Abstract
Although microRNA-21 (miR-21) is emerging as an oncogene and has been shown to target several tumor suppressor genes, including programmed cell death 4 (PDCD4), its precise mechanism of action on cancer stem cells (CSCs) is unclear. Herein, we report that FOLFOX-resistant HCT-116 and HT-29 cells that are enriched in CSCs show a 3- to 7-fold upregulation of pre- and mature miR-21 and downregulation of PDCD4. Likewise, overexpression of miR-21 in HCT-116 cells, achieved through stable transfection, led to the downregulation of PDCD4 and transforming growth factor beta receptor 2 (TGFβR2). In contrast, the levels of β-catenin, TCF/LEF activity and the expression of c-Myc, Cyclin-D, which are increased in CSCs, are also augmented in miR-21 overexpressing colon cancer cells, accompanied by an increased sphere forming ability in vitro and tumor formation in SCID mice. Downregulation of TGFβR2 could be attributed to decreased expression of the receptor as evidenced by reduction in the activity of the luciferase gene construct comprising TGFβR2-3′ untranslated region (UTR) sequence that binds to miR-21. Moreover, we observed that downregulation of miR-21 enhances luciferase-TGFβR2-3′ UTR activity suggesting TGFβR2 as being one of the direct targets of miR-21. Further support is provided by the observation that transfection of TGFβR2 in HCT-116 cells attenuates TCF/LEF luciferase activity, accompanied by decreased expression of β-catenin, c-Myc and Cyclin-D1. Our current data suggest that miR-21 plays an important role in regulating stemness by modulating TGFβR2 signaling in colon cancer cells.
Introduction
Tumor recurrence is one of the most common phenomena in all malignancies and observed in nearly 50% of colorectal cancer, which is the third most common cancer in the USA. This could in part be due to the fact that conventional chemotherapy only targets the rapidly dividing cells that form bulk of the tumor and while chemotherapy can shrink the size of the tumor, the effects are transient and usually do not improve patient’s survival (1). Recent evidence supports the contention that cancer is driven by a small set of self-renewing cells, termed cancer stem cells (CSCs), which are distinct from the bulk of the cells in the tumor. Like normal stem cells, CSCs grow slowly and are more likely to survive chemotherapy than other cells. Hence, proportion of stem cells in the tumor increases after conventional chemotherapy. We have recently reported that exposure of colon cancer HCT-116 or HT-29 cells to FOLFOX that inhibited their growth led to the enrichment of CSC phenotype where Wnt/β-catenin signaling played a critical role in regulating the growth and maintenance of CSCs (2–4); however, the precise molecular mechanism is still unknown.
To that end, we investigated the role of microRNA (miRNA) in colon CSCs. MicroRNAs comprised a broad class of small 19–22 nucleotide long endogenous RNAs that negatively control the expression of target genes by cleaving messenger RNA (mRNA) (5,6) or through translation repression (7). MicroRNA-21 (miR-21) has been found to be overexpressed in most epithelial cancers and therefore believed to play a pivotal role in the progression of many malignancies, including lung, breast, stomach, prostate, colon, brain, head and neck, esophagus and pancreatic cancers (8). Furthermore, studies have shown that knockdown of miR-21 impair growth, induce apoptosis and reduce migration and invasion of cancer cells (9). Additional reports demonstrate that miR-21 counteracts the expression of putative tumor suppressive genes, such as programmed cell death 4 (PDCD4) (10,11), transforming growth factor beta receptor 2 (TGFβR2) (12), phosphatase and tensin (PTEN) (13), maspin (14), NFIB (15), Tropomyosin 1 (TPM1) (16) and reversion-inducing cysteine-rich protein with Kazal motifs (RECK) (17). Based on its tumor promoting functions, miR-21 has been recently referred to as an ‘oncomiR’ (an miRNA with oncogenic properties) (18,19). However, limited information is available concerning the relevance of miR-21 in colon cancer chemoresistance and CSCs. One of the objectives of the current investigation was to examine the role of miR-21 in regulating stemness of colon cancer cells. Furthermore, since miR-21 targets several tumor suppressor genes, we have investigated the interrelationship between miR-21 and TGFβR2.
The transforming growth factor beta (TGFβ) signaling pathway acts through activation of TGFβ-receptor-2 (TGFβR2), which is known to be involved in regulating many cellular processes including growth, differentiation, apoptosis and cellular homeostasis (20). Mutation in TGFβR2 gene appears to increase the risk of developing various cancers including colorectal cancer. It is estimated that 30% of malignant colon tumors have TGFβR2 mutations (21). Recently, Li and Wang have reported that TGFβR2 signaling also affects Wnt/β-catenin signaling pathway (22). Therefore, the primary objective of the current investigation was to examine the role miR-21 in regulating stemness of colon cancer cells. Herein, we report that chemoresistant (CR) colon cancer HCT-116 and HT-29 cells that are highly enriched in CSCs show a marked upregulation of miR-21 accompanied by downregulation of PDCD4. Additionally, we demonstrate that miR-21 induces stemness of colon cancer cells by activating Wnt/β-catenin pathway, which is mediated through downregulation of TGFβR2.
Materials and methods
Cell lines and cell cultures
Human colon cancer HCT-116 and HT-29 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were maintained in Dulbecco's modified Eagle medium (DMEM; 4.5 g/L d-glucose) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Grand Island, NY) and 1% antibiotic/antimycotic in tissue culture flasks in a humidified incubator at 37°C in an atmosphere of 95% air and 5% carbon dioxide. The medium was changed two times a week, and cells were passaged using 0.05% trypsin/ethylenediaminetetraacetic acid (Invitrogen).
Generation of CR colon cancer cells
Unless otherwise stated, 5-Fluorouracil (5-FU)- and oxaliplatin (Ox)-resistant (referred to as CR cells) cells were generated by exposing HCT-116 and HT-29 cells to the combination of 5-FU and Ox at clinically relevant doses and schedules as described previously (2). Briefly, ∼1 × 106 cells were seeded in 100 mm tissue culture dish with DMEM containing 10% FBS and 1% antibiotic/antimycotic for 24 h. The cells were exposed to 5-FU + Ox (25 μM 5-FU and 0.625 μM Ox) for 72 h. The medium was removed and the surviving cells cultured in DMEM containing 10% FBS without the drugs for 3–4 days. The cycle was repeated 12 times. The surviving cells were then passaged and exposed to higher doses of combination of 5-FU + Ox (100 μM 5-FU + 2.5 μM oxaliplatin) for ∼4 weeks. Finally, the surviving cells were maintained in DMEM/10% FBS containing 5-FU + Ox (50 μM 5-FU + 1.25 μM oxaliplatin).
Determination of cellular growth
The growth of parental (P) as well as CR colon cancer HCT-116 and HT-29 cells in response to 5-FU + Ox (250 μM of 5-FU + 6.25 μM oxaliplatin) was assessed by 3-(4,5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide assay as described previously (23).
Western blot analysis
Western blot analysis was performed essentially according to our standard protocol (24,25). Briefly, the cells were solubilized in lysis buffer and the protein concentration was determined by the Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA). The protein were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). The membranes were incubated overnight at 4°C with primary antibodies after blocked. The membranes subsequently washed and incubated with appropriate secondary antibodies. The protein bands were visualized by enhanced chemiluminescence detection system (Amersham, Piscataway, NJ). When appropriate, the membranes were stripped and re-probed with β-actin for verification of loading control.
Isolation of RNA and quantitative polymerase chain reaction analysis
Total RNA was extracted from different cells using RNA-STAT solution (Tel Test, Friendswood, TX) according to the manufacturer's instruction. The total RNA was treated with DNase I and purified with phenol-chloroform. RNA concentration was measured spectrophotometrically at an optical density of 260 nm.
Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) was performed using the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA). Five microliters of complementary DNA (cDNA) products were amplified with SYBR Green Quantitative PCR Master Mix (Applied Biosystems). Polymerase chain reaction (PCR) primers were used as follows: pre-miR-21 forward, 5′-tgagactgatgttgactgttgaa-3′ and reverse, 5′-tgtcagacagcccatcgac-3′; β-actin forward, 5′-cccagcacaatgaagatcaa-3′ and reverse 5′-acatctgctggaaggtggac-3′ and TGFβR2 forward, 5′-aaatggaggcccagaaagat-3′ and reverse, 5′-acttgactgcaccgttgttg-3′. Reactions were carried out in Applied Biosystems 7500 Real-Time PCR System. The conditions for PCR running were as follows: for activating the DNA polymerase, hot start was performed for 10 min at 95°C and then cycling at 95°C for 15 s and 60°C for 1 min for a total of 40 cycles.
Quantitation of miRNA-21
TaqMan microRNA assays were used to quantitate miR-21 in different colon cancer cells according to the manufacturer’s instruction (Applied Biosystems). Briefly, cDNA synthesis was carried out with the TaqMan MicroRNA reverse transcription kit (Applied Biosystems). The miRNA reverse transcription–PCR primers for miR-21, and endogenous control RNU6B were purchased from Applied Biosystems. Real-time qRT–PCR analysis was carried out using Applied Biosystems 7500 Real-time PCR System. The PCR mix containing TaqMan 2× Universal PCR Master Mix were processed as follows: 95°C for 10 min and then 95°C for 15 s, 60°C for 60 s for up to 40 cycles. Signal was collected at the endpoint of every cycle. The gene expression ΔCT values of miRNAs from each sample were calculated by normalizing with internal control RNU6B and relative quantitation values were plotted.
Generation of miR-21 overexpressing HCT-116 cells
pCMV-miR-21 plasmid carrying pre-microRNA-21 and 250–300 nucleotides up- and downstream flanking sequence (OriGene, Rockville, MD) or empty vector DNA alone (pCMV) was transfected into HCT-116 cells by Lipofectamine™ 2000 reagent according to the manufacturer's instructions (Invitrogen Corp., Carlsbad, CA). Several independent sublines (colonies) were obtained over 8–10 weeks of the selection period in the presence of 0.6 mg/ml G418 (Neomycin). Colonies were picked at random and grown as individual cell lines in the presence of 0.4 mg/ml G418. Each cell line was subjected to reverse transcription–PCR analysis to evaluate miR-21 expression.
Formation of colonospheres and extreme limiting dilution analysis
The ability of the miR-21 and control HCT-116 cells to form spheres in suspension was evaluated as described previously (3). Briefly, primary colonospheres were generated by incubating the limited number of pCMV and miR-21 stably transfected HCT-116 (clone 1, pooled) cells at a concentration of 100 cells per 500 μl in serum-free stem cell medium (SCM) containing DMEM/F12 (1:1) supplemented with B27 (Life Technologies, Gaithersburg, MD), 20 ng/ml epidermal growth factor (Sigma, St Louis, MO), 10 ng/ml fibroblast growth factor (Sigma) and antibiotic-anti-mycotic in 24-well plates (Corning, Lowell, MA) for 10 days. The formed colonospheres were centrifuged (1000 r.p.m.), dissociated with 0.05% trypsin/ethylenediaminetetraacetic acid. The single-cell suspension derived from colonospheres that have undergone ≥15 serial passages were used for all experiments.
Self-renewing/regeneration abilities of the sphere, derived from pCMV and miR-21 stably transfected HCT-116 cells, were analyzed for secondary colonospheres formation in the following manner. Primary colonospheres formed over a period of 10 days in SCM containing were collected by centrifugation, dissociated with 0.05% trypsin/ethylenediaminetetraacetic acid and subsequently passed through a 40 μM sieve to obtain single-cell suspensions. An equal number of cells obtained from primary colonospheres culture were plated (100 cells/500 μl in SCM) in ultra low-attachment wells. The secondary colonospheres formed after 5 days were recorded for their number and size by light microscopy.
Extreme limiting dilution analysis (ELDA) was performed essentially according to Hu and Smyth as described by us (3,26). Briefly, single-cell suspension obtained from adherent miR-21 expressing HCT-116 (pooled and clone 1), vector (control) HCT-116 cells and β-catenin small interfering RNA (siRNA)-transfected cells, CR-HCT-116 cells transfected with anti-miR-21 (Invitrogen Corp.) were plated at concentrations of 200, 20 and 2 cells per 100 μl SCM (24 well for each dilution) in 96-well plates and incubated for 5 days. At the end of 5 days, the number of wells showing formation of colonospheres was counted. The frequency of sphere forming cells in a particular cell type was determined using ELDA webtool at http://bioinf.wehi.edu.au/software/elda.
Severe combined immunodeficient mice mice xenografts of miR-21 expressing HCT-116 cells
Four-week-old female ICR/severe combined immunodeficient mice (SCID), obtained from Taconic Laboratory (Germantown, NY) were subcutaneously injected with ∼5.5 × 105 HCT-116 cells that were stably transfected with pCMV/miR-21 or pCMV (control). Tumor measurements were carried out at multiple time points during the experimental period. Mice were weighed and the tumor volume in SCID mice were estimated as follows: volume (cubic millimeters) = (L × W2)/2, where L and W are the tumor length and width (in millimeters), respectively.
siRNA transfection
For the transfection of siRNA into the CR-HCT-116 and miR-21 stably transfected HCT-116 cells cells, Oligofectamine reagent (Invitrogen Corp.) and serum-free Opti-MEM (Invitrogen Corp.) medium were used to prepare transfection complexes according to the manufacturer's instructions. Briefly, the cells were plated in six-well tissue culture plates with normal growth medium and incubated overnight to achieve 40–60% confluence. Next day, the medium was removed, washed twice with serum-free Opti-MEM (Invitrogen Corp.) medium prior to adding the complexes containing non-targeted (control), β-catenin siRNA or TGFβR2 siRNA (Integrated DNA Technologies, Coralville, IA). After 2 days of transfection, the cells were collected and analyzed for protein expression of β-catenin using western blot and for colonosphere formation assay using SCM.
TCF/LEF-dual luciferase assay
The activation of transcription factor TCF/LEF was evaluated by using Cignal TCF/LEF reporter assay kit (SA Biosciences, Frederick, MD) as described previously (3). Briefly, the cells were grown to 25–30% confluence and co-transfected with TCF/LEF reporter constructs along with either non-targeted (control) or β-catenin siRNAs (Integrated DNA Technologies) using SureFECT transfection reagent (SA Biosciences) according to manufacturer's instructions. Two days after transfection TCF/LEF activity reporter assay was performed using a dual-luciferase assay kit (Promega-Biosciences, San Luis Obispo, CA) following the instructions outlined by the manufacturer.
Construction of 3′ untranslated region-luciferase plasmid and reporter assays
A putative miRNA-21-recognition element from the TGFβR2 gene was cloned into the 3′ untranslated region (UTR) of a luciferase miRNA reporter vector (pMIR-REPORT luciferase; Applied Biosystems) using standard molecular biological manipulations (27). The oligonucleotide sequences were designed to carry the HindIII and SpeI sequence at their extremities to allow the plasmid into the HindIII and SpeI sites of pMIR-Report luciferase. The oligonucleotides were: 5′-CTAGTTGACATTGTCATAGGATAAGCTGA-3′, 5′-AGCTTCAGCTTATCCTATGACAATGTCAA-3 (12). For reporter assays, the resultant construct (pMIR-TGFβR2-UTR) and the luciferase reporter vector DNA (pMIR, without 3′ UTR of TGFβR2 insert) were used to transiently transfect the pCMV or miR-21 overexpressing HCT-116 with or without anti-miR-21 (Applied Biosystems) using Lipofectamine™ 2000 reagent. Reporter assays were performed 48 h post-transfection using the luciferase assay system (Promega). The pMIR-REPORT β-gal control plasmid DNA (Applied Biosystems) was co-transfected with the luciferase DNA as internal control and β-galactosidase assays were peformed to normalized the transfection variability.
Overexpression of TGFβR2 in HCT-116 cells
A single-cell suspension of HCT-116 cells was plated in the tissue culture plates to achieve a 90% confluence. Once the 90% confluence is achieved, the adherent cells were transfected using Lipofectamine™ 2000 with plasmid pCMV6-XL4 containing the full-length cDNA of human TGFβR2 (pCMV6-XL4/TGFβR2; OriGene) or empty vector (control) with TCF/LEF Luciferase reporter plasmid in OPTI-MEM medium according to the manufacturer's instruction. After 2 days of transfection, the cells were analyzed for TGFβR2, β-catenin, c-Myc and Cyclin-D1 protein expression by western blot and TCF/LEF Luciferase activity assay.
Statistical analysis
Unless otherwise stated, data are expressed as mean ± SEM. Wherever applicable, the results were analyzed using analysis of variance followed by Fisher’s protected least significant differences or Scheffé’s test. P < 0.05 was designated as the level of significance.
Results
Growth of CR colon cancer cells
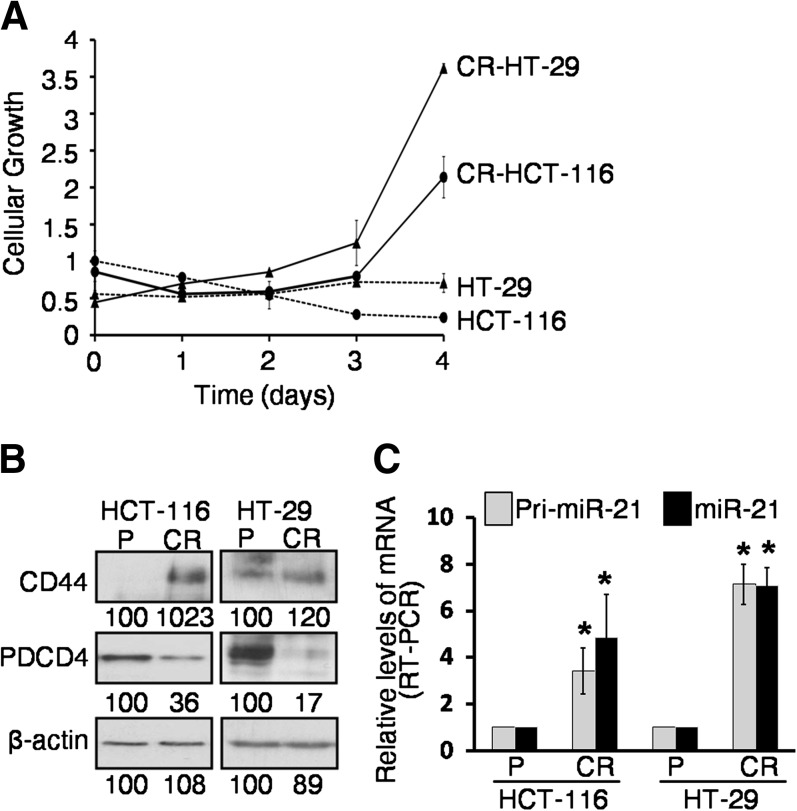
Although FOLFOX [a combination of 5-FU, oxaliplatin and leucovorin] currently remains the backbone of colorectal cancer chemotherapeutics, post-treatment reemergence of cancer has limited its success. To understand the molecular mechanisms for the recurrence of colon cancer, we have generated CR human colon cancer HCT-116 and HT-29 cells lines by exposing them to 5-FU + oxaliplatin at clinically relevant doses and schedules for 4 months. To evaluate the degree of resistance to 5-FU + oxaliplatin of FOLFOX-resistant cells (CR cells), we performed cell viability assay using 250 μM 5-FU + 6.25 μM oxaliplatin (>10-fold the IC50, half maximal inhibitory concentration, of both drugs). We observed that chemotherapeutic combination significantly inhibited the cellular growth of parental HCT-116 and HT-29 cells. In contrast, the same treatment caused a gradual increase in growth of CR colon cancer cells over the 4-day experimental period (Figure 1A).
Fig. 1.
Changes in cellular growth, CSCs markers and miR-21 levels in colon cancer CR and/or parental cells. (A) While the growth of parental (P) colon cancer HCT-116 and HT-29 is inhibited, CR colon cancer cells display induction of the same in response to 5-FU (250 μM) + Ox (6.25 μM) in DMEM/10% FBS over a period of 4 days. (B) Western blot showing stimulation of CSCs marker CD44 and reduction of the pro-apoptotic protein PDCD4 in CR-HCT-116 and CR-HT-29 colon cancer cells, when compared with the corresponding parental (P) cells. β-Actin was used as loading controls. The numbers represent percent of corresponding control normalized to β-actin. (C) Quantitative real-time polymerase chain reaction (RT–PCR) showing upregulation of pri-microRNA-21 and mature microRNA-21 in CR-HCT-116 cells and CR-HT-29 cells, when compared with the parental cells (*P < 0.001).
The next set of experiments was undertaken to examine the potential mechanisms of chemotherapy resistance. In view of the fact that CSCs are resistant to killing by conventional chemotherapy, we hypothesize that CR-HCT-116 and CR-HT-29 cells are enriched in CSCs. Indeed, in both CR-HCT-116 and CR-HT-29 cells, the expression of CD44 (a colon CSC marker), as determined by western blot, was found to be 1023 and 120% higher, respectively, compared with the corresponding parental cells (Figure 1B). This indicates the abundance of CSCs in CR cells. In contrast, the tumor suppressor PDCD4 levels were found to be decreased by 64 and 83% in CR-HCT-116 and CR-HT-29 cells, respectively, compared with the corresponding parental cells (Figure 1B). Additionally, CR-HCT-116 showed 3- to 5-fold increase in the precursor and mature miR-21 levels and a 7-fold increase of the same was noted for CR-HT-29 cells when compared with the corresponding parental cells (Figure 1C) as determined by qRT–PCR.
Overexpression of miR-21 in colon cancer HCT-116 cells increases stemness and the proportion of CSCs
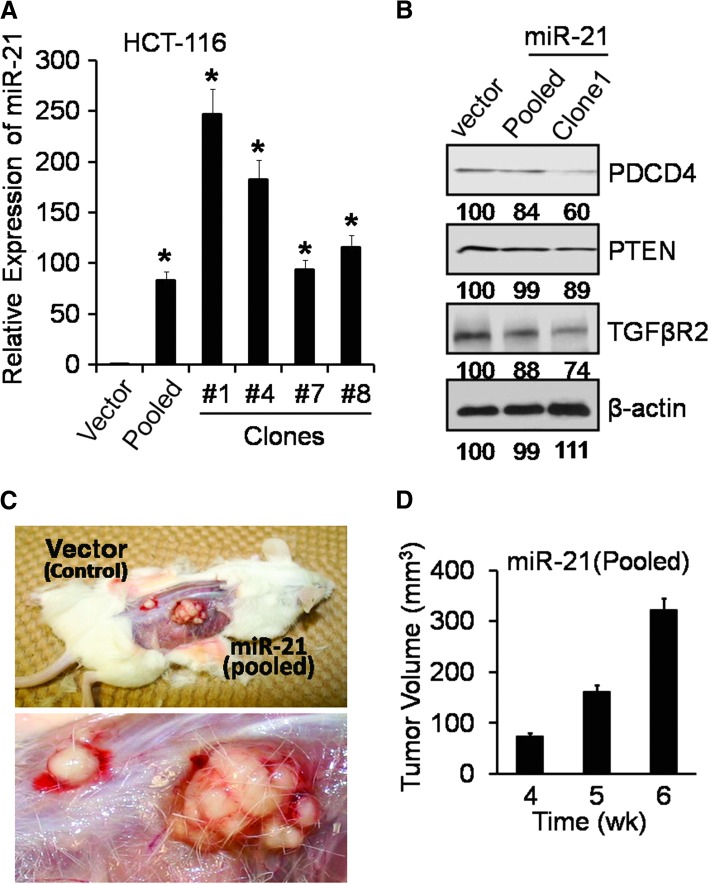
To determine the putative functional properties of miR-21, pCMV/miR-21 plasmid (OriGene) was stably transfected in HCT-116 cells. On the basis of miR-21 expression, as determined by qRT–PCR (real-time PCR) analysis, four miR-21 positive clones were selected. The expression of miR-21 was found to be >240-fold higher in clone 1, compared with empty vector (Figure 2A). The miR-21 overexpressing clone 1 and/or pooled clone were used for subsequent studies. Western blot analysis revealed that the levels of TGFβR2, PDCD4 and PTEN, the targets of miR-21 (10–12), were decreased in miR-21 overexpressing clone 1 as well as in pooled clone as compared with vector-transfected controls (Figure 2B).
Fig. 2.
Overexpression of human miR-21 in colon cancer HCT-116 cells increases tumorigenic potential in SCID mice (A) qRT–PCR showing upregulation of mature miR-21 in HCT-116 clones, derived from the cells stably transfected with pCMV-miR-21 plasmid or the corresponding vector (controls), *P < 0.001. (B) Western blot showing downregulation of TGFβR2, PDCD4 and PTEN by miR-21. β-Actin was used as loading controls. The numbers represent percent of corresponding control normalized to β-actin. (C) Representative photographs showing the anatomical intact xenograft tumor in female SCID mice as observed at the end of 6 weeks of tumor growth after injecting HCT-116 cells stably transfected with pCMV-miR-21 plasmid (pooled) or the corresponding vector (controls), the enlarged xenograft tumors is shown in the lower panel of Figure 2C. (D) Xenograft tumors volume (cubic millimeters) in SCID mice after injecting miR-21 overexpressing HCT-116 cells (pooled); no palpable xenograft tumor was detected after injecting vector (control) transfected HCT-116 cells at corresponding time; Tumor volume = (L × W2)/2; the data represent means of three independent observations.
To determine whether miR-21 may regulate stemness of colon cancer cells, the next set of experiments were carried out. The functional property of cancer stem cells is defined by their ability to form sphere (in vitro) in the serum-free medium containing stem cells growth factors (SCM) when plated in ultra low-attachment plates under extreme limited dilutions (3). To determine whether and to what extent miR-21 overexpression affects the sphere-forming properties of colon cancer cells, HCT-116 cells stably transfected with pCMV/miR-21 (pooled and clone 1) or pCMV vector (control) were subjected to an ELDA. The frequency to form spheroids was found to be 3.5-fold higher for miR-21 overexpressing cells (pooled and clone 1), compared with vector-transfected control (P < 0.001; Table I). Furthermore, the average diameter of spheroids obtained from miR-21 overexpressing clone 1 and pooled clone was found to be 30–40% bigger than the controls. Additionally, we observed a 60–80% increase (P < 0.001) in the self-renewal/re-generation ability of colonospheres in miR-21 overexpressing cells (Table I). Tumorigenic potential of the miR-21 overexpressing cells was further confirmed by generating xenograft tumors in SCID mice. We observed that while the HCT-116 pCMV/miR-21 cells formed palpable tumor within 2 weeks after the first injection, no such tumor could be detected with HCT-116 pCMV (control) cells (Figure 2C and D). The Figure 2C is a representative photograph of anatomically intact xenograft tumor in SCID mice depicting the growth of tumor generated by HCT-116 miR-21 and controls cells. Tumor volumes as calculated after 4 weeks of injection are given in Figure 2D.
Table I.
Sphere-forming frequency, diameter and/or self-renewal ability of cells from colonospheres of stably transfected with pCMV (vector), pCMV/miR-21 (pooled and clone 1) as well as different colon cancer HCT-116 cells where β-catenin or miR-21 is downregulated
| pCMV control | pCMV/miR-21 pooled | pCMV/miR-21 clone 1 | pCMV control β-catenin siRNA | pCMV/miR-21 β-catenin siRNA | CR-HCT-116 scrambled | CR-HCT-116 anti-miR-21 | P value | |
| Sphere-forming frequency (95% CI) | 1/7 | 1/2 | 1/2 | 1/25 | 1/11 | 1/11 | 1/47 | <0.001 |
| Diameter (μm) | 65.8 ± 19.8 | 91.5 ± 28.8 | 86.2 ± 19.8 | — | — | — | — | <0.001 |
| Self-renewal (spheres per 200 cells) | 13 ± 3 | 24 ± 3 | 20 ± 5 | — | — | — | — | <0.001 |
Data are pooled from three independent experiments for each. CI, confidence interval.
Wnt/β-catenin regulates stemness in miR-21 overexpressing colon cancer cells
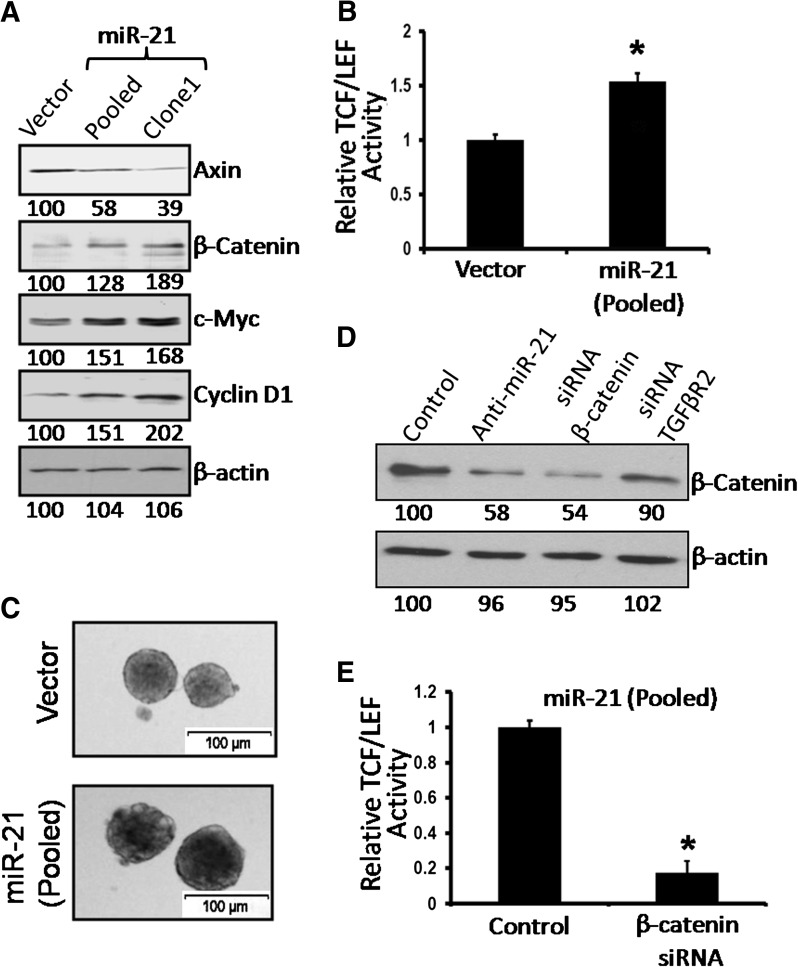
The Wnt/β-catenin signaling pathway is known to be activated in many malignancies, including colorectal cancer (28). This signaling pathway also plays a critical role in regulating the proliferation of CSCs (3). Western blot analysis of miR-21 overexpressing HCT-116 cells showed 28–100% higher expression of total β-catenin, C-Myc and Cyclin-D1 but a 60% reduction in axin levels, compared with the vector-treated control (Figure 3A). These changes were associated with a 50% increase in transcriptional activity of TCF/LEF (Figure 3B) and formation of colonospheres, as evidenced by their increased size (Figure 3C). Our observation of increased TCF/LEF activity together with elevated levels of total β-catenin, c-Myc and Cyclin-D1 and reduction in axin in miR-21 overexpressing HCT-116 cells (pooled and clone 1) suggests activation of Wnt/β-catenin signaling in these cells.
Fig. 3.
miR-21 overexpression activates β-catenin/TCF/LEF activity (A) Western blot showing the expression of different members (axin, β-catenin, c-Myc, Cyclin-D1) of Wnt/β-catenin signaling pathway in miR-21 expressing clones from HCT-116 cells, stably transfected with pCMV-miR-21 plasmid or the corresponding vector. The numbers represent percent of corresponding control normalized to β-actin. (B) miR-21 overexpression resulted in the activation of TCF/LEF activity in the miR-21 pooled clones cells. (C) Representative photographs showing colonospheres formed by the cells from miR-21 expressing clones, derived from HCT-116 cells, stably transfected with pCMV-miR-21 plasmid or the corresponding vector. (D) Western blot showing the expression of β-catenin after downregulation of miR-21, β-catenin and TGFβR2 in the miR-21-overexpressing cells (pooled clones) by anti-miR-21, β-catenin siRNA or TGFβR2 siRNA. (E) Downregulation of β-catenin using β-catenin siRNA inhibits transcriptional activity of TCF/LEF in the clones (pooled) from HCT-116 cells, stably transfected with pCMV-miR-21 plasmid. The corresponding controls were similarly treated with non-targeted siRNA (control). *P < 0.001, compared with the corresponding vector-treated controls.
To further investigate the importance of Wnt/βcatenin signaling in regulating stemness of miR-21 overexpressing cells, we downregulated β-catenin in pCMV/miR-21 stably transfected HCT-116 cells (pooled clones) through transfection of the corresponding siRNA. This resulted in ∼45% reduction in β-catenin levels, compared with the controls, which were transfected with the corresponding vector (NT-siRNA) (Figure 3D). Transfection of anti-miR-21 also downregulated β-catenin by 42% (Figure 3D). Downregulation of β-catenin in miR-21 overexpressing colon cancer cells by the corresponding siRNA resulted in a marked 70–80% reduction in transcriptional activity of TCF/LEF (Figure 3E) and also reduced the sphere forming frequency to 1:11 as opposed to 1:2 observed in miR-21 overexpressing cells (Table I). Interestingly, in CR-HCT-116 cells, where miR-21 is downregulated by anti-miR-21, the sphere-forming frequency was found to be only 1:47 from 1:11, indicating a role for miR-21 in regulating some of the functional properties of CSCs (Table I).
The putative miR-21 binding to TGFβR2 mRNA and activation of Wnt/β-catenin signaling
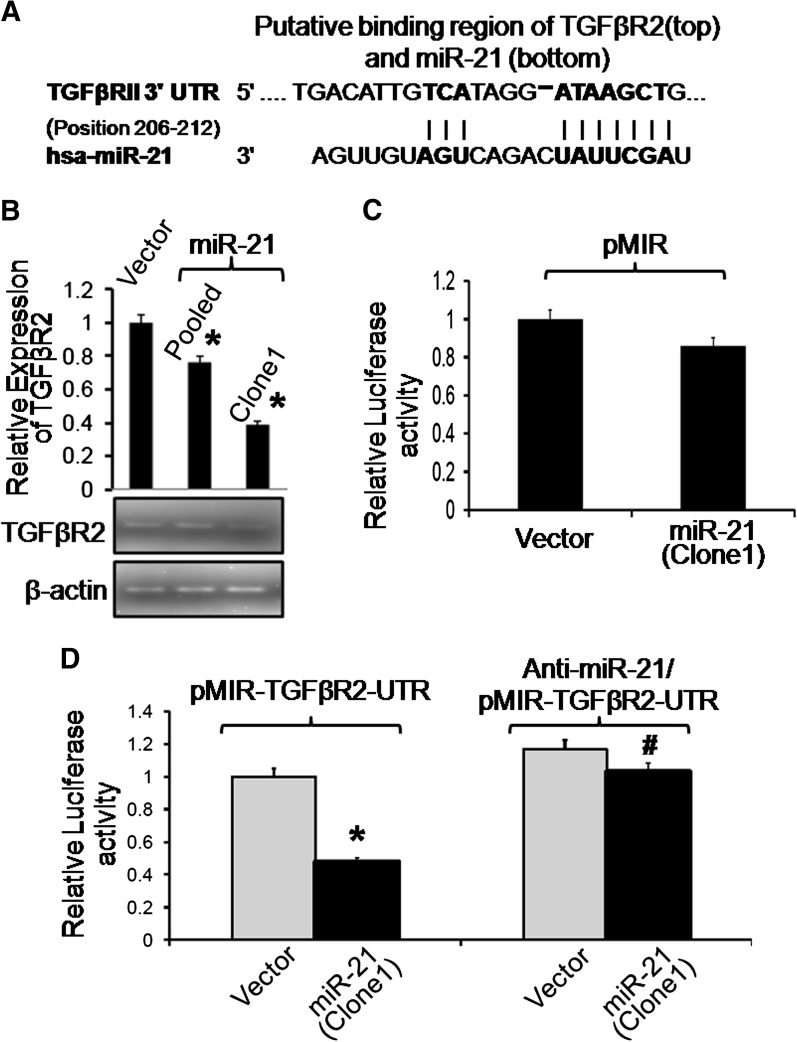
To delineate the mechanism of miR-21 induced stemness by Wnt/β-catenin signaling pathway, we searched miR-21 targets on the web-based resource (http://www.targetscan.org). Interestingly, we found that the putative sites in the 3′ UTR of mRNA of TGFβR2 to be complementary to miR-21 (Figure 4A). It is well known that TGFβR2 is a tumor suppressor (3,29) and TGFβ/TGFβR2 signaling plays important roles in many cellular processes in both adult organism and the developing embryo including growth, differentiation, apoptosis and cellular homeostasis (20). Moreover, recent evidence suggests that TGFβR2 affect Wnt/β-catenin signaling pathway (22). Therefore, we investigated TGFβR2 expression in miR-21 overexpressing clone 1, pooled clones and pCMV-transfected control clone. Reverse transcription–PCR analysis showed a reduction of ∼20 and 60% in mRNA levels of TGFβR2 in pooled clones and clone 1, respectively, compared with the controls (Figure 4B). This was confirmed by western blot analysis which showed a reduction of 12 and 26% TGFβR2 levels in pooled clones and clone 1, respectively, compared with controls (Figure 2B).
Fig. 4.
The putative miR-21 binding to TGFβR2-3′ UTR is responsible for miR-21-mediated downregulation of luciferase activity. (A) A schematic representation of the putative binding sites in 3′ UTR of mRNA of TGFβR2 that was found to be complementary to miR-21 indicating that TGFβR2 might be a target gene of miR-21. (B) Quantitative PCR showing downregulation of human TGFβR2 mRNA in miR-21 (pCMV-miR-21) stably transfected HCT-116 cell clones and control vector (pCMV), (upper panel); and RT–PCR product electrophoresis on agarose gel, 10 μl of the PCR product was electrophoresed on 3% agarose gels containing ethidium bromide, (lower panel). β-Actin as an internal control is also shown. (C) Relative Luciferase activity of pMIR (luciferase reporter without TGFβR2-UTR) in stably transfected miR-21 or pCMV HCT-116 cells. (D) Downregulation of relative luciferase activity of pMIR-TGFβR2-3′ UTR (luciferase reporter with TGFβR2-3′ UTR) in miR-21 overexpressing HCT-116 cells, compared with vector (pCMV) (Left panel), and the recovery following the trasfection of the anti-miR-21 in miR-21 overexpressing HCT-116 cells (clone 1)(right panel). Data represent mean ± SEM (n = 4), *P < 0.01 relative to corresponding control and #P < 0.01 anti-miR-21/pMIR-TGFβR2-3′ UTR relative to corresponding pMIR-TGFβR2-3′ UTR in miR-21 overexpressing cells.
To confirm that the putative complimentary site (as shown in Figure 4A) is the binding site of miR-21 for TGFβR2 regulation, we constructed a Luciferase miRNA expression reporter vector using pMIR-REPORT system (Applied Biosystems). The miR-21 sequence was aligned with 3′ UTR of TGFβR2 (Figure 4A) and inserted to pMIR-report luciferase plasmid HindIII and SpeI site. The resulting construct (pMIR-luciferase-TGFβR2-UTR or parental pMIR-luciferase) was used to transfect miR-21 overexpressing HCT-116 cells (clone 1) and vector (pCMV)-transfected cells. Transfection of the parental luciferase reporter plasmid (pMIR-luciferase, without the TGFβR2 3′ UTR) in miR-21 overexpressing HCT-116 cells (clone 1) produced no significant change in the relative luciferase activity over the control (pCMV) (Figure 4C). However, a significant 50% reduction of luciferase activity was observed in the miR-21 overexpressing cells transfected with pMIR-luciferase-TGFβR2-UTR when compared with the control (pCMV) cells (Figure 4D). The luciferase-TGFβR2-3′ UTR reporter activity was reversed following downregulation miR-21 in overexpressing HCT-116 cells (clone 1) through the transfection of anti-miR-21 (Applied Biosystems) (Figure 4D).
Overexpression of TGFβR2 downregulates β-catenin/TCF/LEF activity
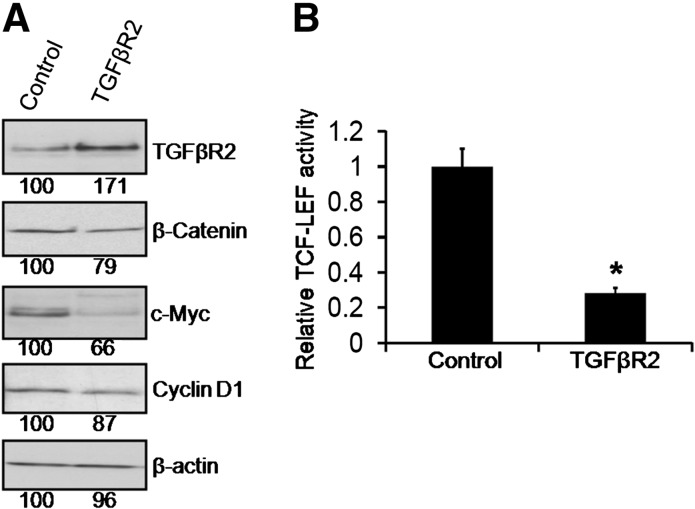
The miR-21 overexpression induced activation of TCF/LEF with concomitant decrease in TGFβR2 expression (both mRNA and protein) indicates a prospective mechanistic pathway for miR-21 mediated modification of TCF/LEF transcription activity. The next set of experiments was carried out to examine the relationship between TGFβR2 and Wnt/β-catenin signaling pathway. This was studied by transfecting the HCT-116 cells with mammalian overexpression vector pCMV6-XL4 containing the full-length cDNA of human TGFβR2 (pCMV6-XL4/TGFβR2; OriGene) or empty vector (control) along with TCF/LEF Luciferase reporter plasmid. After 48 h of transfection, there was a marked increase in the expression of TGFβR2, accompanied by attenuating expression of β-catenin as well as reduction in the levels of Cyclin-D1 and c-Myc, the two major target genes of Wnt/β-catenin signaling in pCMV6-XL4/TGFβR2 transfected cells, when compared with the controls (Figure 5A). Further confirmation of the TGFβR2 induced inhibition of Wnt/β-catenin signaling came from the observation that the relative TCF/LEF activity was reduced by >70% in TGFβR2 overexpressing HCT-116 cells (Figure 5B).
Fig. 5.
Overexpression of TGFβR2 downregulates β-catenin/TCF/LEF activity and downstream target Cyclin-D1 and c-Myc. TCF/LEF Luciferase reporter plasmid with the mammalian overexpression vector pCMV6-XL4 containing the full-length cDNA of human TGFβR2 (TGFβR2) or empty vector (control) were co-transfected into HCT-116 cells for 48 h. (A) Western blot showing TGFβR2, β-cantenin, Cyclin-D1 and c-Myc expression levels in HCT-116 cells after transfecting pCMV6-XL4 containing TGFβR2 (TGFβR2) or empty vector (control). Each lane contained 50 μg of cell lysate protein. β-Actin was used as loading controls. (B) Relative TCF-LEF activity was found to be downregulated following the TGFβR2 overexpression as compared with the corresponding vector-treated control (*P < 0.001).
Discussion
Currently, surgery and chemotherapy are the two major viable strategies exist for the treatment of advanced colorectal cancer. The chemotherapy treatment regimen typically consists of a combination of drugs in order to be more effective. FOLFOX is one regimen for advanced colorectal cancer consists of the combination of 5-FU, leucovorin and oxaliplatin. Despite their synergistic interactions (30) and significant increase in the colorectal cancer patients’ survival rate, FOLFOX fails to eliminate all tumor cells because of intrinsic or extrinsic (acquired) drug resistance, thereby leading to tumor recurrence (31). Our current observation of time-dependent rise in the growth of CR colon cancer HCT-116 and HT-29 cells in response to the combination of 5-FU and oxaliplatin further indicates ineffectiveness of the colon cancer chemotherapeutic to inhibit growth of CR colon cancer cells consistent with the emergence of highly enriched CSCs (2,4). However, the regulatory mechanisms for the development of CSC in response to chemotherapy are unknown.
It is becoming increasingly evident that the miRNAs play important roles in the regulation of proliferation, apoptosis, differentiation, metabolism and invasion (32) thereby affecting normal cell growth and development, leading to a variety of disorders including malignancies (33–35). Recent reports indicate that miRNAs are capable of pleiotropic effects by simultaneously regulating post-transcriptional expression of numerous genes (32), also supporting a role for miRNAs in regulating CSC function which involves a complex interplay between multiple pathways. Indeed, miRNA-21, which promotes cell transformation by targeting the PDCD4 gene (11), has been shown to post-transcriptionally downregulate this protein in colorectal cancer (10). However, the exact role of miRNA-21 in influencing the development of drug resistance and cancer stem cell proliferation is not fully understood. Since colon cancer is a disease driven by the presence of CSCs, which show resistance against various cytotoxic drugs and display activation of Wnt/beta catenin signaling (31,36), the present study was designed to investigate the role miR-21 in regulating the stemness of colon cancer cells.
Previously, we have reported that the newly generated CR HCT-116 and HT-29 cells that are maintained and grown in high doses of 5-FU and oxaliplatin display high proportion of CSCs markers CD44, CD166 and CD133 (2,4). Our current data demonstrate that the CR-HCT-116 and CR-HT-29 cells, in addition to displaying increased expression of CD44, also show an increased expression of premature and mature miR-21 (2,4). Additionally, our data show that CR colon cancer cells, miR-21 overexpression are accompanied by downregulation of the tumor suppressor protein PDCD4. Our results are in direct agreement with those recently reported for breast cancer (37) and glioblastoma (38) cells. Taken together, the results suggest a putative role of miR-21 in regulating the growth of CSCs (39) thereby contributing to the development of chemoresistance.
It is becoming increasingly evident that Wnt/β-catenin pathway, which is activated in many malignancies, including colorectal cancer (40) plays a critical role in regulating the growth and maintenance of CSCs (3). In light of the emerging role of Wnt/β-catenin signaling pathway in the regulation of CSCs, we investigated the influence of miR-21 on the stemness and Wnt signaling pathway by generating miR-21 overexpressing HCT-116 cells through stable transfection. Our results show that miR-21 overexpression results in the activation of Wnt/β-catenin signaling as evident by the reduced levels of axin, increased β-catenin levels as well as induction of TCF/LEF activity and increased protein expression of downstream target genes c-Myc and Cyclin-D1. These findings are important in light of the fact that c-Myc is an oncogene that also regulates the self-renewal processes of CSCs (3,41,42). Furthermore, evidence of miR-21 contributing toward the stemness came from the fact that the cells overexpressing miR-21 show increased size and frequency of formation of spheres. The latter also indicates an increase in the CSCs proliferation. This is further supported by ELDA, an in vitro analysis of the functional frequency of CSCs, and by the observation that miR-21 overexpressing colon cancer cells are able to form tumors in SCID mice at a much shorter time than those by the control cells. Moreover, our observation that downregulation of miR-21 in CR colon cancer cells markedly decreases their sphere-forming ability, a phenomenon similar to that noted for β-catenin-downregulated cells, supports our contention that miR-21 regulates some of the functional properties of colon CSCs. Taken together, these findings indicate that miR-21 induction can lead to an increase in the CSCs proliferation by activation of Wnt/β-catenin signaling and therefore may be responsible for the development of chemoresistance.
Our contention was further supported by the recent clinical findings which showed that overexpression of miR-21 is associated with poor therapeutic response as well as poor prognosis of colon cancer patients (43). Furthermore, recent reports strongly suggest an oncogenic role of miR-21 as it counteracts multiple gene expression including phosphatase and tensin (PTEN), maspin, Tropomyosin 1 (TPM1) and reversion-inducing cysteine-rich protein with Kazal motifs (RECK). It is believed that miR-21 induces cell proliferation and inhibits apoptosis through counteracting the expression of these target genes (44). However, the exact mechanism of miR-21 induced activation of Wnt/β-catenin signaling pathway and its contribution to stemness in colon cancer remains elusive.
In the present report, we show that one of the possible mechanistic pathways of miR-21 actions is mediated through targeting TGFβR2 as evident by the downregulation of mRNA of TGFβR2 in miR-21 overexpressing colon cancer cells. It is well known that TGFβR2 is a tumor suppressor (29) gene and TGFβ/TGFβR2 signaling plays an important role in many cellular processes including growth, differentiation, apoptosis, cellular homeostasis and other cellular functions (20). Our current observation together with what has been reported recently (12) suggests that TGFβR2 is a direct target gene of miR-21 and regulates the stemness by modulating Wnt/β-catenin signaling.
TGFβR2 is pivotal to trigger the signaling pathway induced by TGFβ superfamily ligands as these ligands binds to TGFβR2, which recruits and phosphorylates type I receptor (20). The type I receptor then phosphorylates receptor-regulated SMADs that bind with the downstream complexes and accumulate in the nucleus where they act as transcription factors and participate in the regulation of target gene expression (45). On the other hand, TGFβ can also signal through non-Smad pathways, activate TAK1, ERK, p38, Jun N-terminal protein kinase, Rho GTPase and phosphatidylinositol 3 kinase/AKT pathways (20,46). Studies also indicate that TGFβ and Wnt/β-catenin signaling cross talk in developmental stages as well as during carcinogenesis which can be both cooperative and antagonistic (47–49).
On the basis of the recent observation that 3′ UTR of mRNA of TGFβR2 contains a complimentary sites to miR-21, we speculated that TGFβR2 is the direct target for miR-21. Indeed, our current observations of downregulation of TGFβR2 in miR-21 overexpressing cells, as evidenced by reduction in the activity of luciferase gene construct comprising TGFβR2-3′ UTR sequence that binds to miR-21 and the fact that knockdown of miR-21 enhances luciferase-TGFβR2-3′ UTR activity, strongly suggest that TGFβR2 is one of the direct targets of miR-21. Further support is provided by the fact that upregulation of TGFβR2 in HCT-116 cells decreased β-catenin levels as well as attenuates TCF/LEF luciferase activity, accompanied by downregulation of c-Myc and Cyclin-D1. Taken together, the results suggest that miR-21 is involved in the induction of mRNA cleavage or translational repression of TGFβR2 and may play a role in the activation of Wnt/β-catenin signaling pathway.
The results from the current investigation show that miR-21 overexpression is associated with induction of stemness through downregulation of TGFβR2, a direct target of miR-21 and augmentation of β-catenin TCF/LEF signaling pathway.
Funding
National Institutes of Health/National Institute on Aging (AG014343 to A.P.N.M.); Department of Veterans Affairs to A.P.N.M.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- cDNA
complementary DNA
- CSC
cancer stem cell
- CR
chemoresistant
- DMEM
Dulbecco's modified Eagle medium
- ELDA
extreme limiting dilution analysis
- FBS
fetal bovine serum
- 5-FU
5-fluorouracil
- miR-21
microRNA-21
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- PDCD4
programmed cell death 4
- qRT–PCR
quantitative reverse transcription–polymerase chain reaction
- SCM
stem cell medium
- siRNA, small interfering RNA; TGFβR2
transforming growth factor beta receptor 2
- TGFβ
-
transforming growth factor beta;
UTR, untranslated region
References
- 1.Stockler M, et al. Systematic reviews of chemotherapy and endocrine therapy in metastatic breast cancer. Cancer Treat. Rev. 2000;26:151–168. doi: 10.1053/ctrv.1999.0161. [DOI] [PubMed] [Google Scholar]
- 2.Yu Y, et al. Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Transl. Oncol. 2009;2:321–328. doi: 10.1593/tlo.09193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanwar SS, et al. The Wnt/beta-catenin pathway regulates growth and maintenance of colonospheres. Mol. Cancer. 2010;9:212. doi: 10.1186/1476-4598-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanwar SS, et al. Difluorinated-curcumin (CDF): a novel curcumin analog is a potent inhibitor of colon cancer stem-like cells. Pharm. Res. 2011;28:827–838. doi: 10.1007/s11095-010-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doench JG, et al. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutvagner G, et al. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 7.Olsen PH, et al. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JA, et al. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 10.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 11.Lu Z, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YJ, et al. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- 13.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 15.Fujita S, et al. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J. Biol. Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 17.Gabriely G, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medina PP, et al. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2011;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 19.Ziyan W, et al. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med. Oncol. 2011;28:1469–1474. doi: 10.1007/s12032-010-9563-7. [DOI] [PubMed] [Google Scholar]
- 20.Moustakas A, et al. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 21.Biswas S, et al. Mutational inactivation of TGFBR2 in microsatellite unstable colon cancer arises from the cooperation of genomic instability and the clonal outgrowth of transforming growth factor beta resistant cells. Genes Chromosomes Cancer. 2008;47:95–106. doi: 10.1002/gcc.20511. [DOI] [PubMed] [Google Scholar]
- 22.Li XH, et al. Urothelial transdifferentiation to prostate epithelia is mediated by paracrine TGF-beta signaling. Differentiation. 2009;77:95–102. doi: 10.1016/j.diff.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel BB, et al. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int. J. Cancer. 2008;122:267–273. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 24.Patel BB, et al. Curcumin targets FOLFOX-surviving colon cancer cells via inhibition of EGFRs and IGF-1R. Anticancer Res. 2010;30:319–325. [PMC free article] [PubMed] [Google Scholar]
- 25.Xu H, et al. Epidermal growth factor receptor (EGFR)-related protein inhibits multiple members of the EGFR family in colon and breast cancer cells. Mol. Cancer Ther. 2005;4:435–442. doi: 10.1158/1535-7163.MCT-04-0280. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y, et al. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y, et al. Cloning of a novel EGFR-related peptide: a putative negative regulator of EGFR. Am. J. Physiol. Cell Physiol. 2001;280:C1083–C1089. doi: 10.1152/ajpcell.2001.280.5.C1083. [DOI] [PubMed] [Google Scholar]
- 28.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 29.Munoz NM, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 30.Theile D, et al. Involvement of drug transporters in the synergistic action of FOLFOX combination chemotherapy. Biochem. Pharmacol. 2009;78:1366–1373. doi: 10.1016/j.bcp.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Dean M, et al. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Croce CM, et al. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 34.Hwang HW, et al. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory RI, et al. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 36.Boman BM, et al. Human colon cancer stem cells: a new paradigm in gastrointestinal oncology. J. Clin. Oncol. 2008;26:2828–2838. doi: 10.1200/JCO.2008.17.6941. [DOI] [PubMed] [Google Scholar]
- 37.Bourguignon LY, et al. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J. Biol. Chem. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–18. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 39.Wilson KD, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reya T, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, et al. Increased expression of beta-catenin, phosphorylated glycogen synthase kinase 3beta, cyclin D1, and c-myc in laterally spreading colorectal tumors. J. Histochem. Cytochem. 2009;57:363–371. doi: 10.1369/jhc.2008.953091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krichevsky AM, et al. miR-21: a small multi-faceted RNA. J. Cell. Mol. Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng XH, et al. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell. Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 46.Derynck R, et al. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 47.Labbe E, et al. Transcriptional cooperation between the transforming growth factor-beta and Wnt pathways in mammary and intestinal tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 48.Labbe E, et al. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl Acad. Sci. USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]