Abstract
Mammalian phosphatidylinositol (PI) has a unique fatty acid composition in that 1-stearoyl-2-arachidonoyl species is predominant. This fatty acid composition is formed through fatty acid remodeling by sequential deacylation and reacylation. We recently identified three Caenorhabditis elegans acyltransferases (ACL-8, ACL-9, and ACL-10) that incorporate stearic acid into the sn-1 position of PI. Mammalian LYCAT, which is the closest homolog of ACL-8, ACL-9, and ACL-10, was originally identified as a lysocardiolipin acyltransferase by an in vitro assay and was subsequently reported to possess acyltransferase activity toward various anionic lysophospholipids. However, the in vivo role of mammalian LYCAT in phospholipid fatty acid metabolism has not been well elucidated. In this study, we generated LYCAT-deficient mice and demonstrated that LYCAT determined the fatty acid composition of PI in vivo. LYCAT-deficient mice were outwardly healthy and fertile. In the mice, stearoyl-CoA acyltransferase activity toward the sn-1 position of PI was reduced, and the fatty acid composition of PI, but not those of other major phospholipids, was altered. Furthermore, expression of mouse LYCAT rescued the phenotype of C. elegans acl-8 acl-9 acl-10 triple mutants. Our data indicate that LYCAT is a determinant of PI molecular species and its function is conserved in C. elegans and mammals.
Keywords: Caenorhabditis elegans, lysophosphatidylinositol acyltransferase, fatty acid remodeling, phospholipid, stearic acid, the sn-1 position, lysocardiolipin acyltransferase, epithelial cell division, gas chromatography-mass spectrometry
Phosphatidylinositol (PI) is a relatively minor component of membrane phospholipids but plays important roles in signal transduction through distinct phosphorylated derivatives of the inositol head group (1, 2). Three-fourths or more of membrane PI obtained from mammalian tissues consist of the 1-stearoyl-2-arachidonoyl (18:0/20:4) species (3, 4), which is thought to be formed by a fatty acid remodeling reaction after the de novo synthesis of PI (5–10). The remodeling reaction involves the hydrolysis of a fatty acyl ester bond at the sn-1 or sn-2 position of the newly synthesized PI and subsequent incorporation of the appropriate fatty acid into the position. In an RNA interference (RNAi)-based genetic screen using Caenorhabditis elegans, we identified mboa-7/LPIAT1 as an acyltransferase that selectively incorporates arachidonic acid into the sn-2 position of PI (11). More recently, we demonstrated that C. elegans acl-8, acl-9, and acl-10, which show significant sequence homology to each other, encode acyltransferases that incorporate stearic acid (18:0) into the sn-1 position of PI (12). Stearic acid attached at the sn-1 position of PI was replaced with cis-vaccenic acid (18:1n-7) in acl-8 acl-9 acl-10 triple mutants. The gene product of acl-10, the predominant acyltransferase among acl-8, acl-9, and acl-10, incorporates stearic acid into the sn-1 position of PI in vitro. acl-8 acl-9 acl-10 triple mutants were defective in the asymmetric cell division of epithelial cells. We also showed that acl-8, acl-9, and acl-10 function in the same pathway with ipla-1 (13), a phospholipase A1 that hydrolyzes the fatty acyl chain of PI (12). C. elegans ipla-1 mutants have fatty acid compositions of PI similar to acl-8 acl-9 acl-10 triple mutants. ipla-1 mutants show epithelial cell defects similar to acl-8 acl-9 acl-10 triple mutants. No synergism was observed between the ipla-1 and acl-8 acl-9 acl-10 mutations. These data support a model in which IPLA-1, the gene product of ipla-1, produces the sn-2-acyl lysoPI, which is subsequently reacylated with stearic acid by ACL-8, ACL-9, and ACL-10, gene products of acl-8, acl-9, and acl-10, respectively, in the fatty acid remodeling of the sn-1 position of PI.
ACL-8, ACL-9, and ACL-10 belong to the 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) family (14) (supplementary Table I), in which the members share four conserved AGPAT motifs that are involved in substrate binding and catalysis (15, 16) (supplementary , motif I-IV). In mammals, the AGPAT family consists of at least 16 members. Among these members, lysocardiolipin acyltransferase (LYCAT), also known as LCLAT1 and ALCAT1, is the closest homolog of C. elegans ACL-8, ACL-9, and ACL-10 (supplementary Table I) (14). ACL-8, ACL-9, ACL-10 and mammalian LYCAT possess highly conserved amino acids in the AGPAT motifs, which are unique in LYCAT/ACL-8, -9, -10 subfamily members, but not in other AGPAT family members (supplementary Fig. 1B, amino acids indicated in blue). Acyltransferases with these highly conserved amino acids are evolutionarily conserved in various species including human, zebrafish, and C. elegans, but not in yeast.
Mammalian LYCAT was originally identified as a lysocardiolipin (lysoCL) acyltransferase by an in vitro enzyme assay (17). Thereafter, it has been shown that mammalian LYCAT possesses acyltransferase activity toward the sn-2 position of other anionic lysophospholipids including lysoPI, and lysophosphatidylglycerol (lysoPG) in addition to lysoCL in vitro (18–20). Because C. elegans homologs of LYCAT specifically determines the fatty acyl chain at the sn-1 position of PI in vivo (12), here we examined if LYCAT determines the fatty acid composition of PI in mammals. We generated LYCAT-deficient mice, analyzed the fatty acid composition of phospholipids in the tissues, and found that LYCAT deficiency markedly altered the fatty acid composition of PI, but not that of other phospholipids, in all tissues analyzed. We propose that LYCAT determines the fatty acid composition of PI via fatty acid remodeling and that its function is evolutionarily conserved in C. elegans and mammals.
MATERIALS AND METHODS
Materials
PI and lysoPI from bovine liver, dioleoyl phosphatidylcholine (PC), dioleoyl phosphatidylethanolamine (PE), and 1-palmitoyl-2-oleoyl phosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, AL). Phosphatidylglycerol (PG) from egg yolk was purchased from Sigma-Aldrich (St. Louis, MO). 1,2-Dipalmitoyl PI was purchased from Serdary Research Laboratories (London, ON, Canada). 1,2-Dipalmitoyl phosphatidylinositol monophosphates (PIP1), 1,2-dipalmitoyl phosphatidylinositol bisphosphates (PIP2), and 1,2-dioctanoyl PIP2 were purchased from Cayman Chemical (Ann Arbor, MI). [1-14C]stearoyl-CoA and [1-14C]arachidonoyl-CoA were purchased from American Radiolabeled Chemicals (St. Louis, MO). Rhizopus arrhizus lipase and phospholipase A2 from honey bee venom were purchased from Sigma-Aldrich. DEAE column was purchased from Wako Pure Chemical Industries (Osaka, Japan).
Worm strains
General methods for maintaining C. elegans are described by Brenner (21). The orientation of seam cell division and seam cell lineages were analyzed as previously described (13). The following mutations and transgenes were used: acl-8 acl-9(tm2290), acl-10(tm1045), wIs51[scm::gfp], xhEx3521[dpy-7p::mouse LYCAT; Pges-1::dsREDm] (12).
Preparation of sn-2-acyl lysophospholipids
Each sn-2-acyl-1-lysophospholipid (sn-2-acyl lysophospholipids) was prepared as follows using dioleoyl PC, dioleoyl PE, 1-palmitoyl-2-oleoyl PS, bovine liver PI, or egg yolk PG. Each phospholipid (1 µmol) dissolved in 1 ml of diethyl ether was incubated with 100 µl of 100 mM CaCl2, 600 µl of 50 mM Tris-malate, pH 5.7, and 400 µl of enzyme solution containing 15 mg of Rhizopus arrhizus lipase for 1 h at room temperature while stirring vigorously. After the incubation, the reaction was terminated by adding 1 ml of methanol. Remaining phospholipids and liberated fatty acids were removed by three extractions with 4 ml of diethyl ether-petroleum ether (1:1, v/v). sn-2-acyl lysophospholipid present in the lower layer was extracted by the method of Bligh and Dyer (22) and further purified by TLC on silica gel 60 plates (Merck Biosciences, Darmstadt, Germany) in chloroform-methanol-acetic acid (65:25:13, v/v). The area of silica gel corresponding to sn-2-acyl lysophospholipid was scraped off the plates. sn-2-acyl lysophospholipid was reextracted by methanol and immediately used for acyltransferase assays.
Acyltransferase assay
Mouse tissues were pulverized under liquid nitrogen and homogenized in quadruple volumes (w/v) of SET buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 250 mM sucrose) with protease inhibitors (5 μg/ml pepstatin, leupeptin, and aprotinin). After centrifugation at 2,330 g for 20 min at 4°C, the resulting supernatant was further centrifuged at 105,000 g for 60 min. The resulting pellet (microsomal fraction) was resuspended in homogenizing buffer (50 mM potassium phosphate buffer (pH 7.0) containing 0.15 M KCl, 0.25 M sucrose) and used for the enzyme assay. Acyl-CoA:sn-2-acyl lysophospholipid acyltransferase assay and acyl-CoA:sn-1-acyl lysophospholipid acyltransferase assay were performed essentially as described previously (12), except that the amount of microsomal protein was 10 µg and the incubation temperature was 37°C. Because sn-2-acyl lysophospholipid is known to easily isomerize to sn-1-acyl lysophospholipid, it is possible that the acyl donor is incorporated into sn-1-acyl lysophospholipid and sn-2-acyl lysophospholipid. Thus, an accurate measure of sn-2-acyl lysophospholipid acyltransferase activity could only be obtained by determining the position that had been acylated. To check the positional specificity, the radiolabeled product was treated with bee venom phospholipase A2 (supplementary Fig. II). The distribution of radioactivity between the resultant sn-1-acyl lysophospholipid and free fatty acid was assessed after TLC (12).
Generation of LYCAT-deficient mice
Mouse LYCAT genomic DNA sequences were cloned from a C57BL/6-derived genomic DNA and subcloned into the pPNT vector. The targeting vector contains a PGK-neo cassette with 2.3 kb of LYCAT homologous regions upstream of exon 4 and with 5.9 kb homologous regions downstream of exon 4. The targeting vector was linearized and electroporated into RENKA ES cell lines (TransGenic Inc., Kumamoto, Japan) derived from the C57BL/6 mouse substrain. Stable clones were selected for G418 resistance. Out of 569 G418-resistant clones, three clones were correctly targeted, as confirmed homologous recombination by Southern blot analysis with 5′, 3′, and neo probes. Blastocysts obtained by aggregation methods of ICR morula embryos and three different ES cells to recipient uterus as after 3 days of pseudopregnancy. Chimeric males were mated to C57BL/6 females and offspring animals (F1) were genotyped by Southern blot analysis. F2 homozygous mutant mice were then generated by intercrossing F1 heterozygous males and females. Mice were genotyped by PCR analyses of genomic DNA isolated from tail biopsies. The following primers were used for genotyping: primer 1, 5′-GTG GTA CTG CAC ACC TTT GAT CC-3′; primer 2, 5′-CAA AAT GGC TCT TAT CAT CCT TCC-3′; and primer 3, 5′-GAA CAG AGT ACC TAC ATT TTG AAT GG-3′. The wild-type allele is detected with primers 1 and 2, yielding a 356 bp product, whereas the mutant allele is detected with primers 1 and 3, yielding a 498 bp fragment. The animals used were LYCAT+/+ (wild-type) and LYCAT−/− (homozygous) mice in the same C57BL/6 background between 13 and 20 weeks old. The mice were kept in a temperature-controlled environment with a 12 h light and 12 h dark cycle and received a standard diet and water ad libitum.
Generation of monoclonal antibodies against mouse LYCAT
A recombinant mouse LYCAT (amino acid numbers 78–298 of mouse LYCAT, GenBank accession number NM_001081071) that was expressed and purified by Escherichia coli pCold TF expression system (TaKaRa, Japan) was injected into the hind foot pads of WKY/Izm rat strain by using Freund's complete adjuvant. The enlarged medial iliac lymph nodes were used for cell fusion with mouse myeloma cells, PAI. In the present study, the established monoclonal antibody, named YN1, was used for Western blotting and immunocytochemistry at 1:2,000 and 1:100 dilutions, respectively.
Western blot
Murine tissues were homogenized in quadruple volumes (w/v) of SET buffer with protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 2 µg/ml pepstatin, 2 µg/ml leupeptin, 2 µg/ml aprotinin). After centrifugation at 1,000 g for 10 min at 4°C, the supernatants were used as the total protein extracts. The protein concentrations of samples were determined by the bicinchoninic acid (BCA) assay (Pierce). Each sample (20 µg protein/lane) was subjected to SDS-PAGE and immunoblotting. The following primary antibodies were used: anti-mouse LYCAT monoclonal antibody, anti-mouse LPIAT1 monoclonal antibody, and anti-GAPDH monoclonal antibody (6C5, Calbiochem).
Phospholipid analysis
Lipids of each tissue were extracted by the method of Bligh and Dyer (22). Phospholipids were separated from total lipids by one-dimensional TLC on silica gel 60 plates in chloroform-methanol-acetic acid (65:25:13, v/v). The area of silica gel corresponding to each phospholipid (PC, PE, PG, CL, and PI+PS) was scraped off the plates. The PI+PS fraction was reextracted, separated by TLC in chloroform-methanol-formic acid-water (60:30:7:3, v/v), and the areas of silica gel corresponding to PI and PS were scraped off the plates. Isolated phospholipids were methylated with 2.5% H2SO4 in methanol. The resulting fatty acid methyl esters were then extracted with hexane and subjected to GC-MS analysis as described previously (23). Liquid chromatography-electrospray ionization mass spectrometry (LC-ESIMS) analysis was performed as described previously (12) using 1,2-dipalmitoyl PI (16:0/16:0-PI), as internal standards. Phospholipids in the mouse liver were quantified as follows. Each phospholipid (PC, PE, PI, PS, PG, and CL) was separated from total lipids by TLC as described above. Each lipid spot on the TLC plate was scraped off the plate, and the spots of each phospholipid were then extracted from silica gel by chloroform-methanol-water (1:2:0.8, v/v). The amount of inorganic phosphate in each phospholipid was quantified as described previously (24).
Immunocytochemistry
LYCAT+/+ or LYCAT−/− mouse embryonic fibroblasts (MEFs) were grown on coverslips for 24 h. To achieve mitochondrial staining, MEFs were incubated in the growth medium with MitoTracker Red CM-H2XMRos (Molecular Probes) and fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with PBS, MEFs were permeabilized for 10 min with 0.5% Triton X-100 in PBS. MEFs were then incubated with the anti-mouse LYCAT antibody YN1. As for endoplasmic reticulum (ER) staining, fixed and permeabilized cells were incubated with the rabbit anti-calnexin polyclonal antibody (1:500) (StressGen Biotechnologies, Victoria, BC, Canada) and YN1. Confocal images were obtained using a Zeiss LSM510META confocal microscope system (Carl Zeiss MicroImaging, Thornwood, NY).
Blood biochemistry and measurement of plasma and liver lipid levels
Serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured using a Transaminase CII-test Wako kit (Wako Pure Chemical Industries, Osaka, Japan). Plasma triglyceride and cholesterol levels were measured using a Triglyceride E-test Wako kit and cholesterol E-test Wako kit (Wako Pure Chemical Industries, Osaka, Japan), respectively. To measure hepatic triglyceride contents, livers were homogenized and lipids were extracted by the method of Bligh and Dyer (22) and then suspended in a buffer containing Triton-X100. The triglyceride content was measured using a Triglyceride E-test Wako kit.
Analysis of phosphoinositide molecular species
Livers were weighed and homogenized in 1 ml of HBSS. After addition of 100 pmol of 1,2-dipalmitoyl PIP1 (16:0/16:0-PIP1), 1,2-dipalmitoyl PIP2 (16:0/16:0-PIP2), the homogenates were mixed with 2 ml of methanol. Then, 2 μl of 1 nmol/μl 1,2-dioctanoyl PIP2 (8:0/8:0-PIP2), 1 ml of 2 N HCl, 1 ml of water, 0.3 ml of 1 M NaCl, and 2 ml of chloroform were added, followed by shaking. After centrifugation, the lower layer was collected and 1.5 ml of methanol was added to each sample prior to application to a DEAE column. Column-bound lipids were washed with chloroform-methanol (1:1, v/v) (3 ml) and chloroform-methanol-28% aqueous ammonia-acetic acid (200:100:3:0.9, v/v) (3 ml) in series. Phosphoinositides were then eluted with chloroform-methanol-HCl-water (12:12:1:1, v/v) (1.5 ml). After addition of 0.75 ml of water and 0.1 ml of 1 M NaCl, the solution was shaken and centrifuged to collect the lower layer. After addition of 50 μl of 2 M TMS-diazomethane in hexane, samples were incubated for 10 min at room temperature (25). Then, 10 μl of glacial acetic acid and 0.7 ml of wash solution [chloroform-methanol-water (48:49:3, v/v)] were added to each solution. The solution was shaken and centrifuged to collect the lower layer. Each sample was dried under nitrogen gas and was redissolved in 24 μl of methanol-70% ethylamine (100:0.065, v/v), followed by addition of 8 μl of 1 M ammonium bicarbonate.
The LC-ESIMS/MS analysis was performed by using a TSQ-Vantage (Thermo-Fisher Scientific) with an UltiMate 3000 LC system (Thermo-Fisher Scientific, Waltham, MA) combined with an HTC PAL autosampler (CTC Analytics, Zwingen, Switzerland). The phosphoinositides fractions were separated by a step gradient with mobile phase A (methanol-water-70% ethylamine (20:80:0.13, v/v))/mobile phase B [methanol-water-isopropanol-70% ethylamine (5:5:90:0.13, v/v)] ratios of 90%/10% (0 min), 70%/30% (0–1 min), 10%/90% (1–3 min), 10%/90% (3–15 min), 90%/10% (15–16 min), and 90%/10% (16–22 min). Flow rate was 30 μl/min and the chromatography was performed at room temperature using Waters X-Bridge C8 (3.5 μm, 150 mm × 1.0 mm i.d.) column. Phosphoinositides molecular species were measured by the selected reaction monitoring (SRM) in positive ion mode. The characteristic fragmentation patterns of individual phospholipids were determined by the product ion scan (MS/MS mode). Each molecular species was identified by the LC retention time.
RESULTS
Mouse LYCAT is a functional homolog of C. elegans acl-8, acl-9, and acl-10
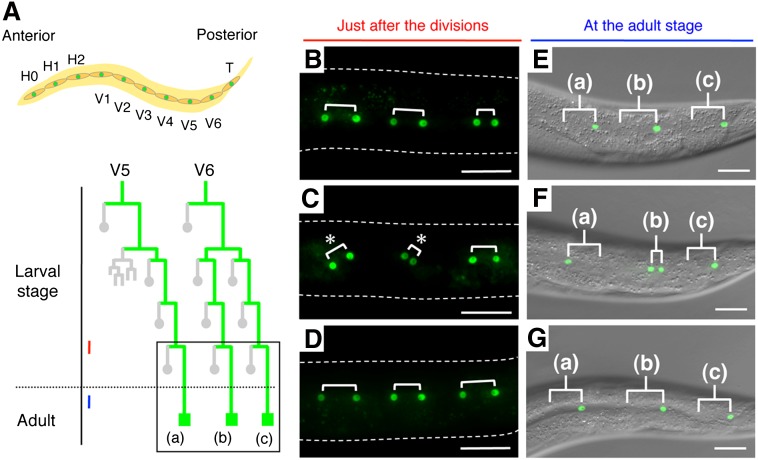
We previously reported that C. elegans acl-8 acl-9 acl-10 triple mutants are defective in the divisions of specialized epithelial cells called seam cells (12). In the wild-type, 10 seam cells (H0-2, V1-6 and T in Fig. 1A) are present on each side of a newly hatched worm and divide asymmetrically in a stem cell-like manner during the larval stage. Both daughter cells just after the division express a seam cell marker, scm::gfp (Fig. 1B). Then, an anterior daughter cell fuses with a major epithelial syncytium and loses the expression of scm::gfp, and a posterior daughter cell assumes the seam cell fate again and continues to express scm::gfp (Fig. 1E). Seam cell division is oriented parallel to the anterior-posterior axis (A-P axis) as judged by scm::gfp, which is expressed in the nuclei of both daughter cells (Fig. 1B). In acl-8 acl-9 acl-10 triple mutants, however, the seam cell division is randomly oriented relative to the A-P axis (Fig. 1C) and the cell-fate determination became symmetric (both daughter cells adopted the seam cell fate) or was reversed (the fates of daughter cells were the opposite of what they are in the wild-type) (Fig. 1F). To determine whether mammalian LYCAT can substitute for acl-8, acl-9, and acl-10 in C. elegans, we expressed mouse LYCAT under the control of the epithelial-specific dpy-7 promoter in acl-8 acl-9 acl-10 triple mutants. Expression of mouse LYCAT restored the parallel orientation of seam cell divisions in acl-8 acl-9 acl-10 triple mutants (Fig. 1D). The angle between the A-P axis and the direction of cell division fell within 10 degrees in most seam cells in LYCAT-expressing mutants. Expression of mouse LYCAT also effectively rescued the defects in cell-fate determination of the daughter cells in acl-8 acl-9 acl-10 triple mutants (Fig. 1G). These results indicate that LYCAT is a functional homolog of C. elegans acl-8, acl-9, and acl-10.
Fig. 1.
Suppression of acl-8 acl-9 acl-10 triple mutant phenotypes by expression of mouse LYCAT. Anterior is toward the left. A, top: Schematic arrangement of seam cells on each side in a newly hatched worm. The ten seam cells are initially present in a line that extends from head to tail along each side of the worm. A, bottom: The postembryonic division pattern of V5 and V6 seam cells. The expression pattern of scm::gfp, which is specifically expressed in the nuclei of seam cells, is indicated in green lines. Gray circles represent anterior daughter cells that differentiate and lose the seam cell fate, and green squares denote seam cells. The box marks the lineages analyzed in this study: V5. pppp lineage (a), V6. papp lineage (b) and V6. pppp lineage (c). Red and blue lines indicate the developmental stages corresponding to those of (B–D) and (E–G), respectively. B–D: Fluorescent images of scm::gfp just after the seam cell divisions. The shapes of the worms are indicated by dotted lines. scm::gfp is expressed in the nuclei of both daughter cells just after the division, indicating the orientation of seam cell division relative to the anterior-posterior axis (A-P axis). Three pairs of daughter cells are shown with brackets. In wild-type worms, all seam cells divide parallel to the A-P axis (B). In acl-8 acl-9 acl-10 triple mutants, the seam cell division is abnormally oriented relative to the A-P axis (C; asterisks). The parallel orientation of seam cell divisions is restored in acl-8 acl-9 acl-10 triple mutants expressing mouse LYCAT under the control of the epithelial-specific dpy-7 promoter (D). E–G: Seam cells at the adult stage visualized by scm::gfp. Merged fluorescence and differential interference contrast images are shown. The letters (a), (b) and (c) correspond to those of (A). E: Wild-type. The anterior daughter cells lose the expression of scm::gfp, and the posterior daughter cells adopt the seam cell fate again and continue to express scm::gfp in all three lineages. In acl-8 acl-9 acl-10 triple mutants, the seam cell division becomes symmetric (both daughter cells adopted the seam cell fate) or is reversed (the fates of daughter cells are the opposite of what they are in the wild-type) (F), and the defects in asymmetric cell-fate determination are rescued by expression of mouse LYCAT (G). Scale bars are 20 μm.
LYCAT deficiency causes reduced acyltransferase activity toward the sn-1 position of PI and PG in mouse liver
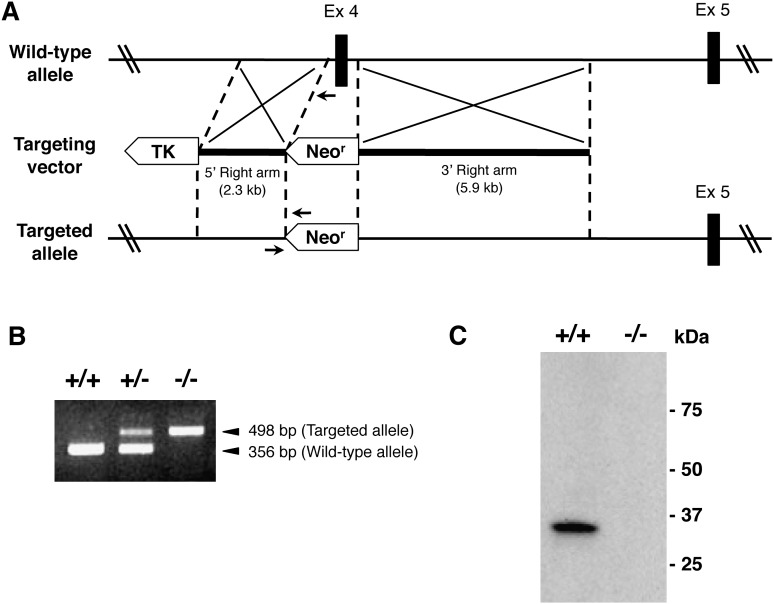
We generated LYCAT-deficient mice by gene targeting. A LYCAT-targeting construct was designed to replace exon 4 encoding the acyltransferase motifs II and III that are conserved in the AGPAT family with a neomycin cassette (Fig. 2A, B) (15, 16). LYCAT protein was not detected in the liver of LYCAT−/− mice (Fig. 2C). LYCAT−/− mice were viable and fertile. They were outwardly healthy and had normal litter sizes. No histological abnormalities were detected in various organs from LYCAT−/− mice by light microscopy. There was no significant change in plasma triglyceride or cholesterol levels (supplementary Fig. III A) or hepatic triglyceride content (supplementary Fig. III B) in LYCAT−/− mice. Moreover, no significant changes were observed in serum markers of liver injury (AST and ALT) (supplementary Fig. III C) or the number of blood cells (leukocytes, platelets, and erythrocytes) (data not shown).
Fig. 2.
Generation of LYCAT-deficient (LYCAT−/−) mice. A: Schematic representation of the LYCAT gene-targeting strategy. Homologous recombination of the targeting vector with the wild-type allele results in the replacement of 0.5-kilobase of the LYCAT genomic sequence containing exon 4 with the neo cassette, deleting the AGPAT motifs II and III that are essential for acyltransferase activity in AGPAT family members (15, 16). A thymidine kinase gene (TK) was used for negative selection of clones with random integration of the targeting vector. B: PCR-based genotyping of genomic DNA from LYCAT+/+, LYCAT+/− and LYCAT−/− mice. The orientation and location of PCR primers are indicated with arrows in (A). C: Western blot analysis using an anti-mouse LYCAT antibody, YN1. Total protein extracts of liver from LYCAT+/+ and LYCAT−/− mice were subjected to SDS-PAGE, blotted onto a PVDF membrane, and stained with YN1.
C. elegans acl-10 shows acyltransferase activity that incorporates stearic acid into the sn-1 position of PI in an in vitro assay (12). To assess the contribution of LYCAT to the acyltransferase activity toward the sn-1 position of phospholipids, we measured [14C]stearoyl-CoA:sn-2-acyl lysophospholipid acyltransferase activity using the liver membrane fraction of LYCAT−/− mice. Because sn-2-acyl lysophospholipid is known to easily isomerize to sn-1-acyl lysophospholipid, it is possible that the [14C]acyl donor is incorporated into both sn-1-acyl lysophospholipid and sn-2-acyl lysophospholipid (supplementary Fig. II). To check the positional specificity, [14C]phospholipid produced after the incubation was treated again with phospholipase A2 (for details, see Materials and Methods). The amount of radioactivity of the resultant lysophospholipid was taken as a measure of acylation at the sn-1 position (sn-2-acyl lysophospholipid acyltransferase activity) and the amount of radioactivity of the free fatty acid was taken as a measure of acylation at the sn-2 position (sn-1-acyl lysophospholipid acyltransferase activity). The sn-1- and sn-2-acyl lysophospholipid acyltransferase activities toward various lysophospholipids in the liver microsomes from wild-type and LYCAT−/− mice are summarized in Table 1. At least 90% of [14C]stearoyl-CoA was incorporated into the sn-1 position of all the lysophospholipids tested (Table 1), indicating that sn-2-acyl lysophospholipid acyltransferase activity was predominant under the present assay conditions. In the liver from LYCAT−/− mice, sn-2-acyl lysoPI acyltransferase (LPIAT) activity was reduced by 64% (from 8.10 ± 0.11 to 2.92 ± 0.22 nmol/min/mg protein) and lysoPG acyltransferase (LPGAT) activity was reduced by 56% (from 10.45 ± 0.05 to 4.57 ± 0.17 nmol/min/mg protein). The sn-2-acyl lysoPE acyltransferase (LPEAT) activity was also reduced, but only slightly. In contrast, sn-2-acyl lysoPC acyltransferase (LPCAT) and lysoPS acyltransferase (LPSAT) activities were unchanged in the liver from LYCAT−/− mice. These results indicate that LYCAT significantly contributes to acyltransferase activity toward the sn-1 position of PI and PG in mouse liver.
TABLE 1.
Acylation of sn-2-acyl lysophospholipid preparations by [14C]stearoyl-CoA in mice liver
| Acyltransferase activity (nmol/min/mg protein) | ||
| Acyl acceptors | sn-1 positiona(sn-2-acyl lysophospholipid acyltransferase activity) | sn-2 positionb(sn-1-acyl lysophospholipid acyltransferase activity) |
| lysoPC | ||
| LYCAT+/+ | 9.87 ± 0.94 | 0.74 ± 0.01 |
| LYCAT−/− | 10.66 ± 0.78 | 0.85 ± 0.05 |
| lysoPE | ||
| LYCAT+/+ | 48.42 ± 1.74 | 2.27 ± 0.03 |
| LYCAT−/− | 41.54 ± 0.09 | 2.15 ± 0.01 |
| lysoPI | ||
| LYCAT+/+ | 8.10 ± 0.11 | 0.15 ± 0.00 |
| LYCAT−/− | 2.92 ± 0.22c | 0.10 ± 0.00c |
| lysoPS | ||
| LYCAT+/+ | 8.62 ± 0.13 | 0.22 ± 0.04 |
| LYCAT−/− | 7.80 ± 0.27 | 0.19 ± 0.00 |
| lysoPG | ||
| LYCAT+/+ | 10.45 ± 0.05 | 0.60 ± 0.07 |
| LYCAT−/− | 4.57 ± 0.17c | 0.26 ± 0.03c |
The acyltransferase reaction mixtures contained 80 µM sn-2-acyl lysophospholipid, 12.5 µM [14C]stearoyl-CoA, and 10 µg of liver microsomal protein in a total volume of 0.8 ml assay buffer. Each 14C-labeled phospholipid produced after the incubation was treated again with phospholipase A2. The distribution of radioactivity between the resultant lysophospholipid and free fatty acid was taken as a measure of acylation at the sn-1 position (sn-2-acyl lysophospholipid acyltransferase activity) (a) and the sn-2 position (sn-1-acyl lysophospholipid acyltransferase activity) (b), respectively. Data represent the mean ± SEM of triplicate measurements. cP<0.01 versus LYCAY+/+.
Furthermore, sn-1-acyl LPIAT activity was reduced by 33% (from 0.15 ± 0.00 to 0.10 ± 0.00 nmol/min/mg protein) and LPGAT activity was reduced by 57% (from 0.60 ± 0.07 to 0.26 ± 0.03 nmol/min/mg protein) (Table 1). This observation is consistent with the previous data that small interfering RNA (siRNA)-mediated knockdown of LYCAT in HeLa cells results in reductions of acyltransferase activities toward the sn-2 position of PI and PG (20). sn-1-acyl LPCAT, LPEAT, and LPSAT activities were not altered significantly in the LYCAT−/− liver (Table 1).
Reduction of acyltransferase activity toward the sn-1 position of PI in various tissues of LYCAT−/− mice
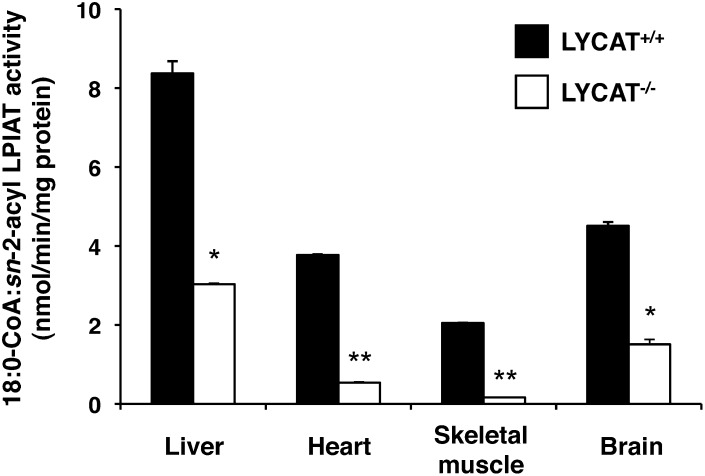
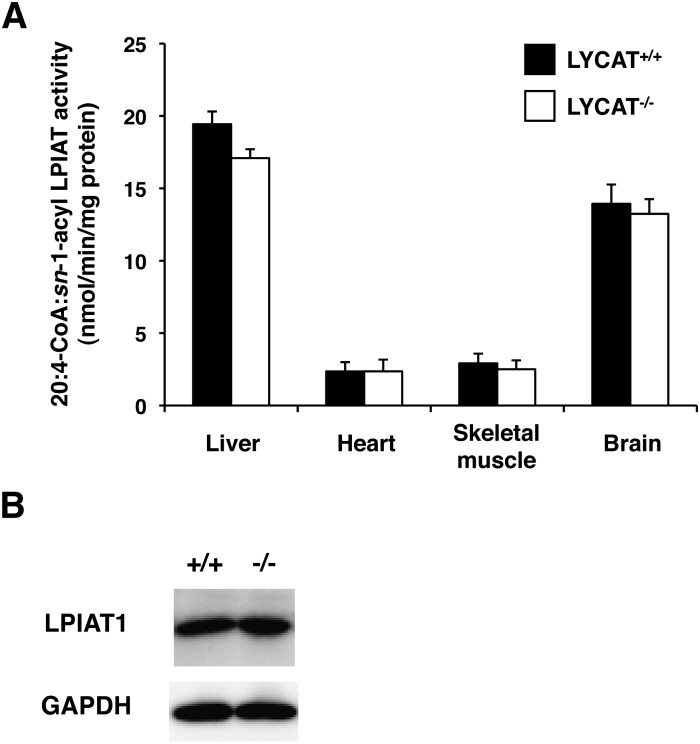
As described above, LYCAT was largely responsible for the acyltransferase activity that incorporates stearic acid into the sn-1 position of PI in the liver (Table 1). We also examined acyltransferase activity toward the sn-1 position of PI in other tissues such as heart, skeletal muscle, and brain. As shown in Fig. 3, a significant portion of the activity was decreased in these tissues from LYCAT−/− mice. Especially in the heart and skeletal muscle, LYCAT was the main acyltransferase that incorporates stearic acid into the sn-1 position of PI.
Fig. 3.
Acyltransferase activity that incorporates stearic acid into the sn-1 position of PI in the membrane fractions from liver, heart, skeletal muscle, and brain. [14C]stearoyl-CoA and sn-2-acyl lysoPI were used as the acyl donor and acceptor, respectively. *P < 0.01, **P < 0.001. Data represent the mean ± SEM of triplicate measurements.
LYCAT deficiency causes a major change in the fatty acid composition of PI
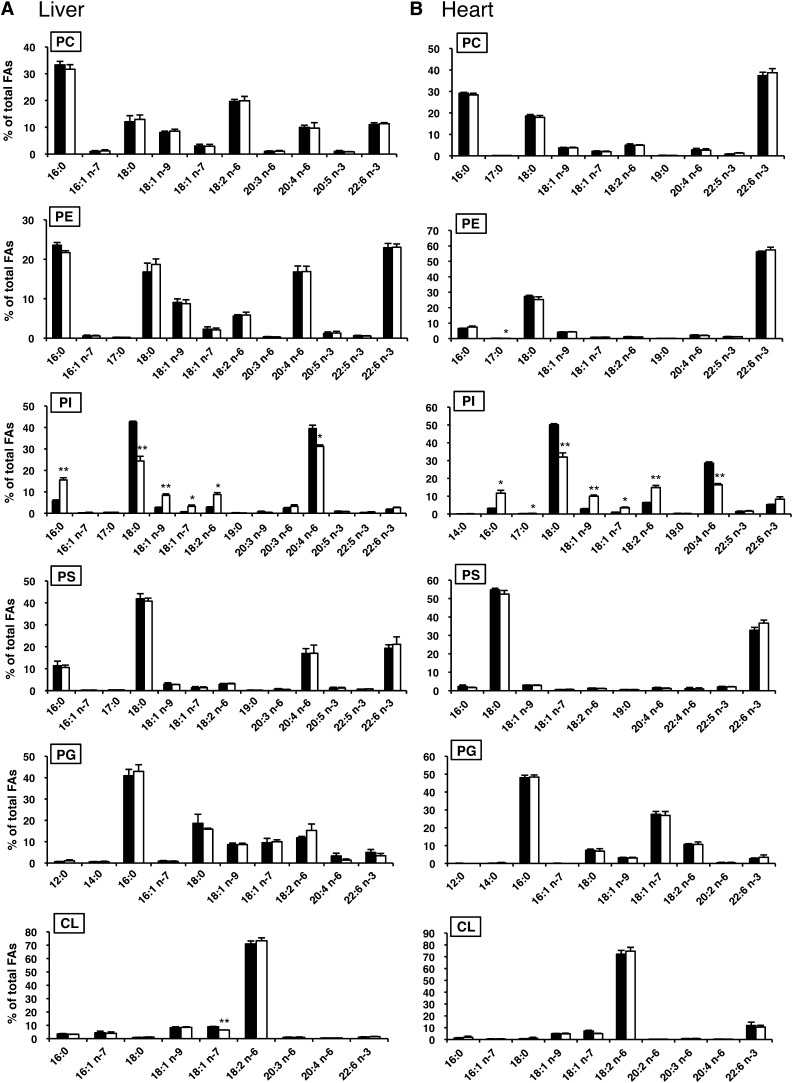
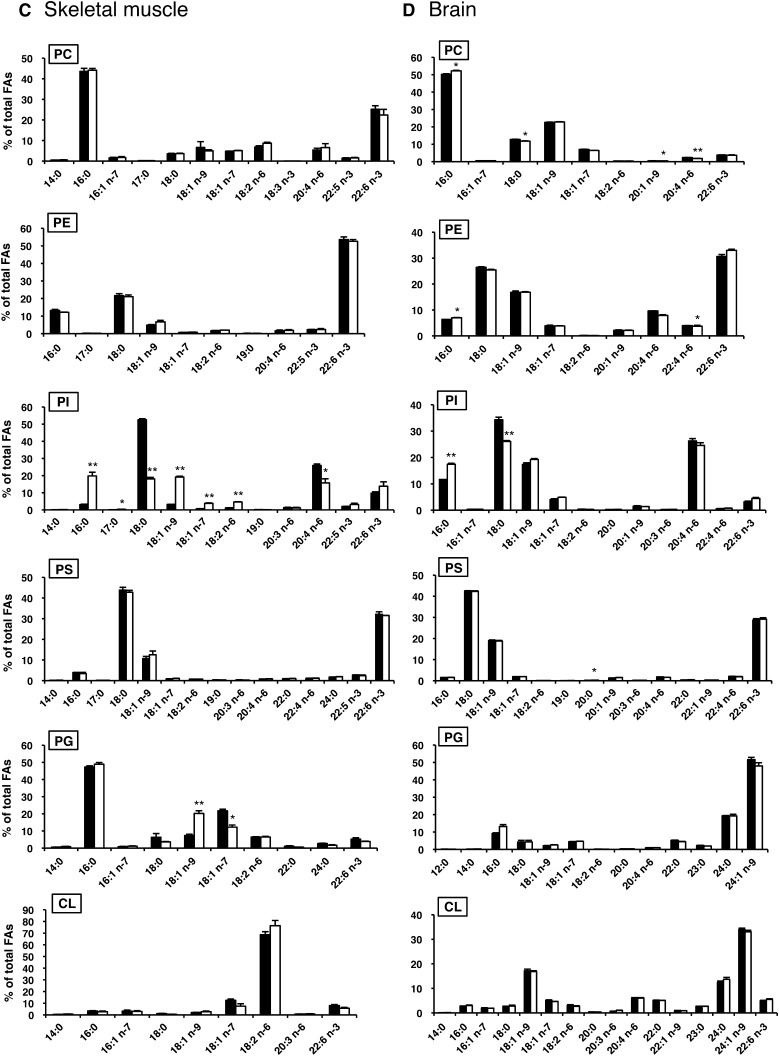
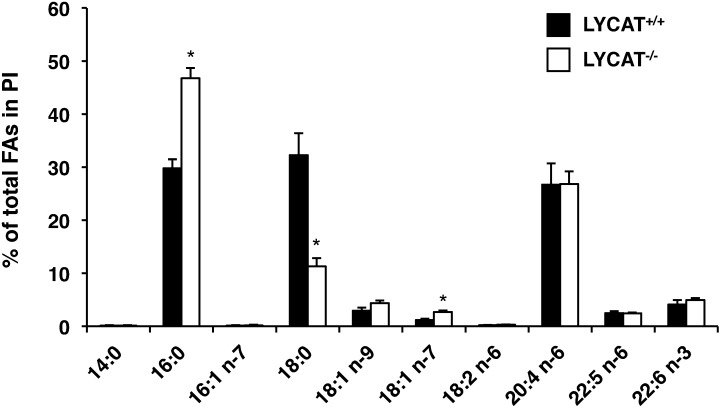
To determine the role of LYCAT in phospholipid metabolism in vivo, we analyzed the fatty acid composition of phospholipids in tissues from LYCAT−/− mice. GC-MS analysis revealed that LYCAT deficiency significantly altered the fatty acid composition of PI in all the tissues examined including liver, heart, skeletal muscle, and brain (Fig. 4). In LYCAT−/− mice, the amounts of stearic acid in PI of the liver, heart, and skeletal muscle were reduced by 42%, 36%, and 66% compared with the amounts in wild-type mice, respectively (Fig. 4A–C). The amount of arachidonic acid, which is a major fatty acid in the sn-2 position of PI, was also reduced in these tissues, though the degree of reduction was less than that of stearic acid. Conversely, the amounts of palmitic acid (16:0), oleic acid (18:1n-9), cis-vaccenic acid (18:1n-7), and linoleic acid (18:2n-6) in PI in these tissues increased. In the brains of LYCAT−/− mice, the change in the fatty acid composition of PI was milder than that in the liver, heart, and skeletal muscle, and the amount of arachidonic acid in PI was not affected (Fig. 4D). Similarly, the amount of arachidonic acid in PI was not affected in the testis of LYCAT−/− mice (Fig. 5). Meanwhile, the amount of stearic acid in PI of the testis was reduced by 66% and the amounts of palmitic acid (16:0) and cis-vaccenic acid (18:1n-7) in PI increased. In contrast to the remarkable change in the fatty acyl chains of PI, no drastic changes were observed in the fatty acid composition of other phospholipids such as PC, PE, PS, PG, and CL (Fig. 4). One exception was a relatively large change in the fatty acid composition of PG in skeletal muscle; i.e., the amount of cis-vaccenic acid (18:1n-7) was reduced and instead, the amount of oleic acid (18:1n-9) increased considerably in LYCAT−/− mice (Fig. 4C). These results indicate that LYCAT mainly determines the fatty acid composition of PI in vivo.
Fig. 4.
Fatty acid compositions of PC, PE, PI, PS, PG and CL in liver, heart, skeletal muscle, and brain. After methyl esterification, fatty acids of each phospholipid were quantified by GC-MS. LYCAT+/+, closed bars; LYCAT−/−, open bars. FAs, fatty acids. *P<0.01, **P<0.001. Data represent the mean ± SEM of triplicate measurements.
Fig. 5.
Fatty acid composition of PI in testis. GC-MS analysis was used to measure the individual fatty acid species. FAs, fatty acids. *P <0.01. Data represent the mean ± SEM of triplicate measurements.
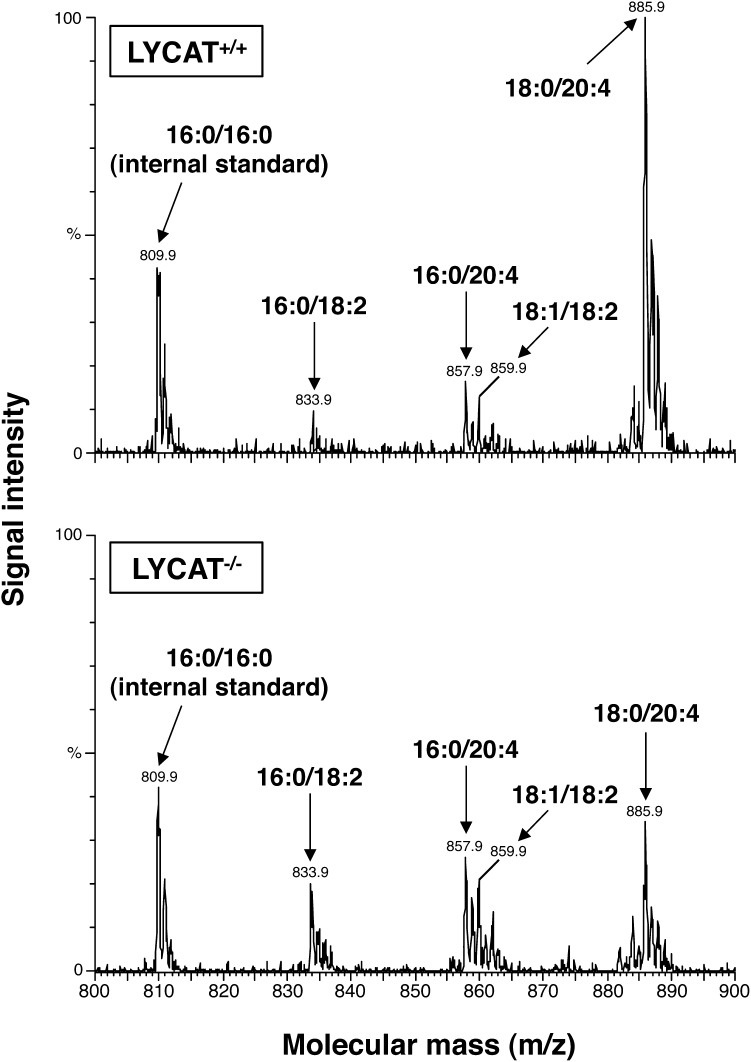
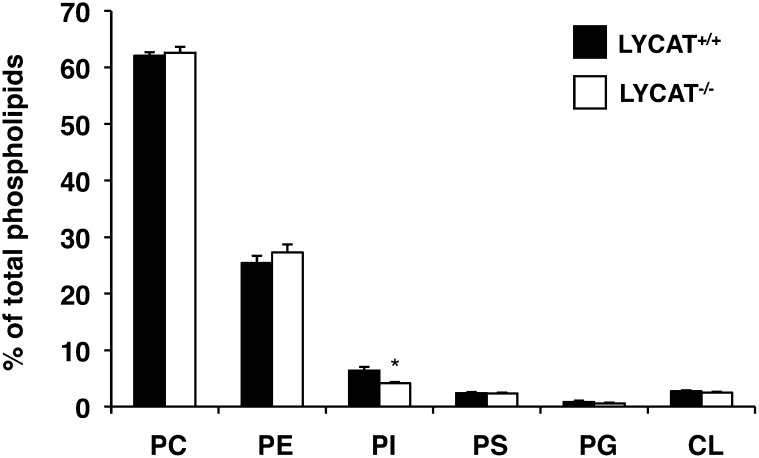
We next analyzed the molecular species of PI in LYCAT−/− mice by LC-ESIMS. In the liver of LYCAT−/− mice, 18:0/20:4-PI, the major molecular species of PI, was significantly decreased, and conversely, 16:0/20:4-PI, 16:0/18:2-PI, and 18:1/18:2-PI were increased compared with those in the liver of wild-type mice (Fig. 6). The ratio of PI to total phospholipids was significantly reduced by 35% compared with the ratio in wild-type mice as judged by quantification of phospholipid phosphorus (Fig. 7). On the other hand, the content and molecular species of PC and PE were not affected in LYCAT−/− mice (Fig. 7 and data not shown). These results confirm the notion that LYCAT plays an important role in determining the molecular species of PI in mice. A deficiency in the reacylation reaction of lysoPI may reduce the PI content.
Fig. 6.
MS analysis of PI molecular species in liver. Negative ionization LC-ESIMS spectra of PI molecular species of LYCAT+/+ (top) and LYCAT−/− mice (bottom) liver. Assigning specific molecular species to m/z values was based on their calculated theoretical monoisotopic masses and verified by MS/MS.
Fig. 7.
Phospholipid composition in liver. The amount of inorganic phosphate in phospholipids was quantified (see Materials and Methods). Data are expressed as phospholipid mol%. *P<0.05. Data represent the mean ± SEM of triplicate measurements.
LYCAT deficiency affects the molecular species of phosphoinositides
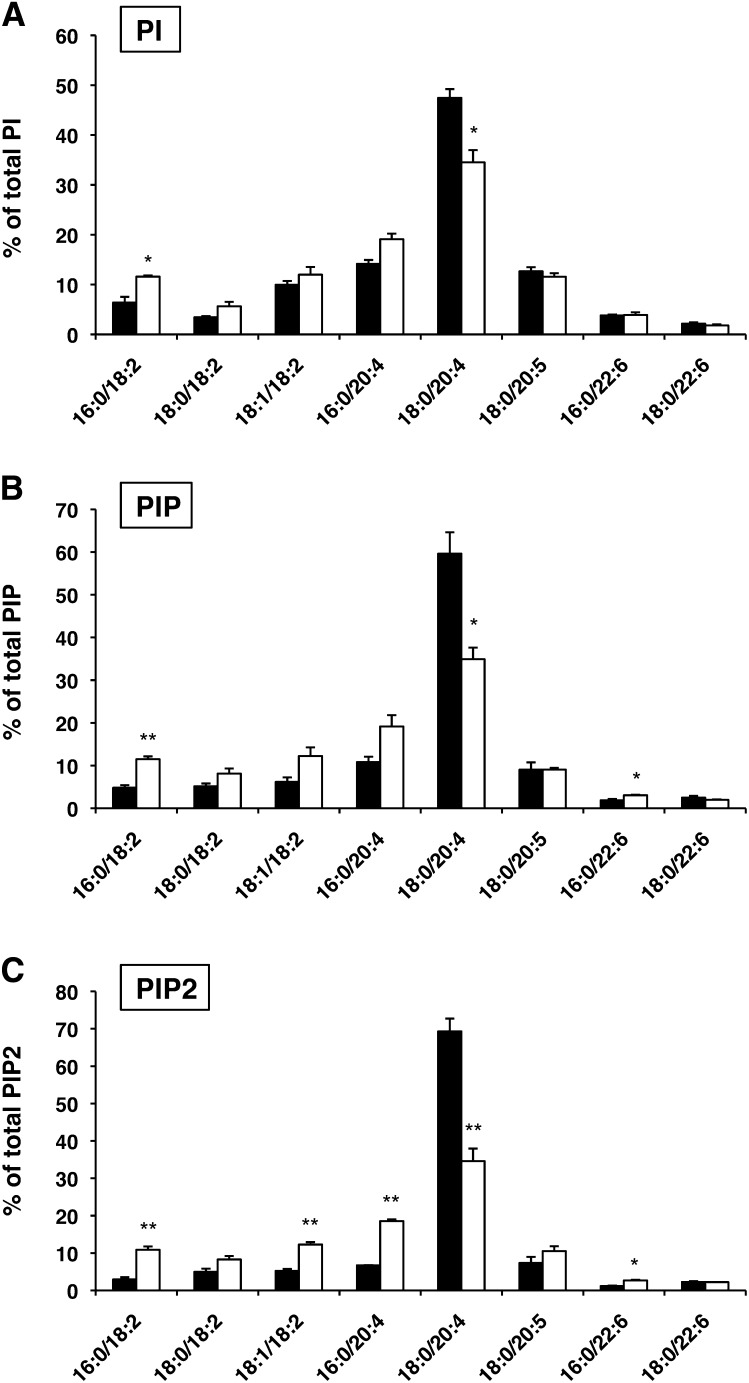
Because PI is a precursor for phosphoinositides (2), we also examined the molecular species of phosphoinositides in the liver of LYCAT−/− mice. For this, we used LC-ESIMS/MS with the selected reaction monitoring (SRM) technique (see Materials and Methods). The amounts of each molecular species in PI, phosphatidylinositol monophosphates (PIP1), or phosphatidylinositol bisphosphates (PIP2) in the liver from wild-type and LYCAT−/− mice are shown in Fig. 8. In PI of the LYCAT−/− liver, the amount of 18:0/20:4 species was reduced by 27% and the amounts of 16:0/18:2, 18:0/18:2, 18:1/18:2, and 16:0/20:4 species increased (Fig. 8A). Similarly, in PIP1 and PIP2, the amounts of 18:0/20:4 species were reduced by 41% and 50%, respectively, and the amounts of 16:0/18:2, 18:0/18:2, 18:1/18:2, 16:0/20:4, and 16:0/22:6 species increased (Fig. 8B, C). These data indicate that LYCAT deficiency affects the molecular species of PIP1 and PIP2 as well as PI.
Fig. 8.
Molecular species composition of PI, PIP1, and PIP2 in liver. Each molecular species of PI, PIP1, and PIP2 were quantified by LC-ESIMS/MS and selected reaction monitoring (SRM), as described in Materials and Methods. LYCAT+/+, closed bars; LYCAT−/−, open bars. *P<0.01, **P<0.001. Data represent the mean ± SEM of triplicate measurements.
LYCAT deficiency does not affect acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI
In the liver, heart, and skeletal muscle of LYCAT−/− mice, the amount of arachidonic acid in PI was also reduced, though the degree of reduction was less than that of stearic acid (Fig. 4A–C). Arachidonic acid is the predominant fatty acid at the sn-2 position of PI in mammals and is incorporated into PI by LPIAT after de novo synthesis (5–8). We first investigated whether LYCAT contributes to acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI in liver, heart, skeletal muscle, and brain. Acyltransferase assays were performed using [14C]arachidonoyl-CoA and sn-1-acyl lysoPI as substrates. As shown in Fig. 9A, [14C]arachidonoyl-CoA: sn-1-acyl LPIAT activity was not altered significantly in all the LYCAT−/− tissues examined. Furthermore, the expression level of LPIAT1, a major acyltransferase that incorporates arachidonic acid into the sn-2 position of PI (11), was not changed in LYCAT−/− liver (Fig. 9B). These results indicate that LYCAT does not contribute to acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI in mouse tissues.
Fig. 9.
Effect of LYCAT deficiency on acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI. A: Acyltransferase activities were measured using the membrane fractions from liver, heart, skeletal muscle, and brain. [14C]arachidonoyl-CoA and sn-1-acyl lysoPI were used as the acyl donor and acceptor, respectively. Data represent the mean ± SEM of triplicate measurements. B: Expression level of LPIAT1 in the liver was measured with Western blot analysis using an anti-mouse LPIAT1 monoclonal antibody. Total protein extracts of liver from LYCAT+/+ and LYCAT−/− mice were subjected to SDS-PAGE, blotted onto a PVDF membrane, and stained with anti-LPIAT1 antibody FT10. GAPDH was used as a loading control.
LYCAT is localized to the ER
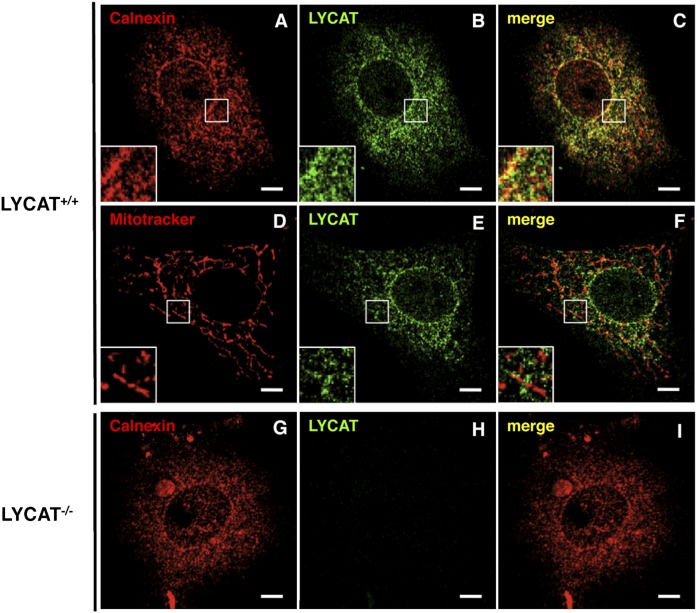
Cao et al. (17) reported that flag-tagged mouse LYCAT transfected into COS-7 cells displayed a perinuclear and punctate pattern that colocalized well with an ER marker but not with a mitochondrial marker. However, using the same type of cells, another study recovered recombinant flag-tagged mouse LYCAT from mitochondria and mitochondria-associated membranes but not from the microsomes (26). In the present study, we established a mouse LYCAT-specific monoclonal antibody YN1 and used it to analyze the subcellular localization of endogenous LYCAT. YN1 did not stain LYCAT−/− MEFs, indicating that YN1 specifically recognized mouse LYCAT (Fig. 10). Wild-type MEFs were costained with MitoTracker Red dye (a mitochondrial selective probe) or were costained with an anti-calnexin antibody that recognizes the calnexin ER-specific protein and the LYCAT antibody. We found that anti-LYCAT staining overlapped substantially with anti-calnexin but not with MitoTracker (Fig. 10), suggesting that endogenous LYCAT is localized to the ER but not to the mitochondria.
Fig. 10.
Subcellular localization of LYCAT in MEFs. MEFs derived from LYCAT+/+ or LYCAT−/− mice were stained with anti-LYCAT antibody YN1 (B, E, H) and anti-calnexin antibody (ER marker, A and G) or MitoTracker (mitochondria marker, D). The corresponding merged images are shown in C, F, I. Note that no signal was detected in LYCAT−/− MEFs when we used YN1 as the first antibody (H), indicating that YN1 specifically stained LYCAT. Scale bars, 10 µm.
DISCUSSION
Mammalian LYCAT is reported to possess acyltransferase activity toward the sn-2 position of anionic lysophospholipids including lysoPI, lysoPG, and lysoCL in vitro (17–20). In this study, we examined the fatty acid composition of each phospholipid from various tissues of LYCAT−/− mice, and showed that LYCAT determines the fatty acid composition of PI in vivo. 1-Stearoyl-2-arachidonoyl (18:0/20:4) PI is a major molecular species of PI in mammals. The amount of stearic acid in PI was greatly reduced in all the tissues examined from LYCAT−/− mice. Acyltransferase activity that incorporates stearic acid into the sn-1 position of PI was also reduced in the tissues from LYCAT−/− mice, whereas acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI was not altered. 1-Stearoyl-2-arachidonoyl PI is thought to be formed through fatty acid remodeling that is regulated by phospholipase A and lysoPI acyltransferase (5–10, 27, 28). Thus, LYCAT should serve as a lysoPI acyltransferase that catalyzes the incorporation of stearic acid into the sn-1 position of PI in the fatty acid remodeling pathway in vivo. Furthermore, mammalian LYCAT can substitute for the C. elegans homologs acl-8, acl-9, and acl-10 because expression of mouse LYCAT efficiently rescued the seam cell defects observed in acl-8 acl-9 acl-10 triple mutants, in which the fatty acid composition of the sn-1 position of PI is selectively altered (12). Our immunocytochemical analysis using MEFs revealed that endogenous LYCAT is mainly localized to the ER, suggesting that the fatty acid remodeling of PI by LYCAT occurs in the ER membrane.
In the membrane fraction of LYCAT−/− mouse liver, acyltransferase activity that incorporates stearic acid into the sn-1 position of PI was reduced to one-third of that in wild-type mice, whereas acyltransferase activities that incorporate stearic acid into the sn-1 position of other phospholipids such as PC, PE, and PS, were not reduced appreciably in the liver of LYCAT−/− mice, which suggests that one or more enzymes other than LYCAT contribute to the activities. In fact, the amounts of stearic acid in phospholipids such as PE and PS were not changed in LYCAT−/− mice. In addition, acyltransferase activity that incorporates stearic acid into the sn-1 position of PI remained in the membrane fraction of LYCAT−/− mice, indicating the existence of other acyltransferase(s) that incorporates stearic acid into the sn-1 position of PI. Acyltransferase activity toward lysoPG was reduced significantly in the membrane fraction of LYCAT−/− mouse liver. Indeed, we observed a relatively large change in the fatty acid composition of PG in the skeletal muscle of LYCAT−/− mice. These results indicate that LYCAT partially determines the fatty acid composition of PG, at least in certain organs.
In this study, we found that the amount of arachidonic acid in PI was also reduced in the liver, heart, and skeletal muscle of LYCAT−/− mice. Acyltransferase activity that incorporates arachidonic acid into the sn-2 position of PI was not reduced in these tissues. Moreover, the expression level of LPIAT1, a major acyltransferase that incorporates arachidonic acid into the sn-2 position of PI (11), was not changed. Thus, the reduction of the amount of arachidonic acid in PI of LYCAT−/− mice may be secondary to the reduction of stearic acid at the sn-1 position of PI. One possible explanation for the reduced arachidonic acid in PI is that LPIAT1 preferentially incorporates arachidonic acid when stearic acid is attached at the sn-1 position of lysoPI. In LYCAT−/− mice, stearic acid attached to PI is replaced by other fatty acids, such as palmitic acid and oleic acid, which may lead to reduced incorporation of arachidonic acid into PI.
Unlike the liver, heart, and skeletal muscle, the brain and testis of LYCAT−/− mice did not show any significant changes in the amount of arachidonic acid in PI. It is interesting to note that these tissues possess CDS1 (29, 30), one isoform of CDP-diacylglycerol synthase, which catalyzes the synthesis of CDP-diacylglycerol, a direct precursor of PI, from phosphatidic acid (PA). CDS1 prefers arachidonic acid-containing PA as a substrate (29) and therefore, CDS1 could have a role in increasing the arachidonic acid content of PI to compensate for the LYCAT deficiency in these tissues.
Recently, Li et al. (26) reported that the fatty acid composition of CL was altered in LYCAT−/− heart under high-fat diet conditions. In their study, however, the fatty acid composition of PI was not presented. The present results show that the fatty acid composition of PI was altered significantly in LYCAT−/− mice, whereas that of CL was less affected under normal diet conditions. So far, we have not observed any apparent abnormalities in LYCAT−/− mice under normal diet conditions. However, it is interesting that insulin-stimulated Akt phosphorylation was significantly enhanced in tissues from LYCAT−/− mice under high-fat diet conditions (26). PI plays important roles in intracellular signaling cascades, such as the PI3K/Akt pathway through distinct phosphorylated derivatives of the inositol head group (1, 2). Further analysis of LYCAT−/− mice should reveal the biological significance of PI molecular species and/or the fatty acid remodeling of PI in mammals.
Supplementary Material
Acknowledgments
The authors thank H. Fukuda for excellent technical assistance. We also thank Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) for strains.
Footnotes
Abbreviations:
- AGPAT
- 1-acylglycerol-3-phosphate O-acyltransferase
- A-P axis
- anterior-posterio axis
- CL
- cardiolipin
- ER
- endoplasmic reticulum
- LPCAT
- lysophosphatidylcholine acyltransferase
- LPEAT
- lysophosphatidylethanolamine acyltransferase
- LPGAT
- lysophosphatidylglycerol acyltransferase
- LPIAT
- lysophosphatidylinositol acyltransferase
- LPSAT
- lysophosphatidylserine acyltransferase
- LYCAT
- lysocardiolipin acyltransferase
- MEF
- mouse embryonic fibroblast
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PIP1
- phosphatidylinositol monophosphates
- PIP2
- phosphatidylinositol bisphosphates
- PS
- phosphatidylserine
- X:Yn-Z
- fatty acid chain of X carbon atoms and Y methylene-interrupted cis bonds (Z indicates the position of the terminal double bond relative to the methyl end of the molecules)
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and three figures.
REFERENCES
- 1.Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443: 651–657. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki T., Takasuga S., Sasaki J., Kofuji S., Eguchi S., Yamazaki M., Suzuki A. 2009. Mammalian phosphoinositide kinases and phosphatases. Prog. Lipid Res. 48: 307–343. [DOI] [PubMed] [Google Scholar]
- 3.Holub B. J., Kuksis A. 1971. Structural and metabolic interrelationships among glycerophosphatides of rat liver in vivo. Can. J. Biochem. 49: 1347–1356. [DOI] [PubMed] [Google Scholar]
- 4.Baker R. R., Thompson W. 1972. Positional distribution and turnover of fatty acids in phosphatidic acid, phosphinositides, phosphatidylcholine and phosphatidylethanolamine in rat brain in vivo. Biochim. Biophys. Acta. 270: 489–503. [DOI] [PubMed] [Google Scholar]
- 5.Akino T., Shimojo T. 1970. On the metabolic heterogeneity of rat liver phosphatidylinositol. Biochim. Biophys. Acta. 210: 343–346. [PubMed] [Google Scholar]
- 6.Holub B. J., Kuksis A. 1971. Differential distribution of orthophosphate- 32P and glycerol-14C among molecular species of phosphatidylinositols of rat liver in vivo. J. Lipid Res. 12: 699–705. [PubMed] [Google Scholar]
- 7.Luthra M. G., Sheltawy A. 1976. The metabolic turnover of molecular species of phosphatidylinositol and its precursor phosphatidic acid in guinea-pig cerebral hemispheres. J. Neurochem. 27: 1501–1511. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa Y., Rüstow B., Rabe H., Kunze D., Waku K. 1989. The de novo synthesis of molecular species of phosphatidylinositol from endogenously labeled CDP diacylglycerol in alveolar macrophage microsomes. Arch. Biochem. Biophys. 268: 559–566. [DOI] [PubMed] [Google Scholar]
- 9.Darnell J. C., Osterman D. G., Saltiel A. R. 1991. Synthesis of phosphatidylinositol in rat liver microsomes is accompanied by the rapid formation of lysophosphatidylinositol. Biochim. Biophys. Acta. 1084: 269–278. [DOI] [PubMed] [Google Scholar]
- 10.Darnell J. C., Osterman D. G., Saltiel A. R. 1991. Fatty acid remodelling of phosphatidylinositol under conditions of de novo synthesis in rat liver microsomes. Biochim. Biophys. Acta. 1084: 279–291. [DOI] [PubMed] [Google Scholar]
- 11.Lee H. C., Inoue T., Imae R., Kono N., Shirae S., Matsuda S., Gengyo-Ando K., Mitani S., Arai H. 2008. Caenorhabditis elegans mboa-7, a member of the MBOAT family, is required for selective incorporation of polyunsaturated fatty acids into phosphatidylinositol. Mol. Biol. Cell. 19: 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imae R., Inoue T., Kimura M., Kanamori T., Tomioka N. H., Kage-Nakadai E., Mitani S., Arai H. 2010. Intracellular phospholipase A1 and acyltransferase, which are involved in Caenorhabditis elegans stem cell divisions, determine the sn-1 fatty acyl chain of phosphatidylinositol. Mol. Biol. Cell. 21: 3114–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanamori T., Inoue T., Sakamoto T., Gengyo-Ando K., Tsujimoto M., Mitani S., Sawa H., Aoki J., Arai H. 2008. β-catenin asymmetry is regulated by PLA1 and retrograde traffic in C. elegans stem cell divisions. EMBO J. 27: 1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shindou H., Hishikawa D., Harayama T., Yuki K., Shimizu T. 2009. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J. Lipid Res. 50(Suppl): S46–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewin T. M., Wang P., Coleman R. A. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38: 5764–5771. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita A., Nakanishi H., Suzuki H., Kamata R., Tanaka K., Waku K., Sugiura T. 2007. Topology of acyltransferase motifs and substrate specificity and accessibility in 1-acyl-sn-glycero-3-phosphate acyltransferase 1. Biochim. Biophys. Acta. 1771: 1202–1215. [DOI] [PubMed] [Google Scholar]
- 17.Cao J., Liu Y., Lockwood J., Burn P., Shi Y. 2004. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279: 31727–31734. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal A. K., Barnes R. I., Garg A. 2006. Functional characterization of human 1-acylglycerol-3-phosphate acyltransferase isoform 8: cloning, tissue distribution, gene structure, and enzymatic activity. Arch. Biochem. Biophys. 449: 64–76. [DOI] [PubMed] [Google Scholar]
- 19.Cao J., Shen W., Chang Z., Shi Y. 2009. ALCAT1 is a polyglycerophospholipid acyltransferase potently regulated by adenine nucleotide and thyroid status. Am. J. Physiol. Endocrinol. Metab. 296: E647–E653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Chen Y. Q., Li S., Konrad R. J., Cao G. 2009. The microsomal cardiolipin remodeling enzyme acyl-CoA lysocardiolipin acyltransferase is an acyltransferase of multiple anionic lysophospholipids. J. Lipid Res. 50: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 23.Ariyama H., Kono N., Matsuda S., Inoue T., Arai H. 2010. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J. Biol. Chem. 285: 22027–22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartlett G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234: 466–468. [PubMed] [Google Scholar]
- 25.Clark J., Anderson K. E., Juvin V., Smith T. S., Karpe F., Wakelam M. J., Stephens L. R., Hawkins P. T. 2011. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods. 8: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Romestaing C., Han X., Li Y., Hao X., Wu Y., Sun C., Liu X., Jefferson L. S., Xiong J., et al. 2010. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metab. 12: 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darnell J. C., Saltiel A. R. 1991. Coenzyme A-dependent, ATP-independent acylation of 2-acyl lysophosphatidylinositol in rat liver microsomes. Biochim. Biophys. Acta. 1084: 292–299. [DOI] [PubMed] [Google Scholar]
- 28.Holub B. J., Piekarski J. 1979. The formation of phosphatidylinositol by acylation of 2-acyl-sn-glycero-3-phosphorylinositol in rat liver microsomes. Lipids. 14: 529–532. [DOI] [PubMed] [Google Scholar]
- 29.Saito S., Goto K., Tonosaki A., Kondo H. 1997. Gene cloning and characterization of CDP-diacylglycerol synthase from rat brain. J. Biol. Chem. 272: 9503–9509. [DOI] [PubMed] [Google Scholar]
- 30.Inglis-Broadgate S. L., Ocaka L., Banerjee R., Gaasenbeek M., Chapple J. P., Cheetham M. E., Clark B. J., Hunt D. M., Halford S. 2005. Isolation and characterization of murine Cds (CDP-diacylglycerol synthase) 1 and 2. Gene. 356: 19–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.