Abstract
Inter-individual DNA methylation variations were frequently hypothesized to alter individual susceptibility to Type 2 Diabetes Mellitus (T2DM). Sequence-influenced methylations were described in T2DM-associated genomic regions, but evidence for direct, sequence-independent association with disease risk is missing. Here, we explore disease-contributing DNA methylation through a stepwise study design: first, a pool-based, genome-scale screen among 1169 case and control individuals revealed an excess of differentially methylated sites in genomic regions that were previously associated with T2DM through genetic studies. Next, in-depth analyses were performed at selected top-ranking regions. A CpG site in the first intron of the FTO gene showed small (3.35%) but significant (P = 0.000021) hypomethylation of cases relative to controls. The effect was independent of the sequence polymorphism in the region and persists among individuals carrying the sequence-risk alleles. The odds of belonging to the T2DM group increased by 6.1% for every 1% decrease in methylation (OR = 1.061, 95% CI: 1.032–1.090), the odds ratio for decrease of 1 standard deviation of methylation (adjusted to gender) was 1.5856 (95% CI: 1.2824–1.9606) and the sensitivity (area under the curve = 0.638, 95% CI: 0.586–0.690; males = 0.675, females = 0.609) was better than that of the strongest known sequence variant. Furthermore, a prospective study in an independent population cohort revealed significant hypomethylation of young individuals that later progressed to T2DM, relative to the individuals who stayed healthy. Further genomic analysis revealed co-localization with gene enhancers and with binding sites for methylation-sensitive transcriptional regulators. The data showed that low methylation level at the analyzed sites is an early marker of T2DM and suggests a novel mechanism by which early-onset, inter-individual methylation variation at isolated non-promoter genomic sites predisposes to T2DM.
INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a common disorder of complex genetic and environmental origin, accounting for >95% of diabetes worldwide. T2DM-associated sequence polymorphisms have been identified in 30 linkage disequilibrium (LD) blocks across the human genome (1,2), but their combined effect sizes explain only a minor fraction of the observed phenotypic diversity among individuals in the human population. Many of these polymorphisms do not directly alter coding or control sequences of T2DM genes, but are merely co-transmitted with putative causative polymorphisms in large LD blocks. The causative polymorphisms and the affected control regions assumed to reside in the LD blocks are still largely unknown, and a large portion of the divergence in individual susceptibility to T2DM remains unexplained.
Inter-individual DNA methylation variations were hypothesized to alter individual susceptibility to T2DM and other common human diseases (3–5). Methylation profiles are typically established during early developmental stages prior to major cell differentiation, and subsequently maintained through cell divisions. Thus, individual-specific, disease-related methylations may appear not only in the affected tissue(s), but across the human body including accessible tissues such as blood. Given the well-documented effect of DNA methylation on chromatin activity and gene-expression patterns, such early-onset methylation variation may predispose to disease development at adulthood, in a manner similar to the effect of DNA sequence variations.
Connections between DNA methylation and gene-expression patterns were traditionally studied at clusters of CpG methylation sites located in gene promoters. However, more recent studies demonstrated that connections between small methylation differences at isolated CpG sites and large differences in gene-expression levels can be found across a substantial portion of the human genome (6–8). Hence, we sought to include in our study not only methylation clusters in gene promoters, but also isolated non-promoter methylation sites.
An essential step towards the elucidation of disease-contributing epigenetic mechanisms is the mapping of disease-associated methylations across the genome through epigenome-wide association studies (EWAS). An excellent review describing the goals, the abilities and the limitations of the EWAS approach was recently published (9). Unfortunately, none of the available mapping technologies allow cost-effective quantitative evaluation of the 28 million CpG methylation sites in the human genome across sufficient numbers of case and control individuals. To overcome this obstacle, here we apply a stepwise study design: first, we utilized a microarray-based assay to evaluate average methylation levels of case and control individuals assembled in DNA pools. This allowed us to discover differentially methylated regions (DMRs) across the genome and analyze whether they are concentrated in specific genomic locations. Following these microarray-based analyses, we applied deep sequencing to evaluate the significance of case–control differentiation at particular methylation sites following multiple hypothesis testing. Finally, we examined various aspects of the association between methylation and the disease through individual-level analyses. This research approach allowed us to conduct an effective screen among hundreds of case and control individuals, and thus to reveal a T2DM-specific methylation pattern that appeared prior to the onset of clinical T2DM manifestations and carried a high effect size on disease risk.
RESULTS
DNA pooling allows accurate assessment of average DNA methylation in large groups of individual genomes (10,11). We have previously showed the effectiveness of DNA pooling in highlighting actual epigenetic patterns over the background of the numerous stochastic inter-individual differences typical for epigenetic marks (12). Here, we applied this pool-based strategy for the mapping of disease-specific patterns shared by many individuals in the ‘case’ pools and distinguish them from the controls. In the first experiment (step i in Fig. 1), 710 T2DM patients and 459 control individuals were assembled in four (two cases and two controls) age-matched DNA pools, as described in Table 1. An additional control pool of young individuals was used to evaluate the possible effect of age. All participants were of Ashkenazi Jews ethnic origin (all four grandparents) who attended various Israeli medical centers. Patients were treated for T2DM for at least 10 years prior to blood collection. Controls were self-reported healthy individuals.
Figure 1.
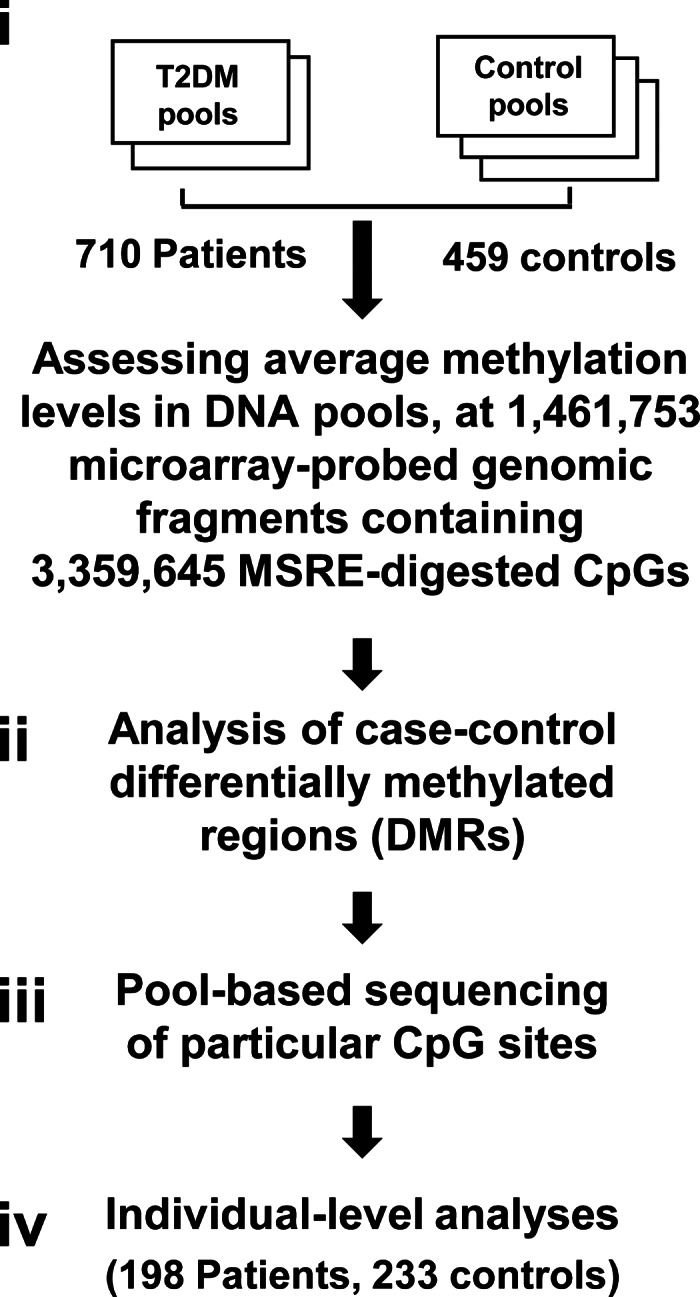
A flowchart of the multistep study design. Pool-based assessments of CpG sites in methyl-sensitive restriction enzyme (MSRE) sites within microarray-probed DNA fragments (i), allowed genome-scale evaluation of differentially methylated regions (DMRs) (ii). Following these steps, deep bisulfite sequencing was utilized to verify particular case–control-differentiating sites (iii). Finally, effect size, sensitivity and mechanistic aspects were explored through individual-level analyses (iv).
Table 1.
Case and control DNA pools that were analyzed in the microarray-based experiment
| Pool | n | Age (average) | Age (SD) | Age (minimum) | Age (maximum) | BMI (average) | BMI (SD) | BMI (minimum) | BMI (maximum) | Males’ fraction |
|---|---|---|---|---|---|---|---|---|---|---|
| T2DM 1 | 361 | 64.51 | 9.99 | 42 | 92 | 30.16 | 5.51 | 19.15 | 62.36 | 0.50 |
| T2DM 2 | 349 | 64.72 | 9.12 | 40 | 89 | 30.32 | 5.63 | 20.76 | 77.63 | 0.48 |
| Control 1 | 169 | 64.40 | 12.52 | 40 | 92 | 28.08 | 5.68 | 16.51 | 55.84 | 0.27 |
| Control 2 | 156 | 65.25 | 13.81 | 33 | 91 | 27.93 | 5.72 | 17.34 | 56.92 | 0.31 |
| Control 3 (young) | 134 | 25.89 | 6.39 | 11 | 41 | 21.98 | 3.84 | 14.90 | 30.84 | 0.77 |
| Total cases | 710 | |||||||||
| Total controls | 459 |
Applying an established microarray-based assay (6,12), we analyzed the methylation levels of each pool at 1 461 753 DNA genomic fragments containing 3 359 645 CpG methylation sites (Supplementary Material, Fig. S1). Following this step, the assessed genomic fragments were ranked according to discrimination between case and control pools, using the silhouette statistic (methods), where the fragment showing the highest separation between case and control was ranked number 1, and the fragment showing the lowest separation ranked 1 461 753. We then searched for case–control DMRs, defined as a 10 kb genomic window which contains at least three top-ranking fragments and is also significantly (P < 0.05 or P < 0.005 for stringent DMRs) enriched by top-ranking fragments following correction for the number of fragments in the window (Fig. 1, step ii).
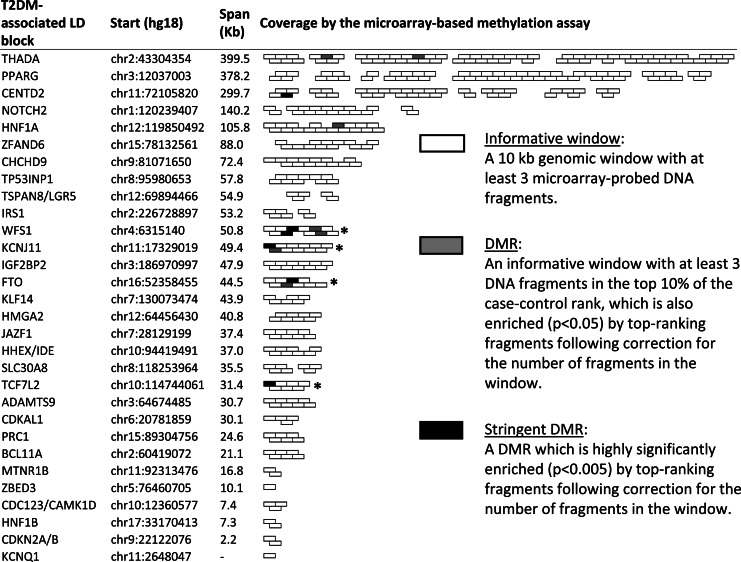
We evaluated the distribution of DMRs across the genome and whether they are concentrated in specific genomic locations. Within the 30 LD blocks which were previously shown to contain T2DM-associated sequence polymorphisms, six stringent DMRs were identified (Fig. 2). In contrast, the average number of stringent DMRs among the informative windows in 10 000 sets of randomly selected similar-sized genomic blocks was 2.23 ± 1.73 (P = 0.034 for the T2DM blocks differing from the genome average as represented by the 10 000 randomly selected sets). Thus, the group of T2DM-associated blocks is significantly enriched with DMRs when compared with the genome. Moreover, a further analysis of the enrichment within blocks showed that four of the T2DM-associated LD blocks were significantly (P < 0.05) enriched by stringent DMRs following correction for block size (marked by asterisks in Fig. 2), compared with an average of 1.05 ± 1.01 enriched blocks in the 10 000 random genomic sets (P = 0.019). Thus, T2DM-associated blocks are not only enriched as a group, but also tend to contain more differential methylations than the average similar-size genomic block. As shown in Supplementary Material, Figure S1, this enrichment is not due to an intrinsic bias of the differential methylation assay towards T2DM-associated blocks. We also analyzed other sets of genomic locations, including the blocks containing metabolic genes that were not directly associated with T2DM, or various gene-ontology terms. These sets did not show enrichment relative to the genome. The control pool of young individuals showed a slight tendency towards higher methylation levels across the genome, compared with the pools of old individuals (Supplementary Material, Fig. S1e). This global methylation difference between young and old pools did not confound the enrichment analysis, as repeating the analysis using only the age-matching pools revealed similar enrichment of DMRs in the associated LD blocks. We conclude that T2DM-associated LD blocks are specifically enriched with case–control DMRs.
Figure 2.
Case–control differentially methylated regions (DMRs) in T2DM-associated genomic blocks. The chromosomal locations and sizes of the 30 established T2DM-associated genomic linkage disequilibrium (LD) blocks, and their coverage by the microarray-based assay are shown. The indicated genes provide the common reference in the literature, and not the complete list of genes residing in these regions. Unshaded boxes indicate the coverage of these regions by microarray-probed 10 kb ‘informative’ windows (50% overlapped). Gray boxes indicate DMRs, and black boxes indicate stringent DMRs. Asterisks indicate LD blocks that were significantly (P < 0.05) enriched by stringent DMRs following correction to the number of informative windows within the block. Note that the indicated genes are only reference to the block identity as given in the study of Voight et al. (2), not necessarily the actual T2DM gene affected by the disease-associated (genetic or epigenetic) polymorphisms in the region.
We further sought to replicate the observed case–control differences at particular CpG sites embedded in the microarray-probed genomic fragments, using a different methylation assay. In this analysis, age-matching pools were used, and similar numbers of men and women were applied (Table 2). Selected top-ranking genomic fragments were PCR-amplified from bisulfite-converted DNA pools, and 93 particular CpG sites embedded in these fragments were sequenced at ultra-deep (>450×) coverage using a genomic sequencer. Following multiple hypothesis testing (q-value), 13 out of the 93 CpG sites, located in six different LD blocks, showed significant (P < 0.05, q < 0.05) case–control differences (Table 3; the results of all 93 sites are given in Supplementary Material, Table S1). We further analyzed methylation differentiation among neighboring CpG sites by assessing the frequency of sequence reads in which all CpGs in a DNA fragment were methylated (complete methylation) out of all reads. The analysis revealed significant case–control differences in four DNA fragments containing more than one CpG (Table 4; the results of all 22 fragments with more than one methylation site are given in Supplementary Material, Table S2). We conclude that case–control differential methylation could be observed within T2DM-associated LD blocks at regional (Fig. 2), local (Table 4) or site-specific (Table 3) levels.
Table 2.
Case and control DNA pools that were analyzed by ultra-deep bisulfite sequencinga
| Pool | n | Age (average) | Age (SD) | Age (minimum) | Age (maximum) | BMI (average) | BMI (SD) | BMI (minimum) | BMI (maximum) | Males’ fraction |
|---|---|---|---|---|---|---|---|---|---|---|
| T2DM 1 | 361 | 64.51 | 9.99 | 42 | 92 | 30.16 | 5.51 | 19.15 | 62.36 | 0.50 |
| T2DM 2 | 349 | 64.72 | 9.12 | 40 | 89 | 30.32 | 5.63 | 20.76 | 77.63 | 0.48 |
| Control 4 | 220 | 65.80 | 10.26 | 40 | 92 | 28.56 | 5.83 | 17.34 | 56.92 | 0.49 |
| Control 5 | 217 | 65.51 | 9.90 | 41 | 91 | 28.14 | 5.90 | 16.51 | 55.84 | 0.51 |
| Total cases | 710 | |||||||||
| Total controls | 437 |
aThe patient pools evaluated in the sequencing experiment are equivalent to the case pools in the microarray experiment. The control pools were reorganized to allow age matching and similar numbers of males and females, utilizing additional controls from a population-representative sample of Ashkenazi Jews (E.L.L).
Table 3.
CpG sites showing significant (P < 0.05) differences between T2DM and control pools following multiple hypothesis testing (q < 0.05). Average methylation levels, number of aligned sequence reads and absolute methylation differences are shown
| Chromosome | Position (hg18) | Nearest gene | Gene relation | Control methylation (%) | T2DM methylation (%) | Control reads | T2DM reads | Methylation difference | P-value | q-value |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 43590864 | THADA | Intron | 95.64 | 94.41 | 3947 | 3957 | 1.23 | 0.0120 | 0.0464 |
| 7 | 28143482 | JAZF1 | Intron | 94.54 | 92.94 | 4305 | 3555 | 1.60 | 0.0034 | 0.0188 |
| 8 | 118257326 | SLC30A8 | 3′-UTR | 85.61 | 83.43 | 5159 | 4888 | 2.18 | 0.0025 | 0.0188 |
| 8 | 118257358 | SLC30A8 | 3′-UTR | 96.12 | 95.07 | 5130 | 4845 | 1.05 | 0.0102 | 0.0428 |
| 8 | 118258573 | SLC30A8 | Down | 61.78 | 65.43 | 3370 | 2930 | 3.65 | 0.0027 | 0.0188 |
| 10 | 114734658 | TCF7L2 | Intron | 88.76 | 86.41 | 4618 | 3755 | 2.34 | 0.0012 | 0.0116 |
| 10 | 114739401 | TCF7L2 | Intron | 99.25 | 98.53 | 4642 | 1768 | 0.72 | 0.0079 | 0.0361 |
| 10 | 114743601 | TCF7L2 | Intron | 94.52 | 92.59 | 3542 | 4777 | 1.93 | 0.0004 | 0.0055 |
| 10 | 114743664 | TCF7L2 | Intron | 98.23 | 96.91 | 3551 | 4788 | 1.32 | 0.0001 | 0.0025 |
| 11 | 2805916 | KCNQ1 | Intron | 90.23 | 92.20 | 2979 | 4180 | 1.97 | 0.0033 | 0.0188 |
| 11 | 2806049 | KCNQ1 | Intron | 88.06 | 90.97 | 2981 | 4187 | 2.92 | 0.0001 | 0.0015 |
| 11 | 2806079 | KCNQ1 | Intron | 93.43 | 95.03 | 2968 | 4168 | 1.60 | 0.0038 | 0.0192 |
| 16 | 52366732 | FTO | Intron | 30.15 | 27.53 | 12180 | 11013 | 2.61 | 1e−5 | 0.0006 |
Table 4.
Significant (P < 0.05, q < 0.05) case–control differences at clusters of neighboring CpG sites. Start and end sites of the sequenced DNA fragments, the number of CpG sites in each fragment and the fraction of sequence reads in which all CpGs in a DNA fragment were methylated (complete methylation) out of all reads are shown
| Chromosome | Start | End | CpG (n) | Nearest gene | CTRL reads | T2DM reads | Complete methylation (%) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CTRL | T2DM | Difference | P-value | q-value | |||||||
| 8 | 118247780 | 118247919 | 2 | SLC30A8 | 4205 | 4048 | 56.81 | 53.88 | 2.93 | 0.0074 | 0.0250 |
| 8 | 118257203 | 118257365 | 2 | SLC30A8 | 5168 | 4895 | 83.39 | 80.75 | 2.64 | 0.0006 | 0.0047 |
| 10 | 114743495 | 114743680 | 7 | TCF7L2 | 3558 | 4795 | 83.62 | 81.04 | 2.57 | 0.0024 | 0.0115 |
| 16 | 52366689 | 52366689 | 2 | FTO | 12185 | 11021 | 29.84 | 27.28 | 2.55 | 2e−5 | 0.0003 |
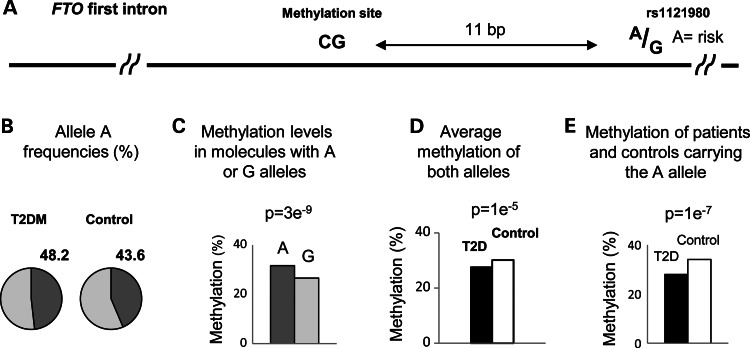
We next selected a CpG site in the first intron of the FTO gene for further in-depth analysis. Figure 3A shows the location of this site, 11 bp upstream of the obesity/T2DM-associated rs1121980 polymorphic sequence. The A allele of rs1121980 is an established risk factor for obesity and T2DM (1,2,13). We analyzed the relationships between these neighboring genetic and epigenetic polymorphisms. In theory, two alternative models could explain the co-association of the adjacent genetic and epigenetic sites with the disease. First, it is possible that the DNA polymorphism affects the methylation level of the neighboring site, a situation that was widely observed in the human genome (14–16). In this case, the association of the methylation level with the disease is secondary to the association between the sequence polymorphism and the disease. The alternative, and the more significant situation, is an independent association with the disease due to disconnected genetic and epigenetic mechanisms that independently affect the function of a control region in which they both reside. We explored these alternative possibilities utilizing pool-based sequencing. Among 23 193 sequenced molecules obtained from the two case and two matching control pools (Table 2), the frequency of the A risk allele was 43.6% in the controls and 48.2% among patients (Fig. 3B). These frequencies are in accordance with the allelic distribution of rs1121980 in the Western European population (17), a population that is genetically related to the Ashkenazi Jews population. In addition, the observed difference in the frequency of the risk allele between cases and controls is in agreement with the reported effect size of sequence polymorphisms in the FTO LD block (1,2). We next analyzed the methylation status of 10 601 molecules carrying the A allele and 12 592 molecules carrying the G allele. The molecules carrying the A risk allele were significantly hypermethylated relative to molecules carrying the G allele (Fig. 3C). This result is in accordance with a recent study report on hypermethylation of a T2DM-associated haplotype in the FTO LD block (18). Therefore, if the entire association with the disease was secondary to the effect of the sequence, we would expect the patient groups, which possess higher frequency of the A allele, to be more methylated than the control group. Instead, T2DM patients were significantly hypomethylated (Table 3 and Fig. 3D). Moreover, among molecules carrying the A allele cases were still hypomethylated relative to controls (Fig. 3E). Thus, the methylation level of this particular site is under the control of an independent mechanism, dictating hypomethylation of the patients in spite of their higher frequency of the nearby A allele (Fig. 3B) and the general tendency of the region towards hypermethylation of the risk alleles. We conclude that T2DM-associated sequences cannot account for the observed case–control methylation difference. As the other disease-associated polymorphisms in the LD block are tightly linked with rs1121980 and with each other, the above conclusion is valid not only for rs1121980 but for the entire block.
Figure 3.
Sequence-independent differential methylation. (A) Map of a DNA fragment from the first intron of the FTO gene, containing a CpG site and a T2DM/obesity-associated polymorphic site. (B–E) The results of ultra-deep sequencing of this fragment among two case and two control pools, allowing co-assessment of genotype and epigenotype differences. (B) Frequencies of the A allele among case and controls pools. (C) Methylation levels of sequenced DNA molecules carrying the A or the G alleles. The P-value of the difference between the case and control pools is indicated. (D) Methylation levels of case and control pools (both alleles). (E) Methylation levels of molecules from the case or the control pools carrying the A allele. The data show that despite the higher frequency of the A risk allele among the cases (B), and the higher methylation level associated with the risk allele (C), cases are more methylated than controls (D), and this trend persists among DNA molecules carrying the A allele (E). Thus, the hypomethylation of the patients is sequence-independent.
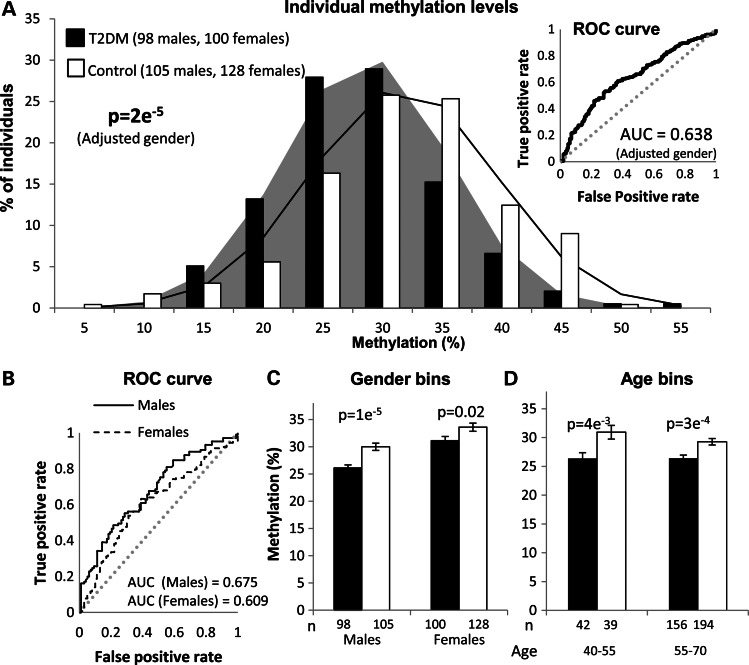
Despite the advantages of the pool-based approach in initial screens, some downstream analyses require individual-level data. We next studied individual methylation levels at the FTO site employing quantitative bisulfite sequencing. Out of the participants in the pool-based sequencing experiment, we randomly selected similar numbers of T2DM and control males and females with low and high body mass index (BMI) at ages 40–70 and analyzed their methylation levels by pyrosequencing. As previously observed in the pool-based experiments, the results showed small (3.35%) but significant (P = 0.000021) hypomethylation of cases relative to controls (Fig. 4A and Supplementary Material, Tables S3 and S4). These results provide further confirmation, at the level of individual analyses, for the association between hypomethylation in this site and T2DM, independent of the general hypermethylation of risk alleles in the FTO block.
Figure 4.
Individual-level analyses of the FTO methylation site. Methylation levels of the methylation site presented in Figure 3 were evaluated in patients and controls by quantitative pyrosequencing. (A) Frequencies and Gaussian expectations of methylation levels among 198 T2DM patients and 233 controls. The P-value of the methylation difference between cases and control following adjustment for the effect of gender is indicated. Insert: Receiver-operating characteristic (ROC) analysis of the same individuals. The area under the curve (AUC) is a measurement of the sensitivity, as expressed by the ratio of true-positive to false-positive rates. The dashed line represents the null hypothesis of no prediction. (B) ROC analyses as above, but for males and females separately. (C) Methylation levels of case and control individuals categorized by gender. (D) Methylation levels of case and control individuals categorized by age.
The odds ratio for 1% lower methylation was 1.061 (95% CI: 1.032–1.090) (i.e. the odds of belonging to the T2DM group increased by 6.1% for every 1% decrease in methylation). Receiver-operating characteristic analysis (insert in Fig. 4A) suggests that methylation level in this single site is more closely related to T2DM (area under the curve (AUC) = 0.638, 95% CI: 0.586–0.690) than the sequence variant with the largest effect identified to date (rs7901695 in the TCF7L2 LD block, AUC = 0.55), or the 18 most established genetic variants combined (AUC = 0.6) (19). Interestingly, men were hypomethylated relative to women, and the effect was stronger in men than in women (P = 0.034 for sex interaction, AUC = 0.675 among men and 0.609 among women; Fig. 4B). Nevertheless, the hypomethylation of cases relative to controls appeared in both genders (Fig. 4C). No significant association with age appeared in the examined age range (P = 0.791 for age interaction) (Supplementary Material, Fig. S2) and cases were discriminated from controls at low and high ages (Fig. 4D). The P-value of case–control differences adjusted to age and gender was 3e−5.
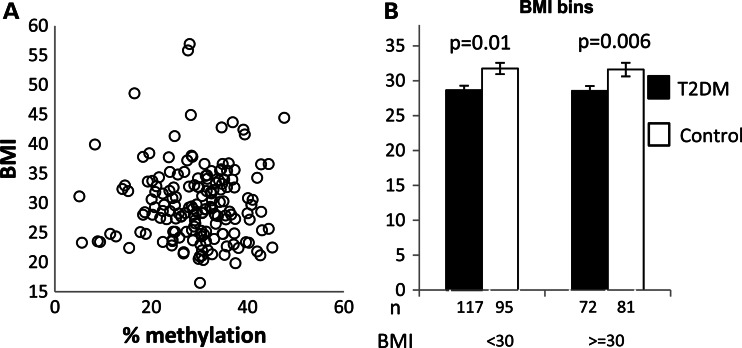
Interestingly, we observed no correlation between methylation and BMI (Fig. 5A; P = 0.954 for BMI interaction), and cases were hypomethylated relative to controls in both obese and non-obese subjects (Fig. 5B). These results suggest that the observed association with T2DM is not mediated through obesity. This is of some surprise, as the association between the FTO alleles and obesity is well established (13). Initially, the effect on T2DM was thought to be fully explained by the known connection between BMI and T2DM risk. This notion was supported by the original analysis of the region (13) and through a set of following studies. However, some other works have reported on residual association with T2DM following adjustment for BMI, in different human populations (20,21). Importantly, a recent large-scale meta-analysis among 41 504 subjects from Scandinavian populations confirmed an effect on T2DM risk, partly independent of the observed effect on BMI (22). Therefore, it was suggested that some of the polymorphisms in the FTO region affect T2DM subsequently to obesity, while others are more directly connected with T2DM possibly through differential effect of polymorphisms in the block among tissues. It appears that the particular epigenetic variation we studied belongs to this second type.
Figure 5.
Case–control differentiation is not due to body mass index BMI. (A) BMI of control individuals as a function of their methylation level. (B) Methylation levels of case and control individuals categorized by BMI. BMI data were available for 365 out of the 431 individuals presented in Figure 4.
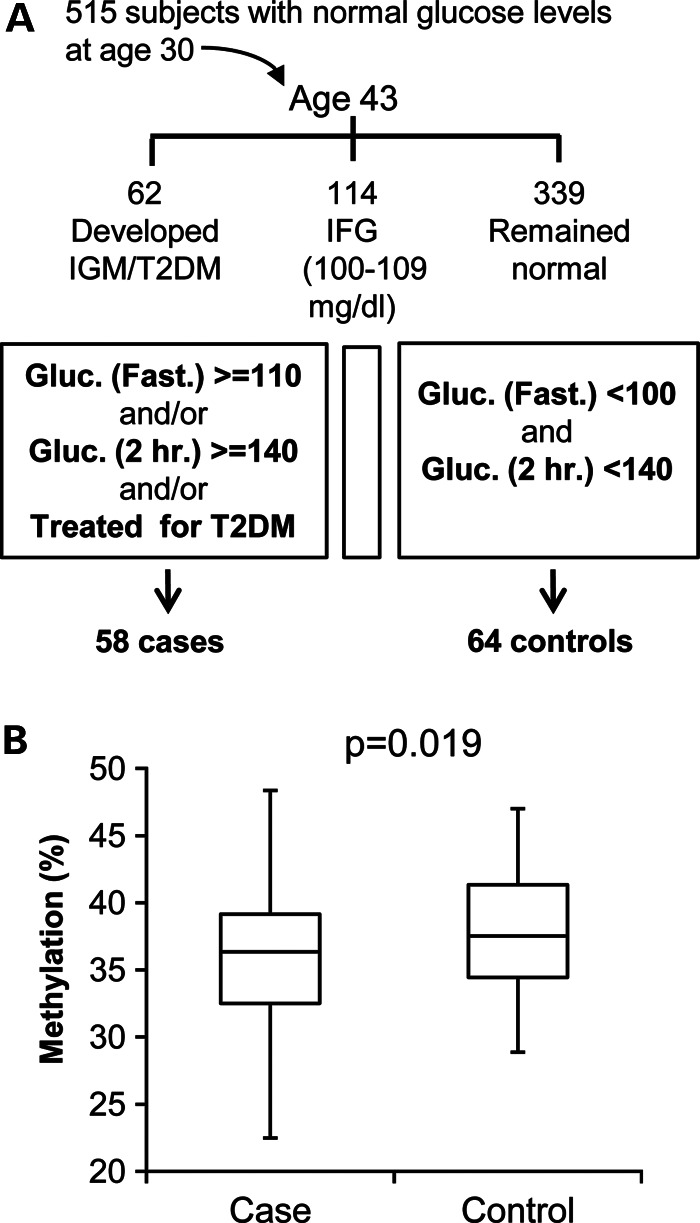
We next sought to replicate the association with the disease in an independent population cohort and at the same time to explore whether the hypomethylation of the cases could be observed prior to the onset of clinical disease manifestations. For this, we took advantage of the Jerusalem LRC longitudinal study (23). Out of 515 initially healthy participants in this study, 62 had developed impaired glucose metabolism (IGM) between ages 30 and 43. We analyzed the methylation levels at age 30 of 58 individuals who developed IGM and of 64 randomly selected control participants who remained free of IGM. During the entire course of this analysis, case and control samples were mixed together and the laboratory was blinded for their identity. The analysis showed that those individuals who progressed to IGM were hypomethylated relative to controls (P = 0.019), already before the appearance of IGM/diabetes (Fig. 6 and Supplementary Material, Tables S5 and S6). Thus, we independently confirmed the initial finding of hypomethylation in T2DM individuals, and additionally showed that low methylation level is not the result of the disease, but rather an early risk factor that predisposes to disease manifestation later in life.
Figure 6.
Abnormal methylation levels precede the onset of disease manifestations. (A) Scheme of the Jerusalem LRC longitudinal (cohort) study: Out of 515 individuals with normal glucose metabolism at age 30, 62 had, by age 43, developed IGM, defined as fasting plasma glucose levels ≥110 mg/dl and/or ≥140 mg/dl 2h after a 75 g oral glucose load, including eight that fulfilled clinical criteria for T2DM; 114 developed impaired fasting glucose (IFG, fasting glucose 100–109 mg/dl); and 339 remained normal (fasting glucose <100 mg/dl and post-challenge <140 mg/dl). (B) Methylation levels at baseline (age 30) of 58 incidence cases of IGM (including seven with diabetes) and 64 randomly selected normal controls. P-value (logistic regression, one-sided) was obtained following adjustment for gender and lymphocyte count.
As the studied DNA samples were obtained from whole blood, we also sought to analyze the potential contribution of differential case–control composition of blood cell types to the observed methylation differences. Using data from the Jerusalem LRC cohort study, we compared methylation levels with differential blood counts. We found that lymphocytes were significantly hypermethylated relative to granulocytes and monocytes (Supplementary Material, Fig. S3). However, no significant differences in blood counts appeared between cases and controls, and adjustment for blood lineage count did not affect the association of methylation with the incidence of IGM/T2DM. In fact, owing to reduced inter-individual variation following this procedure, the hypomethylation of the cases become even more significant (Supplementary Material, Fig. S4 and note Supplementary Material, S1).
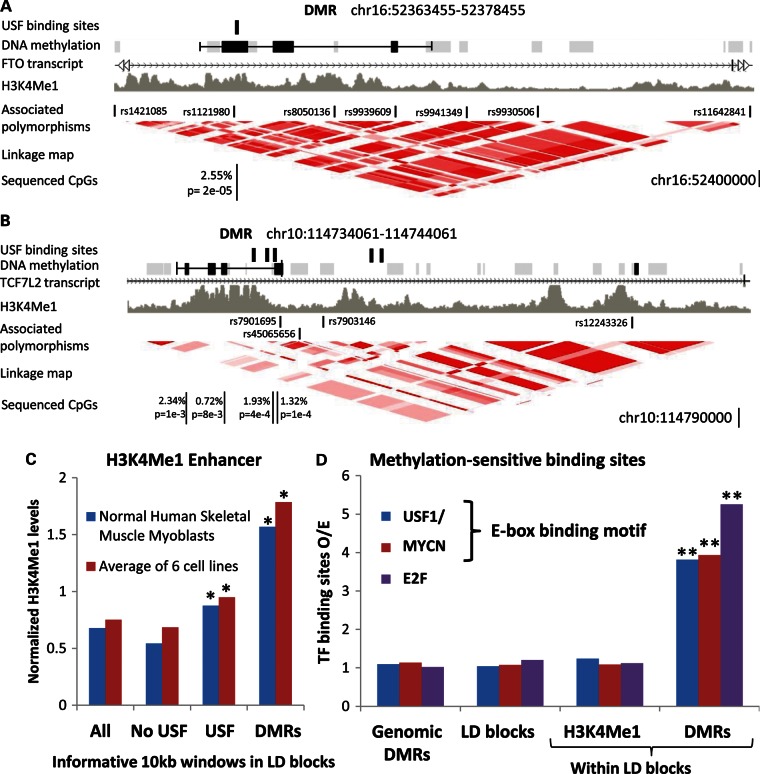
Finally, we explored potential connections between T2DM-associated methylation and gene-control mechanisms. We found that case–control DMRs were co-localized with sub-regions of the LD blocks that carry a chromatin signature of transcription enhancers in various cell types (Fig. 7A–C). This suggests that differential methylations are concentrated at functional gene-controlling regions embedded in the LD blocks. We further asked whether the observed case–control methylation differences may directly affect, or be affected by, binding of transcription factors. Strikingly, we found that the analyzed methylation site in the FTO LD block resides within a perfect binding motif of USF1/2 transcription activators (5′-GTCACGTGTC-3′), which binds these factors in silico and in vivo (deposited data in (24,25)). The USF1/2 factors play an important role in the regulation of glucose–lipid metabolism in response to insulin, and also participate in beta-cell development (26,27). Strikingly, the binding of these factors to their DNA targets is controlled by the methylation level of a single CpG dinucleotide included in their targeted sequence (28,29). Thus, the observed hypomethylation of T2DM patients can modify the binding affinity of methylation-sensitive transcription regulators, which are closely related to the mechanism of T2DM, to a putative regulatory element located within a T2DM-associated genomic region.
Figure 7.
Differentially methylated regions (DMRs) are co-localized with transcription enhancers and binding sites for methylation-sensitive transcription factors. (A) A map of the FTO LD block showing (from top to bottom) USF1/2 binding sites, probed (gray boxes) and differentially methylated (black boxes) DNA fragments, DMRs (black line), H3K4Me1 chromatin marks (deposited data in Mikkelesen et al. (#43)), T2DM-associated polymorphic sequences and a genetic linkage map. CpG sites that showed significant differences between case and control pools (hypomethylation in patients) are also indicated. (B) Same as in panel (A), but for the TCF7L2 block. (C) Average levels of H3K4Me1 enhancer marks in 10 kb windows along the T2DM LD blocks, in windows without or with USF1/2 binding sites, and in DMRs. To allow evaluation on a common scale, the H3K4Me1 levels of six cell lines were normalized to a common mean and variance. (D) The ratio of observed/expected numbers of binding sites (expectation is based on the frequency across the genome) in all genomic DMRs, in LD block windows, in LD block windows enriched with H3K4Me1 marks and in LD block DMRs. The data show that DMRs within the LD blocks are significantly enriched with binding sites for methylation-sensitive transcription factors. The presented data refer to all DMRs and similar enrichments were observed for the stringent DMRs (Supplementary Material, Fig. S5). USF1/2 and MYCN shared the E-box sequence-binding motif (CACGTG), while E2F binds a different core motif (C/GGCGC). USF1/2 were the only methylation-sensitive transcription factors that showed enrichment P-values <0.001 in both DMRs and stringent DMR analyses. Asterisks denote P < 0.05; double asterisks denote P < 0.01.
After making these observations, we asked whether other differentially methylated sites also bind methylation-sensitive transcription factors. We analyzed the genomic distribution of binding motifs for transcription factors with CpG dinucleotides in their binding sequences, which were shown to be affected by methylation. Remarkably, we found that case–control DMRs in the T2DM LD blocks are significantly (P < 0.01) enriched with binding sites for USF1/2, MYCN and E2F transcription factors (Fig. 7D), compared with DMRs across the genome or to informative windows in the T2DM LD blocks. Moreover, this enrichment was not due to a general tendency of the factors to bind enhancer regions, as only the DMRs, but not the other windows enriched with H3KMe1 chromatin marks, were enriched with binding sites for USF1/2 (Fig. 7D). Similar to USF1/2, MYCN and E2F family members bind their target sequences in a methylation-dependent manner (30,31), and might participate in T2DM-related pathways (32). Thus, case–control-differentiating regions are specifically enriched with binding sites for T2DM-related transcription factors, which may be affected by the observed T2DM-associated methylation.
A region few hundreds of basepairs downstream to the analyzed CpG site has been shown to bind the chromatin modulators FOXA1, FOXA2 and HDAC2 in various cell types (HudsonAlpha-deposited data). FOXA1 and FOXA2 bind enhancer regions during early development and regulate chromatin structure through cooperation with histone modifiers and other factors. We analyzed a CpG site located 860 bp downstream to rs1121980, and found that it is tightly correlated with the methylation of the previously analyzed site across individuals and exhibits similar methylation differences between cases and controls. Thus, a tract of case–control-differentiating methylation sites is co-localized with a chromatin region that serves as a cis-acting control element and binds enhancer-specific factors.
Taken together, the results revealed case–control-differentiating methylation in a region that serves as a distant regulator of gene transcription. Whether the gene under control is the FTO itself, one of its neighboring genes, or a more distant gene, is currently unknown and should await further research.
DISCUSSION
Despite intensive research, much of the variance in individual susceptibility to T2DM remains unexplained. Here, we show that sub-regions of the T2DM-associated LD blocks carry a unique epigenetic signature. In contrast to the previously reported sequence-directed methylations in T2DM (18,33), the methylation pattern described here is sequence-independent. Thus, it indicates a new type of inter-individual variation underlying an independent source of human diversity.
As an alternative to complete bisulfite sequencing of many case and control genomes, which is currently impractical, and to the conventionally reduced representation mapping methods, which are strongly biased towards certain fractions of the genome, here we demonstrated the efficiency of a pool-based large-scale screen among hundreds of case and control individuals followed by zooming-in on particular CpG sites. Our microarray-based assay offers fairly unbiased representation of the genome (Supplementary Material, Fig. S1), high throughput capability and sensitive detection of small methylation differences along large regions as demonstrated here (Fig. 2) and in our study of gene-body methylation in fresh human tissues (12). However, the analysis of methylation levels at isolated CpG sites using this assay is somewhat less reliable (Supplementary Material, Fig. S1 in the study of Aran et al. (12)). In our multistep study design, this limitation is compensated by the high fidelity of the pyrosequencing assay (Supplementary Material, Fig. S6).
DNA methylation levels are generally established early in development and are stably maintained thereafter (34). Our finding of abnormal methylation levels in young adulthood prior to the appearance of disease manifestations (Fig. 6) clearly support an early onset methylation risk profile for later IGM and subsequent diabetes. The occurrence of T2DM-related methylation in a tissue (blood) not directly involved in insulin secretion or action further suggests that the observed T2DM-associated methylation were established during early developmental stages, prior to major tissue differentiation.
The origin of predisposing methylation variations awaits further research: both stochastic epigenetic mutations and directed epigenetic programs are possible, and can lead to the observed results. According to the stochastic model, somatic epimutations produce random methylation differences between individuals. Out of this bulk of random variations, those which happen to affect disease pathways are enriched among patients and thus may be captured by our case–control comparative assay.
According to the alternative model of programmed methylations, particular alternations in the epigenetic patterning during early development are associated with elevated disease risk. One interesting possibility is pre-patterning by chromatin-modulating factors. A few specific DNA-binding proteins, referred to as ‘pioneer’ transcription factors, are able to bind naive chromatin during early developmental stages and to regulate chromatin structure and methylation levels through cross-talk with histone modifiers and other factors (35–38). If the binding profile of such factors differs between patients and controls, it could lead to differential case–control methylation. The chromatin immunoprecipitation data showing binding of the pioneering factors, FOXA1 and FOXA2 (HudsonAlpha deposited data), to the DMR are in line with this possibility.
In this study, we established that intronic regions of T2DM-associated genes carry small, but highly significant case–control methylation differences. While these differences are smaller than that generally observed between active and inactive gene promoters, we and others have recently shown that within gene bodies, small methylation differences are tightly correlated with substantial differences in gene-expression levels (6–8). It is also worth noting that over long periods of time, even small expression differences may predispose to a late-onset disease as T2DM. Thus, the observed methylation differences are capable of producing or indicating real expression differences, which may lead to the observed enhanced disease risk.
The identity of the gene (or genes) affected by the polymorphisms (either genetic or epigenetic) in the FTO LD block is yet to be clarified. As cis-acting enhancers can work over large distances, the potential impact on other genes in the area (some of them are reasonable T2DM gene candidates) and even more distant genes should be examined. The targeted tissue is also currently unclear. Given that the FTO gene is expressed in many tissues including in pancreatic isles, skeletal muscles, adipose tissues and other T2DM-related tissues, it would be interesting to evaluate methylation-expression relationships across these tissues. The systematic evaluation of case–control-differentiating sites, provided by this and by similar genome-wide studies, should open the way for downstream experiments aiming at the identification of targeted genes and tissues.
In theory, the binding affinity of transcription factors may be affected by either local alteration of their DNA target sequences or through more global changes in chromatin accessibility. Although it is well known that both genetic and epigenetic variations are capable of doing both, methylation marks were generally assumed to act through the synergistic effect of contiguous methylation marks on global chromatin structures. Our results call for reevaluation of the contribution of isolated methylation sites to disease mechanisms.
Genetic association studies have limited power to map specific functional regions embedded in the LD blocks. We suggest that sub-regions of the LD blocks, characterized by (i) frequent occurrence of differential methylation, (ii) an enhancer-like chromatin structure and (iii) frequent binding sites for methylation-sensitive transcription factors, have a greater probability to include actual control elements than the entire LD blocks, or other genomic regions. Whereas these control regions may be affected by either genetic or epigenetic variations, the generally higher occurrence of epigenetic variations relative to sequence variations suggests that more individuals might be affected by epigenetic rather than by genetic variations.
In conclusion, here we revealed a novel T2DM-specific methylation signature at isolated, non-promoter CpG sites. These methylations alter binding sites of T2DM-related transcription factors, within cis-acting regulation elements. Notably, these findings were obtained by applying a partial representation of the genome in a particular human population. Thus, further high-resolution analyses across various ethnic origins and different environments are likely to uncover many additional differentially methylated sites. The discovery of methylation patterns independent of known T2DM risk factors may improve our ability to predict diabetes risk through combined genetic/epigenetic DNA-based tests, and may lead to a better understanding of T2DM mechanisms.
MATERIALS AND METHODS
Participants and DNA samples
Participants in the pool-based and individual-based cross-sectional case–control studies were Israeli residents of Jewish Ashkenazi origin (four grandparents). Participants in the Jerusalem Lipid Research longitudinal (cohort) study were Jewish residents of Jerusalem of various ethnic origins. T2DM subjects (n = 710) and controls (n = 304) were obtained from the Israel Diabetes Research Group. Additional controls were obtained from the Jerusalem Perinatal Study (39) (n = 155) and from a population-representative sample of Ashkenazi Jews (n = 147) (E.L.L). T2DM subjects were at least 10 years post-diagnosis at the time of DNA collection and were treated at Israeli medical centers. DNA samples were obtained from peripheral white blood cells and were analyzed anonymously. All subjects provided signed informed consent. The study was approved by the Hadassah Medical Center Review Committee, the Shaare Zedek Medical Center Review Committee and the Israeli National Helsinki Committee for Genetic Studies.
DNA pooling
DNA samples were diluted in reduced ethylene diaminetetraacetic acid (EDTA) buffer (10 mm Tris–HCl, 0.5 mm EDTA) to a precise concentration of 50 ± 0.4 ng/µl and equivalent amounts of DNA from the participating individuals were assembled in DNA pools.
Microarray-based methylation assay
The assay was fully described elsewhere (6,12). Briefly, 1 µg of genomic DNA was digested with a cocktail of methylation-sensitive restriction enzymes (MSREs), ethanol-precipitated and re-suspended in tris-EDTA buffer to a concentration of 50 ng/µl. Digested and non-digested DNAs were hybridized to Affymetrix SNP6 microarrays and scanned according to the manufacturer's instructions (www.Affymetrix.com). Hybridization intensities were normalized by applying an invariant set of probed genomic fragments without MSRE sites. Methylation signals, defined as 1-log2 of the ratio between hybridization intensities before/after MSRE treatments, were assigned to each one of the 1 461 753 small (size range 100–3600 bp, average 1106 bp) DNA fragments with at least one MSRE site probed on the Affymetrix SNP6 microarray.
Data filtering
Genomic fragments containing published (SNP db build no. 130) or sequenced polymorphism which may affect data interpretation were excluded from the microarray and sequencing-based analyses.
Ranking of microarray-probed DNA fragments by case–control differentiation
Microarray-probed genomic fragments were ranked according to discrimination between case and control pools, where the fragment showing the highest separation between case and control ranked number 1, and the fragment showing the lowest separation ranked 1 461 753. Ranking was performed applying the silhouette statistic, according to the following formula:
where s(i) is computed for each value, a(i) is the squared Euclidean distance from the ith point (methylation percentage) to the other points in its cluster and b(i) is the average distance from the ith point to points in the other cluster.
Analysis of differentially methylated regions
Sliding 10 kb windows, 50% overlapped, were analyzed along the genome. An informative window was defined as a 10 kb window containing at least three microarray-probed DNA fragments. A DMR was defined as an informative window containing at least three probed DNA fragments in the top 10% of the case–control-differentiating rank, which is also significantly enriched (P < 0.05, or P < 0.005 for stringent DMRs) by top-ranking fragments following correction for the number of encompassing fragments. Sliding windows (50% overlapped) were applied in the DMR screens.
T2DM-associated LD blocks
LD blocks were defined as a region between the two most distant single-nucleotide polymorphisms with r2>0.8 (1000 Genomes Pilot 1) (40) encompassing an established T2D variant.
Bisulfite sequencing
PCR fragments were amplified from bisulfite-treated DNA (EZ-DNA kit, Zymo Research), quantified and sequenced by either 454 FLX Titanium genome sequencer (Roche), or PyroMark Q24 bench-top sequencer (Qiagen). Raw data were analyzed using the BISMA tool as described (41).
Longitudinal study
The Jerusalem Lipid Research Clinic (LRC) study initially examined full cohorts of 17-year-old Jerusalem residents in 1976–1978 (n = 8646), and subsequently at age 28–32 (mean age 30.1 years) examined a random sample of those who remained Jerusalem residents as well as those whose parents experienced an acute myocardial infarction during a 10-year follow-up (n = 1052, response rate 72%) (42). At ages 41–47 (mean 43.2 years), 631 of the 1052 participants were reexamined (71% response among the eligible). Of these, 515 were classified as having normal glucose metabolism at ages 28–32 (i.e. fasting glucose <100 mg/dl and 2 h post-challenge glucose <140 mg/dl); they served as the cohort for the current study. Over the mean 13.1 years of follow-up, 62 cases of IGM/T2DM were identified (Fig. 4), of which 58 were tested for methylation; 339 remained normal (defined as fasting glucose <100 mg/dl and 2 h post-challenge <140 mg/dl); of these 64 were randomly selected for methylation studies. For the methylation determination, samples were encoded and bisulfite-sequenced in 24-well plates containing even numbers of cases and controls. Assay reproducibility was controlled by repeated pyrosequencing analyses of 21 samples (Supplementary Material, Fig. S6a). Cohort quality was evaluated by pyrosequencing duplicated samples of 11 (10%) of the participants in the longitudinal study (Supplementary Material, Fig. S6b). The laboratory was blinded to this process. Decoding and analysis were performed after all plates had been sequenced.
Control for confounding effect of blood lineages
Lineage-specific methylation levels were estimated using linear least-squares analysis with upper and lower bounds (0–100%). The analysis was performed on 110 participants of the Jerusalem LRC study with full blood counts and methylation assessments.
Analysis of in silico binding affinities
Matrices of transcription factors’ binding sites were obtained from TRANSFAC database. In silico binding affinities were determined using the FIMO software from the MEME suite. A binding site was defined using a threshold of P-value <1e−5.
Data mining
H3K4Me1 level in normal human skeletal muscle and in other five different cell types were obtained from the Broad Histone Chip-Seq (43), and normalized to a common mean and variance. Genetic linkage data (CEU population) obtained from the Hapmap project (40).
Statistical analyses
The methylation data and P-values presented in Figures 4–6 and in Supplementary Material, S2, S3 and S4 were adjusted to gender, except for the gender bins in Figure 4C. The data and P-values presented in Figure 6 were additionally adjusted for lymphocyte percentage. Correlations were evaluated by applying linear Pearson's correlation. Differences between distributions were analyzed by the Kolmogorov–Smirnov (K–S) test. Adjustments and interactions were analyzed using binomial logistic regression. Case–control methylation differences were analyzed by either two-proportion z-tests for pools, paired two-sided t-tests for individuals or as odds ratios estimated from binomial logistic regression models fitted for individual-level analyses. False discovery rates associated with multiple hypothesis testing were estimated using the Benjamini and Hochberg procedure (44) or by q-values (45). Standard error of the mean (SEM = SD/sqrt(n)) are presented in bar graphs.
Data deposition
Raw microarray hybridization outputs and processed methylation signals are available through the GEO public database (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE33032.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the United States-Israel Binational Science Foundation (to J.D.K.), the Breast Cancer Research Foundation and The Israeli HPR (to E.L.L.), the Russell Berry Foundation/D Cure (to B.G.), the Israel Cancer Research Fund (to A.H.), the NIH research grant R01HL088884 (to Y.F.) and the Israel Science Foundation (to A.H., to Y.F. and to J.D.K.).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Howard Cedar and Yuval Dor for helpful discussions and comments. We also thank the additional members of the Israel Diabetes Research Group who played active roles in this research: Itamar Raz, Ardon Rubenstein, Ilana Harman-Boehm, Joseph Cohen, Oscar Minuchin, Yair Yerushalmi, Andreas Buchs, Anat Tsur and Clara Norymberg; and Ronit Sinnreich and Nehama Goldberger who contributed to the Jerusalem LRC Longitudinal Study data used in this research.
REFERENCES
- 1.Prokopenko I., McCarthy M.I., Lindgren C.M. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. doi:10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. doi:10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornsson H.T., Fallin M.D., Feinberg A.P. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi: 10.1016/j.tig.2004.06.009. doi:10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. doi:10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 5.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. doi:10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 6.Hellman A., Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. doi:10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 7.Ball M.P., Li J.B., Gao Y., Lee J.H., LeProust E.M., Park I.H., Xie B., Daley G.Q., Church G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. doi:10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. doi:10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakyan V.K., Down T.A., Balding D.J., Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12:529–541. doi: 10.1038/nrg3000. doi:10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Docherty S.J., Davis O.S., Haworth C.M., Plomin R., Mill J. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenet. Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. doi:10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Docherty S.J., Davis O.S., Haworth C.M., Plomin R., Mill J. DNA methylation profiling using bisulfite-based epityping of pooled genomic DNA. Methods. 2010;52:255–258. doi: 10.1016/j.ymeth.2010.06.017. doi:10.1016/j.ymeth.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 12.Aran D., Toperoff G., Rosenberg M., Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. doi:10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 13.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. doi:10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E., et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. doi:10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 15.Hellman A., Chess A. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenet. Chromatin. 2010;3:11. doi: 10.1186/1756-8935-3-11. doi:10.1186/1756-8935-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schalkwyk L.C., Meaburn E.L., Smith R., Dempster E.L., Jeffries A.R., Davies M.N., Plomin R., Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am. J. Hum. Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. doi:10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. doi:10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell C.G., Finer S., Lindgren C.M., Wilson G.A., Rakyan V.K., Teschendorff A.E., Akan P., Stupka E., Down T.A., Prokopenko I., et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5:e14040. doi: 10.1371/journal.pone.0014040. doi:10.1371/journal.pone.0014040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lango H., Palmer C.N., Morris A.D., Zeggini E., Hattersley A.T., McCarthy M.I., Frayling T.M., Weedon M.N. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. doi:10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herder C., Rathmann W., Strassburger K., Finner H., Grallert H., Huth C., Meisinger C., Gieger C., Martin S., Giani G., et al. Variants of the PPARG, IGF2BP2, CDKAL1, HHEX, and TCF7L2 genes confer risk of type 2 diabetes independently of BMI in the German KORA studies. Horm. Metab. Res. 2008;40:722. doi: 10.1055/s-2008-1078730. doi:10.1055/s-2008-1078730. [DOI] [PubMed] [Google Scholar]
- 21.Bressler J., Kao W.H., Pankow J.S., Boerwinkle E. Risk of type 2 diabetes and obesity is differentially associated with variation in FTO in whites and African-Americans in the ARIC study. PLoS One. 2010;5:E10521. doi: 10.1371/journal.pone.0010521. doi:10.1371/journal.pone.0010521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertel J.K., Johansson S., Sonestedt E., Jonsson A., Lie R.T., Platou C.G., Nilsson P.M., Rukh G., Midthjell K., Hveem K., et al. FTO, Type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41 504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60:1637–1644. doi: 10.2337/db10-1340. doi:10.2337/db10-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kark J.D., Sinnreich R., Leitersdorf E., Friedlander Y., Shpitzen S., Luc G. Taq1B CETP polymorphism, plasma CETP, lipoproteins, apolipoproteins and sex differences in a Jewish population sample characterized by low HDL-cholesterol. Atherosclerosis. 2000;151:509–518. doi: 10.1016/s0021-9150(99)00408-6. doi:10.1016/S0021-9150(99)00408-6. [DOI] [PubMed] [Google Scholar]
- 24.Wingender E., Dietze P., Karas H., Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. doi:10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. doi:10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 26.Vallet V.S., Casado M., Henrion A.A., Bucchini D., Raymondjean M., Kahn A., Vaulont S. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J. Biol. Chem. 1998;273:20175–20179. doi: 10.1074/jbc.273.32.20175. doi:10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 27.van Deursen D., Jansen H., Verhoeven A.J. Glucose increases hepatic lipase expression in HepG2 liver cells through upregulation of upstream stimulatory factors 1 and 2. Diabetologia. 2008;51:2078–2087. doi: 10.1007/s00125-008-1125-6. doi:10.1007/s00125-008-1125-6. [DOI] [PubMed] [Google Scholar]
- 28.Wong R.H., Chang I., Hudak C.S., Hyun S., Kwan H.Y., Sul H.S. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–1072. doi: 10.1016/j.cell.2008.12.040. doi:10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watt F., Molloy P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. doi:10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 30.Prendergast G.C., Ziff E.B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. doi:10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 31.Campanero M.R., Armstrong M.I., Flemington E.K. CpG methylation as a mechanism for the regulation of E2F activity. Proc. Natl Acad. Sci. USA. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. doi:10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyblom H.K., Bugliani M., Fung E., Boggi U., Zubarev R., Marchetti P., Bergsten P. Apoptotic, regenerative, and immune-related signaling in human islets from type 2 diabetes individuals. J. Proteome Res. 2009;8:5650–5656. doi: 10.1021/pr9006816. doi:10.1021/pr9006816. [DOI] [PubMed] [Google Scholar]
- 33.Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K.T., Frigge M.L., et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. doi:10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. doi:10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 35.Xu J., Pope S.D., Jazirehi A.R., Attema J.L., Papathanasiou P., Watts J.A., Zaret K.S., Weissman I.L., Smale S.T. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. doi:10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu C.R., Cole P.A., Meyers D.J., Kormish J., Dent S., Zaret K.S. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–966. doi: 10.1126/science.1202845. doi:10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sérandour A.A., Avner S., Percevault F., Demay F., Bizot M., Lucchetti-Miganeh C., Barloy-Hubler F., Brown M., Lupien M., Metivier R., et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. doi:10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Schug J., Tuteja G., White P., Kaestner K.H. The nucleosome map of the mammalian liver. Nat. Struct. Mol. Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. doi:10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedlander Y., Manor O., Paltiel O., Meiner V., Sharon N., Calderon R., Hochner H., Sagy Y., Avgil M., Harlap S., et al. Birth weight of offspring, maternal pre-pregnancy characteristics, and mortality of mothers: the Jerusalem perinatal study cohort. Ann. Epidemiol. 2009;19:112–117. doi: 10.1016/j.annepidem.2008.11.002. doi:10.1016/j.annepidem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durbin R.M., Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. doi:10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohde C., Zhang Y., Reinhardt R., Jeltsch A. BISMA–fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinform. 2010;11:230. doi: 10.1186/1471-2105-11-230. doi:10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kark J.D., Sinnreich R., Rosenberg I.H., Jacques P.F., Selhub J. Plasma homocysteine and parental myocardial infarction in young adults in Jerusalem. Circulation. 2002;105:2725–2729. doi: 10.1161/01.cir.0000017360.99531.26. doi:10.1161/01.CIR.0000017360.99531.26. [DOI] [PubMed] [Google Scholar]
- 43.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. doi:10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
- 45.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. doi:10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.