Abstract
Endoplasmic Reticulum (ER) stress refers to a condition of accumulation of unfolded or misfolded proteins in the ER lumen. A variety of biochemical stimuli or pathophysiologic conditions can directly or indirectly induce ER stress, leading to activation of an ER-originated adaptive signaling response called Unfolded Protein Response (UPR). Recent studies demonstrated that ER stress and UPR signaling are critically involved in the initiation and progression of many diseases, such as metabolic disease, cardiovascular disease, neurodegenerative disease, and cancer. In this study, we show that ER stress induced by pharmacologic reagents, including tunicamycin (TM) and thapsigargin (Tg), promotes hepatic lipogenesis and lipid droplet formation. Using quantitative gene expression analysis, we identified 3 groups of key lipogenic regulators or enzymes that are inducible by pharmacological ER stress in a human hepatoma cell line Huh-7. These ER stress-inducible lipogenic factors include: 1) lipogenic trans-activators including CCAAT/ enhancer binding protein alpha (C/EBPα), peroxisome proliferator-activated receptor gamma (PPARγ), PPARγ coacti-vator 1-alpha (PGC1α), and Liver X receptor alpha (LXRα); 2) components of lipid droplets including fat-specific protein 27 (FSP27), adipose differentiation related protein (ADRP), fat-inducing transcript 2 (FIT2), and adipocyte lipid-binding protein (AP2); 3) key enzymes involved in de novo lipogenesis including acetyl-CoA carboxylase 1 (ACC1) and stearoyl-CoA desaturase-1 (SCD1). Supporting the role of pharmacologic ER stress in up-regulating de novo lipogenesis, TM or Tg treatment significantly increased accumulation of cytosolic lipid droplet formation in the hepatocytes. Moreover, we showed that forced expression of an activated form of X-box binding protein 1 (XBP1), a potent UPR trans-activator, can dramatically increase expression of PPARγ and C/EBPα in Huh-7 cells. The identification of ER stress-inducible lipogenic regulators provides important insights into the molecular basis by which acute ER stress promotes de novo lipogenesis. In summary, the findings from this study have important implication in understanding the link between ER stress and metabolic disease.
Keywords: Endoplasmic reticulum (ER) stress, hepatic lipogenesis, lipid droplet formation
Introduction
In eukaryotic cells, the ER is the site of folding of membrane and secreted proteins, synthesis of lipids and sterols, and storage of free calcium [1, 2]. As a protein-folding compartment, the ER is exquisitely sensitive to alterations in homeo-stasis, and provides stringent quality control systems to ensure that only correctly folded proteins transit to the Golgi and unfolded or mis-folded proteins are retained in the ER and ultimately degraded. A number of biochemical stimuli and physiological and pathological processes, such as perturbation in calcium homeo-stasis, elevated secretory protein synthesis, and expression of misfolded proteins, can disrupt ER homeostasis, impose stress to the ER, and subsequently lead to accumulation of unfolded or misfolded proteins in the ER lumen. To cope with accumulation of unfolded or misfolded proteins in the ER, the cell has evolved highly specific signaling pathways called the unfolded protein response (UPR) to reduce the amount of new proteins translocated into the ER lumen, increase retrotranslocation and degradation of ER-localized proteins, and bolster the protein-folding capacity and secretion potential of the ER [2]. The UPR is orchestrated by transcrip-tional activation of multiple genes mediated by the protein kinase/endoribonuclease IRE1 (inositol-requiring 1) and the b-ZiP transcription factor ATF6 (activating transcription factor 6), and a general decrease in translation initiation and the selective translation of specific mRNAs mediated by the protein kinase PERK (double-strand RNA-activated kinase-like ER kinase) [1-4].
Liver is a major organ responsible for lipid and glucose metabolism. Dysregulation of hepatic lipid metabolism is closely associated with the initiation and progression of metabolic syndrome. Recent studies suggest that ER stress response plays important roles in maintaining lipid homeostasis [5-9]. The UPR branches through IRE1α and/or ATF6 is required to prevent hepatic steatosis upon acute ER stress [6, 7, 10]. It has also been shown that the IRE1α/ XBP1 UPR branch is activated by the dietary high-carbohydrate and controls the expression of lipogenic enzymes, such as ACC2, DGAT2 and SCD1, that are essential for fatty acid and cholesterol biosynthesis [11]. Moreover, the UPR pathway through PERK/eIF2a was documented to be required for the expression of lipogenic genes and the development of hepatic steatosis [9]. Together, these observations suggest that ER stress and the UPR signaling are critically involved in regulating hepatic lipid metabolism.
In this study, we utilized two structurally-unrelated ER stress-inducing reagents, tunicamycin (TM) and Thapsigargin (Tg), to induce pharmacologic ER stress in Huh-7, a human hepatoma cell line that maintains key features of hepatic lipid metabolism [12]. Through this approach, we confirmed the effect of pharmacologic ER stress in up-regulating de novo lipogenesis and lipid droplet formation. Importantly, we have identified a subset of genes encoding key lipogenic trans-activators and enzymes, which are inducible by acute ER stress. The results from this study provide important insights into ER stress-induced hepatic lipogenesis
Materials and methods
Materials
Chemicals were purchased from Sigma unless indicated otherwise. Synthetic oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Antibodies against XBP1, C/EBPα, and PPARγ were from Santa Cruz Biotechnologies, Inc (Santa Cruz, CA). Anti-bodies against GAPDH and p-actin were purchased from Sigma (St. Louis, MO). Tunicamycin was from Sigma. BODIPY staining kit was purchased from Invitrogen. Human hepatoma cell line Huh-7 was kindly provided by Drs. Christopher M. Schonhoff (Tufts University Cummings School of Veterinary Medicine).
Huh-7 cell culture and TM and Tg treatment
HuH-7 cells were cultured at 37 °C and 5% C02 in DMEM containing high glucose (25 mM) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, and 10% heat-inactivated fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Huh-7 cells at 70% confluency were treated with tunicamycin (TM; 5, 10, and 20 μg/ml) or thapsigargin (Tg; 0.5, 1, and 1.5 μM) or vehicle PBS for 6, 12, and 24 hrs.
BODIPY staining of lipid droplets
Cells were washed with PBS, fixed with 3% formaldehyde for 15 min, and stained with BODIPY 493/503 (Invitrogen, stock concentration 1mg/ ml, working solution 1:1000 dilution) for 15min at room temperature. Cells were then mounted with Prolong gold anti-fade reagent (Invitrogen) followed by washing in PBS for 3 times.
Western Blot and IP-Western blot Analyses
To determine expression levels of XBP1, PPARγ, C/EBPα, and GAPDH, total cell lysates were prepared from cultured Huh-7 cells using NP-40 lysis as previously described [13]. Denatured proteins were separated by SDS-PAGE on 10% Tris-glycine polyacrylamide gels and transferred to a 0.45-mm PVDF membrane (GE Healthcare). Membrane-bound antibodies were detected by an enhanced chemiluminescence detection reagent (GE Healthcare).
Recombinant adenoviral infection
Huh-7 cells at 60% confluency were infected by recombinant adenovirus expressing GFP or an activated form of XBP1 protein at an MOI of 100 for 48 hours before cell lysates were collected for Western blot analysis. Adenovirus expressing spliced XBP1 was kindly provided by Dr. Umut Ozcan (Harvard University) [14]. Recombinant adenovirus expressing GFP was kindly provided by Dr. Jiande Lin (University of Michigan).
Quantitative real-time RT-PCR analysis
For real-time PCR analysis, the reaction mixture containing cDNA template, primers, and SYBR Green PCR Master Mix (Invitrogen) was run in a 7500 Fast Real-time PCR System (Applied Bio-systems, Carlsbad, CA). The real-time PCR primer sequences used in this study are described in supplemental information. Fold changes of mRNA levels were determined after normalization to internal control β-actin RNA levels.
Statistics analysis
Experimental results are shown as mean ± STDEV (for variation between experiments). The mean values for biochemical data from the experimental groups were compared by a paired or unpaired, 2-tailed Student's t test. Statistical tests with P < 0.05 were considered significant.
Results
Pharmacologic ER stress induced by TM or Tg promotes lipid droplet formation
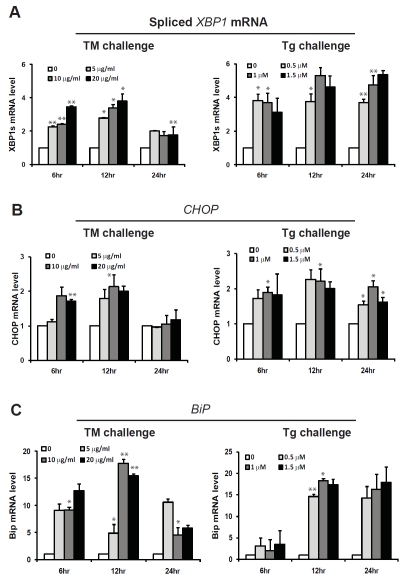
To study the effect of pharmacologic ER stress on hepatic lipid metabolism, we challenged a human hepatoma cell line, Huh-7, with two structurally-unrelated ER stress-inducing drugs, tunicamycin (TM) and Thapsigargin (Tg). Huh-7 is a human hepatocellular carcinoma cell line that has been used for studying hepatic lipid metabolism [12]. TM is a bacterial nucleoside antibiotic that can block N-linked glycoproteins and cause accumulation of unfolded or mis-folded proteins in the ER [15]. Tg is a specific inhibitor of intracellular SERCA-type Ca2+ pumps present in the sarcoplasmic/ER [16, 17]. Tg treatment can disrupt ER calcium homeostasis, leading to accumulation of unfolded or mis-folded proteins in the ER lumen. Both TM and Tg have been routinely used as experimental tools to induce pharmacologic ER stress [18]. To delineate gene expression profiles in hepatic lipid metabolism upon pharmacologic ER stress challenge, Huh-7 cells were treated with TM at doses ranging from 5 to 20 μg/ml or Tg at doses ranging from 0.5 to 1.5 μM. The time intervals for each treatment were 6, 12, and 24 hours. Quantitative real-time RT-PCR analysis indicated that expression of the UPR target mRNA or genes, including spliced Xbp1 mRNA, Bip, and Chop, was increased upon TM or Tg treatment in a dose-and time-dependent manner (Figure 1). This result suggests that TM and Tg can efficiently induce ER stress and activation of the UPR signaling in Huh-7 cells. Note that the levels of the spliced XBP1, CHOP, and BiP mRNAs in Huh-7 cells under different doses of TM treatment were comparable at 24 hours post TM treatment (Figure 1A-C), suggesting that Huh-7 cells can adapt to ER stress at the late stage of TM treatment.
Figure 1.
Quantitative real-time RT-PCR analysis of the mRNAs encoding spliced XBP1 (A), CHOP (B), and BiP (C) in Huh-7 cells. Total RNAs were isolated from Huh-7 cells treated with TM (5, 10, and 20 μg/ml) or Tg (0.5, 1, and 1.5 μM) for 6, 12 and 24 hrs. Fold changes of mRNA are shown by comparing to the vehicle-treated control. Each bar denotes mean ± SEM (n= 3). * p<0.05; ** p<0.01.
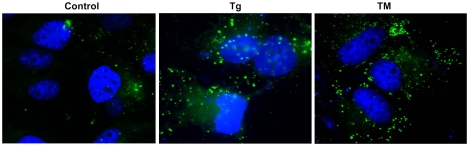
Next, we evaluated the impact of pharmacologic ER stress in de novo hepatic lipogenesis, a key lipid synthesis progress that is tightly regulated by multiple layers of metabolic and stress signals [19, 20]. We examined the production of cytosolic lipid droplets, a major indicator of de novo lipogenesis, in the Huh-7 cells upon TM or Tg challenge. Production of cytosolic lipid droplets, as indicated by Bodipy staining, was significantly increased in the Huh-7 cells after 6 hours of TM or Tg treatment, compared to that after vehicle treatment (Figure 2). This result suggests that pharmacologic ER stress induced by TM or Tg can promote hepatic lipid droplet formation.
Figure 2.
Bodipy staining of lipid droplets in Huh-7 cells. Huh-7 cells were treated with TM (10 μg/ml) or Tg (1 μM) for 6 hrs and then stained with Bodipy for lipid droplets. Magnification: 630 x.
Challenge of TM or TG up-regulates expression of the genes encoding key lipogenic trans-activators
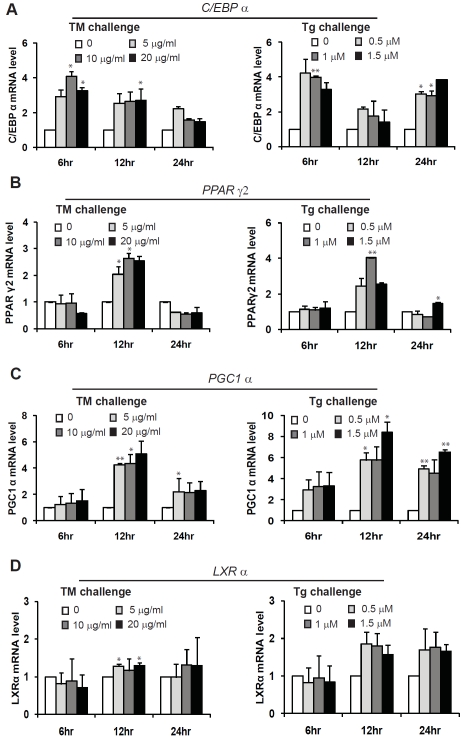
To understand the mechanism by which pharmacologic ER stress promotes lipid droplet formation, we first tested whether TM or Tg can up-regulate trans-activators in de novo lipogenesis. Huh-7 cells were treated with different doses of TM or Tg for a time course from 6, 12, to 24 hours. Quantitative real-time RT-PCR analysis was performed with the TM or Tg-treated Huh-7 cells to determine ER stress-inducible target genes in trans-activation of de novo lipogenesis. Among known lipogenic trans-activators we examined, expression of the genes encoding lipogenic trans-activators including CCAAT/ enhancer binding protein alpha (C/EBPα), per-oxisome proliferator-activated receptor gamma 2 (PPARγ2), PPARγ coactivator 1-alpha (PGC1α), and Liver X receptor alpha (LXRα) was significantly increased in the Huh-7 cells under the treatment of TM or Tg (Figure 3A-D). Although all these genes are inducible by TM or Tg, the expression dynamics of these genes were different upon TM or Tg challenge. Expression of the C/EBPα, LXRα, and PGC1α genes was inducible by TM or Tg in the time windows from 6 to 24 hours post treatment (Figure 3A, C and D). However, expression of the gene encoding PPARγ2 was only inducible at 12 hours after TM or Tg treatment (Figure 3B). Both TM and Tg challenge failed to increase PPARγ2 expression at either early 6 hours or late 24 hours post treatment. It has been documented that PPARγ2, which is usually expressed in adipose tissue, is inducible in steatotic livers, and contributes to increased de novo lipogenesis [19, 21-23]. Our data suggest that pharmacologic ER stress can induce expression of PPARγ2 in hepatocytes that may contribute to ER stress-induced lipogenesis and lipid droplet formation.
Figure 3.
Quantitative real-time RT-PCR analysis of the mRNAs encoding key lipogenic trans-activators, including C/EBPα (A), PPARγ2 (B), PGC1α (C), and LXRα (D), in Huh-7 cells. Total RNAs were isolated from Huh-7 cells treated with TM (5, 10, and 20 μg/ml) or Tg (0.5, 1, and 1.5 μM) for 6, 12 and 24 hrs. Fold changes of mRNA are shown by comparing to the vehicle-treated control. Each bar denotes mean ± SEM (n= 3). * p<0.05; ** p<0.01.
TM or TG treatment promotes expression of genes encoding key enzymes in lipid droplet formation and triglyceride synthesis
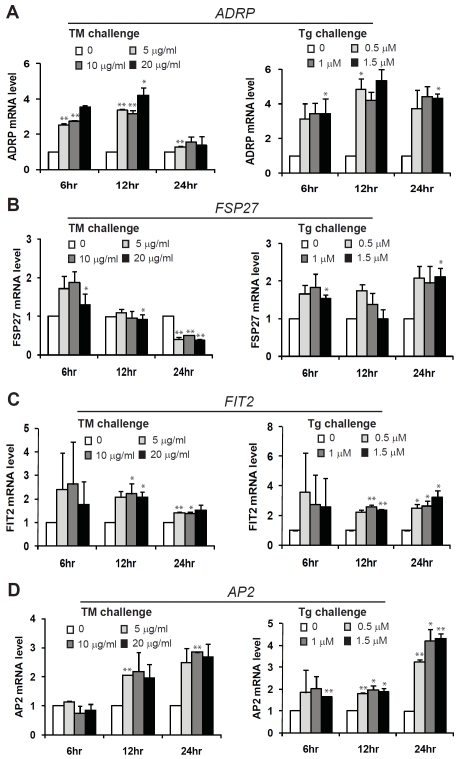
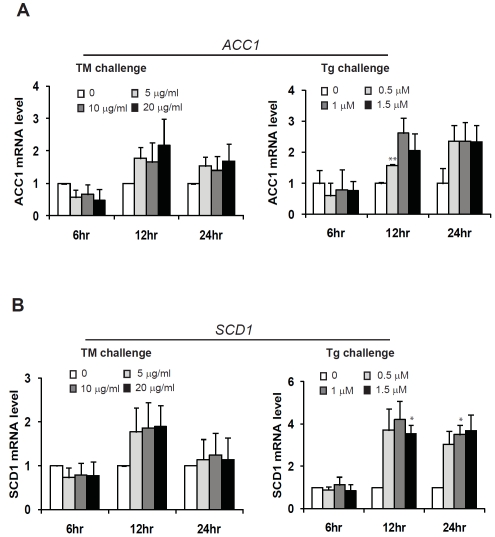
We extended our effort in understanding pharmacologic ER stress-induced hepatic lipogenesis by identifying ER stress-inducible target genes in lipid droplet formation and triglyceride synthesis [19]. Through quantitative real-time RT-PCR analysis, we found that expression of the genes encoding key factors in lipid droplet formation, including fat-specific protein 27 (FSP27), adipose differentiation related protein (ADRP), fat-inducing transcript 2 (FIT2), and adi-pocyte lipid-binding protein (AP2), was increased in Huh-7 cells challenged with TM or Tg (Figure 4). All these genes were inducible by TM or Tg at 6 hours post treatment. However, expression of ADRP and FSP27 was reduced in Huh-7 cells at 24 hours post TM treatment (Figure 4A-B). The expression patterns of ADRP and FSP27 were similar to those of the classic ER stress targets including XBP1, CHOP, and BiP (Figure 1A-C), implying that an ER stress-associated negative feedback regulation may exist for expression of the ADRP and FSP27 genes (Figure 4). In contrast, expression of AP2, a protein factor involved in lipid transport and storage in lipogenesis and lipolysis [24], was increased in response to TM or Tg treatment from 6 to 24 hours (Figure 4D), suggesting a prominent regulation of AP2 gene expression by ER stress. Moreover, we identified two key enzymes required for triglyceride synthesis, ace-tyl-CoA carboxylase 1 (ACC1), and stearoyl-CoA desaturase-1 (SCD1), were inducible by TM or Tg in Huh-7 cells in a dose- and time-dependent manner (Figure 5). Because triglyceride is the core component of lipid droplet, increased expression of key enzymes or protein regulators in triglyceride synthesis and lipid droplet formation may account for ER stress-induced lipid droplet formation.
Figure 4.
Quantitative real-time RT-PCR analysis of the mRNAs encoding protein factors in lipid droplet formation, including ADRP (A), FSP27 (B), FIT2 (C), and AP2 (D), in Huh-7 cells. Total RNAs were isolated from Huh-7 cells treated with TM (5, 10, and 20 μg/ml) or Tg (0.5, 1, and 1.5 μM) for 6, 12 and 24 hrs. Fold changes of mRNA are shown by comparing to the vehicle-treated control. Each bar denotes mean ± SEM (n= 3). * p<0.05; ** p<0.01.
Figure 5.
Quantitative real-time RT-PCR analysis of the mRNAs encoding key enzymes in triglyceride synthesis, including ACC1 (A) and SCD1 (B), in Huh-7 cells. Total RNAs were isolated from Huh-7 cells treated with TM (5, 10, and 20 μg/ml) orTg(O.5, 1, and 1.5 μM) for 6, 12 and 24 hrs. Fold changes of mRNA are shown by comparing to the vehicle-treated control. Each bar denotes mean ± SEM (n= 3). * p<0.05; ** p<0.01.
Activated XBP1 increases expression of PPARγ and C/EBPα in hepatoma cells
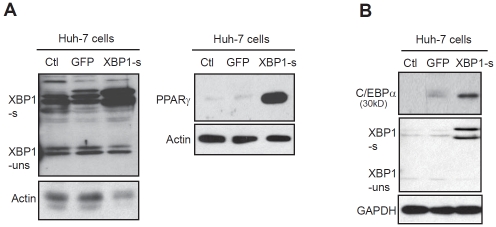
To verify the role of the UPR signaling in regulating expression of lipogenic genes, we forcibly expressed an activated form of human XBP1, an ER stress-inducible transcription factor, in Huh-7 cells by utilizing an adenoviral-based over-expression system. As a control, Huh-7 cells were infected by an adenovirus over-expressing GFP. Under ER stress, the UPR transducer IREα is activated to function as an RNase that splices the mRNA encoding X-box binding protein 1 (XBP1) [25-27]. The spliced XBP1 mRNA, but not the unspliced XBP1 mRNA, encodes an activated transcription factor that potently activates the UPR target genes. Through Western blot analysis, we confirmed that expression levels of the activated form of XBP1 protein (encoded by the spliced XBP1 mRNA), but not the inactivated XBP1 protein, were significantly increased in Huh-7 cells infected by the adenovirus expressing the activated form of human XBP1 (Figure 6A-B). Consistent with the gene expression analysis, Huh-7 cells expressing the activated XBP1 produced much higher levels of PPARγ and C/EBPα, two key lipogenic trans-activators, compared to those expressing GFP (Figure 6A-B). These results confirm the role of the UPR in activating hepatic lipogenesis. It should be noted that over-expression of the activated XBP1 has mild or no effects on activating expression of other lipogenic regulators or enzymes that are inducible by ER stress challenge (data not shown), suggesting that ER stress may regulate the ER stress-inducible lipogenic factors through the other UPR branches. Nevertheless, our study indentified two ER stress-inducible lipogenic trans-activators, C/EBPα and PPARγ, that are under the regulation of the IRE1α/XBP1-mediated UPR pathway.
Figure 6.
Western blot analysis of expression levels of PPARγ2 (A) and C/EBPα (B) proteins in Huh-7 cells. Huh-7 cells were infected with an adenovirus expressing an activated form of human XBP1 protein or GFP control. Levels of (3-actin or GAPDH were determined as a loading control. XBP1-s, the activated form of XBP1 protein encoded by the spliced XBP1 mRNA; XBP1-uns, the inactivated XBP1 protein encoded by the un-spliced XBP1 mRNA.
Discussion
In this study, we demonstrated that pharmacologic ER stress, induced by two structurally-unrelated ER stress-inducing reagents TM and Tg, can promote de novo lipogenesis in the hepatoma cell line Huh-7. Both lipid droplet phenotype and gene expression profile further validated the effect of pharmacologic ER stress in promoting lipogenesis and lipid droplet formation (Figures 2-5). Importantly, we identified three groups of ER stress-inducible regulators and enzymes in de novo lipogenesis (Figures 3-5). In particular, we demonstrated that the ER stress-inducible lipogenic trans-activators, C/EBPα and PPARγ are regulated by the UPR trans -activator XBP1 (Figure 6). These results have important implications in the understanding of the upstream signals that facilitate de novo lipogenesis.
The UPR signaling is an adaptive response that protects cells from ER stress [28]. The UPR signaling mediated through IRE1α/XBP1, ATF6, and PERK/eIF2a reprograms transcription and translation of stressed cells, leading to alterations in cell physiology that helps the stressed cells adapt to ER stress. However, when ER stress gets more severe or prolonged, the same UPR signaling can activate cell death programs to remove the stressed cells. Lipid droplet is a dynamic organelle composed of a monolayer phospholipid embedded with numerous proteins without trans-membrane spanning domains, and a hydrophobic core that contains triglycerides and sterol esters [29]. Under normal physiological conditions, hepatic lipid droplets are important to maintain lipid and energy homeostasis at the cellular and organismal levels. As a defense response to acute liver injuries, accumulation of lipid droplets is increased in the liver of animal models [6, 7, 30]. Our study suggests that the UPR-regulated de novo lipogenesis and accumulation of cytosolic lipid droplets may be parts of the protective response of liver hepatocytes to pharmacologic ER stress. On the other hand, excessive accumulation of lipid droplets is closely associated with the development of metabolic disease [31]. If ER stress-induced lipid droplet accumulation cannot be resolved, prolonged hepatic lipid droplet accumulation may result in metabolic deterioration. This is consistent with the dual roles of the UPR in mediating survival and death signals in the context of cell pathophysiology.
Our work demonstrated that pharmacologic ER stress represents a strong stimulus that triggers de novo lipogenesis and lipid storage. In addition to TM or Tg, many pharmaceutical drugs, for example, clinically-used anti-cancer drug Bortezomib, are strong inducers of pharmacologic ER stress [7, 32, 33]. Although the mechanisms involved in its anticancer activity are still being elucidated, Bortezomib has been shown to cause the accumulation of misfolded proteins in the ER by inhibiting the 26S protea-some activity and subsequent ER-associated protein degradation machinery [34-36]. Previously we demonstrated that Bortezomib induces pharmacologic ER stress, causes hepatic steatosis, and increases hepatotoxicity in an animal model [7]. Our work here confirmed that pharmaceuticals that directly or indirectly induce ER stress in vivo may have side-effects on induction of hepatic steatosis by promoting lipogenesis and lipid deposition.
In summary, our study provides mechanistic evidence that pharmacologic ER stress and its associated UPR signaling can directly regulate hepatic lipid metabolism by stimulating lipogenesis and lipid droplet accumulation. The identification of the ER stress-inducible lipogenic regulators and enzymes provides important insights into the molecular link between ER stress and lipid metabolism. Additional investigations need to be done in the future in order to delineate the regulation of these individual ER stress-inducible targets by the UPR branches. Nevertheless, the findings from this study significantly contribute to our understanding of pathophysiological roles of ER stress and the UPR as well as potential side effects of ER stress-inducing clinically-used drugs.
Acknowledgments
Portions of this work were supported by American Heart Association Grants 0635423Z and 09GRNT2280479 (KZ), National Institutes of Health (NIH) grants DK090313 and ES017829 (KZ), and the Department of Defense Breast Cancer Program grants BC095179P1 (to KZ and ZY).
Glossary
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- TM
tunicamycin
- Tg
thapsi-gargin
- PPARγ
peroxisome proliferator-activated receptor gamma
- C/EBPα
CCAAT/enhancer binding protein alpha
- XBP1
X-box binding protein 1
Conflict of Interest
None.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 4.Dorner AJ, Wasley LC, Kaufman RJ. Increased synthesis of secreted proteins induces expression of glucose regulated proteins in butyrate treated CHO cells. Journal of Biological Chemistry. 1989;264:20602–20607. [PubMed] [Google Scholar]
- 5.Zhang C, Wang G, Zheng Z, Maddipati KR, Zhang X, Dyson G, Williams P, Duncan SA, Kaufman RJ, Zhang K. ER-tethered transcription factor crebh regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress. Hepatology. 2011 doi: 10.1002/hep.24783. doi: 10.1002/hep.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS, Hanson RW, Choi HS, Koo SH. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 13.Laing S, Wang G, Briazova T, Zhang C, Wang A, Zheng Z, Gow A, Chen AF, Rajagopalan S, Chen LC, Sun Q, Zhang K. Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues. Am J Physiol Cell Physiol. 2010;299:C736–749. doi: 10.1152/ajpcell.00529.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, Ueki K, Ozcan U. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 16:429–437. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King IA, Tabiowo A. Effect of tunicamycin on epidermal glycoprotein and glycosaminogly-can synthesis in vitro. Biochem J. 1981;198:331–338. doi: 10.1042/bj1980331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson TR, Patterson SI, Thastrup 0, Hanley MR. A novel tumour promoter, thapsigar-gin, transiently increases cytoplasmic free Ca2+ without generation of inositol phosphates in NG115-401L neuronal cells. Biochem J. 1988;253:81–86. doi: 10.1042/bj2530081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sagara Y, Inesi G. Inhibition of the sar-coplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. J Biol Chem. 1991;266:13503–13506. [PubMed] [Google Scholar]
- 18.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes. Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 19.Musso G, Gambino R, Cassader M. Recent insights into hepatic lipid metabolism in nonalcoholic fatty liver disease (NAFLD) Prog Lipid Res. 2009;48:1–26. doi: 10.1016/j.plipres.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J Clin Invest. 2008;118:829–838. doi: 10.1172/JCI34275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubuquoy L, Dharancy S, Nutten S, Pettersson S, Auwerx J, Desreumaux P. Role of per-oxisome proliferator-activated receptor gamma and retinoid X receptor heterodimer in hepatogastroenterological diseases. Lancet. 2002;360:1410–1418. doi: 10.1016/S0140-6736(02)11395-X. [DOI] [PubMed] [Google Scholar]
- 22.Schadinger SE, Bucher NL, Schreiber BM, Farmer SR. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepato-cytes. Am J Physiol Endocrinol Metab. 2005;288:E1195–1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki T, Shiraishi S, Kishimoto K, Miura S, Ezaki 0. An increase in liver PPARgamma2 is an initial event to induce fatty liver in response to a diet high in butter: PPARgamma2 knockdown improves fatty liver induced by high-saturated fat. J Nutr Biochem. 2010 doi: 10.1016/j.jnutbio.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins: a family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 26.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 27.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 28.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11:R446–449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Jhaveri R, Huang J, Qi Y, Diehl AM. Endoplasmic reticulum stress, hepatocyte CDld and NKT cell abnormalities in murine fatty livers. Lab Invest. 2007;87:927–937. doi: 10.1038/labinvest.3700603. [DOI] [PubMed] [Google Scholar]
- 31.Thomas SE, Dalton LE, Daly ML, Malzer E, Marciniak SJ. Diabetes as a disease of endoplasmic reticulum stress. Diabetes Metab Res Rev. 2010;26:611–621. doi: 10.1002/dmrr.1132. [DOI] [PubMed] [Google Scholar]
- 32.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 33.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R, Siegel D, Orlowski RZ, Kuter D, Limentani SA, Lee S, Hideshima T, Esseltine DL, Kauffman M, Adams J, Schenkein DP, Anderson KC. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 34.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fribley A, Zeng Q, Wang CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24:9695–9704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nawrocki ST, Carew JS, Jr, Dunner K, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]