Abstract
Post-translational modifications (PTMs) play a crucial role during biogenesis of many transmembrane proteins. Previously, it had not been possible to evaluate PTMs in cystic fibrosis transmembrane conductance regulator (CFTR), the epithelial ion channel responsible for cystic fibrosis, because of difficulty obtaining sufficient amounts of purified protein. We recently used an inducible overexpression strategy to generate recombinant CFTR protein at levels suitable for purification and detailed analysis. Using liquid chromatography (LC) tandem and multiple reaction ion monitoring (MRM) mass spectrometry, we identified specific sites of PTMs, including palmitoylation, phosphorylation, methylation and possible ubiquitination. Many of these covalent CFTR modifications have not been described previously, but are likely to influence key and clinically important molecular processes including protein maturation, gating and the mechanisms underlying certain mutations associated with disease.
Keywords: CFTR, mass spectrometry, palmitoylation, post-translational modification, protein purification
Introduction
The cystic fibrosis transmembrane conductance regulator (CFTR) mediates transport of chloride and bicarbonate across apical membranes of epithelial cells. Mutations in CFTR lead to cystic fibrosis (CF), a multi-organ disease of exocrine secretion in the lung, pancreas, intestine, liver and other glands. Mature CFTR is post-translationally processed in the endoplasmic reticulum (ER) and Golgi apparatus and subsequently trafficked to the plasma membrane where it is recycled from early endosomes or marked by ubiquitin for proteolysis (Ward et al., 1995; Sharma et al., 2004). The most common mutation in CFTR, F508del, accounts for ∼70% of defective alleles in the USA (Rowe et al., 2005) and is targeted for ER-associated degradation (ERAD) by mechanisms including covalent side chain attachment (Younger et al., 2006). Identifying the specific amino acids in CFTR that serve as substrates for post-translational modification (PTM) is of considerable and topical interest. In other membrane proteins, palmitoylation, phosphorylation or ubiquitination govern key aspects of biogenesis such as ER to Golgi transit, routing to the proteosome and stability at the cell surface. Reversible PTMs are also well described, including ‘switch’-type mechanisms that underlie membrane protein maturation and activity (Xue et al., 2004; Hunter, 2007; Lin et al., 2009).
F508del CFTR is typically ubiquitinated and routed to ERAD but can be induced to reach the cell surface following low-temperature cell growth or pharmacologic manipulation. Once F508del CFTR is redirected to the plasma membrane, it is recognized by quality control in the early endosome such that surface recycling is obviated and ubiquitination is enhanced (Sharma et al., 2004). In addition to ubiquitination, other PTMs govern biogenesis and function of both wild-type and mutant CFTR (Glozman et al., 2009; Hegedus et al., 2009; Wang et al., 2010). Immature CFTR is N-glycosylated at positions 894 and 900, followed by complex sugar attachment and trafficking to the plasma membrane. Glycosylation is not required for channel activity but has been suggested as a possible contributor to surface stability of mature CFTR by decreasing turnover rate (Chang et al., 2008). The unphosphorylated regulatory domain (R domain) of CFTR acts as a channel inhibitor and establishes contacts with the first nucleotide binding domain (NBD1) that block dimerization with NBD2. Disordered regions associated with NBD1 (regulatory insertion [RI] and regulatory extension [RE]) undergo a conformational shift due to phosphorylation that elicits NBD1/NBD2 dimerization and reorientation of transmembrane regions, leading to an open channel (Lewis et al., 2005; Kanelis et al., 2010). A number of specific phosphorylation sites that control CFTR gating have been described previously by site-directed mutagenesis or mass spectrometry (Townsend et al., 1996; Pasyk et al., 2009).
Identifying PTMs with biological significance could lead to new therapeutic strategies for CF. For example, certain small molecules (among a class of ‘potentiators’) overcome functional defects in CFTR by enhancing phosphate attachment to the R-domain (Pyle et al., 2011). The mechanisms by which phosphorylation sites are influenced by these compounds are not known, but are highly relevant to CF drug development, including agents currently being tested in the clinic. Although technologies for investigating post-translational CFTR modification are available, difficulty in obtaining sufficient amounts of purified protein has stalled progress in this area. In recombinant overexpression systems, CFTR degradation by the proteosome has impeded studies of dynamic folding, crystal structure and biochemical analysis (Rosenberg et al., 2004). We recently developed an inducible overexpression system that yields quantities of recombinant CFTR suitable for purification and analysis by mass spectrometry. In the present study, we describe a proteomic survey of full-length CFTR that revealed numerous and specific PTMs likely to play a role in maturation and activation, including a class of lipid attachments not previously reported for the CFTR gene product.
Materials and Methods
Cell culture
GnTI− HEK293S cells (a gift from Dr Khorana, MIT, Cambridge, MA, USA) lack N-acetylglucosaminyltransferase I, resulting in the absence of complex N-glycans (Reeves et al., 2002). The defect allows large-scale preparation of homogenous, minimally glycosylated (Man5GlcNAc2) membrane proteins. GnTI− HEK293S cells were further modified to become doxycyline inducible by stable expression of the M2 protein (modified tet-on transactivator) (Urlinger et al., 2000). The GnTI− HEK293S cell line with this modification was cultured in DMEM-F12 with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA).
Expression of CFTR in mammalian cells
Recombinant CFTR protein was produced in GnTI− HEK293S cells using a tet-on gene expression strategy (Welman et al., 2007). The approach was designed to limit toxicity associated with constitutive protein overexpression. Briefly, a nucleotide sequence comprising human CFTR, fused in-frame with the tobacco etch virus (TEV) protease cleavage site (Kapust et al., 2001) and a 10× histidine tag, was generated by polymerase chain reaction and cloned into an HIV-1-based lentiviral vector under control of the TRE promoter, generating P2231/TRE-CFTR.TEV.His-IRES-EGFP. Other design features of the vector included an internal ribosomal entry site (IRES) downstream of CFTR followed by the enhanced green fluorescence protein (EGFP) open reading frame (Fig. 1A). The integrity of the P2231 vector, including the CFTR-TEV-His coding sequence, was confirmed by nucleotide sequencing. The vector was packaged and pseudotyped with the amphotropic VSV-G envelope glycoprotein as described previously (Burns et al., 1993; Zufferey et al., 1998). To enable doxycycline (dox)-inducible CFTR expression in the GnTI− HEK293S cell line (Reeves et al., 2002), the cells were modified by transduction with a blasticidin resistance gene-containing lentiviral vector (referred to as No. P2800) constructed to constitutively express the M2 reverse transcriptional transactivator (rtTA) (Urlinger et al., 2000) under control of an EF1α promoter (designated HEK293S.M2). This cell line was then transduced with P2231/TRE-CFTR.TEV.His-IRES-EGFP. Taking advantage of EGFP expression from a bicistronic mRNA, cells were induced with doxycycline (1 µg/ml), and 24 h later high-expression-positive cells were sorted into 48-well plates (one cell per well) using a FACSAria (BD Biosciences, Franklin Lakes, NJ, USA). The single-cell clonal cultures were expanded and analyzed for basal and doxycycline-inducible CFTR expression, multiplication kinetics and stability of inducible CFTR protein. One of the clonal cultures exhibiting favorable characteristics, designated D099/GnTI− HEK293S.M2-CFTR.His, was grown in batch using a 10-l bioreactor (BIOFLO 310, New Brunswick Scientific Co, Inc., Edison, NJ, USA) to produce sufficient quantities of CFTR for purification and analysis of PTM. At near-peak cell culture density (∼3 × 106 cells/ml), medium (CDM HEK293, Hyclone, South Logan, UT, USA) was supplemented with 1.0 µg/ml of doxycycline. The cells were harvested 24–48 h later and analyzed for EGFP expression as a surrogate and quality control measure for CFTR protein expression.
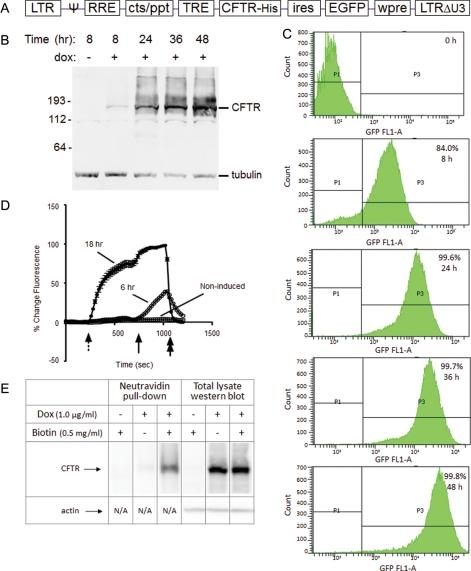
Fig. 1.
CFTR expression in GnTI− HEK293S cells. (A) The genetic structure of the P2231 lentiviral vector-based CFTR expression cassette is depicted. (B, C) Exogenous protein (CFTR and EGFP) expression in the D099 clonal cell line was analyzed by western blot (B) and flow cytometry (C) prior to and after (8, 24, 36 and 48 h) induction with dox (1 µg/ml). (D) Functionality of the recombinant CFTR protein was analyzed by the halide-sensitive fluorescent dye, SPQ. D099 cells that were either non-induced or induced with dox for 6 or 18 h, respectively, were analyzed. The dotted arrow indicates the addition of NaNO3 dequench buffer, followed by the solid arrow indicating addition of forskolin (20 µM) to maximally activate CFTR. NaI quenching buffer is resumed at the end of the experiment as a control (double arrow). (E) Surface localization of CFTR was analyzed by cell surface biotinylation. Biotinylated lysate (1500 µg) was used for the NeutrAvidin pull-down, and 30 µg lysate reserved for western blot analysis.
Surface biotinylation of CFTR
Cells were grown until ∼80% confluent and induced with doxycycline (1 µg/ml) for 4 h. Surface proteins were labeled with 0.5 mg/ml biotin (EZ-Link NHS-SS-Biotin, Thermo Scientific, Pittsburgh, PA, USA) in phosphate-buffered saline with calcium and magnesium for 20 min at 4°C. Cells were rinsed in Tris-buffered saline and lysed in Triton buffer (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris-Cl) + 1% protease inhibitors with EDTA (Thermo Scientific Halt Protease Inhibitor Cocktail, Thermo Scientific). NeutrAvidin Agarose Resin (Thermo Scientific) was blocked with HEK293 lysate (no CFTR expressed) prior to pull-down. Biotinylated lysate (1500 µg) was rotated overnight at 4°C with 15 µl of blocked NeutrAvidin Agarose Resin. Bound protein was examined by western blot analysis using 10B6.2 antibody (CFTR NBD1).
Functional analysis of CFTR
Cells were seeded onto Vectabond®-treated glass coverslips coated with 5 µg/cm2 human placental extracellular matrix and grown until ∼80% confluent. Immediately prior to study, cells were hypotonically loaded with halide-quenched dye (6-methoxy-N-(3-sulfopropyl)quinolinium, SPQ—10 mM, Molecular Probes Inc., Eugene, OR, USA) for 10 min and then placed in a quenching NaI buffer. CFTR robustly conducts iodide in addition to chloride, HCO3− and several other anions, allowing the use of iodide quench as a measure of channel activity. Cells were mounted in a specially designed perfusion chamber and studied using an inverted microscope (excitation at 340 nm, emission at >410 nm), a PTI imaging system (Monmouth Junction, NJ, USA) and a Hamamatsu camera (Bridgewater, NJ, USA) as previously described (Rowe et al., 2007). Baseline fluorescence was measured in isotonic NaI buffer followed by perfusion with isotonic dequench buffer (NaNO3 replaced NaI) as indicated. The perfusate was then switched to dequench buffer plus agonist and requenched with NaI at the end of the experiment. Fluorescence was normalized for each cell to its baseline value and increases shown as percent above basal (quenched) fluorescence.
Purification of CFTR
Cells were lysed in a 25 mM phosphate buffer (pH 7.2) containing 20% glycerol, 100 mM NaCl, 50 mM arginine, 50 mM glutamic acid and 1 mM ATP with 2 mM MgCl2 and EDTA Complete Protease Inhibitor (Roche Applied Science, Bavaria, Germany) using a French press. The lysates were pelleted at 4000 rpm (3313 × g) followed by ultra-centrifugation at 40 000 rpm (186 000 × g) for 6–18 h. The membrane fraction was collected and solubilized using a dounce homogenizer and 25 mM phosphate buffer as above, with 150 mM NaCl and 1% Fos-choline 14 (Anatrace, Maumee, OH, USA). The extraction mixture was pelleted at 20 000 rpm (48 384 × g) in a JA 25.1 rotor, and the supernatant diluted to a final Fos-choline 14 concentration of 0.6 mM (5× CMC) by adding 25 mM phosphate (pH 7.2) containing 20% glycerol, 100 mM NaCl, 50 mM arginine, 50 mM glutamic acid and 1 mM ATP. Pre-washed nickel resin (1 ml per billion cells starting material) was placed on a rotating platform at 4°C overnight. Lysate and resin were centrifuged at 800 rpm (150 × g) in a swinging bucket rotor (Sorval, RTH-750) at 4°C and unbound supernatant decanted. The resin was applied to a column and washed in 25 mM phosphate (pH 7.2) containing 20% glycerol, 100 mM NaCl, 50 mM arginine, 50 mM glutamic acid, 1 mM ATP and 5× CMC Fos-choline 14. Elution was achieved by gravity using step gradients and two washes (in buffer containing 500 mM and 1 M imidazole) or an Akta chromatography system (GE Healthcare, Piscataway, NJ, USA) with 1 M imidazole. Fractions were dot blotted, pooled and concentrated to 10 ml or less, followed by size exclusion chromatography using elution buffer (40 mM Tris-HCl (pH 7), 150 mM NaCl, 2 mM MgCl, 15% glycerol, 0.1% DDM (dodecylmaltoside)) without imidazole. Pooled fractions were run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) for western blotting and Coomassie Blue staining. Total protein concentration was evaluated by precipitation BCA assay. An alternative purification protocol with DDM was also utilized (Rosenberg et al., 2004).
Protein isolation for mass spectrometry
Following protein electrophoresis, Coomassie Blue-stained plugs were excised using the Digilab ProPic automated sample picker (Holliston, MA, USA). Each plug was destained with washes of 100 mM ammonium bicarbonate (ABC, 50%)/acetonitrile (50%). Samples were incubated at 60°C for 60 min in 20 mM dithiothreitol dissolved in 50 mM ABC (for cysteine reduction) and then at room temperature in 55 mM iodoacetamide in 50 mM ABC for free cysteine alkylation. After several washes with 100 mM ABC, samples were dried in a Savant SpeedVac (Thermo Scientific, Pittsburgh, PA, USA).
Proteolytic hydrolysis of CFTR
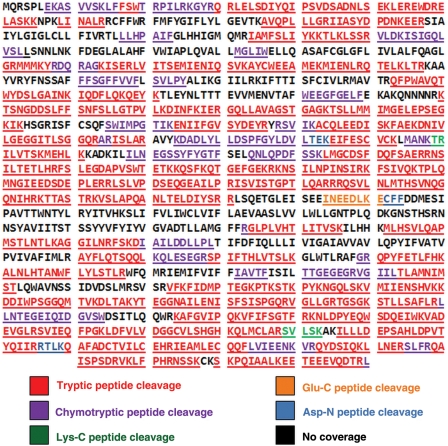
Trypsin (mass spectrometry grade; Promega, Madison, WI, USA) was used for enzymatic digestion at 37°C for 16 h. Peptide solution extraction was performed by removing the incubation supernatant and washing the gel pieces twice with 20 µl of a 50/50 solution of 5% formic acid and acetonitrile for 30 min. Supernatants were combined and dried as above and resuspended in 40 µl of 0.1% formic acid. Other proteolytic digests utilized chymotrypsin, Lys-C, Glu-C and Asp-N. The use of multiple proteases and enzymatic cocktails for protein digestion led to ∼72% coverage of the overall CFTR polypeptide.
Mass spectrometry analysis of CFTR peptides
Aliquots (5–10 µL) of each digestion were loaded onto a 5 mm × 100 µm i.d. C18 reverse-phase cartridge at 20 µl/min using a PAL robot (Leap Technologies, Carrboro, NC, USA). After washing each cartridge for 5 min with 0.1% formic acid in ddH2O, the bound peptides were flushed onto a 22 cm × 100 µm i.d. C18 reverse-phase pulled tip analytical column with a 35 min linear 5–50% acetonitrile gradient in 0.1% formic acid at 500 nl/min using an Eksigent nanopump (Eksigent Technologies, Dublin, CA, USA). The column was washed with 90% acetonitrile–0.1% formic acid for 15 min and then re-equilibrated with 5% acetonitrile–0.1% formic acid for 24 min. Eluted peptides were passed directly from the tip into a modified MicroIonSpray interface of an Applied Biosystems/MDS SCIEX (Concorde, Ontario, Canada) 4000 Qtrap mass spectrometer. IonSpray voltage was set at 2600 V and declustering potential at 60 V with ionspray and curtain gases 12 and 5 psi, respectively (interface heater temperature set at 160°C). Eluted peptides were subjected to a survey mass spectrometry (MS) scan to determine the two most intense ions. A second scan (for enhanced resolution) was next used to determine the charge state of selected ions. Finally, enhanced product ion scans were carried out to obtain the tandem mass spectra of the selected parent ions (with the declustering potential raised to 100 V) over the range from m/z 400 to 1500. Spectra were centroided and de-isotoped by Analyst Software, version 1.42 (Applied Biosystems). The tandem mass spectrometry data were processed to provide potential peptide identifications to the known CFTR sequence used in this study with an in-house MASCOT search engine (Matrix Science, London, England—July 2010). Parameters were set against the NCBInr Homo sapiens protein database and one missed protease cleavage site. The precursor mass tolerance was set to 1.0 Da and the MS/MS tolerance to 0.6 Da. The average error for all spectra was ≤ 150 ppm. Possible modified peptides on the CFTR construct were evaluated by allowing for variable modifications using the MASCOT Server as well as Protein Pilot (AB SCIEX, Foster City, CA, USA). MS/MS spectra were subjected to de novo sequencing.
Multiple reaction ion monitoring
Following SDS–PAGE, multiple reaction ion monitoring (MRM) was used to assess the presence or absence of modifications of interest on peptides at a specific m/z value. Parent molecular ions were passed into an electrospray interface, filtered and collided with neutral gas in the quadrupole of a 4000 Qtrap mass spectrometer. Particular sequence-dependent fragment ions were then selectively filtered in the third quadrupole and measured. Since one peptide at a time was analyzed (for 20–30 ms) and peaks were ∼10 s in width, 30–50 peptides monitored each second (∼10 data points) were sufficient to construct and accurately measure the area under an emerging peak. The strategy offers attomole to femtomole sensitivity for detecting peptide fragments and has previously been used to quantify endogenously expressed CFTR in intact cells (HT-29, colonic; Jiang et al., 2010).
Results
CFTR is expressed in epithelial cells at low levels, and purification of sufficient amounts of the native protein for biophysical studies has been difficult due to rapid degradation of the gene product. In this report, our experimental strategy was to establish a cell line that expressed recombinant CFTR under tight transcriptional control of the TRE promoter (tet-on) using N-acetylglucosaminyltransferase I-deficient (GnTI−) HEK293S cells. The GnTI− deficiency results in reduced levels of complex N-glycans, allowing for large-scale preparation of homogenous, minimally glycosylated (Man5GlcNAc2) CFTR suitable for studies such as crystallization (Reeves et al., 2002). The molecular expression employed a lentiviral vector (designated P2231) comprising a bicistronic open reading frame that encoded CFTR and EGFP separated by an IRES (Fig. 1A). The HEK293S.M2 cell line (containing the M2 reverse transcriptional transactivator; Urlinger et al., 2000) was transduced with the VSV-G pseudotyped P2231 vector, and expression-positive clonal cultures derived by sorting individual GFP + cells (24 h post-dox induction) into wells of a 48-well plate. The clonal cell line (designated D099) exhibited characteristics that appeared most favorable among many clones that were screened: neither CFTR nor EGFP was detected in the absence of dox. However, expression of both was markedly up-regulated after dox induction (99.8%; Fig. 1B and D). Notably, by 24 h post dox induction, the shift to EGFP/CFTR-positive cells was nearly quantitative (Fig. 1C). As expected, due to the nature of the GnTI− cell line, the glycan-deficient CFTR protein exhibited greater mobility in SDS–PAGE gels (MW ∼140 kDa, Fig. 1B).
Expression of CFTR in this cell model leads to surface localized protein (confirmed by functional studies and surface biotinylation (Fig. 1D and E)), together with CFTR en route to degradation (via ERAD). CFTR PTMs present in either pathway (maturation or degradation) are therefore represented. The CFTR ERAD mechanism (Ward and Kopito, 1994; Ward et al., 1995) is known to be regulated by multiple PTMs and is therefore of considerable interest with respect to the present findings. Cells overexpressing the CF protein have commonly been used for studies of CFTR biology, including identification of molecular subcategories of disease, CFTR-binding partners and cellular quality control, ion channel regulation and therapeutic drug discovery (Rowe et al., 2005; Wang et al., 2006; Okiyoneda et al., 2010). Numerous earlier studies, including in vivo trials of emerging CFTR modulators in CF subjects, indicate the relevance of recombinant overexpression systems for studies of CFTR biogenesis, including identification of PTMs (Van Goor et al., 2009; Accurso et al., 2010; Ostedgaard et al., 2011).
Channel activity of the recombinant CFTR protein was analyzed by the halide-based fluorescence SPQ assay. The D099/GnTI− HEK293S.M2-CFTR-His cell line was cultured either with or without dox (1 µg/ml) for 6 or 18 h prior to analysis. Compared with the HEK293S.M2 parental cell line, there was much greater halide efflux from D099 cells detected as early as 6 h post-induction and peaking at 18 h (Fig. 1D). Notably, channel activity was also observed prior to forskolin stimulation and subsequent to halide-free buffer exchange (at 200 s), due to overexpression of constitutively active CFTR. The results demonstrate that glycan-restricted CFTR protein exhibits substantial anion conductance at the cell surface. Consistent with this interpretation, cell surface biotinylation analysis of dox-induced D099 cells indicated abundant CFTR protein in the plasma membrane compartment (Fig. 1E). These biological characteristics were stable over at least 10 serial passages at the 10 l/30–60 × 109 cells scale, which is attributable to tight control over CFTR protein expression.
CFTR was purified by affinity chromatography and high-performance liquid chromatography. An example of the final material is shown in Fig. 2 and indicates a level of purity (as judged by Coomassie Blue stain and western blotting) suitable for proteomic analysis. Following electrophoresis, gel bands were excised and solubilized for proteolytic digestion. To obtain optimal peptide coverage, enzymatic digestions were repeated using multiple proteases (trypsin, chymotrypsin, Glu-C, Lys-C and Asp-N) individually or in combination. Resulting peptides were analyzed by liquid chromatography (LC) tandem mass spectrometry (LC-MS/MS; overall CFTR peptide coverage is summarized in Fig. 3). The data were processed by the MASCOT search engine and revealed PTMs at numerous CFTR positions, including three previously undescribed phosphorylation sites (T717, S1444, S1456) in addition to six CFTR phosphorylation sites reported earlier (S660, S686, S700, S712, S737, S795; Fig. 4, Supplementary Fig. S1). Evidence for each of these sites was provided by the neutral loss of H3PO4 (−98), i.e., the phosphorylated Ser residue appeared in the MS/MS spectrum as a mass increase of 69, not 80, due to the loss of water in addition to the phosphate group.
Fig. 2.
CFTR purified from a solubilized membrane fraction. Elution from a nickel resin (1 ml resin per billion cells starting material) was followed by concentration with an Akta chromatography system (Materials and Methods). Coomassie Blue staining is shown in (A). Western blotting (B) with 1104 antibody (raised against the R domain of CFTR) confirmed that the major band in the Coomassie Blue-stained gel is CFTR.
Fig. 3.
CFTR coverage using proteolytic hydrolysis. Purified full-length CFTR was fragmented using multiple enzymes including trypsin, chymotrypsin, Lys-C, Glu-C and Asp-N. Cleavage by trypsin is shown in areas of protease overlap. Amino acid sequence (MS/MS) verified CFTR as the single polypeptide digested.
Fig. 4.
Mass spectrometry analysis. Summary of PTMs identified in CFTR: Phosphorylation: S660, S686, S700, S712, T717, S737, S795, S1444, S1456. Ubiquitination: K688. Palmitoylation: C524, C1395. Methylation: K698, N699, Q744, T757. Gray letters represent residues not covered by proteolysis. Note: All modifications were identified by MS/MS except for palmitoylation of C524 (identified by MRM-MS).
Also identified was a putative site of ubiquitination (Fig. 4, Supplementary Fig. S2). The tryptic peptide containing K688 (QSFK*QTGEFGEKR) was found to have a mass increase of 128 Da. MS/MS of the triply charged molecular peptide resulted in unmodified y-ions (y1, y2, y4–9), confining the modification(s) to the N-terminal region S686-K688. However, because the b4 ion (m/z 619.3) has an increase in mass of 128 Da, it may represent a composite of more than one modification. For example, the residue at which an additional 14 Da occurs could be S686, which would convert the hydroxyl group into a methoxy group. This could arise, for example, from chemical displacement of a phospho group (S686 is a site of phosphorylation) or even a sulfo group prior to MS/MS analysis.
MS/MS identified multiple sites of methylation (K698, N699, Q744, T757 – see Fig. 4, Supplementary Fig. S3). Additional methylated peptides were detected but the sites of methylation could not be confirmed (Supplementary Table S1). CFTR methylation has not been described previously, but may be of considerable interest with regard to the growing appreciation of this PTM as a multifaceted regulator of proteins other than histones (Tolstykh et al., 2000; Friesen et al., 2001; Sprung et al., 2008; Subramanian et al., 2008; An et al., 2009; Osna et al., 2010; Parry and Ward, 2010).
In addition, LC-MS/MS established a palmitoyl modification, a 16-carbon saturated fatty acid, at C1395 (Fig. 4, Supplementary Fig. S4A). This modification is known to regulate critical aspects of biogenesis in other membrane proteins with parallels to CFTR processing and activity. Since this is a topical PTM not previously reported for CFTR and likely to be of considerable mechanistic interest, we extended our mass spectrometry-based investigation. Using MRM-MS, which offers higher sensitivity for specific peptides of interest, we were able to identify one additional palmitoylated CFTR residue, C524 (Fig. 4, Supplementary Fig. S4B and C).
Discussion
Purification of complex integral membrane proteins for biochemical and structural analysis represents a major barrier to performing detailed studies of membrane protein biochemistry. Approaches such as cell-free expression systems, prokaryotic technologies, baculovirus expression and yeast systems (e.g. Pichia) have been successful in specific settings, but can be limited by issues such as cellular toxicities associated with recombinant polypeptide overexpression, differences in membrane lipid composition, aggresome or inclusion body sequestration and aberrant folding pathways present in non-mammalian cells. In the present experiments, we utilized a robust and novel lentiviral strategy combined with inducible protein expression and concomitant reporter gene detection to generate high levels of CFTR in multiple cellular compartments, including functional ion channels in the plasma membrane. To our knowledge, CFTR purification of the scale and purity described here has not been reported previously (Rosenberg et al., 2004). This method allowed the first comprehensive study of CFTR PTM, leading to the identification of a number of novel PTMs. The same overall approach could be applied to other integral membrane proteins expressed in mammalian cells.
Palmitoylation is a covalent, typically reversible PTM known to govern various aspects of membrane protein biogenesis of particular relevance to the mechanisms that underlie CF. In numerous membrane proteins, including ion channels and an ABC transporter in the same gene family as CFTR, palmitoylation has been shown to mediate maturation beyond the ER, plasma membrane localization, ion channel phosphorylation, recycling from the cell surface and protein activation (Lam et al., 2006; Kinlough et al., 2006; Jindal et al., 2008; Singaraja et al., 2009; Mueller et al., 2010). Because the common F508del mutation in CF disrupts many of these same processes, identification of specific sites for palmitoylation may provide new insight into trafficking and function of both wild type and commonly observed mutant forms of CFTR. Similarly, the finding of multiple CFTR methylation sites suggests a previously unappreciated regulatory pathway relevant to CFTR biogenesis and/or ion channel activity. Protein methylation is well recognized for its epigenetic role during histone modification, and there is also growing appreciation of the ways methyl group PTMs may influence processes relevant to CFTR pathogenicity, including protein localization, protein–protein interactions or stability in the plasma membrane (Ciorba et al., 1997; Stadtman and Levine, 2003; Xing et al., 2008; Egorova et al., 2010; Hsu et al., 2011).
In many eukaryotic proteins, multiple PTMs interact dynamically, and different side chain attachments may compete for binding to the same residue (so-called post-translational ‘switches’; Acconcia et al., 2009; Lin et al., 2009). Based on findings presented here, a number of positions near each other in CFTR accommodate two distinct side chain modifications. For example, ubiquitination regulates F508del CFTR degradation and is associated with retrieval from the plasma membrane and proteosomal targeting (Sharma et al., 2004). The putative site of CFTR ubiquitination described above (K688) is in close proximity to a well-described position for phosphorylation (S686). If ubiquitination at this position is confirmed by direct labeling, the results could provide a biochemical explanation for diminished sensitivity of F508del CFTR to PKA. Further mass spectrometry, mutagenesis and functional studies will be necessary to definitively assign K688 as ubiquitinated and to determine whether an expected triply charged ion m/z of 327.5 occurs due to a 14 Da modification on Q685 or S686. Identifying the specific residue(s) of CFTR ubiquitination is of considerable interest, would contribute to studies of CFTR ERAD and could improve understanding of (and help optimize) small-molecule ERAD inhibitors designed to act synergistically with ‘correctors’ that augment processing of F508del CFTR.
In summary, results presented here describe the first application of comprehensive analysis of PTMs within CFTR. Protein samples used for mass spectrometry were obtained after recombinant overexpression and represent both properly folded and unfolded configurations. Distinguishing among CFTR modifications specific to the ER, Golgi, cell surface, during retrograde translocation, etc. will be necessary to fully characterize the relevant pathways, including their mechanistic significance. Mass spectrometry technologies such as MRM (shown above) permit specific residues to be queried even when present at very low (attomole to femtomole) concentrations. Such techniques necessitate pre-identification of residues known to be substrates for PTM and can be guided by the results shown in Fig. 4. For example, MRM will allow mature, properly folded CFTR to be tested with respect to specific palmitate or ubiquitin attachments. The present findings also describe a new perspective on CFTR processing defects attributable to the common F508del mutation, and suggest novel therapeutic targets (e.g. those involving palmitoylation) that have not been available previously. These methods for high-level CFTR production and PTM analysis therefore provide a means by which CFTR can be better characterized and understood in the future.
Supplementary Data
Funding
This work was supported by the Cystic Fibrosis Foundation (R464-CR07) and the National Institutes of Health (P30 DK072482 to E.J.S; 1K23DK075788 and 1R03DK084110 to SMR). The expression and purification of CFTR protein was supported by Cystic Fibrosis Foundation (DELUCA05XX0) and the Genetically Defined Microbe and Expression Core of the University of Alabama at Birmingham Mucosal HIV and Immunobiology Center (NIH DK64400). The mass spectrometer used in this study was purchased with funds from a National Institutes of Health/National Center for Research Resources Shared Instrumentation Grant (S10 RR019231). Funds for the operation of the Targeted Metabolomics and Proteomics Laboratory come in part from the University of Alabama at Birmingham Center for Nutrient-Gene Interaction (U54 CA100949), the Purdue-University of Alabama at Birmingham Botanicals Center for Age-Related Disease (P50AT00477), the University of Alabama at Birmingham O'Brien Acute Kidney Injury Center (P30DK079337), the University of Alabama at Birmingham Skin Disease Research Center (P30AR50948) and the University of Alabama at Birmingham Lung Health Center.
Supplementary Material
Acknowledgements
We thank Mrs J. Mott and Ms C. Owens for assistance with preparation of the manuscript.
References
- Acconcia F., Sigismund S., Polo S. Exp. Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Accurso F.J., Rowe S.M., Clancy J.P., et al. N. Engl. J. Med. 2010;363:1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Yun M., Park Y.G., Park G.H. Electrophoresis. 2009;30:2412–2421. doi: 10.1002/elps.200800772. [DOI] [PubMed] [Google Scholar]
- Burns J.C., Friedmann T., Driever W., Burrascano M., Yee J.K. Proc. Natl Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X.B., Mengos A., Hou Y.X., Cui L., Jensen T.J., Aleksandrov A., Riordan J.R., Gentzsch M. J. Cell. Sci. 2008;121:2814–2823. doi: 10.1242/jcs.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciorba M.A., Heinemann S.H., Weissbach H., Brot N., Hoshi T. Proc. Natl Acad. Sci. USA. 1997;94:9932–9937. doi: 10.1073/pnas.94.18.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova K.S., Olenkina O.M., Olenina L.V. Biochemistry (Mosc). 2010;75:535–548. doi: 10.1134/s0006297910050019. [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Massenet S., Paushkin S., Wyce A., Dreyfuss G. Mol. Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Glozman R., Okiyoneda T., Mulvihill C.M., Rini J.M., Barriere H., Lukacs G.L. J. Cell. Biol. 2009;184:847–862. doi: 10.1083/jcb.200808124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus T., Aleksandrov A., Mengos A., Cui L., Jensen T.J., Riordan J.R. Biochim. Biophys. Acta. 2009;1788:1341–1349. doi: 10.1016/j.bbamem.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Hsu J.M., Chen C.T., Chou C.K., et al. Nat. Cell Biol. 2011;13:174–181. doi: 10.1038/ncb2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jiang H., Ramos A.A., Yao X. Anal. Chem. 2010;82:336–342. doi: 10.1021/ac902028f. [DOI] [PubMed] [Google Scholar]
- Jindal H.K., Folco E.J., Liu G.X., Koren G. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H2012–2021. doi: 10.1152/ajpheart.01374.2007. [DOI] [PubMed] [Google Scholar]
- Kanelis V., Hudson R.P., Thibodeau P.H., Thomas P.J., Forman-Kay J.D. EMBO J. 2010;29:263–77. doi: 10.1038/emboj.2009.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R.B., Tozser J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., Waugh D.S. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Kinlough C.L., McMahan R.J., Poland P.A., et al. J. Biol. Chem. 2006;281:12112–12122. doi: 10.1074/jbc.M512996200. [DOI] [PubMed] [Google Scholar]
- Lam K.K., Davey M., Sun B., Roth A.F., Davis N.G., Conibear E. J. Cell. Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis H.A., Zhao X., Wang C., et al. J. Biol. Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- Lin D.T., Makino Y., Sharma K., Hayashi T., Neve R., Takamiya K., Huganir R.L. Nat. Neurosci. 2009;12:879–887. doi: 10.1038/nn.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller G.M., Maarouf A.B., Kinlough C.L., Sheng N., Kashlan O.B., Okumura S., Luthy S., Kleyman T.R., Hughey R.P. J. Biol. Chem. 2010;285:30453–30462. doi: 10.1074/jbc.M110.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Barrière H., Bagdány M., Rabeh W.M., Du K., Höhfeld J., Young J.C., Lukacs G.L. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osna N.A., White R.L., Donohue T.M., Jr, Beard M.R., Tuma D.J., Kharbanda K.K. Biochem. Biophys. Res. Commun. 2010;391:1291–1296. doi: 10.1016/j.bbrc.2009.12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard L.S., Meyerholz D.K., Chen J.H., et al. Sci. Transl. Med. 2011;3:74ra24. doi: 10.1126/scitranslmed.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry R.V., Ward S.G. Trends Immunol. 2010;31:164–169. doi: 10.1016/j.it.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Pasyk S., Taylor P., Ramjeesingh M., Moran M., Bear C.E. 2009. Abstract, North American Cystic Fibrosis Conference, 2009.
- Pyle L., Ehrhardt A., Mitchell L.H., et al. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011;4:L587–L597. doi: 10.1152/ajplung.00465.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.J., Callewaert N., Contreras R., Khorana H.G. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M.F., Kamis A.B., Aleksandrov L.A., Ford R.C., Riordan J.R. J. Biol. Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- Rowe S.M., Miller S., Sorscher E.J. N. Engl. J. Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Rowe S.M., Varga K., Rab A., Bebok Z., Byram K., Li Y., Sorscher E.J., Clancy J.P. Am. J. Respir. Cell. Mol. Biol. 2007;37:347–356. doi: 10.1165/rcmb.2006-0176OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Pampinella F., Nemes C., et al. J. Cell. Biol. 2004;164:923–933. doi: 10.1083/jcb.200312018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja R.R., Kang M.H., Vaid K., et al. Circ. Res. 2009;105:138–147. doi: 10.1161/CIRCRESAHA.108.193011. [DOI] [PubMed] [Google Scholar]
- Sprung R., Chen Y., Zhang K., Cheng D., Zhang T., Peng J., Zhao Y. J. Proteome Res. 2008;7:1001–1006. doi: 10.1021/pr0705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman E.R., Levine R.L. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- Subramanian K., Jia D., Kapoor-Vazirani P., Powell D.R., Collins R.E., Sharma D., Peng J., Cheng X., Vertino P.M. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstykh T., Lee J., Vafai S., Stock J.B. EMBO J. 2000;19:5682–5691. doi: 10.1093/emboj/19.21.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend R.R., Lipniunas P.H., Tulk B.M., Verkman A.S. Protein Sci. 1996;5:1865–1873. doi: 10.1002/pro.5560050912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlinger S., Baron U., Thellmann M., Hasan M.T., Bujard H., Hillen W. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F., Hadida S., Grootenhuis P.D., et al. Proc. Natl. Acad. Sci. USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Venable J., LaPointe P., et al. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Wang W., Wu J., Bernard K., Li G., Wang G., Bevensee M.O., Kirk K.L. Proc Natl Acad Sci U S A. 2010;107:3888–3893. doi: 10.1073/pnas.0913001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Kopito R.R. J. Biol. Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Ward C.L., Omura S., Kopito R.R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Welman A., Barraclough J., Dive C. Translational Oncogenomics. 2007;2:17–33. [PMC free article] [PubMed] [Google Scholar]
- Xing G., Zhang J., Chen Y., Zhao Y. J Proteome Res. 2008;7:4603–4608. doi: 10.1021/pr800456q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Gollapalli D.R., Maiti P., Jahng W.J., Rando R.R. Cell. 2004;117:761–771. doi: 10.1016/j.cell.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Younger J.M., Chen L., Ren H.Y., Rosser M.F., Turnbull E.L., Fan C.Y., Patterson C., Cyr D.M. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.