Abstract
A number of amyloidogenic variants of apoA-I have been discovered but most have not been analyzed. Previously, we showed that the G26R mutation of apoA-I leads to increased β-strand structure, increased N-terminal protease susceptibility, and increased fibril formation after several days of incubation. In vivo, this and other variants mutated in the N-terminal domain (residues 26 to ∼90) lead to renal and hepatic accumulation. In contrast, several mutations identified within residues 170 to 178 lead to cardiac, laryngeal, and cutaneous protein deposition. Here, we describe the structural changes in the fibrillogenic variant L178H. Like G26R, the initial structure of the protein exhibits altered tertiary conformation relative to wild-type protein along with decreased stability and an altered lipid binding profile. However, in contrast to G26R, L178H undergoes an increase in helical structure upon incubation at 37°C with a half time (t1/2) of about 12 days. Upon prolonged incubation, the L178H mutant forms fibrils of a diameter of 10 nm that ranges in length from 30 to 120 nm. These results show that apoA-I, known for its dynamic properties, has the ability to form multiple fibrillar conformations, which may play a role in the tissue-specific deposition of the individual variants.
Keywords: high density lipoprotein, apoA-I, fibril formation, amyloid, protein stability
Amyloid protein causes and/or exacerbates a variety of diseases, including Alzheimer's, Huntington's, Parkinson's, and systemic amyloidoses. It is increasingly clear that a wide variety of proteins can form amyloid structures under certain conditions. Understanding the causes behind amyloid formation and the structures of amyloid proteins is critical to preventing or alleviating symptoms in these diseases. In the majority of cases, amyloid protein develops from an increase in β-strand content leading to fibrils possessing cross-β structure.
ApoA-I is the major protein component of HDL and has a central role in cholesterol metabolism. As such, apoA-I/HDL possesses significant anti-atherogenic properties, making it a good candidate for treatment of cardiovascular disease. Both animal models and clinical trials provide striking evidence for the regression of atherosclerotic plaque upon treatment with HDL discs containing apoA-I [reviewed in (1)]. Amyloid-forming variants of apoA-I are found to accumulate in different tissues and wild-type (WT) apoA-I is present in atherosclerotic lesions as well as tissues (2–4). This presents a point of concern for HLD-based infusion therapies as repeated infusions could result in vascular deposition of excess A-I and thereby counter the therapeutic aim.
The variants of apoA-I with an increased propensity to form amyloid fibrils include both point mutations and deletions. Point mutations cluster in the amino-terminus (residues 1–100) and in residues 173–178 (5). Amyloid protein from these variants is deposited in a tissue-specific manner but the basis of tissue specificity is not known. We reported the first analysis of the structure of an amyloid apoA-I, G26R (apoA-IIOWA) (6). This variant exhibits increased β-strand content, N-terminal protease sensitivity, and binding to amyloid-detecting dyes as well as altered lipid-binding properties. We postulated that the conversion to amyloid structure results from the positioning of two adjacent arginine residues, favoring β-strand extension from residue 26 to 57. Acquisition of additional β-strand either causes or is accompanied by destabilization of the N-terminal region, which increases amyloid propensity. This analysis remains the only study to date of the properties of a full-length amyloid variant. A number of questions remain to be answered concerning apoA-I amyloidosis, including the effect of mutation position in the two clusters on structure and amyloid properties, the relationship between mutation position and tissue deposition, and the influence of in vivo factors on amyloid formation. To address the first of these issues, which will help determine whether a universal mechanism underlies amyloid formation, we elected to examine the structure and in vitro amyloid propensity of the L178H variant. This mutation is associated with severe cardiac and laryngeal amyloidosis as well as skin lesions (7); since this first report involving position 178, additional studies have linked C-terminal cluster mutations with cardiac and laryngeal deposition, including a recent report involving a L178P variant (8). Here, we report that L178H exhibits structural alterations similar to those observed for G26R and forms fibrils with dimensions similar to those formed by other fibrillogenic proteins. However, in contrast to G26R, L178H exhibits increased helical content, which is reflected in a lack of thioflavin T (ThT) binding. To our knowledge, this is the first report of a full-length apoA-I protein that forms fibrils with predominantly helical structure.
METHODS
Production of recombinant protein
A bacterial expression system consisting of pNFXex plasmid in Escherichia coli strain BL21(DE3) pLysS cells (Invitrogen) was used to prepare the apoA-I proteins, as previously described (9, 10). Primer-directed PCR mutagenesis was used to create the L178H mutation. The mutation was verified by dideoxy automated fluorescent sequencing. In brief, human apoA-I proteins were purified from bacterial cell lysate by immobilized metal affinity chromatography (HiTrap columns, GE Healthcare, Waukesha, WI) under denaturing conditions (3 M guanidine-HCl), extensively washed with PBS (20 mM phosphate, 500 mM NaCl), pH 7.4, supplemented with protease inhibitors (Sigma #P2714), and then eluted by increasing concentrations of imidazole. Refolded protein was dialyzed into PBS, pH 7.4, 150 mM NaCl, concentrated with 30 kDa molecular weight cut-off Amicon Ultra centrifugal filter devices (Millipore), and stored at 4°C with or without protease inhibitor cocktail (Sigma, # P8849) prior to use. The time-course experiments were synchronized to be initiated the same day (day 0) or the day after (day 1) purification. Protein purity was confirmed by SDS-PAGE with Coomassie blue staining and concentrations determined by the MicroBCA assay kit (Pierce), using BSA as a standard, or by absorbance of proteins diluted into guanidine using an extinction coefficient of 1.13 cm2/mg. Proteins were periodically checked for proteolytic degradation by SDS-PAGE; only those samples showing intact protein were used.
CD spectroscopy
Circular dichroism (CD) measurements were performed on a Jasco J-810 spectropolarimeter equipped with a Jasco CDF-426S Peltier set to 25°C. For thermal stability experiments, spectra were obtained from 25°C to 80°C with 2.5°C increments and with a thermal equilibration time of 10 s. ApoA-I was diluted to 0.4 mg/ml in PBS (final concentration was 25 mM phosphate, 25 mM NaCl, pH 7.4), placed in a 0.1 mm quartz cuvette and, after extensive purging with nitrogen, scanned in the region 180 to 260 nm (scan speed was 20 nm/min). Averages of five scans were baseline-subtracted (PBS buffer; 25 mM phosphate, 25 mM NaCl) and the α-helical content was calculated from the molar ellipticity at 222 nm as previously described (11). The Boltzmann function within the Origin 8.1 software (OriginLab, Northampton, MA) was used to fit the helical content values of the time course to a sigmoidal fit curve.
Denaturation
Equilibrium denaturation was carried out on samples using guanidine (Gn-HCl) and urea. Stock solutions of 8 M urea and Gn-HCl were prepared from their respective ultra pure solid crystals. Protein was incubated for at least 24 h at 4°C at 0.1 mg/ml prior to addition of denaturant (final protein concentration = 0.05 mg/ml). Incubated protein samples were combined in triplicate with denaturant by dilution from 8 M stock solutions and stored at 4°C for a minimum of 24 h prior to data collection. Changes in tryptophan fluorescence were recorded using a Shimadzu RF5301 fluorometer with excitation at 295 nm and excitation and emission slit widths of 5 and 3 nm, respectively. The unfolding transition was monitored by recording the wavelength at maximum fluorescence intensity (WMI) or maximum wavelength (λmax).
The WMIs of triplicate dilutions of each sample were averaged and plotted against the denaturant concentration. The resulting curves exhibited characteristics of a sigmoidal function and were fitted with the Hill equation. Assuming a two-state model (folded and unfolded), the fraction of unfolded protein (fD) can be calculated using:
where ymax is the maximum WMI. The equilibrium constant (KD) for the process can be calculated as KD = fD/(1/fD), yielding the free energy of denaturation ΔGD = -RTlnKD. D1/2, m, and ΔGDo values were obtained by plotting ΔGD versus denaturant concentration to give the linear equation (12) ΔGD = ΔGDo − m[denaturant], where ΔGD° is the free energy of protein folding in water (0 M denaturant), and m the cooperativity of denaturation. In most cases, the values obtained were averages of data from three separate protein expressions.
Dye-binding assays
ThT binding assay.
L178H and WT (1 mg/ml) were incubated at 37°C and diluted to 0.25 mg/ml with PBS at time of use. A total of 182 μl of diluted protein was incubated for 5 min in the dark with 1.82 ml of a ThT (5 μM)/glycine (50 mM) solution (ThT stock: 1 mM stored in the dark at 4°C. Glycine buffer stock: 0.1M at pH 8.5 stored at 4°C). ThT fluorescence was then measured using a VICTOR3 Multilabel Plate Counter (PerkinElmer, Waltham, MA) spectrofluorometer at an excitation of 450 nm, with excitation and emission slit widths of 10 nm.
1-Anilinonaphthalene-8-sulfonic acid (ANS) binding assay.
Protein solutions (50 μg/ml in PBS) were incubated with 250 μM ANS (>100 molar excess) in a total volume of 2 ml. Samples were excited at 395 nm with 5 nm excitation and emission slit widths. Emission was recorded from 410 nm to 560 nm, which corresponds to the blue-shifted emission spectra of ANS bound to protein (13).
Limited proteolysis
Following brief dialysis to remove protease inhibitors, protein (20 μg) was treated with 1:2000 molar ratio of high purity chymotrypsin (Calbiochem #230834) for the indicated periods of time. Previous studies employing Staphylococcus aureus V8 protease (Glu-C specificity) and trypsin on WT apoA-I gave the same results obtained using chymotrypsin; therefore, chymotrypsin was used here as the representative protease (6, 14, 15). Reactions were stopped with protease inhibitor cocktail (Sigma #P2714) followed by addition of SDS loading buffer. Samples were stored at −20°C until analysis with SDS-PAGE. Selected bands were blotted onto polyvinylidene fluoride and submitted to the Glycoprotein Core facility at University of Texas Medical Branch for N-terminal sequence analysis.
Electron microscopy
Protein samples incubated at 37°C for 30 days were diluted and analyzed by negative stain electron microscopy as described previously (16). Five microliter aliquots were adsorbed onto carbon-coated grids for 1 min, washed with two drops of water, and stained on two drops of 0.75% uranyl formate. The grids were rendered hydrophilic by glow discharge at low pressure in air. Specimens were observed in a JEOL JEM 1230 electron microscope operated at 80 kV accelerating voltage, and images were recorded with a Gatan Multiscan 791 CCD camera.
Formation of reconstituted HDL (rHDL)
Lyophilized dimyristoylphosphatidylcholine (DMPC, Avanti Polar Lipids) was dissolved in 3:1 chloroform-methanol by thorough vortexing and the solvent was totally evaporated by overnight incubation under a stream of nitrogen gas. The phospholipids were then dissolved in PBS by vortexing and the lipid suspension was extruded through a 100 nm polycarbonate membrane using the LiposoFast system (Avestin) to create unilamellar vesicles. ApoA-I WT and L178H were incubated at physiological temperature (37°C) and at the transition temperature of the lipid (24°C) with unilamellar DMPC at a molar ratio of protein to phospholipid of 1:100 and a protein concentration of 0.4 mg/ml. Samples were taken at 0, 1, 3, 6, 9, 24 h and at 4 days and stored at −80°C with 10% w/v sucrose until analysis by blue native-PAGE using the NativePAGE Novex Bis-Tris Gels System (Invitrogen) according to the manufacturer's instructions. The lipid binding capacity was evaluated by a spectrophotometric lipid clearance assay as described previously (6). Briefly, unilamellar DMPC vesicles were prepared as described above and placed in a 96-well plate, where lipid-free apoA-I proteins were added at a 1:100 protein to lipid ratio (mol/mol). The spectral changes (at 600 nm) were monitored over time as phospholipid molecules were used in the formation of smaller protein-lipid complexes with a consequent clearance of larger DMPC vesicles. Although the lipid binding of apolipoproteins is known to be a complex process involving several intermediate steps, this assay is accepted in the field as an approximate measure of the lipid-binding capability of an apolipoprotein (17).
RESULTS
L178H has increased exposed hydrophobic surface area and an altered amino-terminus
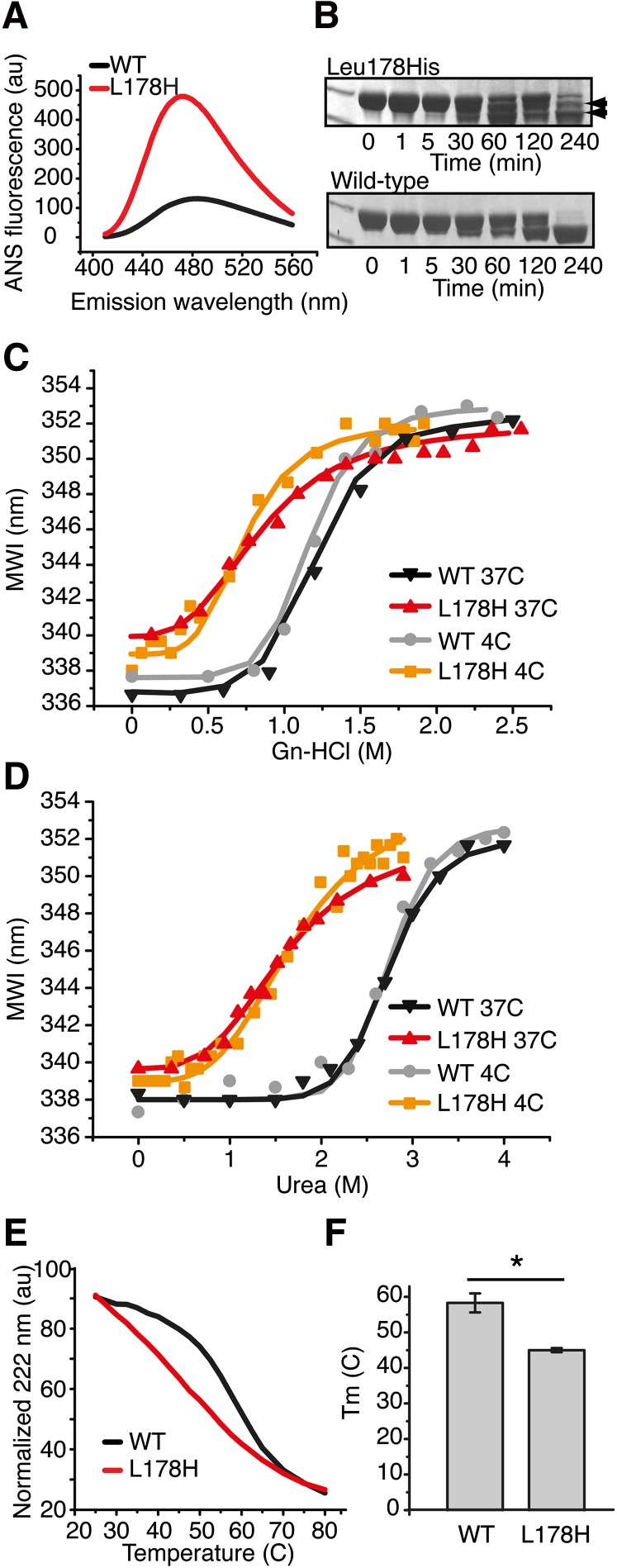
Aggregation of proteins, as in amyloid formation, is often brought about by increased exposure of hydrophobic regions. To determine the surface hydrophobicity of L178H, the dye ANS was used. WT protein binds a significant amount of ANS compared with a protein of similar molecular weight such as carbonic anhydrase. We compared the ANS binding of both G26R and L178H to WT apoA-I. L178H exhibit about a 4-fold increase in binding ANS (Fig. 1A) and a 2-fold increase for G26R (not shown), indicating that both mutations alter apoA-I structure in a manner that increases the fraction of residues accessible to bind hydrophobic dyes.
Fig. 1.
L178H exhibits greater hydrophobic exposure, altered structural organization, and decreased structure stability compared with WT apoA-I. A: ANS binding. Wild-type and L178H proteins (50 µg/ml) were incubated with excess ANS (ratio 1:150) followed by reading of the blue-shifted emission spectra between 410 to 560 nm. B: Limited proteolysis of L178H and wild-type apoA-I proteins was used to assay structure accessibility. ApoA-I proteins (20 µg) were incubated in the presence of chymotrypsin (1:2000 molar ratio) at 37°C for the indicated times. Top panel, L178H; bottom panel, WT. Edman degradation assay of the fragmented mutant apoA-I (indicated with arrows) was used to identify the specific cleavage sites (see Table 1). C, D: Increasing concentrations of urea (C) and guanidine (D) were used in intrinsic fluorescence experiments to compare the stability of L178H with wild-type apoA-I. E: A temperature gradient ranging from 25°C to 80°C combined with CD measurements was used to analyze the thermal stability. F: Melting point values (Tm) for WT and L178H proteins. Bars are averaged Tm values (n = 3). * P < 0.05. WT, wild-type apoA-I protein; L178H, L178H apoA-I protein.
We previously demonstrated that the G26R mutation leads to increased protease sensitivity in the N terminus, presumably due to an altered structure triggered by the increased β structure in residues 26–50 (6). To determine the effect of the L178H mutation on protease sensitivity, we subjected the protein to limited proteolysis using chymotrypsin (Fig. 1B; Table 1). Like G26R, L178H exhibits a marked increase in protease sensitivity in the N terminus with major cleavage occurring at F57. However, an additional cleavage was observed at residue 18 in L178H that we did not detect under the same conditions in G26R. We also did not detect in L178H a fragment observed in both WT and G26R that is produced by cleavage at the extreme carboxy-terminus, near residue 225. These results suggest that although both G26R and L178H have increased N-terminal sensitivity, some structural differences exist between the two variants.
TABLE 1.
Proteolytic cleavage sites (chymotrypsin) of L178H, WT and G26R apoA-I
| apoA-I variant | Proteolytic cleavage sitesa | Fragmentsb |
| L178Hc | W8, Y18, F57 | D9-D243, V19-D243, S58-D243 |
| WTd | Y192, F229 | M1-Y192, M1-F229 |
| G26Rd | Y18, F57 | V19-D243, S58-D243 |
Determined by Edman degradation of blotted SDS-PAGE fragments.
Based on the molecular estimate from SDS-PAGE.
This work.
From ref (6).
L178H has decreased stability
Many proteins convert to an amyloidogenic state when they are destabilized in some way. Equilibrium solvent denaturation using either guanidine or urea was used to examine the stability of L178H. WT incubated at 4°C for one day prior to addition of denaturant yielded D1/2 (D1/2 is the concentration of denaturant at which the protein is half folded) values for guanidine and urea of 1.23 ± 0.02 M and 2.81 ± 0.05 M, respectively, similar to values reported elsewhere in the literature, where samples were also incubated at 4°C (Fig. 1C D, Table 2). Likewise, ΔGDo and cooperativity (m) values were nearly identical to those reported previously. After incubation at 4°C for one day, L178H exhibited lower stability and loss of cooperativity (Fig. 1C, D), reflected by substantially lower ΔGDo and D1/2 values and less pronounced sigmoidality of the curves. Due to the low protein concentration used in these studies (0.05 mg/ml), we believe the denaturation data show that the monomeric structure of L178H is both less stable and less cooperative at 4°C than monomeric WT protein. There was little difference between the two denaturants as the same reductions in thermodynamic parameters for L178H versus WT (∼40%, 60%, and 40% for D1/2, ΔGDo, and m, respectively) were observed (Table 2). A similar difference in stability between WT and L178H was observed for proteins incubated at 37°C for one day, with L178H being much less stable than WT. Measurement of the proteins’ thermal stabilities using CD measurements yielded a similar result with Tm values of 45 ± 0.6°C for L178H and 58 ± 2.7°C for WT and reduced cooperativity in L178H (Fig. 1E, F). Thus, L178H, like most amyloid proteins, has substantially reduced stability.
TABLE 2.
Denaturation parameters for the folding of apoA-I variants.a
| D1/2 (M) | ΔGD (kcal/mol) | m | Ref. | |
| Urea | ||||
| WT, 4°Ca | 2.81 ± 0.05 | 6.16 ± 0.19 | 2.19 ± 0.11 | this work |
| 2.7 | 7.07 ± 0.65 | 2.57 ± 0.24 | (37) | |
| 3.0 ± 0.2 | 6.2 ± 0.2 | 2.0 ± 0.1 | (12) | |
| WT, 37°Ca | 2.82 ± 0.07 | 5.39 ± 0.33 | 1.91 ± 0.08 | this work |
| L178H, 4°C | 1.74 ± 0.08 | 2.55 ± 0.07 | 1.45 ± 0.07 | this work |
| L178H, 37°C | 1.76 ± 0.13 | 2.36 ± 0.14 | 1.34 ± 0.06 | this work |
| Gn-HCl | ||||
| WT, 4°C | 1.23 ± 0.02 | 4.74 ± 0.38 | 3.85 ± 0.34 | this work |
| 1.0 | 4.6 ± 0.3 | 4.6 | (38) | |
| 1.05 ± 0.13 | 3.1 ± 0.2 | 3.0 | (39) | |
| WT, 37°Cb | 1.22 | 3.82 | 3.14 | this work |
| L178H, 4°Cb | 1.08 ± 0.07 | 1.93 ± 0.14 | 1.76 ± 0.06 | this work |
| L178H, 37°C | 0.90 ± 0.02 | 1.55 ± 0.27 | 1.74 ± 0.31 | this work |
Unless otherwise indicated, data are averages from triplicate incubation samples of three different protein preparations.
Triplicate samples of two different protein preparations.
L178H has reduced lipid-binding ability
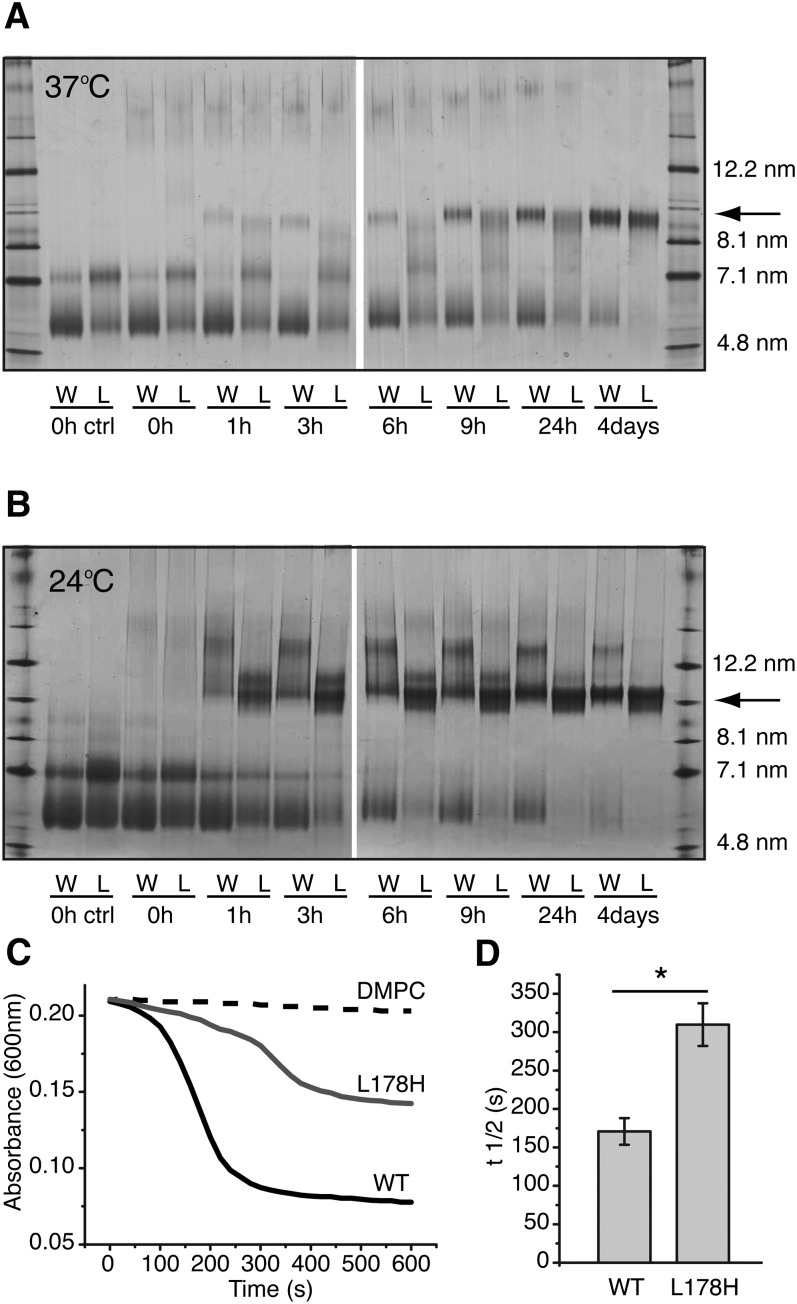
The majority of plasma apoA-I, more than 95%, is bound to lipid. At present, it is not known whether lipid-free or lipid-bound protein is responsible for amyloid formation in vivo. To examine the effect of the L178H mutation on lipid binding, synthetic rHDL was prepared and analyzed by non-denaturing gradient gel electrophoresis. DMPC lipids were incubated with purified apoA-I proteins (at a molar ratio of 1:100, protein to lipid) at 37°C (physiological temperature) and at 24°C (the transition temperature of the lipid) for up to 4 days. As can be seen in Fig. 2A, at physiological temperature, the mutant protein forms recombinant HDL with an estimated diameter of 9.6 nm to a similar extent (at 4 days) as the WT protein but this particle develops at a slower rate. Moreover, there is an apparent difference in the intermediate structures of the remodeling process during the first day of incubation. At these time points, fewer 9.6 nm rHDL particles of the L178H mutant are formed and several less defined particle sizes can instead be seen. Also of note is that a larger proportion of the mutant protein is oligomeric in the lipid-free state (0 h samples and control). Similarly to rHDL formed at physiological temperature, rHDL particles of similar sizes are formed for both proteins after incubation at transition temperature for 4 days (Fig. 2B). At this temperature, however, the formation of rHDL of the L178H mutant is faster than at 37°C and involves fewer intermediate structures. This may reflect a lower degree of self-association of the L178H mutants at this temperature as self-association in the apo-state is known to decrease lipid-binding (18).
Fig. 2.
Lipid-binding properties of L178H variant. Incubation of apoA-I with DMPC results in time-dependent formation of rHDL. A, B: ApoA-I WT and L178H (0.38 mg/ml) were incubated with DMPC (molar ratio 1:100) at 37°C (A) and 24°C (B) for the indicated times and then stored at −80°C with 10% sucrose until analysis. The samples were analyzed by Blue Native-PAGE on 4-16% polyacrylamide gels. Stoke's diameters of reference proteins are shown to the right. Arrows indicate the formed rHDL particles with an estimated diameter of 9.6 nm. C, D: A lipid clearance assay was used to make a quantitative comparison of the lipid-binding rate between wild-type and L178H proteins. Proteins (0.4 mg/ml) were mixed with DMPC (protein to lipid molar ratio of 1:100) at time 0 followed by spectrophotometric measurement at 600 nm. Bars are averaged t1/2 values (n = 3). * P < 0.05. W, wild-type; L, L178H.
The lipid-binding activity was also measured in a lipid clearance assay. WT and L178H proteins were mixed with preformed 100 nm DMPC lipid vesicles followed by spectrophotometric reading of the clearance of DMPC vesicles (Fig. 2C). The L178H mutant showed a markedly decreased rate of lipid binding with a t1/2 of ∼300 s, which is almost 2-fold higher than that observed for the WT protein (t1/2 ∼170 s) (Fig. 2D). These studies indicate that the L178H variant is capable of forming rHDL with a diameter of 9.6 nm, however, at a slower rate at 37°C and with variable specific intermediate protein-lipid complexes.
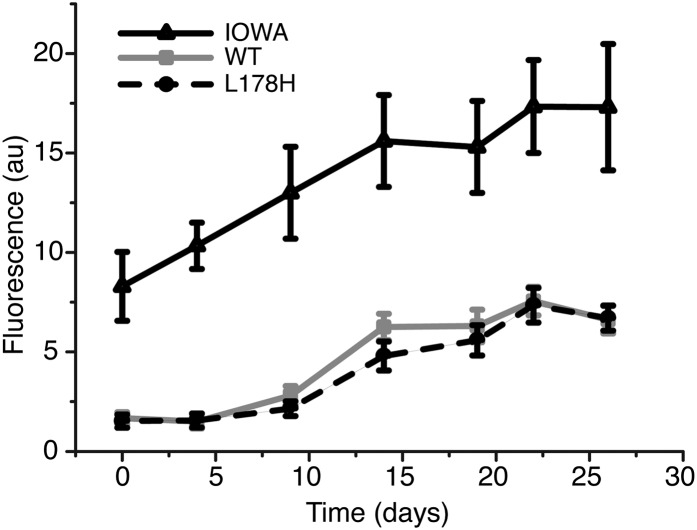
L178H does not bind ThT
The presence of amyloid cross-β structure can be assessed by binding to the amyloidophilic dye ThT. The binding of L178H to ThT was assessed and compared with WT and to G26R as a positive control (Fig. 3). WT protein exhibited a smaller increase in ThT binding, with the amount and rate of increase similar to our previous observation for WT (6). A similar low increase in ThT binding was observed for L178H over the 26 days of incubation at 37°C. These results are in contrast to G26R, which had higher ThT binding than WT and L178H from the first measurement recorded (day 0). This suggests that L178H, in contrast to the G26R variant, does not form the typical β-structure present in amyloids.
Fig. 3.
ThT binding experiment indicates that the L178H variant does not form amyloids. ThT was used to determine the time dependent formation of β-sheet containing amyloids. Proteins were incubated at 37°C for the indicated times followed by incubation with ThT and spectrofluorometric reading. The G26R variant, which is known to form amyloids, was used as a positive control. n = 3 for each data point.
L178H forms fibrils with helical structure
To determine whether L178H protein forms aggregates over a similar time period, the solubility of the protein was examined (not shown). Protein was incubated for 4 weeks at either 4 or 37°C. The solubility of both proteins decreased over time when incubated at 37°C, to a greater extent with L178H than WT (not shown).
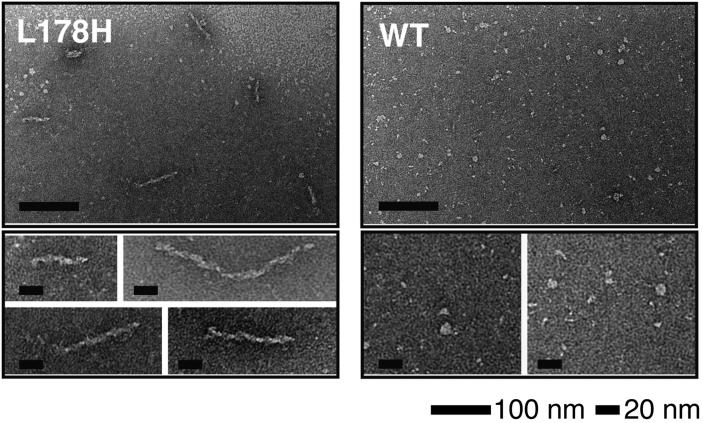
L178H and WT solutions were examined by negative stain electron microscopy to determine the ultra structure of the formed aggregates (Fig. 4). Fibrillar structures ranging in length from 30 to 120 nm with a 10 nm diameter were observed for L178H incubated at 37°C for 30 days. Under the same conditions, WT solutions did not produce fibrils, although small spherical aggregates were observed. The studies show that the L178H form insoluble aggregates with a fibril morphology partly consistent with the characteristics of fibril-forming proteins. However, only shorter fibrils were observed.
Fig. 4.
Electron microscopy analysis shows fibril formation of L178H. Mutant and wild-type protein (both at 0.4 mg/ml) were incubated in tris-buffer saline, pH 7.4, at 37°C for 4 weeks followed by negative stain electron microscopy analysis. The L178H mutant proteins formed elongated fibrils with lengths in the range of 30 to 120 nm (left panels). Wild-type apoA-I protein incubated at identical conditions is less prone to form higher order structures (right panels). Long and short scale bars are 100 nm and 20 nm, respectively.
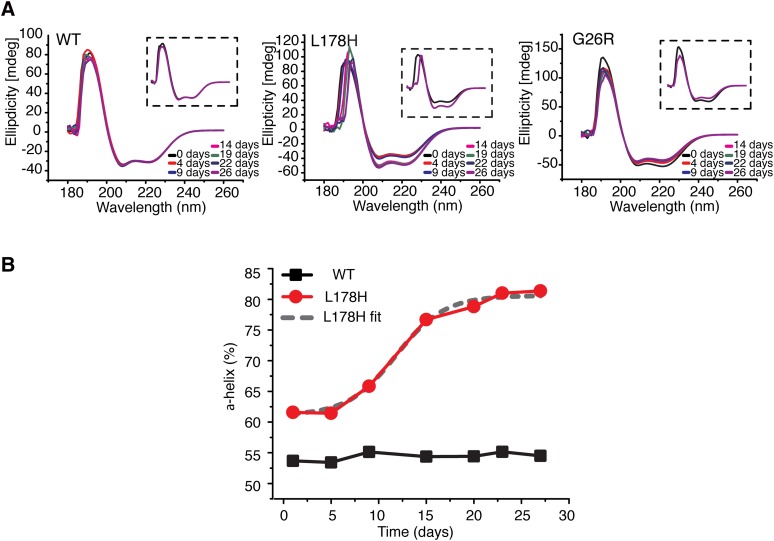
The hallmark feature of amyloid proteins is the increase in β-strand content. To determine the secondary structural changes in L178H, CD spectra were collected and compared with WT and G26R proteins (Fig. 5). Spectra were collected for a 4 week period on samples incubated at 37°C. The helical content of the proteins during the incubation was estimated from their molar ellipticities at 222 nm. The initial helical content of the L178H was slightly elevated (∼62% α-helix) as compared with the WT protein, which exhibited a helical content (∼55%) in agreement with previous estimations by use of CD spectroscopy (19, 20). During the incubation, L178H showed a slight increase in helix content at day 9 with an additional larger increase at day 14; little change was observed following day 14 up to 26 days of incubation. The change in helix content was fit to a sigmoidal curve indicating cooperativity in the structural transition to the fibrillar organization. The t1/2 of the structural conversion of the L178H into largely helical structure was about 12 days. The change in the CD signal could be influenced by self-association, because a modest increase in helix content occurs upon self-association (18), about 10% increase between 0.1 and 1.0 mg/ml. However, the increase observed here (in a 0.2 mg/ml solution) is much higher. Furthermore, over these same time periods and under the same conditions, WT showed no change in secondary structure whereas G26R showed a decrease in helix content. This analysis therefore shows a time-dependent increase in helical structure that appears to be L178H specific.
Fig. 5.
Structural transitions of apoA-I proteins. CD spectroscopy was used to analyze secondary structure changes over time. A: Spectra (average of 5 readings) of WT (left), L178H (middle), and G26R (right) apoA-I proteins (0.2 mg/ml) were obtained (180 nm to 260 nm) at indicated times of incubation at 37°C. Insets show the initial (black spectra) and final (purple spectra) spectra of the respective apoA-I proteins. B: The relative α-helical structure content of the L178H (red) and WT (black) apoA-I proteins. The t1/2 value for the structural transition is ∼12 days (see Boltzmann fit of the L178H graph; dashed gray line).
DISCUSSION
Sixteen of the ∼70 known apoA-I variants are known to be amyloidogenic (21). Most of these have point mutations in the N-terminal 100 amino acids of apoA-I, although several cluster in the short span 173–178. In either location, the in vivo amyloid material is composed primarily of a proteolytic N-terminal fragment of about 90 residues, commonly 1–93. We reported the first analysis of a full-length amyloid mutant, G26R, and showed the mutation triggers secondary structure conversion with an increase in β-strand structure (6). Our current analyses of the fibrillogenic L178H apoA-I variant show that mutations in the C-terminal cluster can per se (and independently of other macromolecular factors) trigger fibril formation. Notably, despite the apparent presence of formed fibrils as shown by negative stain electron microscopy, our studies show no significant increase in ThT binding of the L178H apoA-I variant, nor an increase in β-strand structure but rather a large increase in helical structure. The apparent ordered aggregates of this particular mutant thus represent an alternate mode of fibril formation distinct from that of G26R.
Although amyloid fibrils have historically been defined as containing increased β-structure, evidence for the role of helical structure in amyloid formation and in fibrils associated with amyloid deposits is accumulating. For example, helical-containing assemblies as transient intermediate structures in the fibrillization process have been described for the apoE protein (22) and the Abeta peptide in Alzheimer's disease (23). Furthermore, helix formation may be a key requirement in the development of amyloid from intrinsically disordered proteins (24).
Several studies involving apoA-I peptides link α-helical conformation to fibril formation (25–27) and formation of helical intermediates has been postulated as a key step in the formation of β-amyloid structure (26). The most common species present in in vivo amyloid deposits is an ∼9 kDa N-terminal fragment. Di Gaetano et al. (26) analyzed recombinant 1–93 and demonstrated formation of helical structure prior to aggregation into fibrils, particularly under acidic conditions. However, the end fibrillar product contained β-amyloid structure. These authors suggested that helical intermediates precede the formation of cross-β structure, and that stabilization of helix through lipid-binding can slow or prevent the formation of β-amyloid (26, 27).
Other studies involving apoA-I peptides demonstrate the presence of helical structure within fibrils. Explanted fibrils of the amyloidogenic variant L174S from cardiac tissue were examined and found to contain both cross-β and residual helical structure (28). The reflections in the fiber diffraction pattern supported coiled-coil helical structure with the helix axis aligned perpendicular to the long axis of the fibril. Similarly, X-ray fiber diffraction of a series of synthetic peptides based on a single repeat of the 18A sequence (29) interspersed with turn sequences derived from human apoA-I demonstrated the formation of helical fibrils at physiological pHs in which the helix axis of the peptides is perpendicular to the fibril axis (25). In this case, only helical secondary structure was present.
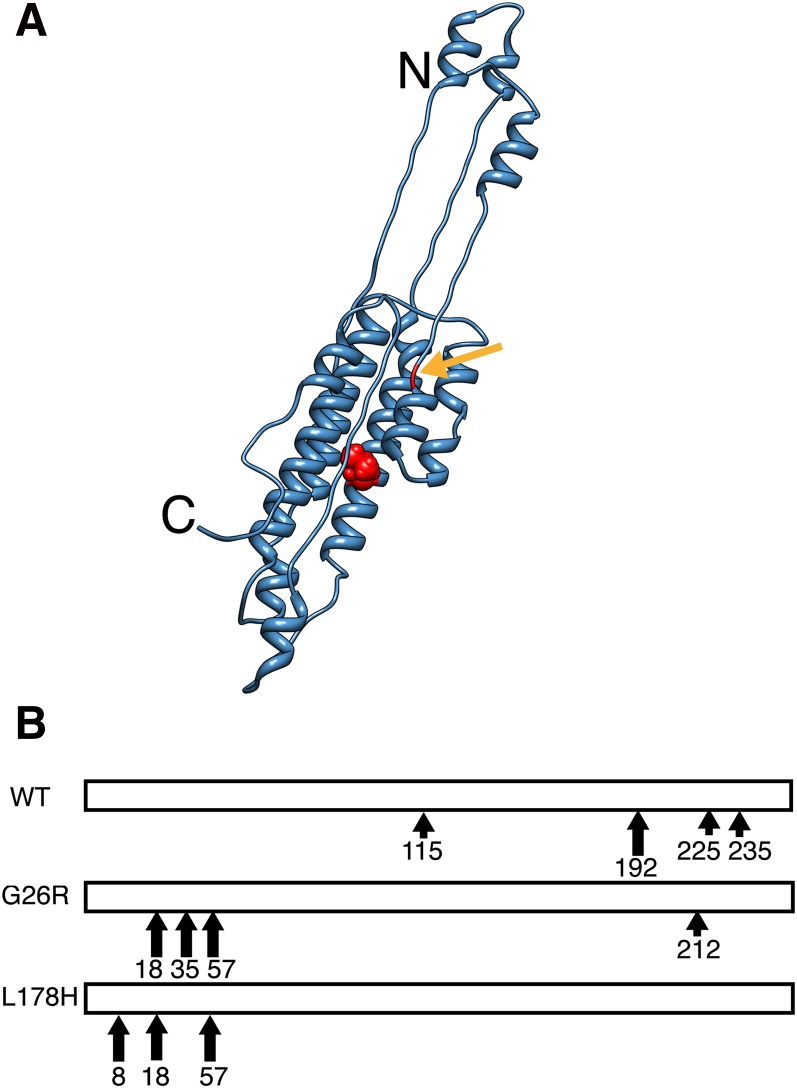
What underlies the differences in secondary structure in L178H versus G26R? L178H is an “outside” mutation,occurring outside the 1–90 segment that is found in most apoA-I amyloid deposits in vivo, whereas G26R is an “inside” mutation. However, both mutations are in the N-terminal helix bundle domain comprising residues 1–190 (20, 30, 31) (Fig. 6A). Both mutations also create adjacent positive charges (G26R results in R26,R27; L178H results in R177,H178). The two mutations produced similar initial structures as evidenced by increased ANS binding, increased N-terminal protease sensitivity, and decreased lipid binding ability. However, incubation over a relatively short time leads to an increase in helix content in L178H versus a decrease in helix content with increase in β structure in G26R. The changes in secondary structure in both proteins leads to aggregated forms with elongated fibrils being evident in L178H samples after 4 weeks of incubation and the formation of annular protofilaments within 1 week of incubation in G26R samples. The differences in these two variants may be due to the ability of the mutation to promote helical versus β structure over time. Interestingly, the L178H mutation actually reduces the predicted helix propensity compared with G26R (32), suggesting that the secondary structure that develops in the fibrils is strongly influenced by the mutations’ effect on the tertiary, rather than the secondary, structure. This is supported by the greatly reduced stability of L178H compared with WT proteins. How this effect on tertiary structure is manifested is unclear because the initial structures exhibit similar destabilization of their extreme amino-termini (Fig. 6B), which may result from the relatively close proximity of the two mutations within the N-terminal helix bundle domain (Fig. 6A). N-terminal destabilization may be a common theme underlying all amyloid apoA-I mutations, whether they be “inside” or “outside” mutations. An examination of the full-length structures and stabilities of all amyloid apoA-I variants will determine if this is the case.
Fig. 6.
Similar effect of “inside” and “outside” mutations on lipid-free structure. A: The side-chain of residue 178 is shown as a red sphere in the monomeric (blue), lipid-free apoA-I model structure [modified from (40)]. Arrow indicates residue 26 in the lipid-free structure. B, Comparison of the proteolytic cleavage sites in WT, G26R and L178H. Large and small arrows indicate major and minor cleavage sites, respectively.
Other plasma factors may also contribute to the fibrillogenesis of apoA-I. Transthyretin, a plasma transporter of retinol and thyroxine also known to be associated with HDL (33), colocalizes with L178H apoA-I in skin fibrils (7). This protein has proteolytic activity and cleaves WT apoA-I after phenylalanine 225 (34). A potential role of transthyretin in the generation of fibrillogenic apoA-I fragments has thus been proposed (35). In our limited proteolysis analyses, we previously reported cleavage near residue F225 for the WT protein (F225 and F229 using chymotrypsin) and in the G26R mutant (E223 using V8 protease) but here do not observe the same for the L178H mutant using similar conditions (Fig. 6B) (6). As transthyretin is regarded as a chymotrypsin-like protease with similar sensitivity to protease inhibitors as chymotrypsin (34), this finding may suggest that this position of the L178H variant could be less accessible for cleavage by the transthyretin protein. Indeed, a significant portion of the apoA-I protein found in the skin fibrils corresponds to full-length apoA-I (7). Finally, transthyretin is per se amyloidogenic (36), and it is not yet clear whether transthyretin and L178H apoA-I indeed form molecular complexes, or if the colocalization is due to other factors such as cell and tissue preferences.
Further analysis on the influence of transthyretins on the fibrillogenesis of the L178H apoA-I variant will be of great importance to understanding the interplay of the proteins in the formation of fibrils. In addition, more careful analysis of aggregation kinetics and fibril morphologies between G26R and L178H, analysis of full-length structures of “inside” versus “outside” fibrillogenic mutants, and the relationship between mutation position and type of fibril formed as well as tissue localization will provide mechanistic details that will be of clinical relevance.
Note added in proof
Since the acceptance of this paper, the authors have had a paper accepted (40) that better reflects the model described in Fig. 6. The original accepted version of this paper cited ref. 10 and did not include ref. 40.
Acknowledgments
The authors thank Maria Baumgarten for skillful work and Rita Wallén for help with electron microscopy. The authors also thank Dr. John C. Voss (University of California) for careful reading and constructive comments on the manuscript.
Footnotes
Abbreviations:
- ANS
- 1-Anilinonaphthalene-8-sulfonic acid
- CD
- circular dichroism
- DMPC
- dimyristoylphosphatidylcholine
- D1/2
- the concentration of denaturant at which the protein is half folded
- G26R
- glycine to arginine mutation at residue 26
- Gn-HCl
- guanidine
- L178H
- leucine to histidine mutation at residue 178
- rHDL
- reconstituted HDL
- ThT
- thioflavin T
- t1/2
- half time
- WMI
- wavelength at maximum fluorescence intensity
- WT
- wild-type
This work was supported by grants from the Swedish Research Council (522-2008-3724, 7480), the Wenner-Gren Foundation, the Petrus and Augusta Hedlund Foundation, the Crafoord Foundation, the Carl Trygger Foundation for Scientific Research, the Magnus Bergvall Foundation, the Greta and Johan Kocks Foundation, by an equipment grant from the Knut and Alice Wallenberg Foundation, and by internal awards programs at California State University, Sacramento.
REFERENCES
- 1.Tardif J. C. 2010. Emerging high-density lipoprotein infusion therapies: fulfilling the promise of epidemiology? J. Clin. Lipidol. 4: 399–404. [DOI] [PubMed] [Google Scholar]
- 2.Westermark P., Mucchiano G., Marthin T., Johnson K. H., Sletten K. 1995. Apolipoprotein A1-derived amyloid in human aortic atherosclerotic plaques. Am. J. Pathol. 147: 1186–1192. [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnato C., Thumar J., Mayya V., Hwang S. I., Zebroski H., Claffey K. P., Haudenschild C., Eng J. K., Lundgren D. H., Han D. K. 2007. Proteomics analysis of human coronary atherosclerotic plaque: a feasibility study of direct tissue proteomics by liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteomics. 6: 1088–1102. [DOI] [PubMed] [Google Scholar]
- 4.Lepedda A. J., Cigliano A., Cherchi G. M., Spirito R., Maggioni M., Carta F., Turrini F., Edelstein C., Scanu A. M., Formato M. 2009. A proteomic approach to differentiate histologically classified stable and unstable plaques from human carotid arteries. Atherosclerosis. 203: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreola A., Bellotti V., Giorgetti S., Mangione P., Obici L., Stoppini M., Torres J., Monzani E., Merlini G., Sunde M. 2003. Conformational switching and fibrillogenesis in the amyloidogenic fragment of apolipoprotein a-I. J. Biol. Chem. 278: 2444–2451. [DOI] [PubMed] [Google Scholar]
- 6.Lagerstedt J. O., Cavigiolio G., Roberts L. M., Hong H. S., Jin L. W., Fitzgerald P. G., Oda M. N., Voss J. C. 2007. Mapping the structural transition in an amyloidogenic apolipoprotein A-I. Biochemistry. 46: 9693–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Sousa M. M., Vital C., Ostler D., Fernandes R., Pouget-Abadie J., Carles D., Saraiva M. J. 2000. Apolipoprotein AI and transthyretin as components of amyloid fibrils in a kindred with apoAI Leu178His amyloidosis. Am. J. Pathol. 156: 1911–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazenberg A. J., Dikkers F. G., Hawkins P. N., Bijzet J., Rowczenio D., Gilbertson J., Posthumus M. D., Leijsma M. K., Hazenberg B. P. 2009. Laryngeal presentation of systemic apolipoprotein A-I-derived amyloidosis. Laryngoscope. 119: 608–615. [DOI] [PubMed] [Google Scholar]
- 9.Ryan R. O., Forte T. M., Oda M. N. 2003. Optimized bacterial expression of human apolipoprotein A-I. Protein Expr. Purif. 27: 98–103. [DOI] [PubMed] [Google Scholar]
- 10.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. 2007. Electron paramagnetic resonance spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apoA-I in solution. J. Biol. Chem. 282: 9143–9149. [DOI] [PubMed] [Google Scholar]
- 11.Morrow J. A., Segall M. L., Lund-Katz S., Phillips M. C., Knapp M., Rupp B., Weisgraber K. H. 2000. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 39: 11657–11666. [DOI] [PubMed] [Google Scholar]
- 12.Koyama M., Tanaka M., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2009. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 48: 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastukhov A. V., Ropson I. J. 2003. Fluorescent dyes as probes to study lipid-binding proteins. Proteins. 53: 607–615. [DOI] [PubMed] [Google Scholar]
- 14.Ji Y., Jonas A. 1995. Properties of an N-terminal proteolytic fragment of apolipoprotein AI in solution and in reconstituted high density lipoproteins. J. Biol. Chem. 270: 11290–11297. [DOI] [PubMed] [Google Scholar]
- 15.Roberts L. M., Ray M. J., Shih T. W., Hayden E., Reader M. M., Brouillette C. G. 1997. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apo A-I. Biochemistry. 36: 7615–7624. [DOI] [PubMed] [Google Scholar]
- 16.Engel J., Furthmayr H. 1987. Electron microscopy and other physical methods for the characterization of extracellular matrix components: laminin, fibronectin, collagen IV, collagen VI, and proteoglycans. Methods Enzymol. 145: 3–78. [DOI] [PubMed] [Google Scholar]
- 17.Panagotopulos S. E., Witting S. R., Horace E. M., Hui D. Y., Maiorano J. N., Davidson W. S. 2002. The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 277: 39477–39484. [DOI] [PubMed] [Google Scholar]
- 18.Jayaraman S., Abe-Dohmae S., Yokoyama S., Cavigiolio G. 2011. Impact of self-association on function of apolipoprotein A-I. J. Biol. Chem. 286: 35610–35623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brubaker G., Peng D. Q., Somerlot B., Abdollahian D. J., Smith J. D. 2006. Apolipoprotein A-I lysine modification: effects on helical content, lipid binding and cholesterol acceptor activity. Biochim. Biophys. Acta. 1761: 64–72. [DOI] [PubMed] [Google Scholar]
- 20.Silva R. A., Hilliard G. M., Fang J., Macha S., Davidson W. S. 2005. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 44: 2759–2769. [DOI] [PubMed] [Google Scholar]
- 21.Zannis V. I., Chroni A., Krieger M. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. (Berl). 84: 276–294. [DOI] [PubMed] [Google Scholar]
- 22.Hatters D. M., Zhong N., Rutenber E., Weisgraber K. H. 2006. Amino-terminal domain stability mediates apolipoprotein E aggregation into neurotoxic fibrils. J. Mol. Biol. 361: 932–944. [DOI] [PubMed] [Google Scholar]
- 23.Kirkitadze M. D., Condron M. M., Teplow D. B. 2001. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J. Mol. Biol. 312: 1103–1119. [DOI] [PubMed] [Google Scholar]
- 24.Abedini A., Raleigh D. P. 2009. A role for helical intermediates in amyloid formation by natively unfolded polypeptides? Phys. Biol. 6: 015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazar K. L., Miller-Auer H., Getz G. S., Orgel J. P., Meredith S. C. 2005. Helix-turn-helix peptides that form alpha-helical fibrils: turn sequences drive fibril structure. Biochemistry. 44: 12681–12689. [DOI] [PubMed] [Google Scholar]
- 26.Di Gaetano S., Guglielmi F., Arciello A., Mangione P., Monti M., Pagnozzi D., Raimondi S., Giorgetti S., Orru S., Canale C., et al. 2006. Recombinant amyloidogenic domain of ApoA-I: analysis of its fibrillogenic potential. Biochem. Biophys. Res. Commun. 351: 223–228. [DOI] [PubMed] [Google Scholar]
- 27.Monti D. M., Guglielmi F., Monti M., Cozzolino F., Torrassa S., Relini A., Pucci P., Arciello A., Piccoli R. 2010. Effects of a lipid environment on the fibrillogenic pathway of the N-terminal polypeptide of human apolipoprotein A-I, responsible for in vivo amyloid fibril formation. Eur. Biophys. J. 39: 1289–1299. [DOI] [PubMed] [Google Scholar]
- 28.Mangione P., Sunde M., Giorgetti S., Stoppini M., Esposito G., Gianelli L., Obici L., Asti L., Andreola A., Viglino P., et al. 2001. Amyloid fibrils derived from the apolipoprotein A1 Leu174Ser variant contain elements of ordered helical structure. Protein Sci. 10: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anantharamaiah G. M., Jones J. L., Brouillette C. G., Schmidt C. F., Chung B. H., Hughes T. A., Bhown A. S., Segrest J. P. 1985. Studies of synthetic peptide analogs of the amphipathic helix. Structure of complexes with dimyristoyl phosphatidylcholine. J. Biol. Chem. 260: 10248–10255. [PubMed] [Google Scholar]
- 30.Rogers D. P., Roberts L. M., Lebowitz J., Engler J. A., Brouillette C. G. 1998. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 37: 945–955. [DOI] [PubMed] [Google Scholar]
- 31.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obici L., Franceschini G., Calabresi L., Giorgetti S., Stoppini M., Merlini G., Bellotti V. 2006. Structure, function and amyloidogenic propensity of apolipoprotein A-I. Amyloid. 13: 191–205. [DOI] [PubMed] [Google Scholar]
- 33.Sousa M. M., Berglund L., Saraiva M. J. 2000. Transthyretin in high density lipoproteins: association with apolipoprotein A-I. J. Lipid Res. 41: 58–65. [PubMed] [Google Scholar]
- 34.Liz M. A., Faro C. J., Saraiva M. J., Sousa M. M. 2004. Transthyretin, a new cryptic protease. J. Biol. Chem. 279: 21431–21438. [DOI] [PubMed] [Google Scholar]
- 35.Liz M. A., Gomes C. M., Saraiva M. J., Sousa M. M. 2007. ApoA-I cleaved by transthyretin has reduced ability to promote cholesterol efflux and increased amyloidogenicity. J. Lipid Res. 48: 2385–2395. [DOI] [PubMed] [Google Scholar]
- 36.Westermark P., Sletten K., Johansson B., Cornwell G. G., 3rd 1990. Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc. Natl. Acad. Sci. USA. 87: 2843–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers D. P., Roberts L. M., Lebowitz J., Datta G., Anantharamaiah G. M., Engler J. A., Brouillette C. G. 1998. The lipid-free structure of apolipoprotein A-I: effects of amino-terminal deletions. Biochemistry. 37: 11714–11725. [DOI] [PubMed] [Google Scholar]
- 38.Alexander E. T., Tanaka M., Kono M., Saito H., Rader D. J., Phillips M. C. 2009. Structural and functional consequences of the Milano mutation (R173C) in human apolipoprotein A-I. J. Lipid Res. 50: 1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka M., Dhanasekaran P., Nguyen D., Nickel M., Takechi Y., Lund-Katz S., Phillips M. C., Saito H. 2011. Influence of N-terminal helix bundle stability on the lipid-binding properties of human apolipoprotein A-I. Biochim. Biophys. Acta. 1811: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagerstedt J. O., Budamagunta M. S., Liu G. S., DeValle N. C., Voss J. C., Oda M. N. 2011. The “Beta-Clasp” model of apolipoprotein A-I - a lipid-free solution structure determined by electron paramagnetic resonance spectroscopy. Biochim. Biophys. Acta. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]