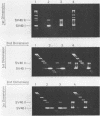
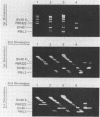
Abstract
A method that provides an easy, rapid, and reproducible way for separating closed circular DNA species from linear DNA and nicked circles is described. The method is based on the difference in mobility of form I (supercoiled) DNA and form II (nicked circles), and the differential mobility of relaxed circular DNA in the presence and absence of ethidium bromide (EtdBr). It can be used for detection or for purification of plasmid, episomal, or viral DNA from the bulk of cellular DNA, or from other DNA mixtures.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3760–3764. doi: 10.1073/pnas.76.8.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. F., Lowy D. R., Dvoretzky I., Howley P. M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci U S A. 1981 May;78(5):2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Mizusawa H., Kakefuda T. Unwinding of double-stranded DNA helix by dehydration. Proc Natl Acad Sci U S A. 1981 May;78(5):2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Modak S. P., Beard P. Analysis of DNA double- and single-strand breaks by two dimensional electrophoresis: action of micrococcal nuclease on chromatin and DNA, and degradation in vivo of lens fiber chromatin. Nucleic Acids Res. 1980 Jun 25;8(12):2665–2678. doi: 10.1093/nar/8.12.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Morgan A. R. The sense of naturally occurring superhelices and the unwinding angle of intercalated ethidium. J Mol Biol. 1975 Jan 5;91(1):1–13. doi: 10.1016/0022-2836(75)90368-x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980 Aug;21(1):103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]