Abstract
Serum gamma-glutamyl transferase (GGT) activity is a marker of liver disease which is also prospectively associated with the risk of all-cause mortality, cardiovascular disease, type 2 diabetes and cancers. We have discovered novel loci affecting GGT in a genome-wide association study (rs1497406 in an intergenic region of chromosome 1, P = 3.9 × 10−8; rs944002 in C14orf73 on chromosome 14, P = 4.7 × 10−13; rs340005 in RORA on chromosome 15, P = 2.4 × 10−8), and a highly significant heterogeneity between adult and adolescent results at the GGT1 locus on chromosome 22 (maximum PHET = 5.6 × 10−12 at rs6519520). Pathway analysis of significant and suggestive single-nucleotide polymorphism associations showed significant overlap between genes affecting GGT and those affecting common metabolic and inflammatory diseases, and identified the hepatic nuclear factor (HNF) family as controllers of a network of genes affecting GGT. Our results reinforce the disease associations of GGT and demonstrate that control by the GGT1 locus varies with age.
INTRODUCTION
Gamma-glutamyl transferase (GGT) is a membrane-bound enzyme which transfers glutamyl groups linked through the gamma-carboxylic acid from peptides such as glutathione to acceptors. In humans, the active enzyme is coded by GGT1 on chromosome 22. Its main physiological function is to make cysteine available for regeneration of intracellular glutathione and hence to protect against oxidative stress (1).
Measurement of GGT activity in serum is used clinically as a liver function test and a biomarker for excessive alcohol consumption (2), but there is also strong evidence of prospective associations between GGT activity and all-cause mortality, cardiovascular disease, type 2 diabetes and cancers. For example, a study of >0.25 million people followed for a median of 7.6 years (3) showed a relative risk of death from any cause of 2.0 when the top 15% of participants were compared against the bottom 37% for GGT at entry. Relative risk was similar for cancer-related and non-cancer deaths. For hepatobiliary mortality in general and for hepatoma, these relative risks were 15- and 18-fold, respectively, while for the much more common cardiovascular deaths the ratio was 1.6. Other studies have examined disease incidence rather than mortality, comparing rates of incident cardiovascular disease or type 2 diabetes by GGT at enrolment. For cardiovascular disease (coronary heart disease or stroke), the relative risk per unit increase in the natural logarithm of GGT estimated from meta-analysis was 1.53, or 1.34 after adjustment for potential confounders (4). Meta-analysis of studies on diabetes, contrasting participants in the bottom quartile against the top quartile of GGT at entry, showed a relative risk after covariate adjustment of 2.94 (5). The activity of other enzymes used as liver function tests [alanine aminotransferase (ALT), aspartate aminotransferase (AST)] has shown similar epidemiological associations with mortality or morbidity but generally to a lesser degree.
Given this biological and epidemiological information about GGT, identification of polymorphisms, genes and pathways influencing serum GGT should contribute to our understanding of the response to oxidative stress and the causes of cardiovascular and metabolic risk. Significant allelic associations have been reported for single-nucleotide polymorphisms (SNPs) near the GGT1 gene on chromosome 22 and HNF1A on chromosome 12 (6–8). GGT1 codes for the active form of GGT and SNP effects in this chromosome 22 region presumably regulate gene expression. The effects of the other known locus are more diverse, as significant associations at HNF1A have been reported for many phenotypes. Apart from the known association with GGT, there are associations with serum C-reactive protein (CRP) (9), low-density lipoprotein (LDL) cholesterol and coronary artery disease (10) and type 2 diabetes (11). HNF1A codes for a regulator of the expression of multiple genes in the liver and also in the pancreatic islet cells. As well as the reported allelic association with type 2 diabetes, mutations throughout HNF1A cause maturity-onset diabetes in youth type 3 (MIM 600496), primarily through impaired insulin secretion (12).
As with many quantitative traits, the heritability of serum GGT activity [50–60% in adults (13,14)] is substantially greater than can be accounted for by identified and significant SNP associations. We previously found heritability for GGT of ∼70% in adolescents (15), with evidence for some genetic effects common to ages 12, 14 and 16 and others which change with age. There is also evidence for genetic correlation between serum GGT, ALT and AST activity (13).
We have conducted a genome-wide association study (GWAS) for serum GGT activity in 12 526 adults and adolescents. Our aims were to identify novel loci, test whether they affect other markers of liver function (ALT, AST) or only GGT, and compare allelic effects between adults and adolescents. Using both significant and suggestive SNP associations, we searched for overlap between genes or pathways affecting GGT and those affecting cardiovascular or diabetic risk.
RESULTS
We tested for SNP associations of serum GGT activity in two Australian cohorts, of adults and adolescents, and compared the adolescent results with the larger adult data set for replication and heterogeneity testing. This was followed by confirmation of age heterogeneity at one locus using additional adult and adolescent cohorts, and exploration of the disease associations of the significant or suggestive SNPs.
Allelic associations
Five genome-wide-significant associations (P < 5 × 10−8) were found for GGT in either the adult or the combined adult and adolescent data (Table 1). There were three novel associations, with rs1497406 (P = 3.9 × 10−8) in an intergenic region at 16 Mbp on chromosome 1; rs944002 (P = 2.3 × 10−12) at 103 Mbp on chromosome 14 in C14orf73 and rs340005 (P = 2.4 × 10−8) at 61 Mbp on chromosome 15 in RORA. The previously reported associations for GGT with HNF1A on chromosome 12 and GGT1 on chromosome 22 were confirmed. Plots for chromosome 1, 12, 14 and 15 loci are shown in Supplementary Material, Figure S1, and the more complex results for chromosome 22 are presented below. A list of SNPs with P ≤ 10−5 for GGT in the combined adult and adolescent data is provided in Supplementary Material, Table S1. Further exploration of our results using gene-based testing (16) showed only three significant regions. These covered HNF1A, OASL and C12orf43 on chromosome 12, C14orf73 on chromosome 14 and C22orf36 and GGT1 on chromosome 22 (Supplementary Material, Table S2), consistent with the SNP association results.
Table 1.
GGT results for adults and adolescents separately and together, based on the most significant SNP for allelic association from each locus with any SNP with P < 5 × 10−8 in either group or in the combined analysis
| SNP | Chr | Bp (NCBI B36, dbSNP b126) | Gene | Minor (effect) allele | Adults |
Adolescents |
Meta-analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Beta | SE | P-value | MAF | Beta | SE | P-value | PHet | Beta | SE | PAssoc | |||||
| rs1497406 | 1 | 16,377,907 | – | T | 0.41 | −0.084 | 0.016 | 2.50E−07 | 0.41 | −0.056 | 0.031 | 0.077 | 0.422 | −0.078 | 0.014 | 3.93E−08 |
| rs1169288 | 12 | 119,901,033 | HNF1A | G | 0.31 | −0.126 | 0.017 | 4.30E−13 | 0.32 | −0.155 | 0.033 | 2.2E−06 | 0.435 | −0.132 | 0.015 | 2.33E−18 |
| rs944002 | 14 | 102,642,568 | C14orf73 | G | 0.22 | 0.135 | 0.020 | 1.50E−11 | 0.21 | 0.105 | 0.039 | 0.0064 | 0.494 | 0.129 | 0.018 | 4.66E−13 |
| rs340005 | 15 | 58,665,322 | RORA | G | 0.36 | −0.080 | 0.017 | 1.30E−06 | 0.37 | −0.097 | 0.032 | 0.0022 | 0.639 | −0.084 | 0.015 | 2.43E−08 |
| rs5751901 | 22 | 23,322,266 | GGT1 | C | 0.32 | 0.117 | 0.017 | 8.70E−12 | 0.32 | −0.105 | 0.033 | 0.0015 | 2.23E−09 | – | – | – |
All SNPs in this Table were imputed, with R2> 0.6 and the best-guess genotypes were used in the association analysis. Betas are estimated as the per-allele effect of the minor allele. For the meta-analysis data, PHet is the probability of the observed heterogeneity between adult and adolescent allelic associations occurring by chance and PAssoc is the probability for the allelic associations.
We compared results for additional liver function tests, ALT and AST. There was no overlap between our genome-wide-significant results for GGT and nominally significant results for ALT or AST, or vice versa. An intergenic region on chromosome 7 showed suggestive effects on all three enzyme activities, with a block of SNPs showing lowest P-values of 1.3 × 10−6 for GGT, 8.3 × 10−5 for ALT and 1.2 × 10−6 for AST (Supplementary Material, Fig. S2). The reported association for AST on chromosome 10, in MRC1/MRC1L1 (8), was confirmed (rs691461, P = 7.3 × 10−15), and that between ALT and PNPLA3 (7) showed P = 0.0010 at rs2281135 in our data. In view of the associations between GGT activity and cardiometabolic risk factors, we also tested whether the SNPs showing significant associations with GGT had effects on BMI, lipids, glucose and CRP. Results are shown in Supplementary Material, Table S3; rs1169288 in HNF1A showed the expected associations with CRP and LDL-C but the other GGT-related SNPs had no significant effects (after adjusting for multiple testing) on the tested phenotypes.
Adult–adolescent heterogeneity
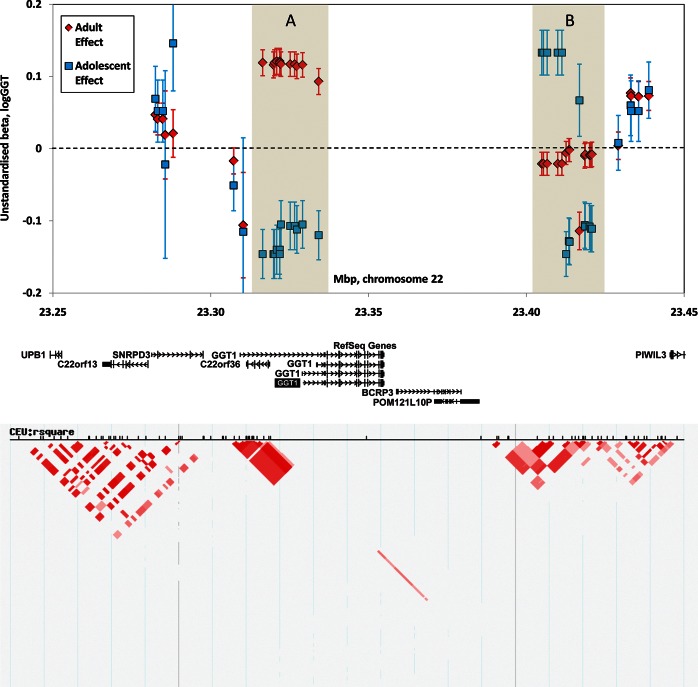
Comparison of our adult and adolescent results from the discovery studies showed consistent allelic effects for all the significant loci except GGT1 (Table 1). This locus showed contrasting allelic effects (Fig. 1, Supplementary Material, Fig. S3), with maximum adult-adolescent heterogeneity of P = 5.6 × 10−12 at rs6519520. The adult SNP associations are significant at 6 × 10−11–8.7 × 10−12 at 11 SNPs in the block labelled A in Figure 1, where the adolescent data show P-values down to 1.4 × 10−5 but in the opposite direction. In the block labelled B in Figure 1, the association P-values for the adolescent data are again highly suggestive (minimum P in this region 3.1 × 10−6), while the adult data show only one SNP with P < 0.10 (rs17661237, P = 9.7 × 10−6) and this SNP is not significant in the adolescents. Details are given in Supplementary Material, Table S1.
Figure 1.
Comparison of the magnitude and direction of allelic effects on GGT activity at the GGT1 locus in adults (red diamonds) and adolescents (blue squares). Gene locations are shown below the main panel, with the linkage disequilibrium (R2) between SNPs below that. Points shown are the unstandardized betas for the minor-allele effect on log-transformed GGT activity in an additive model, and bars show standard errors. Between rs5760485 at 23 316 473 bp and rs2001180 at 23 334 206 bp (shaded region A), the allelic effects are in opposite directions in adults and adolescents. Between rs6004233 at 23 404 863 bp and rs5760590 at 23 420 768 bp (shaded region B), the allelic effects in adults are negligible but those for adolescents are similar to those for the 23.316–23.334 Mbp region.
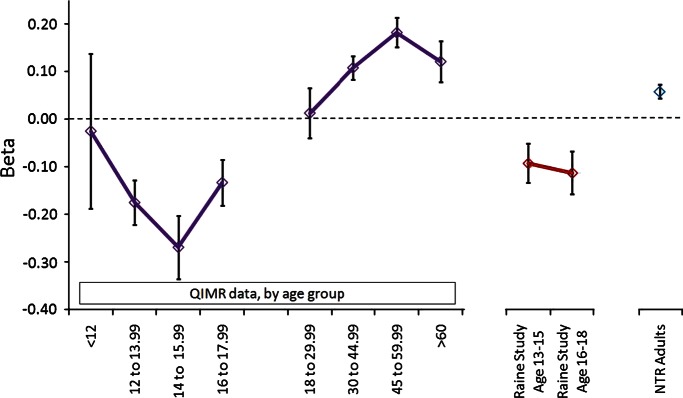
We replicated this GGT1 × age effect in independent samples from the Western Australian Pregnancy (Raine) Cohort Study (adolescents in Perth, Australia) and the Netherlands Twin Registry (adults). Subjects in these additional studies had GGT results which were similar to those in the adult and adolescent discovery cohorts (see Supplementary Material, Table S4). In the replication, we concentrated on rs6519519, which was genotyped in the QIMR data. The outcome is shown in Table 2 and Figure 2, confirming the opposite allelic effects for this SNP at the GGT1 locus in adults and adolescents.
Table 2.
Comparison of allelic effects for rs6519519 at the GGT1 locus between adults and adolescents
| Adult studies |
Adolescent studies |
Comparison | |||||
|---|---|---|---|---|---|---|---|
| β | SE (β) | P-value | β | SE (β) | P-value | PHet | |
| GGT1 | |||||||
| rs6519519 (QIMR) | 0.121 | 0.018 | 1.50E−11 | −0.140 | 0.034 | 3.40E−05 | 1.67E−11 |
| rs6519519 (NTR) | 0.059 | 0.015 | 1.14E−04 | – | – | – | – |
| rs6519519 (Raine, age 13–15) | – | – | – | −0.092 | 0.041 | 0.027 | – |
| rs6519519 (raine, age 16–18) | – | – | – | −0.112 | 0.045 | 0.013 | – |
Betas are reported as the per-allele effect of the minor allele (T) on log10(GGT).
Figure 2.
Allelic effects at rs6519519 on log10GGT, by age group; QIMR data by age, and results from replication in RAINE adolescent and NTR adult studies. Error bars show SE for each estimated beta.
Disease and pathway analysis
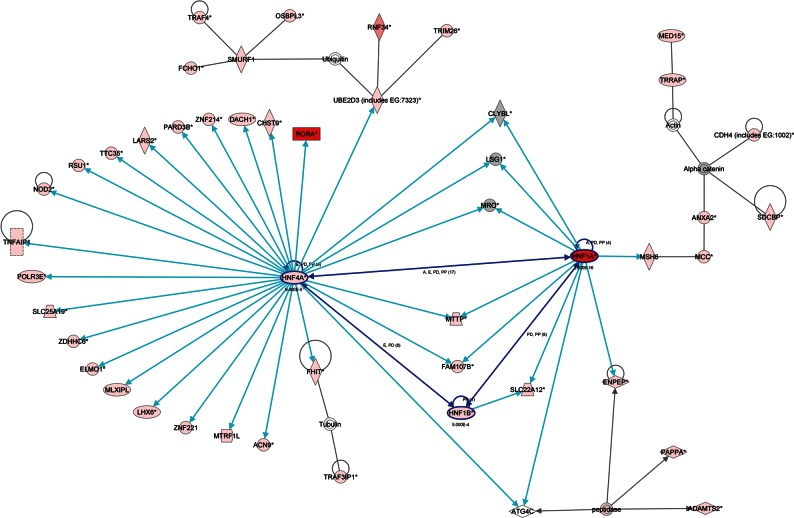
There was substantial overlap between genes containing significant or suggestive SNPs for GGT and genes showing documented disease associations. For 321 genes identified by Ingenuity Pathway Analysis from the list of SNPs with P < 0.001 for GGT, an excess of disease associations was found for the categories ‘coronary artery disease’, ‘hypertension’, ‘non-insulin-dependent diabetes’, ‘insulin-dependent diabetes’, ‘rheumatoid arthritis’, ‘Crohn's disease’, ‘inflammatory bowel disease’, ‘progressive motor neuropathy’ and ‘amyotrophic lateral sclerosis’, all at P < 10−10 (by Fisher's exact test). The conditions and genes identified are summarized in Supplementary Material, Table S5. Varying the list of SNPs by raising the P-value required for inclusion to 10−2 produced more significant P-values for these disease associations, and showed P-values <10−10 for additional neurodegenerative and psychiatric conditions. The pathway analysis at P < 0.001 also showed networks of genes in which variation is associated with GGT activity, connected by previously documented effects (mainly effects of one gene product on the expression of another gene). The most striking of these networks is shown in Figure 3.
Figure 3.
Network of genes containing SNPs affecting GGT at P < 0.001 (pink blocks) or P < 5 × 10−8 (red blocks), emphasizing HNF connections. (Details of the genes in this network, the most significant SNPs in each gene and their P-values are given in Supplementary Material, Table S7.)
DISCUSSION
Two main points arise from our results. First, GGT activity has been shown in many prospective studies to have important disease associations, including cardiovascular disease and type 2 diabetes. We and others have previously reported significant phenotypic correlations with quantitative traits or biomarkers defining dyslipidaemia, obesity and metabolic syndrome, confirmed in our current data. Its allelic associations should be considered in the light of these, and we do find that the set of significant and suggestive SNPs for GGT in our data overlaps strongly with genes implicated by previous studies in complex diseases and with pathways related to them. Secondly, there is a remarkable difference in allelic effects of GGT1 SNPs on GGT activity at different ages. Developmental changes in SNPs' effects are potentially common and biologically important, but there are few specific examples for disease-related phenotypes in humans.
Allelic associations
We replicated previously reported associations between GGT activity and SNPs in GGT1 and HNF1A. These are discussed in more detail, in relation to the age heterogeneity and the network analysis, below.
Of the novel loci, that on chromosome 1 is intergenic, between EPHA2 and ARHGEF19, and shows some linkage disequilibrium with each, but neither gene shows obvious connections with GGT. We examined the ENCODE data (at http://genome.ucsc.edu/ENCODE/) for the lead SNP in this region, rs1497406, and found evidence of transcription factor binding (including HNF4A, central to the network of genes shown in Figure 3) spanning this location. This SNP is also within a region showing ‘histone marks’ (evidence for promoter activity) in several cell lines, including the liver-derived HepG2. Functional studies of this region in relation to GGT and other phenotypes reported to show associations with members of the HNF gene family may clarify the role of this locus.
The significant SNPs on chromosome 14 cover C14orf73 and include two non-synonymous coding SNPs (rs2297067, Arg77His and rs10131298, Leu185His) with potential functional effects. The protein from C14orf73 (or EXOC3L4) was also known as SEC6-like protein, and SEC6 is a component of the exocyst involved in insulin granule secretion (17). This provides a potential connection between control of glucose homeostasis and GGT activity, another parallel to the known phenotypic associations.
The chromosome 15 locus contains RORA, a retinoid-related orphan receptor. Knockout mouse studies have implicated RORA in control of high-density lipoprotein and triglycerides, obesity and accumulation of triglyceride in the liver, indicating a potential connection with GGT as a marker of hepatic steatosis (18).
Age heterogeneity
SNPs in and near GGT1 are known to be associated with serum GGT activity (6–8), but the age effect means that this relationship is more complex than previously thought. This interaction is an unexpected result, although we have previously found evidence from a longitudinal twin study that genetic effects on GGT activity differ with age across adolescence (15). GGT activity is known to change with age and also differs between men and women; in our data, the sex difference becomes apparent at about age 14 and there is a substantial increase in mean GGT between the ages of 18 and 30 (see Supplementary Material, Table S4). Similar differences between mean GGT in adolescents and adults were present in the replication cohorts. We cannot be more precise about the age when this change occurs because our adult cohort contained very few people aged <30. Among adult men, GGT changes little with age; the values in adult women increase with age but remain lower than those for men. As for other biochemical phenotypes which change with age, existing knowledge does little more than describe the changes which occur and the mechanisms are poorly understood, but the existence of age-by-genotype effects for serum GGT adds to the complexity and emphasizes genetic causes of differences between people.
Detailed examination shows there are two sets of SNPs (either side of GGT1) which show subtly different age effects. For one group of SNPs, the direction of the allelic effect is opposite in adults and adolescents, and for the other group effects are present only in the adolescents (Fig. 1). The block of SNPs which show the greatest age × allelic effects includes rs5751901, which has been shown to affect expression of GGT1, GGT2 and GGTLA4 in human liver tissue (19); and rs6519519, which affects expression of the same genes in lymphoblastoid cell lines (20). The second block, which shows effects on GGT activity in adolescents but not adults, is situated between GGT1 and PIWIL3 (see Fig. 1 and Supplementary Material, Fig. S3), and there appears to be no published information about effects within this block on GGT expression.
Out of the GGT and GGT-related gene family, only GGT1 has been shown to produce an active enzyme (21), although GGT protein, GGT1 transcripts and other GGT genes have physiological functions independent of enzymatic activity (22,23). Multiple promoters give rise to at least four human GGT1 transcripts, differing in the 5′UTR but leading to the same protein; the SNPs with opposite effects in adults and adolescents cover the region containing these promoters. Alternative splicing of GGT mRNA is associated with tissue-specific expression and developmental regulation (24). It is likely that the observed differences in SNP effects on the GGT phenotype between adolescents and adults, like the changes in expression during embryonic development, are connected with differences in the mRNAs and the control of splicing.
Results from the genome-wide assessment of heterogeneity between adult and adolescent SNP associations showed an excess of heterogeneity P-values in the range 10−3–10−5 (Supplementary Material, Fig. S4). This suggests that other loci also change their effects on GGT activity with age. A group of SNPs in the CELF2 (CUGBP, Elav-like family member 2; CUGBP2) gene show heterogeneity P-values of 8.8 × 10−7, and this gene has biological plausibility for trans effects on GGT expression. The CUG-binding proteins coded by this gene family bind to specific RNA sequences and control both alternative splicing and mRNA translation and stability (25).
Disease and pathway associations
Although only five loci met genome-wide significance criteria for GGT, many other variants with smaller effect sizes or lower minor-allele frequencies (MAFs) must contribute to the heritability of this phenotype. Gene- and pathway-based analyses can aid our understanding of these. Results from the ingenuity analysis showed substantial overlap between genes containing these SNPs and genes showing documented disease associations. An excess of disease associations was found for cardiovascular disease and hypertension, both type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes, inflammatory conditions (rheumatoid arthritis, Crohn's disease, inflammatory bowel disease) and rather unexpectedly for progressive motor neuropathy and amyotrophic lateral sclerosis. All these were significant at P < 10−10 when SNPs showing P < 0.001 for GGT were included, and became stronger when the list of SNPs was expanded to P < 0.01 for GGT. In view of the known phenotypic associations between GGT and cardiovascular diseases and diabetes, some of these gene overlaps are consistent with expectations but those with inflammatory and neurological conditions are novel. The association with amyotrophic lateral sclerosis may be because of the role of oxidative stress in that condition (26,27) and the function of GGT in replenishing glutathione.
This pathway analysis also showed networks of genes in which variation is associated with GGT activity, connected by previously documented effects (mainly effects of one gene product on the expression of another gene). Figure 3 emphasizes the existence of a group of genes whose variation affects GGT activity and which are controlled by or connected with the Hepatic Nuclear Factor family. Multiple genes, which are controlled by HNF1a, HNF4a or both, contain SNPs which are associated with GGT activity at P < 0.001, and SNPs in both HNF4A and (even more strongly) HNF1A have apparently direct effects on GGT. This reinforces the phenotypic associations between serum GGT and obesity and inflammation, while the SNP-disease associations are consistent with the predictive validity of serum GGT for diabetes, cardiovascular disease and overall mortality.
Limitations
As with other GWASs, only the genetic variants which are tagged by the SNPs included in the genotyping process can be tested for association with the phenotype. Those which are not in linkage disequilibrium with tagging SNPs are not included, and those with low MAF require large samples to give adequate power even if they are typed or imputed. However, this limitation does not lead to false-positive associations. Our study focused on people of European descent, and variants which are monomorphic in Europeans but polymorphic in other populations [such as rs671 in ALDH2 (8)] would not be detected. The age comparison of the effects of SNPs at GGT1 is based on independent cohorts, and comparisons based on longitudinal measurements of GGT or on within-family comparisons such as parent-offspring data would be desirable. However, the subjects in the four cohorts (three Australian and one from the Netherlands) had similar GGT results and each was filtered on the basis on ancestry-informative markers to include only people of European descent.
Conclusions
Serum GGT activity is an independent predictor of morbidity and mortality, and increases in obesity and metabolic syndrome. These associations are strengthened by our discovery that SNPs affecting GGT, even at suggestive rather than genome-wide-significant levels, include a highly significant excess of those affecting cardiovascular disease and diabetes. The direction of association between SNPs at the GGT1 locus (and potentially elsewhere) and serum GGT activity changes with age, and differences in allelic effects on GGT between health and disease deserve further study.
SUBJECTS AND METHODS
Discovery cohorts
The Adult studies conducted by Queensland Institute of Medical Research (QIMR) comprised:
a study of genetic effects on risk factors for common psychiatric and metabolic diseases (1993–96 Adult Study), described in ref. (13). All participants were Australian adult twins, aged 29–92 years, not selected for any disease. Enzyme activity in serum was measured on 3060 participants.
a study of anxiety and depression, drawn from participants in our previous twin studies and selecting for pairs either highly concordant or highly discordant for neuroticism score (28). A total of 2351 participants in this study had enzyme measurements.
a study on genetic risk factors for endometriosis (29), from which 3688 participants had enzyme measurements; and
a twin-family study of alcohol and nicotine dependence and metabolic risk factors for alcoholic liver disease. Enzyme measurements were available from 9527 participants.
From the combined Adult Studies, 17 071 people had GGT and other liver enzyme results, some from more than one study. Of them, 9971 had both enzyme results and GWAS genotyping.
The Brisbane Adolescent Twins Study, described in (30), focused on cognition, risk factors for melanoma and metabolic risk factors in adolescence. A total of 3023 twins or their siblings had biochemical results from participation in these studies between 1992 and 2006, mostly on more than one occasion. Of them, 2555 had both enzyme results and GWAS genotyping.
Each of the studies was approved by the QIMR Ethics Review Committee and the participants (and for those aged <18, their parent or guardian) gave informed consent.
Information on participants' characteristics is shown in Supplementary Material, Table S6. There was no overlap of individuals between the Adolescent and Adult studies. Where multiple measurements were available for any participant, the mean value was used in the genetic association analysis.
Serum was separated from clotted blood samples and stored at −70°C until analysed. For the 1993–96 Adult Study, GGT, ALT, AST, lipids and glucose were measured with standard clinical laboratory methods on a Hitachi 747 analyser. For later studies, these phenotypes, and also CRP, were measured on either a Roche 917 or Modular P analyser (Roche Diagnostics, Castle Hill, NSW, Australia). Enzyme activity results were log-transformed, and adjusted for sex, age, age2, sex × age and sex × age2 in the association analyses.
Genotyping was performed on DNA extracted from blood samples by standard methods. Genome-wide SNP typing was performed using Illumina 317K, 370K or 610K chips at CIDR, deCode Genetics or the Finnish Genome Centre. Data cleaning and analysis involved checks for quality of SNPs and of samples, with exclusions as previously described (31,32). SNPs were included in the analyses if they had Hardy–Weinberg equilibrium (HWE) test P ≥ 10−6, MAF ≥ 1%, call-rate ≥ 0.95 and the mean value of BeadStudio GenCall score ≥ 0.7. We used principal components analysis (EIGENSTRAT) of the genotyping data to exclude subjects of non-European ancestry. We used a set of ∼275 000 autosomal SNPs that were common to Australian samples, HapMap 3 (11 global populations) and 5 Northern European (Denmark, Finland, the Netherlands, the UK and Sweden) populations from the GenomEUtwin Consortium. We excluded 277 individuals (1.7% of the total genotyped individuals) who were more than 6 standard deviations from the mean of principal components (PCs) 1 and 2 derived from the European populations.
Imputation of non-typed HapMap SNPs was carried out using MACH (http://www.sph.umich.edu/csg/abecasis/mach/index.html). To avoid bias due to both missingness and SNP density, we used a set of SNPs common to the genotyping chips (n = 274 604). The imputation procedure, and the set of SNPs used, was the same for both the adult and the adolescent data. The imputed genotypes were used for the association analyses if they met the same conditions as mentioned for the genotyped SNPs as well as an imputation quality score, R-sq-hat (the squared correlation between imputed and true genotypes) of 0.3 or greater. The SNPs which showed significant associations with GGT had R-sq-hat values between 0.65 and 0.98. A total of 2 380 486 SNPs were used for the association analyses. The imputed genotypes were used for all SNPs including those originally genotyped. For the locus at GGT1 which showed heterogeneity of allelic effects between adults and adolescents, we checked the allelic association results for rs6519519 (Table 2) by repeating the analysis using the original (pre-imputation) genotypes and found essentially identical results.
Prior to association analyses, the phenotypes were adjusted for the effects of sex, age, age × age, sex × age and sex × age × age. We tested each SNP for association with the phenotype under an additive model using a family-based score test implemented in Merlin (33). Both within- and between-family components were used to produce a total association estimate. We meta-analysed the SNP–phenotype associations from the adult and adolescent cohorts by weighting the effect size estimates using the inverse of the corresponding standard errors in Metal (34). The heterogeneity between the estimates was assessed using Cochran's Q statistic implemented in Metal. A genome-wide significance level of 5 × 10−8 was used for both allelic association and heterogeneity P-values, and results are reported (Table 1) for the most significant SNP at each locus with P-values below this threshold either in the meta-analysis of adult and adolescent data or in the adults or adolescents separately. There was one locus with genome-wide-significant heterogeneity, and we tested for replication at one SNP (rs6519519) at this locus, so the significance threshold used for the replication data was P < 0.05.
Replication cohorts
The adult replication data came from individuals who took part in the Netherlands Twin Registry (NTR) Biobank study conducted between January 2004 and July 2008 (35). This study collected blood from 9530 adult twins and their family members from 3477 families who participate in the longitudinal studies of the NTR. At the time of analysis, genome-wide SNP data were available in 4201 individuals, including 1428 individuals who are part of a monozygotic (MZ) twin pair. If both twins from an MZ pair had valid GGT data, the average value was taken. The final sample size for this analysis was 3225 individuals, 63.3% women and mean age 47.4 years (SD = 14.1). GGT was determined in heparin plasma using the Vitros assay (Johnson & Johnson, Rochester, USA). The mean for log10GGT was 1.43 (SD = 0.23).
Genotype data were available for individuals who participated in at least one of the following six genotyping projects of the NTR: the GAIN-MDD study (1864 participants, who were mainly controls for MDD; Perlegen/Affymetrix 660K SNP chip) (36), the NTR2 study (1335 unrelated individuals unselected for phenotype; Illumina 660K), the GenomEUtwin study (287 female MZ twins, unselected for phenotype; Illumina 310K), the MDD2000 study (26 cases for MDD; Affymetrix 600K), an ADHD study (155 subjects from families with high or low scoring offspring; Affymetrix 1M) and a subsample from the GAIN-MDD study (96 controls) who were genotyped for a second time on Illumina 660K. The majority were genotyped as part of the GAIN-MDD or NTR2 studies (1576 and 1282 individuals, respectively, out of 3225).
Individuals' genotype data were checked for European ancestry, gender inconsistencies, Mendelian errors and level of heterozygosity. Other checks including HWE, MAFs, SNP and sample call rates were conducted in each sample independently and in the combined sample. Samples typed twice and allele frequencies were used to verify the alignment of strands between chips. After that, imputation of all HAPMAP phase II SNPs in all genotyped individuals was conducted using the HAPMAP II build 36 release 24 CEU sample as a reference panel. rs6519519 genotypes were extracted for the replication analyses. Because imputation was run in the entire sample, the genotype data were imputed in all individuals, even when this SNP was genotyped on one of the chips. The r-squared of the SNP was 0.87.
The adolescent replication data came from individuals who took part in the Western Australian Pregnancy (Raine) cohort. Between 1989 and 1991, 2900 pregnant women were recruited prior to 18-week gestation into a randomized controlled trial to evaluate the effects of repeated ultrasound in pregnancy (37–39). Recruitment predominantly took place at King Edward Memorial Hospital (Perth, Western Australia). Their children have been comprehensively phenotyped from birth to 21 years of age (average ages of 1, 2, 3, 6, 8, 10, 14, 17 and currently 21) by the Raine research team. Data collection included questionnaires completed by the child's primary carer and by the adolescent from age 14, physical assessments by trained assessors at all follow-up years and blood collection from the year 14 and 17 follow-ups. The study was conducted with appropriate institutional ethics approval, and written informed consent was obtained from a parent/guardian, and from the adolescent from year 17. The cohort has been shown to be representative of the population presenting to the antenatal tertiary referral centre in Western Australia.
Serum was separated from clotted blood and stored at −70°C until analysis. Serum GGT was analysed on an Architect c16000 Analyser (Abbott Diagnostics, Abbott Laboratories, Abbott Park, IL 60064, USA) using reagents from Abbott Diagnostics. Log-transformed GGT z-scores from the 14- and 17-year follow-ups were used. Of the 2868 individuals who participated in the childhood follow-ups, 1377 individuals had GGT levels measured at the 14-year follow-up and 1268 at the 17-year follow-up. A subset of those individuals were selected for analysis based on singleton birth, having no siblings in the study, Caucasian ethnicity, no congenital abnormalities and relevant genotype data. This led to 1149 individuals with genotype and GGT data at the 14-year follow-up and 1046 individuals at the 17-year follow-up, which are the final sample sizes for the analysis. The mean age of the participants at each follow-up was 14.1 (SD = 0.20; minimum = 13.0, maximum = 15.1) and 17.1 (SD = 0.25; minimum = 16.0, maximum = 18.3).
Individual genotype data were extracted from the genome-wide Illumina 660 Quad Array. Briefly, the genotyping was performed on the Illumina BeadArray Reader at the Centre for Applied Genomics (Toronto, Ontario, Canada). Individuals' data were checked for gender inconsistencies, level of heterozygosity and relatedness with other individuals in the sample. SNPs were removed through genotyping QC if they had a call rate of <95%, had a Hardy–Weinberg P-value <5.7 × 10−7 or had a MAF <1%. All analyses were performed using R version 2.11.1. Allelic association for rs6519519 was performed by multivariate linear regression on gender and SNP allele count.
Disease and pathway analysis
We submitted all SNPs showing associations with GGT at P < 0.001 (n = 3073) for analysis using Ingenuity Pathway Analysis® (IPA; Ingenuity Systems, Inc., Redwood City, CA, USA). This identifies genes within 10 kbp of the SNPs, and then tests for associations between these genes and known biological pathways or previously reported disease associations. The IPA database contains multiple types of information including published associations between genes and diseases. In some cases, the data are more extensive than that published in the original papers, so the system can make use of suggestive associations as well as the published significant ones. The sources used are cited in the Ingenuity output. IPA provides statistics on associations between diseases (e.g. non-insulin-dependent diabetes) and the genes containing significant or suggestive SNPs, based on a standard 2 × 2 contingency table. The four cells comprise the number of genes identified in the association analysis, categorized as associated with the disease/not associated with the disease, and the number in the set of all genes in the database, again categorized as associated with the disease/not associated with the disease. The statistical test used is a one-tailed Fisher's exact test (one-tailed because we are interested in over-representation of genes associated with the disease). As well as the P-values for disease association, the number of molecules (genes) in the submitted list which have been associated with the disease, and the sources of that information, are reported.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
The Australian studies at QIMR were supported by NIH grants AA07535, AA07728, AA13320, AA13321, AA13326, AA14041, AA11998, AA17688, DA012854, DA019951; by grants from the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498); from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921); and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). Genotyping at CIDR was supported by grant AA13320 to the late Richard Todd, PhD, MD. R.P.M., S.E.M., D.R.N., and G.W.M. are supported by the National Health and Medical Research Council (NHMRC) Fellowship Scheme. The authors gratefully acknowledge the NH&MRC for their long-term contribution to funding the Raine Study over the last 20 years and also the following Institutions for providing funding for Core Management of the Raine Study: The University of Western Australia; Raine Medical Research Foundation; UWA Faculty of Medicine, Dentistry and Health Sciences; The Telethon Institute for Child Health Research; and the Women and Infants Research Foundation. The authors gratefully acknowledge the assistance of the Western Australian DNA Bank (National Health and Medical Research Council of Australia National Enabling Facility). The authors also acknowledge the support of the National Health and Medical Research Council of Australia (Grant ID 572613 and ID 003209) and the Canadian Institutes of Health Research (Grant ID 166067). For the Netherlands studies, funding was obtained from the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW) Grants 904-61-090; 985-10-002; 904-61-193; 480-04-004; 400-05-717; 911-09-032; Addiction-31160008; Spinozapremie (SPI 56-464-14192); CMSB: Center for Medical Systems Biology (NWO Genomics); NBIC/BioAssist/RK/2008.024; BBMRI-NL: Biobanking and Biomolecular Resources Research Infrastructure; the VU University: Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); the European Science Foundation (ESF): EU/QLRT-2001-01254; European Community's Seventh Framework Program (FP7/2007-2013): ENGAGE (HEALTH-F4-2007-201413); the European Science Council (ERC) Genetics of Mental Illness (230374); Rutgers University Cell and DNA Repository cooperative agreement (NIH U24 MH068457-06); Collaborative study of the genetics of DZ twinning (NIH R01D0042157-01A); and the Genetic Association Information Network, a public–private partnership between the NIH and Pfizer Inc., Affymetrix Inc. and Abbott Laboratories. M.H.M.M. is financially supported by NWO (VENI-grant 016-115-035).
Supplementary Material
ACKNOWLEDGEMENTS
For the Raine study, the authors are grateful to the Raine Foundation, to the study participants and their families, and to the research staff for cohort coordination and data collection. We gratefully acknowledge the assistance of the Telethon Institute for Child Health Research and the Raine Medical Research Foundation of the University of Western Australia.
REFERENCES
- 1.Zhang H., Forman H.J., Choi J. Gamma-glutamyl transpeptidase in glutathione biosynthesis. Methods Enzymol. 2005;401:468–483. doi: 10.1016/S0076-6879(05)01028-1. doi:10.1016/S0076-6879(05)01028-1. [DOI] [PubMed] [Google Scholar]
- 2.Whitfield J.B. Gamma glutamyl transferase. Crit. Rev. Clin. Lab Sci. 2001;38:263–355. doi: 10.1080/20014091084227. doi:10.1080/20014091084227. [DOI] [PubMed] [Google Scholar]
- 3.Kazemi-Shirazi L., Endler G., Winkler S., Schickbauer T., Wagner O., Marsik C. Gamma glutamyltransferase and long-term survival: is it just the liver? Clin. Chem. 2007;53:940–946. doi: 10.1373/clinchem.2006.081620. doi:10.1373/clinchem.2006.081620. [DOI] [PubMed] [Google Scholar]
- 4.Fraser A., Harris R., Sattar N., Ebrahim S., Smith G.D., Lawlor D.A. Gamma-glutamyltransferase is associated with incident vascular events independently of alcohol intake: analysis of the British Women's Heart and Health Study and Meta-Analysis. Arterioscler. Thromb. Vasc. Biol. 2007;27:2729–2735. doi: 10.1161/ATVBAHA.107.152298. doi:10.1161/ATVBAHA.107.152298. [DOI] [PubMed] [Google Scholar]
- 5.Fraser A., Harris R., Sattar N., Ebrahim S., Davey Smith G., Lawlor D.A. Alanine aminotransferase, gamma-glutamyltransferase, and incident diabetes: the British Women's Heart and Health Study and meta-analysis. Diabetes Care. 2009;32:741–750. doi: 10.2337/dc08-1870. doi:10.2337/dc08-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. doi:10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X., Waterworth D., Perry J.R., Lim N., Song K., Chambers J.C., Zhang W., Vollenweider P., Stirnadel H., Johnson T., et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. doi:10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. doi:10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 9.Reiner A.P., Barber M.J., Guan Y., Ridker P.M., Lange L.A., Chasman D.I., Walston J.D., Cooper G.M., Jenny N.S., Rieder M.J., et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. doi:10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. doi:10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamagata K. Regulation of pancreatic beta-cell function by the HNF transcription network: lessons from maturity-onset diabetes of the young (MODY) Endocr. J. 2003;50:491–499. doi: 10.1507/endocrj.50.491. doi:10.1507/endocrj.50.491. [DOI] [PubMed] [Google Scholar]
- 13.Whitfield J.B., Zhu G., Nestler J.E., Heath A.C., Martin N.G. Genetic covariation between serum gamma-glutamyltransferase activity and cardiovascular risk factors. Clin. Chem. 2002;48:1426–1431. [PubMed] [Google Scholar]
- 14.Bathum L., Petersen H.C., Rosholm J.U., Hyltoft P.P., Vaupel J., Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: results from a population-based Danish twin study. Clin. Chem. 2001;47:81–87. [PubMed] [Google Scholar]
- 15.Middelberg R.P., Medland S.E., Martin N.G., Whitfield J.B. A longitudinal genetic study of uric acid and liver enzymes in adolescent twins. Twin Res. Hum. Genet. 2007;10:757–764. doi: 10.1375/twin.10.5.757. doi:10.1375/twin.10.5.757. [DOI] [PubMed] [Google Scholar]
- 16.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. doi:10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., Liu Y., Adamson C.L., Valdez G., Guo W., Hsu S.C. The mammalian exocyst, a complex required for exocytosis, inhibits tubulin polymerization. J. Biol. Chem. 2004;279:35958–35966. doi: 10.1074/jbc.M313778200. doi:10.1074/jbc.M313778200. [DOI] [PubMed] [Google Scholar]
- 18.Jetten A.M. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. doi:10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. doi:10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 21.Heisterkamp N., Groffen J., Warburton D., Sneddon T.P. The human gamma-glutamyltransferase gene family. Hum. Genet. 2008;123:321–332. doi: 10.1007/s00439-008-0487-7. doi:10.1007/s00439-008-0487-7. [DOI] [PubMed] [Google Scholar]
- 22.Joyce-Brady M., Jean J.C., Hughey R.P. gamma -glutamyltransferase and its isoform mediate an endoplasmic reticulum stress response. J. Biol. Chem. 2001;276:9468–9477. doi: 10.1074/jbc.M004352200. doi:10.1074/jbc.M004352200. [DOI] [PubMed] [Google Scholar]
- 23.Niida S., Kawahara M., Ishizuka Y., Ikeda Y., Kondo T., Hibi T., Suzuki Y., Ikeda K., Taniguchi N. Gamma-glutamyl transpeptidase stimulates receptor activator of nuclear factor-kappaB ligand expression independent of its enzymatic activity and serves as a pathological bone-resorbing factor. J. Biol. Chem. 2004;279:5752–5756. doi: 10.1074/jbc.M311905200. doi:10.1074/jbc.M311905200. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda Y., Taniguchi N. Gene expression of gamma-glutamyltranspeptidase. Methods Enzymol. 2005;401:408–425. doi: 10.1016/S0076-6879(05)01025-6. doi:10.1016/S0076-6879(05)01025-6. [DOI] [PubMed] [Google Scholar]
- 25.Vlasova I.A., Bohjanen P.R. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi L., Ke Y., Luo C., Gozal D., Liu R. Depletion of reduced glutathione enhances motor neuron degeneration in vitro and in vivo. Neuroscience. 2007;144:991–1003. doi: 10.1016/j.neuroscience.2006.09.064. doi:10.1016/j.neuroscience.2006.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyderman H., Hutson P.G., Matusica D., Rogers M.L., Rush R.A. The human G93A-superoxide dismutase-1 mutation, mitochondrial glutathione and apoptotic cell death. Neurochem. Res. 2009;34:1847–1856. doi: 10.1007/s11064-009-9974-z. doi:10.1007/s11064-009-9974-z. [DOI] [PubMed] [Google Scholar]
- 28.Kirk K.M., Birley A.J., Statham D.J., Haddon B., Lake R.I., Andrews J.G., Martin N.G. Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Res. 2000;3:299–309. doi: 10.1375/136905200320565274. doi:10.1375/136905200320565274. [DOI] [PubMed] [Google Scholar]
- 29.Treloar S.A., Wicks J., Nyholt D.R., Montgomery G.W., Bahlo M., Smith V., Dawson G., Mackay I.J., Weeks D.E., Bennett S.T., et al. Genomewide linkage study in 1176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am. J. Hum. Genet. 2005;77:365–376. doi: 10.1086/432960. doi:10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright M.J., Martin N.G. Brisbane Adolescent Twin Study: outline of study methods and research projects. Aust. J. Psychol. 2004;56:65–78. doi:10.1080/00049530410001734865. [Google Scholar]
- 31.Medland S.E., Nyholt D.R., Painter J.N., McEvoy B.P., McRae A.F., Zhu G., Gordon S.D., Ferreira M.A., Wright M.J., Henders A.K., et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am. J. Hum. Genet. 2009;85:750–755. doi: 10.1016/j.ajhg.2009.10.009. doi:10.1016/j.ajhg.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benyamin B., Middelberg R.P., Lind P.A., Valle A.M., Gordon S., Nyholt D.R., Medland S.E., Henders A.K., Heath A.C., Madden P.A., et al. GWAS of butyrylcholinesterase activity identifies four novel loci, independent effects within BCHE, and secondary associations with metabolic risk factors. Hum. Mol. Genet. 2011;20:4504–4514. doi: 10.1093/hmg/ddr375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W.M., Abecasis G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. doi:10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willemsen G., de Geus E.J., Bartels M., van Beijsterveldt C.E., Brooks A.I., Estourgie-van Burk G.F., Fugman D.A., Hoekstra C., Hottenga J.J., Kluft K., et al. The Netherlands Twin Register biobank: a resource for genetic epidemiological studies. Twin Res. Hum. Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231. doi:10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- 36.Boomsma D.I., Willemsen G., Sullivan P.F., Heutink P., Meijer P., Sondervan D., Kluft C., Smit G., Nolen W.A., Zitman F.G., et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur. J. Hum. Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. doi:10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 37.Newnham J.P., Evans S.F., Michael C.A., Stanley F.J., Landau L.I. Effects of frequent ultrasound during pregnancy: a randomised controlled trial. Lancet. 1993;342:887–891. doi: 10.1016/0140-6736(93)91944-h. doi:10.1016/0140-6736(93)91944-H. [DOI] [PubMed] [Google Scholar]
- 38.Evans S., Newnham J., MacDonald W., Hall C. Characterisation of the possible effect on birthweight following frequent prenatal ultrasound examinations. Early Hum. Dev. 1996;45:203–214. doi: 10.1016/0378-3782(96)01728-8. doi:10.1016/0378-3782(96)01728-8. [DOI] [PubMed] [Google Scholar]
- 39.Williams L.A., Evans S.F., Newnham J.P. Prospective cohort study of factors influencing the relative weights of the placenta and the newborn infant. BMJ. 1997;314:1864–1868. doi: 10.1136/bmj.314.7098.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.