival is not compromised. J. Biol. Chem. 2001;276:17301–17306. doi: 10.1074/jbc.M010450200. [DOI] [PubMed] [Google Scholar]
- 46.Vitolo O.V., Sant'Angelo A., Costanzo V., Battaglia F., Arancio O., Shelanski M. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc. Natl Acad. Sci. USA. 2002;99:13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saura C.A., Choi S.Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana Rao B.S., Chattarji S., Kelleher R.J., 3rd, Kandel E.R., Duff K., et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- 48.Caccamo A., Maldonado M.A., Bokov A.F., Majumder S., Oddo S. CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2010;107:22687–22692. doi: 10.1073/pnas.1012851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oddo S., Caccamo A., Kitazawa M., Tseng B.P., LaFerla F.M. Amyloid deposition precedes tangle formation in a triple transgenic model of Alzheimer's disease. Neurobiol. Aging. 2003;24:1063–1070. doi: 10.1016/j.neurobiolaging.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Oddo S., Caccamo A., Shepherd J.D., Murphy M.P., Golde T.E., Kayed R., Metherate R., Mattson M.P., Akbari Y., LaFerla F.M. Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 51.Espana J., Valero J., Minano-Molina A.J., Masgrau R., Martin E., Guardia-Laguarta C., Lleo A., Gimenez-Llort L., Rodriguez-Alvarez J., Saura C.A. beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 2010;30:9402–9410. doi: 10.1523/JNEUROSCI.2154-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flammer J.R., Popova K.N., Pflum M.K. Cyclic AMP response element-binding protein (CREB) and CAAT/enhancer-binding protein beta (C/EBPbeta) bind chimeric DNA sites with high affinity. Biochemistry. 2006;45:9615–9623. doi: 10.1021/bi052521a. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X., Odom D.T., Koo S.H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl Acad. Sci. USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seamon K.B., Padgett W., Daly J.W. Forskolin: unique diterpene activator of adenylate cyclase in membranes and in intact cells. Proc. Natl Acad. Sci. USA. 1981;78:3363–3367. doi: 10.1073/pnas.78.6.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger F., Lau C., Dahlmann M., Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 56.Mayer P.R., Huang N., Dewey C.M., Dries D.R., Zhang H., Yu G. Expression, localization, and biochemical characterization of nicotinamide mononucleotide adenylyltransferase 2. J. Biol. Chem. 2010;285:40387–40396. doi: 10.1074/jbc.M110.178913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruzalegui F.H., Kapiloff M.S., Morfin J.P., Kemp B.E., Rosenfeld M.G., Means A.R. Regulation of intrasteric inhibition of the multifunctional calcium/calmodulin-dependent protein kinase. Proc. Natl Acad. Sci. USA. 1992;89:12127–12131. doi: 10.1073/pnas.89.24.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarty D.M., Young S.M., Jr, Samulski R.J. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- 59.Grieger J.C., Choi V.W., Samulski R.J. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 2006;1:1412–1428. doi: 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- 60.Kugler S., Hahnewald R., Garrido M., Reiss J. Long-term rescue of a lethal inherited disease by adeno-associated virus-mediated gene transfer in a mouse model of molybdenum-cofactor deficiency. Am. J. Hum. Genet. 2007;80:291–297. doi: 10.1086/511281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burger C., Nash K., Mandel R.J. Recombinant adeno-associated viral vectors in the nervous system. Hum. Gene Ther. 2005;16:781–791. doi: 10.1089/hum.2005.16.781. [DOI] [PubMed] [Google Scholar]
- 62.Tenenbaum L., Chtarto A., Lehtonen E., Velu T., Brotchi J., Levivier M. Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med. 2004;6(Suppl. 1):S212–S222. doi: 10.1002/jgm.506. [DOI] [PubMed] [Google Scholar]
- 63.Benito E., Barco A. CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Alberini C.M. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonze B.E., Ginty D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 66.Bito H., Takemoto-Kimura S. Ca(2+)/CREB/CBP-dependent gene regulation: a shared mechanism critical in long-term synaptic plasticity and neuronal survival. Cell Calcium. 2003;34:425–430. doi: 10.1016/s0143-4160(03)00140-4. [DOI] [PubMed] [Google Scholar]
- 67.Mantamadiotis T., Lemberger T., Bleckmann S.C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J., et al. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 68.Valor L.M., Jancic D., Lujan R., Barco A. Ultrastructural and transcriptional profiling of neuropathological misregulation of CREB function. Cell Death Differ. 2010;17:1636–1644. doi: 10.1038/cdd.2010.40. [DOI] [PubMed] [Google Scholar]
- 69.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto-Sasaki M., Ozawa H., Saito T., Rosler M., Riederer P. Impaired phosphorylation of cyclic AMP response element binding protein in the hippocampus of dementia of the Alzheimer type. Brain Res. 1999;824:300–303. doi: 10.1016/s0006-8993(99)01220-2. [DOI] [PubMed] [Google Scholar]
- 71.Smith D.L., Pozueta J., Gong B., Arancio O., Shelanski M. Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc. Natl Acad. Sci. USA. 2009;106:16877–16882. doi: 10.1073/pnas.0908706106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong B., Cao Z., Zheng P., Vitolo O.V., Liu S., Staniszewski A., Moolman D., Zhang H., Shelanski M., Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 73.Martin-Loeches M., Molina V., Munoz F., Hinojosa J.A., Reig S., Desco M., Benito C., Sanz J., Gabiri A., Sarramea F., et al. P300 amplitude as a possible correlate of frontal degeneration in schizophrenia. Schizophr. Res. 2001;49:121–128. doi: 10.1016/s0920-9964(00)00125-0. [DOI] [PubMed] [Google Scholar]
- 74.Rouaux C., Jokic N., Mbebi C., Boutillier S., Loeffler J.P., Boutillier A.L. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu H.Y., Hudry E., Hashimoto T., Kuchibhotla K., Rozkalne A., Fan Z., Spires-Jones T., Xie H., Arbel-Ornath M., Grosskreutz C.L., et al. Amyloid beta induces the morphological neurodegenerative triad of spine loss, dendritic simplification, and neuritic dystrophies through calcineurin activation. J. Neurosci. 2010;30:2636–2649. doi: 10.1523/JNEUROSCI.4456-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kogel D., Peters M., Konig H.G., Hashemi S.M., Bui N.T., Arolt V., Rothermundt M., Prehn J.H. S100B potently activates p65/c-Rel transcriptional complexes in hippocampal neurons: Clinical implications for the role of S100B in excitotoxic brain injury. Neuroscience. 2004;127:913–920. doi: 10.1016/j.neuroscience.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 77.Ittner L.M., Ke Y.D., Delerue F., Bi M., Gladbach A., van Eersel J., Wolfing H., Chieng B.C., Christie M.J., Napier I.A., et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 78.Zempel H., Thies E., Mandelkow E., Mandelkow E.M. Abeta oligomers cause localized Ca(2+) elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J. Neurosci. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thompson L.T., Moskal J.R., Disterhoft J.F. Hippocampus-dependent learning facilitated by a monoclonal antibody or D-cycloserine. Nature. 1992;359:638–641. doi: 10.1038/359638a0. [DOI] [PubMed] [Google Scholar]
- 80.Flood J.F., Morley J.E., Lanthorn T.H. Effect on memory processing by D-cycloserine, an agonist of the NMDA/glycine receptor. Eur. J. Pharmacol. 1992;221:249–254. doi: 10.1016/0014-2999(92)90709-d. [DOI] [PubMed] [Google Scholar]
- 81.Coleman M.P., Freeman M.R. Wallerian degeneration, wld(s), and nmnat. Annu. Rev. Neurosci. 2010;33:245–267. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lunn E.R., Perry V.H., Brown M.C., Rosen H., Gordon S. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur. J. Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 83.Conforti L., Tarlton A., Mack T.G., Mi W., Buckmaster E.A., Wagner D., Perry V.H., Coleman M.P. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc. Natl Acad. Sci. USA. 2000;97:11377–11382. doi: 10.1073/pnas.97.21.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mack T.G., Reiner M., Beirowski B., Mi W., Emanuelli M., Wagner D., Thomson D., Gillingwater T., Court F., Conforti L., et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 85.Gillingwater T.H., Haley J.E., Ribchester R.R., Horsburgh K. Neuroprotection after transient global cerebral ischemia in Wld(s) mutant mice. J. Cereb. Blood Flow Metab. 2004;24:62–66. doi: 10.1097/01.WCB.0000095798.98378.34. [DOI] [PubMed] [Google Scholar]
- 86.Sajadi A., Schneider B.L., Aebischer P. Wlds-mediated protection of dopaminergic fibers in an animal model of Parkinson disease. Curr. Biol. 2004;14:326–330. doi: 10.1016/j.cub.2004.01.053. [DOI] [PubMed] [Google Scholar]
- 87.Mi W., Beirowski B., Gillingwater T.H., Adalbert R., Wagner D., Grumme D., Osaka H., Conforti L., Arnhold S., Addicks K., et al. The slow Wallerian degeneration gene, WldS, inhibits axonal spheroid pathology in gracile axonal dystrophy mice. Brain. 2005;128:405–416. doi: 10.1093/brain/awh368. [DOI] [PubMed] [Google Scholar]
- 88.MacDonald J.M., Beach M.G., Porpiglia E., Sheehan A.E., Watts R.J., Freeman M.R. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 89.Zhai R.G., Rizzi M., Garavaglia S. Nicotinamide/nicotinic acid mononucleotide adenylyltransferase, new insights into an ancient enzyme. Cell Mol. Life Sci. 2009;66:2805–2818. doi: 10.1007/s00018-009-0047-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan T., Feng Y., Zheng J., Ge X., Zhang Y., Wu D., Zhao J., Zhai Q. Nmnat2 delays axon degeneration in superior cervical ganglia dependent on its NAD synthesis activity. Neurochem. Int. 2010;56:101–106. doi: 10.1016/j.neuint.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 91.Pollak N., Dolle C., Ziegler M. The power to reduce: pyridine nucleotides—small molecules with a multitude of functions. Biochem. J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.David D.C., Hauptmann S., Scherping I., Schuessel K., Keil U., Rizzu P., Ravid R., Drose S., Brandt U., Muller W.E., et al. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J. Biol. Chem. 2005;280:23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- 93.Rhein V., Song X., Wiesner A., Ittner L.M., Baysang G., Meier F., Ozmen L., Bluethmann H., Drose S., Brandt U., et al. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc. Natl Acad. Sci. USA. 2009;106:20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perreault S., Bousquet O., Lauzon M., Paiement J., Leclerc N. Increased association between rough endoplasmic reticulum membranes and mitochondria in transgenic mice that express P301L tau. J. Neuropathol. Exp. Neurol. 2009;68:503–514. doi: 10.1097/NEN.0b013e3181a1fc49. [DOI] [PubMed] [Google Scholar]
- 95.Samulski R.J., Chang L.S., Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Philpott N.J., Gomos J., Berns K.I., Falck-Pedersen E. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl Acad. Sci. USA. 2002;99:12381–12385. doi: 10.1073/pnas.182430299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grimm D., Kay M.A., Kleinschmidt J.A. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 98.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 99.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 100.Chen C., Cheng X., Dieter M.Z., Tanaka Y., Klaassen C.D. Activation of cAMP-dependent signaling pathway induces mouse organic anion transporting polypeptide 2 expression. Mol. Pharmacol. 2007;71:1159–1164. doi: 10.1124/mol.106.030841. [DOI] [PubMed] [Google Scholar]
- 101.Lee T.I., Johnstone S.E., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.