Abstract
Sphingolipids are not only important components of membranes but also have functions in protein trafficking and intracellular signaling. The LCB1 gene encodes a subunit of the serine palmitoyltransferase, which is responsible for the first step of sphingolipid synthesis. Here, we show that activation of the unfolded protein response (UPR) can restore normal ceramide levels and viability in yeast cells with a conditional defect in LCB1. Dependence on UPR was demonstrated by showing the HAC1-dependence of the suppression. A similar induction of ceramides by UPR seems to take place in mammalian cells. In rat pancreatic INS-1E cells, UPR activation induces the transcription of the CerS6 gene, which encodes a ceramide synthase. This correlates with the specific accumulation of ceramide with a C16 fatty acyl chain upon UPR activation. Therefore, our study reveals a novel connection between UPR induction and ceramide synthesis that seems to be conserved between yeast and mammalian cells.
Keywords: sphingolipids, serine palmitoyltransferase, endoplasmic reticulum stress, CerS6, fatty acid hydroxylation
The sphingolipid biosynthesis pathways in yeast and mammals are conserved up to the synthesis of dihydroceramide (1, 2). In yeast, ceramides are synthesized using mainly C26 acyl-CoA by two redundant proteins (3, 4), Lag1p and Lac1p, whereas in mammalian cells, six ceramide synthases (CerS1 through CerS6) have specific fatty acyl CoA chain length preferences (5). The sphingolipid synthesis pathway starts with the condensation of palmitoyl-CoA and serine carried out by the enzyme serine palmitoyltransferase (decarboxylating) (SPT). In yeast, one of the subunits of this enzyme is encoded by LCB1, an essential gene (6). A temperature-sensitive mutant in this gene was isolated originally as a mutant defective in the internalization step of endocytosis (7). When the corresponding gene was isolated, the temperature-sensitive allele was named lcb1-100 and studies showed that sphingoid bases play a crucial role in the internalization step of endocytosis, because addition of stereoisomers of sphinganine that cannot be converted to dihydroceramide could restore endocytosis (8). The strain shows reduced sphingolipid synthesis at permissive temperature, which is accentuated at nonpermissive temperature. Subsequently, the lcb1-100 allele has been used to investigate the role of sphingolipid biosynthesis in the heat shock response (9–11), to differentiate between the contribution of de novo and degradative pathways in sphingolipid function (12), and to study trafficking and function of solute transporters (13–15), the role of sphingolipids in membrane domain formation (16), and the intracellular transport of glycosylphosphosphatidylinositol (GPI)-anchored proteins (17, 18).
De novo synthesis of sphingolipids was first shown to be required for the transport of GPI-anchored proteins from the endoplasmic reticulum (ER) to the Golgi compartment by discovery that myriocin (ISP-1), an inhibitor of SPT, rapidly inhibits this pathway (19). Subsequently, it was shown that the lcb1-100 mutant is defective in the same transport step (17). Another set of proteins that are required for the transport of GPI-anchored proteins to the Golgi in yeast and mammalian cells is the p24 family (20–23). Mutations of members of the p24 complex in yeast, emp24 and erv25 for instance, have been shown to induce the unfolded protein response (UPR) (23–25).
The UPR is a pathway activated to protect cells when misfolded proteins accumulate in the ER. Many components of this signaling cascade were first discovered in yeast (26, 27). The HAC1 gene has been identified as an essential transcription factor required for the activation of UPR response (26). Genome-wide studies have identified a number of proteins that are either upregulated or downregulated in cells due to the accumulation of unfolded proteins in the ER (28, 29). This connects the activation of UPR with many other pathways than just the regulation of ER resident proteins and their refolding or degradation.
Recently, evidence has surfaced for an involvement of ceramide synthases in the activation of UPR response. The downregulation of CerS2-affected ceramide homeostasis leading to an increase in C16 ceramide levels, probably resulting from upregulation of CerS5 and CerS6 mRNAs. It also led to a series of physiological responses, including induction of UPR (30). Other lipids have also been implicated in the induction of UPR response. The upregulation of sphingosine-1-phosphate was shown to induce UPR (31). One mammalian cell line in which UPR and the effect of lipids have been best studied is INS-1E cells. These rat insulinoma-derived cells constitute a widely used β-cell surrogate and have been cloned into a stable cell line (32). It has been shown that p24 proteins are required in this cell line for insulin biosynthesis and secretion (33).
To investigate the interaction between the roles of ceramide and p24 proteins in GPI-anchored protein transport, we created a double mutant, emp24 lcb1-100, and analyzed its phenotype. We expected to see a defect equivalent to the more severe of the two mutants, lcb1-100. Surprisingly, this was not the case; rather, emp24 seemed to suppress the defect of the lcb1-100 mutation. The mechanistic explanation for this uncovers a novel connection between UPR induction and ceramide synthesis that seems to be conserved in INS-1E insulinoma cells.
MATERIAL AND METHODS
Strains and reagents
The strains used in this study were the following: RH2888 (MATa his3 leu2 lys2 trp1 ura3 can1 bar1), which corresponds to our lab background wild-type, RH3948 (MATa lcb1-100 his4 leu2 lys2 ura3), RH3949 (MATa lcb1-100 emp24Δ::KanMx his4 leu2 lys2 ura3), RH3950 (MATα emp24Δ::KanMx his4 leu2 lys2 ura3), RH7307 (MATa hac1Δ::KanMx his3 leu2 trp1 ura3), RH7333 (MATa lcb1-100 hac1Δ::KanMx his3 leu2 ura3 bar1), RH6974 (MATα TDH3::FLAG-MmCerS5::TRP1 lac1Δ::ADE2 lag1Δ::HIS3 leu2 ura3 lys2 can1 bar1), and RH6975 (MATa TDH3::FLAG-MmCerS6::TRP1 lac1Δ::ADE2 lag1Δ::HIS3 leu2 ura3 lys2 can1 bar1). The wild-type strain corresponding to the cbr1Δ::KanMx mutation is BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the mutant strain was made by the Saccharomyces cerevisiae. deletion consortium (34). All mutations were constructed using standard gene disruption procedures, with complete removal of open reading frames in the RH2888 strain background. Double and triple mutants were obtained by genetic crosses. Synthetic N-terminally FLAG epitope-tagged open reading frames corresponding to mouse CerS5 and CerS6 were purchased from GeneArt (Germany) as yeast codon optimized open reading frames, subcloned for expression under control of a TDH3 promoter and inserted into the genome to construct stable cell lines. Sequences are available upon request.
For CerS assay in vivo and in vitro, strains RH2888, RH6974, and RH6975 were grown in rich medium to a final concentration of 1 OD600/ml. Rich medium (20 g/l glucose, 20 g/l peptone, and 10 g/l yeast extract with adenine, uracil, and tryptophan at 40 mg/l) was used in all experiments. Aureobasidin A was purchased from Takara Shuzo (Shiga, Japan) and myriocin from Sigma (St. Louis, MO).
Pulse-chase analysis and dilution plating
Yeast strains were grown overnight in SDYE and pulse-labeled for 5 min with 35S methionine and cysteine (Expre35S35S; NEN-Dupont) and chased for the indicated times in presence of methionine, cysteine, and ammonium sulfate. Cells were extracted, Gas1p immunoprecipitated, and analyzed as described (19).
For the plating assays, yeast were grown to stationary phase and diluted to 107 cells per ml in water, and 10-fold serial dilutions were prepared and pinned onto agar plates containing rich medium with or without the indicated additives. Each experiment was repeated at least three times and the pictures shown are representative of the results found.
Cell culture
INS-1E cells (32) were routinely cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, 2 mM glutamine, 10 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin. This medium contains 11.2 mM glucose; for test conditions, glucose was supplemented to 30 mM or DTT to 10 mM as appropriate.
Lipid extraction
For yeast in vivo assay, lipids were extracted from 50 ml of cells as described below. For the extraction of yeast lipids, internal standards were added to the samples [10 µg each of Cer (d18:1,17:0) and GlcCer (d18:1,8:0), Avanti Polar Lipids], which were then extracted using an ethanol-ether based solvent and base-treated with monomethylamine as described (35). Lipids were desalted with butanol and analyzed by mass spectrometry.
Mammalian cells were scraped in 2 × 800 µl water and internal standards were added [0.5 µg of Cer (d18:1,17:0) and 0.1 µg GlcCer (d18:1,8:0)]. Three milliliters chloroform-methanol 2:1 were added to the pooled solvent, samples were sonicated for 5 min and incubated at 50°C for 30 min. One milliliter chloroform and 1 ml water were added, vortexed, and the whole extract was centrifuged at 3,200 g, 4°C for 5 min. The upper phase was discarded and lower phase was dried, base-treated, and desalted as described above.
ESI-MS analysis
For the quantification of yeast and mammalian extracts, lipids were introduced into a Varian 320MS triple quadrupole in LC mode by direct infusion in methanol-chloroform-water (7/2/1, containing 2 mM ammonium acetate) (36). To quantify lipid species, multiple ion monitoring was used for each species (35). Data were collected from at least three independent samples with triplicate measurements. The recoveries and amounts of different inositolphosphorylceramide (IPC) species were calculated relative to the GlcCer (d18:1, 8:0) or to Cer (d18:1, 17:0) standard according to a standard curve. Results are presented as arbitrary units detected because absolute amounts could not be determined without an identical internal standard. The mass spectrometry measurements for the cbr1 mutant and corresponding wild-type were performed using a TSQ Vantage mass spectrometer equipped with an Advion Nanomate for direct infusion in positive and negative modes. First, data from the mass spectrometry measurements were converted from the proprietary formats (.xms and .raw files for Varian and Thermo machines, respectively) to a list of intensities for each transition. Second, signals were computed, automatically assigned to lipid species, and fitted to standard curves to deduce their quantity (Qlip) in the sample, knowing the quantity of standard injected before the extraction (Qstd_injected) and the quantities of each lipid and related standard as detected by the machine.
These quantities of each lipid (Qlip_computed) and the associated standard (Qstd_computed) are computed from the respective measured signals and the linear region of standard curves.
where a and b are the coefficients of the linear portion of the standard curve used for a given lipid.
Finally, for each lipid species, we combined the results from technical replicates.
In vitro assay of dihydroceramide synthase
Five milliliters of cells grown to 1 OD600/ml were harvested and spheroplasted as described. Supernatant was centrifuged and resuspended in 200 µl TE 10 mM Tris.HCl 1 mM EDTA (pH 7.5) with protease inhibitors and homogenized using a 20 gauge syringe. Protein concentrations were determined by Bradford (BioRad Protein Assay) according to manufacturer's protocol. Samples were quickly frozen in liquid nitrogen and kept at −20°C. Homogenates were assayed as previously described (11) using 3H-sphinganine and C16 acyl-CoAs, in a reaction of 100 μl, containing BSA. The reaction products were isolated and dihydroceramide was separated by thin layer chromatography (solvent, CHCl3:CH3OH, 100:7) and exposed to a phosphoimager screen. Images were quantified using ImageQuant (PerkinElmer).
RNA extraction and quantitative RT-PCR
Total RNA was extracted from INS-1E cells with the Qiagen RNEasy Mini kit according to the manufacturer's instructions. Contaminating DNA was removed by subsequent digestion with DNase I (Ambion/Applied Biosystems). RNA was converted to cDNA using SuperScript II (Invitrogen). All primers were designed using Primer Express software (Applied Biosystems). Real-time PCR was carried out on an ABI 7000 Sequence Detection system (Applied Biosystems) with the SYBR Green reagent (Roche) for quantification of product fluorescence using the standard curve method. Amplifications were performed in duplicate for four independent experiments and the values normalized to the housekeeping gene cyclophilin D. Test reactions were performed on DNase I treated RNA to ensure that no product was amplified from contaminating genomic DNA. Gene names and primers used can be found in Table 1.
TABLE 1.
Quantitative RT-PCR primer sequences
| Gene | Symbol | Forward primer | Reverse primer |
| Acetyl CoA carboxylase α | acc | caacgccttcacaccacctt | tcatcaaagatcctgacgaaatctt |
| ATP citrate lyase | acly | aggcagcattgcaaacttca | ggaccctggtaatctcgaattg |
| Fatty acid synthase | fasn | ggacatggtcacagacgatgac | gtcgaacttggacagatccttca |
| Ketosphinganine reductase | kdsr | tctcccgcaaacccaaac | tctgcccgagcgtctactg |
| Ceramide synthase 5 | cers5 | catcagtttctcctacatcaacaaca | gtcggcggagtcatgca |
| Ceramide synthase 6 | cers6 | cccttggccttctgcatct | ggcacatggtttggctatga |
| Serine palmitoyltransferase | spt | caccgagcactatgggatca | cgagcgcattctccatgtta |
| Neutral sphingomyelinase | nsm | ccggatgcacactacttcagaa | ggattgggtgtctggagaaca |
| Glucose-regulated protein 78 | bip | cacgtccaacccggagaac | ttccaagtgcgtccgatga |
| C/EBP homologous protein | chop | ggaaagtggcacagcttgct | ctggtcaggcgctcgatt |
Statistical analysis
All results are representative of at least three independent experiments. All statistical analysis was performed using Student's t test and differences between pairs were considered significant for P < 0.05 (*).
RESULTS
Deletion of EMP24 partially rescues the GPI-anchored protein transport defect and growth at nonpermissive temperature of the lcb1-100 mutant
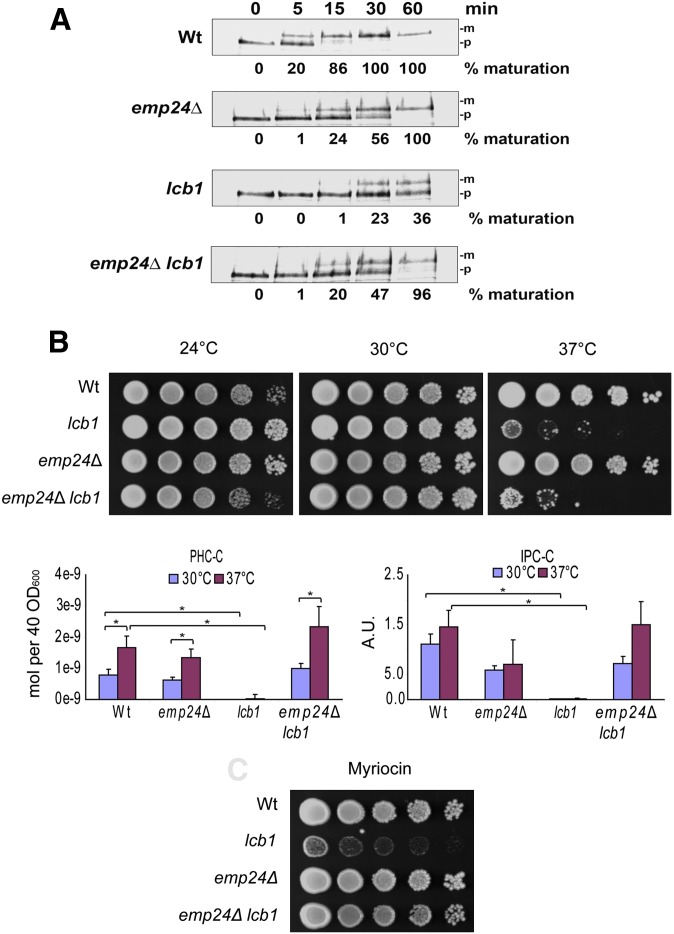
As described above, mutations in either sphingolipid biosynthesis or in members of the p24 family have a strong effect on the transport of GPI-anchored proteins from the ER to the Golgi compartment (17, 19, 20). The transport of GPI-anchored proteins is slower in the emp24 mutant than in the wild-type cells but eventually reaches completion after 60 min chase. The lcb1-100 mutant has a stronger phenotype and at the end of the chase period, only 36% of the GPI-anchored protein Gas1p is matured. In order to examine a possible interaction between these two requirements, we constructed a double mutant, emp24 lcb1-100, and measured the rate of maturation of Gas1p as a measure of transport to the Golgi compartment. We expected the transport defect to be similar to that of the lcb1-100 mutant at 37°C because it has the more severe phenotype. Surprisingly, the transport defect was virtually identical to that in the emp24 mutant (Fig. 1A). The lcb1-100 allele grew normally at 30°C but the strain was unviable at 37°C. Consistent with the suppression of the GPI-anchored protein trafficking defect, the growth defect of lcb1-100 was also partially suppressed by the emp24 mutation and the double mutant grew better at 30°C than the lcb1-100 single mutant (Fig. 1B). The emp24 mutation was unable to suppress the growth defect of lcb1-100 at 37°C.
Fig. 1.
Deletion of EMP24 partially rescues the lcb1-100 phenotype. A: Pulse-chase analysis of the maturation of the GPI-anchored protein, Gas1p, in wild-type RH2888 (Wt), lcb1-100 cells, RH3948 (lcb1), emp24Δ cells, RH3950 (emp24Δ), and double mutant cells RH3949 (emp24Δ lcb1-100). The precursor form (p) of Gas1p is generated in the ER and maturation (m) occurs upon arrival in the Golgi compartment. The percentage of mature form is quantified. B: Serial dilutions of wild-type and mutant cells were spotted on YPD plates and incubated at the indicated temperatures for 3 days. Quantification by ESI-MS of the major ceramide and IPC species produced in each strain after incubation overnight at 30°C with or without a 1 h shift to 37°C before harvesting. Asterisks indicate P < 0.05 for the pairwise combinations designated. C: Dilution series of strains were spotted as in (B) onto plates containing 0.5µg/ml of myriocin, grown at 30°C. Plates shown are representative of at least three replicates per condition.
To understand the suppression effect, we first investigated whether the sphingolipid levels were affected by introduction of the emp24 mutation. We grew the single and double mutants at 30°C, divided them in two, and shifted one aliquot to 37°C for 1 h. Lipids were extracted and analyzed by ESI-MS. Strikingly, the double mutant had normal levels of sphingolipids when grown at 30°C, providing an explanation for the suppression. The lcb1-100 strain had almost no detectable t18:0/26:0 phytoceramide C (PHC-C, the major ceramide species in yeast) and t18:0/26:0 inositolphytoceramide C (IPC-C, the major inositolceramide). However, the double mutant had normal levels of both lipids when grown at 30°C and even when heat-shocked for 1 h at 37°C (Fig. 1C).
We also tested the behavior of both strains in the presence of myriocin, a compound that inhibits the activity of SPT. In the presence of sublethal concentrations of myriocin, the wild-type and double mutant grew normally whereas the lcb1-100 mutant grew poorly (Fig. 1D).
Activation of the UPR pathway by addition of DTT rescues the lcb1-100 temperature sensitivity
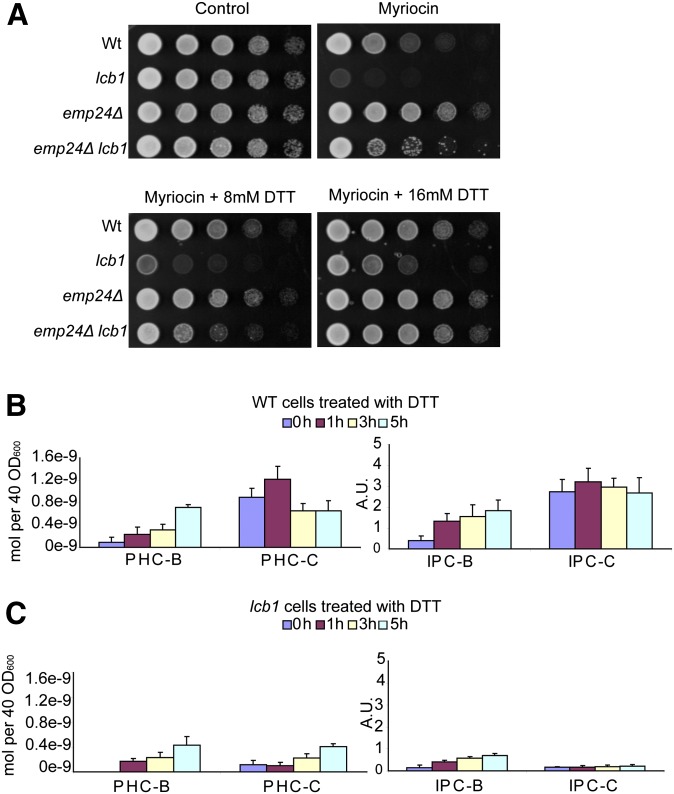
One possible explanation for the mechanism of suppression by the emp24Δ mutation is that the UPR pathway, which is also induced by the mutation, regulates the ceramide synthesis pathway, leading to an increase in SPT activity. To test this hypothesis, we plated the four strains on rich solid medium containing myriocin to inhibit SPT and added increasing amounts of the UPR inducer DTT (28). DTT is a strong reducing agent, which prevents protein disulfide formation. Treatment with DTT partially rescued the lcb1-100 mutant sensitivity to the sublethal dose of myriocin with DTT giving a concentration dependent improvement in growth (Fig. 2A). When the effect of DTT on sphingolipids was analyzed, we found that the total amounts of PHC and IPC were increased (Table 2). This increase was more prominent on lipids without a hydroxylation on the fatty acyl moiety (PHC-B and IPC-B), which are not normally found in high amounts (Fig. 2B, C).
Fig. 2.
Activation of the UPR pathway by addition of DTT rescues the lcb1-100 phenotype. A: Serial dilutions of wild-type and mutant strains were prepared and spotted as in Fig. 1 onto plates containing myriocin. Increasing amounts of DTT were added to induce UPR response. B, C: ESI-MS identification and quantification of the major ceramides and IPCs of wild type (WT) (B) and lcb1-100 (C) strains. Error bars represent the SD of three analyzed extracts.
TABLE 2.
Total ceramides found in WT, lcb1, and hac1Δ strains as determined from data presented in Figs. 2 and 3.
| Total ceramides(hours after DTT treatment) |
||||
| 0 h | 1 h | 3 h | 5 h | |
| WT | 9.84E+01 | 1.46E+02 (*) | 9.67E+01 | 1.37E+02 (*) |
| lcb1-100 | 1.19E+01 | 2.65E+01 (*) | 4.57E+01 (*) | 8.35E+01 (*) |
| hac1Δ | 1.42E+02 | 8.03E+01 | 9.25E+01 | 1.30E+02 |
Amounts are given in nanomoles per 40 OD600 units of cells. Asterisks indicate P < 0.05 relative to 0 h in the same strain.
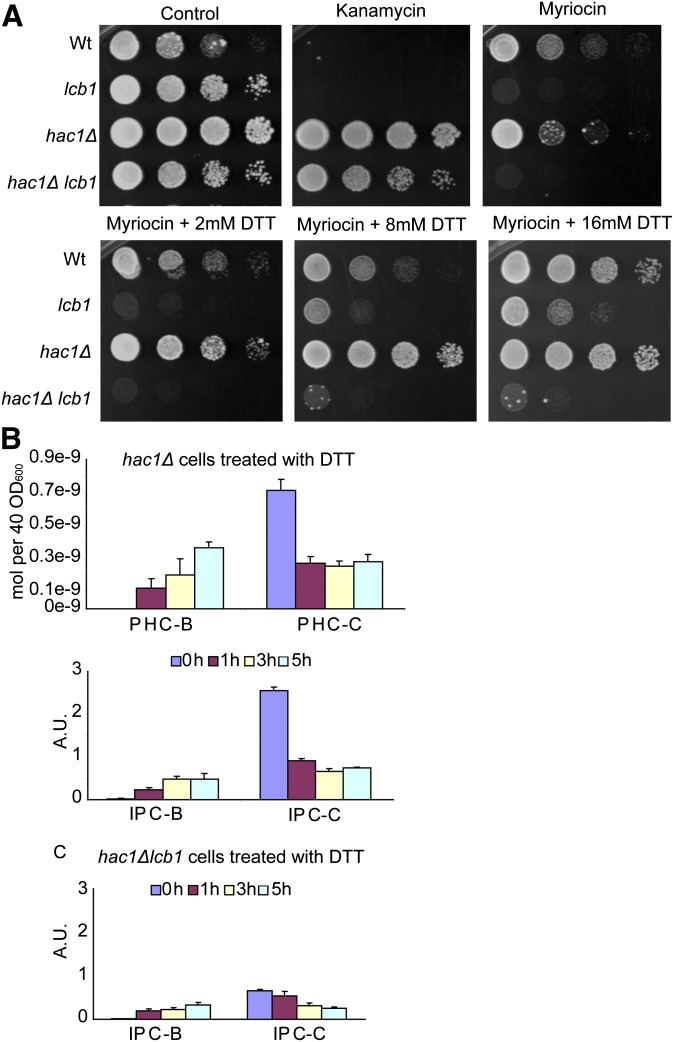
Deletion of HAC1 prevents DTT rescue of mutant strains
To prove that activation of the UPR was responsible for the rescue of lcb1-100 sensitivity to myriocin, we blocked UPR induction by introducing the hac1Δ mutation into the lcb1-100 strain (Fig. 3A). HAC1 encodes a transcription factor that is necessary to induce UPR (26). In the hac1Δ lcb1-100 strain, the addition of DTT was unable to rescue growth in the presence of sublethal doses of myriocin. Sphingolipid analysis also revealed that there was no increase in total amounts of ceramides or IPCs in the hac1Δ mutant (Table 2), even though we still found a decrease in fatty acid hydroxylation in the sphingolipids, suggesting that the hydroxylation effect does not pass through the UPR pathway and might be more direct (Fig. 3B). In the hac1Δ lcb1-100 double mutant, no ceramides were detected, a fact that can be explained by the low amounts of ceramide found in the lcb1-100 mutant and the inability to increase the IPCs due to the hac1 mutation (Fig. 3C).
Fig. 3.
Deletion of HAC1 prevents DTT rescue of mutant strains. A: Serial dilutions of wild-type, lcb1-100, hac1, and lcb1-100 hac1 strains were prepared and spotted on plates as in Fig. 2. Cells were plated on control YPD plates, with kanamycin (as a marker for hac1 deletion), and YPD plus myriocin with increasing amounts of DTT. All plates were grown at 30°C and plates shown are representative of three replicates per condition. B, C: ESI-MS identification and quantification of the major ceramides and IPCs of hac1 (B) and of IPCs of lcb1-100 hac1 (C) strains. Error bars represent the SD of three analyzed extracts.
The specific increase in the amount of B forms of ceramides and sphingolipids in the presence of DTT, which did not depend upon UPR, was intriguing. We reasoned that this could be due to reduction of an essential factor that is required for fatty acid hydroxylation. In contrast to the Sur2p hydroxylase, which acts on the sphingoid base (37), the Scs7p hydroxylase, working on the fatty acid, contains a cytochrome b5 domain (38). However, no cytochrome b reductase that is specific for this hydroxylation has been identified. We examined the lipid profile of a mutant in a candidate gene for this function, CBR1, which encodes a cytochrome b5 reductase homolog (39). The cbr1Δ mutant accumulates much higher amounts of the B forms of ceramides and sphingolipids than wild-type cells (Table 3). We conclude that Cbr1p encodes a cytochrome b5 reductase that donates electrons for the hydroxylation of fatty acids in sphingolipids. A redox hydroxylation reaction is a likely candidate for inhibition by DTT and could be a trivial explanation for accumulation of B rather than C forms of sphingolipids. It is likely that there are other genes with a redundant cytochrome b5 reductase activity, which could explain why the effect of the cbr1 mutation is not complete.
TABLE 3.
Phytoceramide and IPC species in wild-type and cbr1 mutant yeast
| Strain | PHC-B | PHC-C | IPC-B | IPC-C |
| Wild-type | 0.5% | 99.5% | 0.25% | 99.75% |
| cbr1 | 15.2% | 84.8% | 14.1% | 85.9% |
Ceramide and IPC species were quantified by mass spectrometry and the percentage of total amounts that represent the B (non hydroxylated on the fatty acyl chain) or C (hydroxylated on the fatty acyl chain) species are shown.
In mammalian cells induction of the UPR pathway leads to an increase in C16 ceramide but not in sphingomyelin
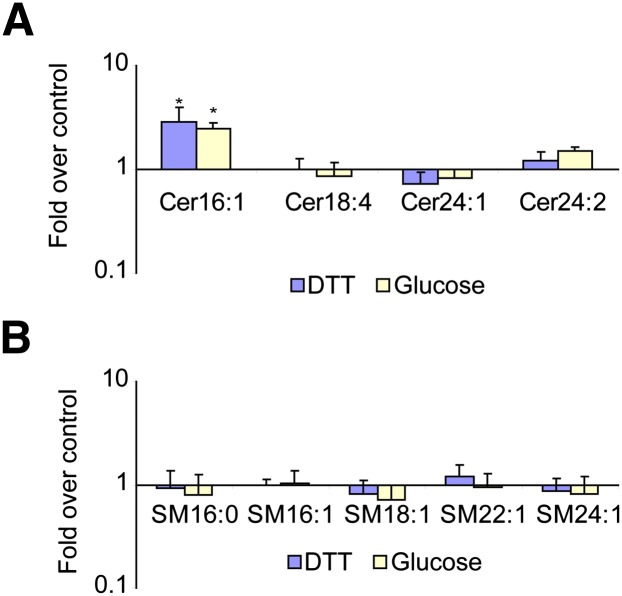
From the experiments above, it seems clear that induction of UPR increases the production of ceramide and sphingolipids. As ceramides are important signaling molecules in mammals and are thought to play roles in many processes related to cell survival, we thought it important to determine whether ceramides are also induced in a mammalian cell culture system. Insulin secreting cells are prone to ER stress, as insulin must be processed and folded in the ER lumen. Disruption of protein folding, for example, with denaturing agents such as DTT, promptly induces the ER stress response. High glucose may also induce some branches of the UPR, although conflicting reports have been published (40–43). Moreover, high glucose levels have been shown to induce chronic stresses (44). For these studies, we used the rat insulinoma INS-1E cells. This cell line was designed as a model for the study of diabetes and the induction of the insulin biosynthesis and secretory pathway (32, 45). One can induce ER stress by a glucose overload and UPR by various reagents affecting protein folding and disulfide formation, including DTT. Therefore, we treated the cells independently with either DTT or a high-glucose medium for 4 and 72 h respectively, and analyzed the sphingolipids of the treated and untreated cells. Treatment with either glucose or DTT caused a specific increase in ceramides containing palmitate as the fatty acid moiety but had a much lesser effect on other ceramide species (Fig. 4A). We also did not observe any differences in the levels of sphingomyelins (Fig. 4B). Therefore, the two types of ER stress inducers have in common the increase in C16-ceramide.
Fig. 4.
In mammalian cells, induction of the UPR pathway leads to an increase in C16 ceramide but not in sphingomyelin. INS-1E rat insulinoma cells were treated with either 10 mM DTT (4 h) or 30 mM glucose (72 h) and harvested for lipid extraction. Different chain-length ceramides (A) and sphingomyelins (B) were measured. Error bars represent the SD of three analyzed extracts. Asterisks indicate P < 0.05 relative to untreated control. For Cer16:1, ceramide levels measured were 500 ± 5 nmol/mg of protein in the untreated control condition.
Gene expression during ER stress
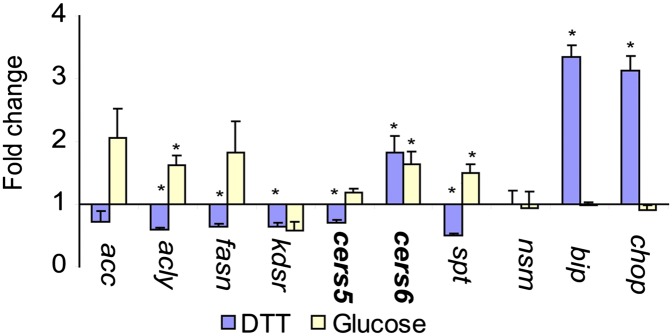
Next, we wanted to know whether the increase in C16 ceramide was the result of the increased expression of specific genes or a posttranscriptional event (5). We grew INS-1E cells as above in the presence of high glucose or DTT and used RT-PCR to analyze the variation in expression of selected genes that comprise fatty acid and sphingolipid biosynthesis pathways. This included the two ceramide synthases known to be responsible for the synthesis of C16 sphingolipids, namely CerS5 and CerS6. As expected, glucose induced genes involved in fatty acid biosynthesis but not in the UPR pathway and DTT induced genes in the UPR pathway but not in fatty acid biosynthesis. Interestingly, both stimuli induced a 1.5- to 2-fold increase in expression of CerS6 mRNA (Fig. 5), suggesting that the C16 ceramide accumulation seen in both the DTT and glucose treatments could have been generated by the CerS6 gene. One of the subunits of the first enzyme in sphingolipid biosynthesis, SPT was induced only by glucose and not by DTT, behaving in a similar manner to the fatty acid synthase genes.
Fig. 5.
Effects of UPR activation in INS-1E cells. A: Real time RT-PCR analysis of genes involved in lipid biosynthesis under UPR induction. Cells were treated with the same compounds and for the same length of time as in Fig. 4 and RNA was harvested for quantification. Full gene names and primers can be found in Table 2. Asterisks indicate P < 0.05 relative to untreated control.
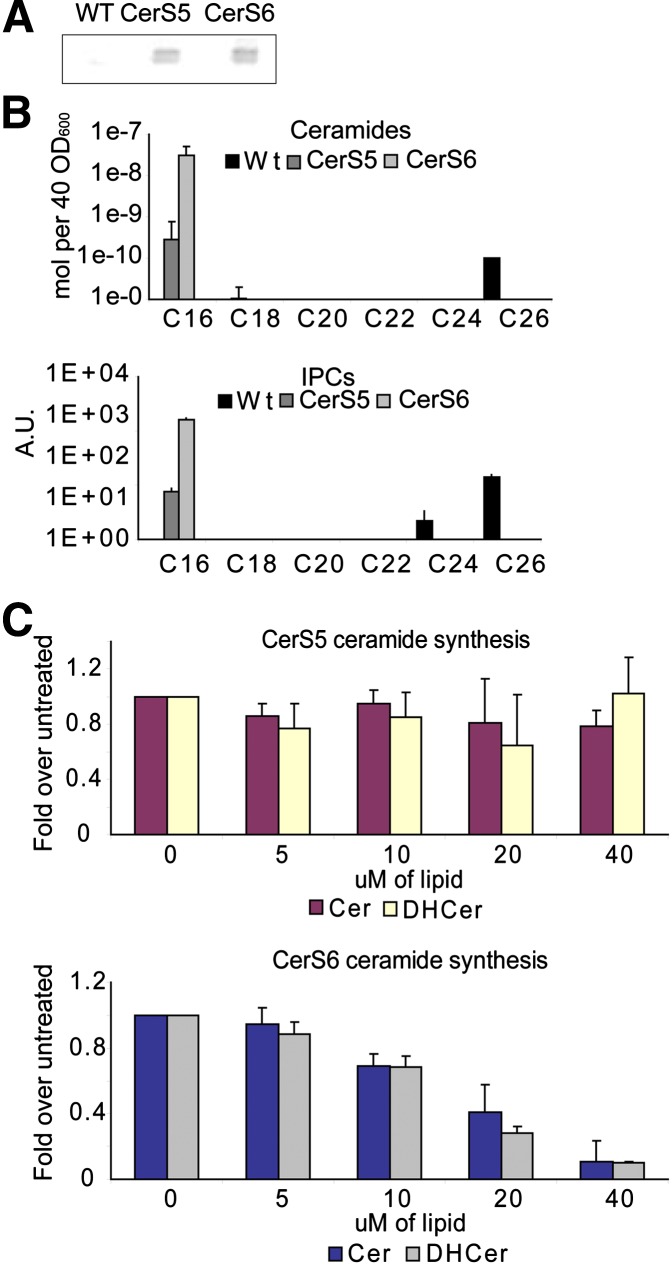
To determine whether we could find any intrinsic differences in the properties of the CerS5 and CerS6 enzymes, we expressed the two genes heterologously using a yeast system. An engineered yeast strain was constructed that had both of its endogenous ceramide synthases deleted (lag1Δ lac1Δ) and yeast codon optimized synthetic open reading frames of either mouse CerS5 and CerS6 with an additional FLAG epitope at the N-terminus, cloned behind the TDH3 promoter inserted into the yeast genome, creating stable cell lines. Lipid analysis by ESI-MS showed that both enzymes retain their specificity in yeast and have a very strong preference for C16 fatty acids (Fig. 6B). Under our conditions, the CerS6-expressing strain accumulated ∼100 times more C16 ceramide than the CerS5 expressing strain and also much more total ceramide than the wild-type strain even though CerS5 and CerS6 protein levels are similar to each other by Western blotting (Fig. 6A). Consequentially, there are also large differences in the amounts of IPC. What is striking is the extremely high activity of the CerS6 in yeast. This could be explained by a higher specific activity or a lack of feedback inhibition. To check this, we performed an in vitro enzyme assay on yeast homogenates from the CerS5 and CerS6 strains. The overall in vitro activity of CerS6 was at least 1,000 times higher than the activity of CerS5 because we had to use 1,000 more homogenate in the in vitro assay enzyme assay from the CerS5 strain in order to obtain signals on the phosphorimager after 3 days, with the CerS6 signal still being stronger (data not shown). Next, we titrated into the in vitro assay the reaction product, C16-dihydroceramide, or the product of the next step in mammalian cells, C16 ceramide. Only CerS6 activity was inhibited by products, not CerS5 (Fig. 6C). Therefore, CerS6 has an apparently much higher specific activity under these conditions and this cannot be explained by a lack of feedback inhibition.
Fig. 6.
CerS5 and CerS6 have similar specificity but different specific activity and regulation. A: WT yeast or yeast strains with their endogenous ceramide synthases (lag1 lac1) deleted and replaced by either CerS5 or CerS6 were grown to OD600=1, harvested, and proteins extracted for Western blotting with anti-FLAG antibodies. Approximately equal amounts of CerS5 and CerS6 were expressed. B: Cells were grown as in (A) and lipids were extracted and analyzed by ESI-MS. C: The same strains were homogenized and subjected to an in vitro ceramide synthase assay with 3H-sphinganine and palmitoyl-CoA and an increasing concentration of C16 dihydroceramide or ceramide. One thousand times higher concentration of homogenate was used for the CerS5 assay and the signal was still weaker than that for CerS6.
DISCUSSION
The major finding of this study was that ceramide levels are substantially increased upon induction of the UPR pathway in both yeast and pancreatic INS-1E cells. The initial discovery of the increase in ceramide synthesis came from studies of a specific double mutant defective in SPT and the p24 family member, Emp24p, which is required for GPI-anchored protein exit from the ER and therefore induces the UPR pathway when absent. In the single SPT mutant, ceramide levels are decreased, which leads to an accumulation of GPI-anchored proteins in the ER. In the emp24 mutant, there is a similar phenotype although it is caused by a different defect, namely the absence of the cargo adaptor. In the double mutant, the absence of Emp24p leads to an increase in ceramides that is capable of partially rescuing GPI transport. The rescue is not complete because the increase in ceramide cannot compensate for the lack of the cargo adaptor Emp24p.Therefore, the partial restoration of cell growth at 30°C is most likely not due to the correct transport of GPI-AP proteins but instead due to UPR activation and increased ceramide levels.
To provide an independent demonstration of the effects of UPR on ceramide levels, we used chemical modulators of the two pathways, myriocin and DTT. Myriocin inhibition of ceramide biosynthesis could be suppressed by titration of DTT, which induces UPR. The yeast model system therefore allowed us to show that activation of the UPR pathway leads to an increased viability in cells with a disrupted sphingolipid biosynthesis pathway and a resulting restoration of sphingolipid levels. Specific disruption of UPR activation by hac1 deletion blocked the restoration of ceramide levels and viability. In INS-1E pancreatic cells, the activation of UPR stress also led to an increase in ceramides, specifically those with C16 fatty acids. We were not able to determine whether disruption of the UPR pathway in INS-1E cells also blocks the induction of ceramides with C16 fatty acids because disruption of this pathway is toxic in this cell line (46). Another stimulus that induces another type of ER stress, excess glucose in the medium, also led to an increase in C16 ceramide levels even though it did not induce the UPR pathway (42, 43). Real-time PCR showed that the increase in C16-ceramide is coordinated with the increased expression of ceramide synthase CerS6. We analyzed the properties of the two CerS genes in yeast and found that CerS6 is substantially more active, yielding at least 100 times more ceramide in vivo and we showed that only CerS6 is feedback-inhibited in vitro in this heterologous system. The high specific activity of CerS6 coupled with feedback inhibition could help to explain the transient increases in ceramides that are often seen under stress conditions or other stimulation where it has been proposed to play important roles in activating direct targets in signaling (47, 48). In contrast to our findings in INS-1E cells, thapsigargin-induced ER stress was previously associated with stimulated ceramide production due to increased neutral sphingomyelinase activity in INS832/13 cells (49). It may be speculated that the latter cells are more prone to ER stress-related ceramide production, as they, in contrast to INS-1E cells, express human insulin in addition to the two allelic rat insulins.
In most yeast strains, the major sphingolipids have a fatty acyl chain of 26 carbons with a hydroxylation at the 4 position on the sphingoid base and one on the fatty acid moiety (called t18:0/26:0 PHC-C). Low levels of sphingolipids with either 24 carbons (t18:0/24:0) or nonhydroxylated moieties (PHC-B) are also encountered. In the lcb1-100 emp24 double mutant, we observed an increase in the major species with very little change in distribution among the different species. On the other hand, whenever we stimulated UPR using DTT, we found an increase in PHC-B. This difference in lipid species found might be attributed to the inhibition of fatty acid hydroxylation by DTT. The sphingoid base hydroxylase Sur2p uses molecular oxygen and NADPH to hydroxylate the sphingoid base, whereas the redox energy is thought to come from a cytochrome b5 reductase for the fatty acid hydroxylation because the enzyme Scs7p has a cytochrome b5 domain. Consistent with this hypothesis, we have shown that the CBR1 gene encoding a cytochrome b5 reductase plays a role in fatty acid hydroxylation of sphingolipids, making it the likely target of DTT.
An increase in lipid synthesis due to induction of UPR has previously been seen and is explained as a means to proliferate the ER (50, 51). Ceramides might also be important in ER proliferation, but they also may act as regulators of signaling pathways (52). Our study does not address the issue of where the stimulation of ceramide synthesis takes place, but a previous study has shown that activation of UPR in yeast upregulates the LCB1 mRNA (28), providing a plausible explanation for how activation of UPR can restore ceramide biosynthesis in cells with the conditionally defective lcb1-100 allele or in presence of myriocin. Recently, a novel regulation of SPT by the ORM proteins in yeast and mammalian cells has been described (53, 54). The phosphorylation status of the ORM proteins regulates their interaction with SPT and SPT activity. It is possible that ORM protein amounts or phosphorylation status might play a role in the UPR-dependent upregulation of ceramide levels.
The induction of ceramide accumulation by the UPR is not restricted to yeast cells. Interestingly, in INS-1E cells, which have a diversity of ceramide fatty acid chain lengths, only the C16 ceramides are substantially upregulated. The upregulation correlates with the elevated expression of CerS6, a ceramide synthase that produces this ceramide species when expressed heterologously in yeast. This suggests that CerS5 and CerS6 may play different roles in INS-1E cells. CerS5, which in our assay does not seem to be feedback regulated, may account for the basal synthesis of C16-ceramides, whereas CerS6, which is induced by UPR and glucose, would provide a rapid increase in C16 ceramides that is either transient or could reach a plateau at a higher level due to product inhibition.
INS-1E cells are insulinoma cells that are models for the study of pancreatic β cells and their impaired function in diabetes. Free fatty acids have been shown to induce cell death by activation of UPR (40) although not all of its components were increased during treatment (55). Although the UPR pathway was first found to be a mechanism for the cell to survive ER stress, recent evidence has shown that when cells can no longer cope with the stress that originated the response, this same pathway can promote cell death (56, 57). We would predict that inducing UPR at the same time as providing excess palmitate, the substrate for SPT, could lead to a large overproduction of C16 ceramide and aberrant cellular signaling. If the transient increase in ceramide is an adaptive response to protect the cells from ER stress by proliferation of the ER, too much ceramide may be detrimental through activation of other signaling pathways such as apoptosis. In mammalian cells, the UPR response is more complex than in yeast. There are distinct UPR-promoting proteins termed PERK and IRE1. When PERK signaling was activated, cells stopped proliferating and became apoptotic whereas the activation of IRE1 promoted cell proliferation (58). If this is the case, the activation of CerS6 could be either protective or apoptotic and could be an important feature in determining cell fate. Further studies will be necessary to determine which UPR activators affect ceramide synthesis. The finding that both glucose and UPR activate ceramide synthesis raises the possibility that combinations of glucose stress and UPR might cause an overproduction of C16 ceramide that might have a role in driving cells into apoptosis (59). It is possible that ceramide accumulation through stress responses and diet may play an important role in metabolic diseases like type 2 diabetes.
Acknowledgments
The authors thank members of the Wollheim and Riezman labs for helpful advice and comments throughout this work and on the manuscript and Maya Schuldiner for providing strains.
Footnotes
Abbreviations:
- ER
- endoplasmic reticulum
- GPI
- glycosylphosphosphatidylinositol
- IPC
- inositolphosphorylceramide
- PHC
- phytoceramide
- SPT
- serine palmitoyltransferase
- UPR
- unfolded protein response
This work was partially funded by grants from SystemsX.ch (H.R.) evaluated by the Swiss National Science Foundation, the Swiss National Science Foundation (C.B.W. and H.R.), and by the European Foundation for the Study of Diabetes/Lilly grant (C.B.W.). The authors have no competing interests.
REFERENCES
- 1.Futerman A. H., Riezman H. 2005. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15: 312–318. [DOI] [PubMed] [Google Scholar]
- 2.Dickson R. C. 2008. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 49: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. 2001. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20: 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallée B., Riezman H. 2005. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 24: 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stiban J., Tidhar R., Futerman A. H. 2010. Ceramide synthases: roles in cell physiology and signaling. Adv. Exp. Med. Biol. 688: 60–71. [DOI] [PubMed] [Google Scholar]
- 6.Buede R., Rinker-Schaffer C., Pinto W. J., Lester R. L., Dickson R. C. 1991. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173: 4325–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munn A. L., Riezman H. 1994. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanolari B., Friant S., Funato K., Sutterlin C., Stevenson B. J., Riezman H. 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19: 2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friant S., Meier K. D., Riezman H. 2003. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. EMBO J. 22: 3783–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier K. D., Deloche O., Kajiwara K., Funato K., Riezman H. 2006. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol. Biol. Cell. 17: 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowart L. A., Okamoto Y., Pinto F. R., Gandy J. L., Almeida J. S., Hannun Y. A. 2003. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J. Biol. Chem. 278: 30328–30338. [DOI] [PubMed] [Google Scholar]
- 12.Cowart L. A., Okamoto Y., Lu X., Hannun Y. A. 2006. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem. J. 393: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bocer T., Zarubica A., Roussel A., Flis K., Trombik T., Goffeau A., Ulaszewski S., Chimini G. 2011. The mammalian ABC transporter ABCA1 induces lipid-dependent drug sensitivity in yeast. Biochim. Biophys. Acta. doi:10.1016/j.bbalip.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Dupré S., Haguenauer-Tsapis R. 2003. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic. 4: 83–96. [DOI] [PubMed] [Google Scholar]
- 15.Hearn J. D., Lester R. L., Dickson R. C. 2003. The uracil transporter Fur4p associates with lipid rafts. J. Biol. Chem. 278: 3679–3686. [DOI] [PubMed] [Google Scholar]
- 16.Bagnat M., Keranen S., Shevchenko A., Simons K. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 97: 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sütterlin C., Doering T. L., Schimmoller F., Schroder S., Riezman H. 1997. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci. 110: 2703–2714. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe R., Funato K., Venkataraman K., Futerman A. H., Riezman H. 2002. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J. Biol. Chem. 277: 49538–49544. [DOI] [PubMed] [Google Scholar]
- 19.Horvath A., Sutterlin C., Manning-Krieg U., Movva N. R., Riezman H. 1994. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 13: 3687–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schimmöller F., Singer-Kruger B., Schroder S., Kruger U., Barlowe C., Riezman H. 1995. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 14: 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., Kinoshita T. 2011. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takida S., Maeda Y., Kinoshita T. 2008. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 409: 555–562. [DOI] [PubMed] [Google Scholar]
- 23.Castillon G. A., Aguilera-Romero A., Manzano-Lopez J., Epstein S., Kajiwara K., Funato K., Watanabe R., Riezman H., Muniz M. 2011. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell. 22: 2924–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belden W. J., Barlowe C. 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 12: 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., et al. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323: 1693–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J. S., Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 87: 391–404. [DOI] [PubMed] [Google Scholar]
- 27.Kimata Y., Kimata Y. I., Shimizu Y., Abe H., Farcasanu I. C., Takeuchi M., Rose M. D., Kohno K. 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell. 14: 2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101: 249–258. [DOI] [PubMed] [Google Scholar]
- 29.Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8: 519–529. [DOI] [PubMed] [Google Scholar]
- 30.Spassieva S. D., Mullen T. D., Townsend D. M., Obeid L. M. 2009. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 424: 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lépine S., Allegood J. C., Park M., Dent P., Milstien S., Spiegel S. 2011. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 18: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merglen A., Theander S., Rubi B., Chaffard G., Wollheim C. B., Maechler P. 2004. Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology. 145: 667–678. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L., Volchuk A. 2010. p24 family type 1 transmembrane proteins are required for insulin biosynthesis and secretion in pancreatic beta-cells. FEBS Lett. 584: 2298–2304. [DOI] [PubMed] [Google Scholar]
- 34.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418: 387–391. [DOI] [PubMed] [Google Scholar]
- 35.Guan X. L., Riezman I., Wenk M. R., Riezman H. 2010. Yeast lipid analysis and quantification by mass spectrometry. Methods Enzymol. 470: 369–391. [DOI] [PubMed] [Google Scholar]
- 36.Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., Fornallaz-Mulhauser M., Hengartner M. O., Gomez M., Riezman H., Martinou J. C. 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science. 324: 381–384. [DOI] [PubMed] [Google Scholar]
- 37.Haak D., Gable K., Beeler T., Dunn T. 1997. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272: 29704–29710. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell A. G., Martin C. E. 1997. Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the alpha-hydroxylation of sphingolipid-associated very long chain fatty acids. J. Biol. Chem. 272: 28281–28288. [DOI] [PubMed] [Google Scholar]
- 39.Csukai M., Murray M., Orr E. 1994. Isolation and complete sequence of CBR, a gene encoding a putative cytochrome b reductase in Saccharomyces cerevisiae. Eur. J. Biochem.219: 441–448. [DOI] [PubMed]
- 40.Wang H., Kouri G., Wollheim C. B. 2005. ER stress and SREBP-1 activation are implicated in beta-cell glucolipotoxicity. J. Cell Sci. 118: 3905–3915. [DOI] [PubMed] [Google Scholar]
- 41.Elouil H., Bensellam M., Guiot Y., Vander Mierde D., Pascal S. M., Schuit F. C., Jonas J. C. 2007. Acute nutrient regulation of the unfolded protein response and integrated stress response in cultured rat pancreatic islets. Diabetologia. 50: 1442–1452. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L., Lai E., Teodoro T., Volchuk A. 2009. GRP78, but not protein-disulfide isomerase, partially reverses hyperglycemia-induced inhibition of insulin synthesis and secretion in pancreatic {beta}-cells. J. Biol. Chem. 284: 5289–5298. [DOI] [PubMed] [Google Scholar]
- 43.Eizirik D. L., Cardozo A. K., Cnop M. 2008. The role for endoplasmic reticulum stress in diabetes mellitus. Endocr. Rev. 29: 42–61. [DOI] [PubMed] [Google Scholar]
- 44.Hou Z. Q., Li H. L., Gao L., Pan L., Zhao J. J., Li G. W. 2008. Involvement of chronic stresses in rat islet and INS-1 cell glucotoxicity induced by intermittent high glucose. Mol. Cell. Endocrinol. 291: 71–78. [DOI] [PubMed] [Google Scholar]
- 45.Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. 1992. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 130: 167–178. [DOI] [PubMed] [Google Scholar]
- 46.Kirkpatrick C. L., Wiederkehr A., Baquie M., Akhmedov D., Wang H., Gauthier B. R., Akerman I., Ishihara H., Ferrer J., Wollheim C. B. 2011. Hepatic nuclear factor 1alpha (HNF1alpha) dysfunction down-regulates X-box-binding protein 1 (XBP1) and sensitizes beta-cells to endoplasmic reticulum stress. J. Biol. Chem. 286: 32300–32312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta N., Nodzenski E., Khodarev N. N., Yu J., Khorasani L., Beckett M. A., Kufe D. W., Weichselbaum R. R. 2001. Angiostatin effects on endothelial cells mediated by ceramide and RhoA. EMBO Rep. 2: 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolova-Karakashian M. N., Rozenova K. A. 2010. Ceramide in stress response. Adv. Exp. Med. Biol. 688: 86–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei X., Zhang S., Bohrer A., Bao S., Song H., Ramanadham S. 2007. The group VIA calcium-independent phospholipase A2 participates in ER stress-induced INS-1 insulinoma cell apoptosis by promoting ceramide generation via hydrolysis of sphingomyelins by neutral sphingomyelinase. Biochemistry. 46: 10170–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sriburi R., Jackowski S., Mori K., Brewer J. W. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ron D., Hampton R. Y. 2004. Membrane biogenesis and the unfolded protein response. J. Cell Biol. 167: 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mousley C. J., Tyeryar K., Ile K. E., Schaaf G., Brost R. L., Boone C., Guan X., Wenk M. R., Bankaitis V. A. 2008. Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Mol. Biol. Cell. 19: 4785–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han S., Lone M. A., Schneiter R., Chang A. 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA. 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. 2006. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 147: 3398–3407. [DOI] [PubMed] [Google Scholar]
- 56.Malhotra J. D., Kaufman R. J. 2011. ER stress and its functional link to mitochondria: role in cell survival and death. Cold Spring Harb. Perspect. Biol. doi: 10.1101/cshperspect.a004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., Wang S., Malhotra J., Hassler J. R., Back S. H., Wang G., Chang L., Xu W., Miao H., Leonardi R., et al. 2011. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 30: 1357–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin J. H., Li H., Zhang Y., Ron D., Walter P. 2009. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS ONE. 4: e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettus B. J., Chalfant C. E., Hannun Y. A. 2002. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta. 1585: 114–125. [DOI] [PubMed] [Google Scholar]