Abstract
We hypothesized that reduction/loss of very long chain PUFAs (VLC-PUFAs) due to mutations in the ELOngase of very long chain fatty acid-4 (ELOVL4) protein contributes to retinal degeneration in autosomal dominant Stargardt-like macular dystrophy (STGD3) and age-related macular degeneration; hence, increasing VLC-PUFA in the retina of these patients could provide some therapeutic benefits. Thus, we tested the efficiency of elongation of C20-C22 PUFA by the ELOVL4 protein to determine which substrates are the best precursors for biosynthesis of VLC-PUFA. The ELOVL4 protein was expressed in pheochromocytoma cells, while green fluorescent protein-expressing and nontransduced cells served as controls. The cells were treated with 20:5n3, 22:6n3, and 20:4n6, either individually or in equal combinations. Both transduced and control cells internalized and elongated the supplemented FAs to C22-C26 precursors. Only ELOVL4-expressing cells synthesized C28-C38 VLC-PUFA from these precursors. In general, 20:5n3 was more efficiently elongated to VLC-PUFA in the ELOVL4-expressing cells, regardless of whether it was in combination with 22:6n3 or with 20:4n6. In each FA treatment group, C34 and C36 VLC-PUFAs were the predominant VLC-PUFAs in the ELOVL4-expressing cells. In summary, 20:5n3, followed by 20:4n6, seems to be the best precursor for boosting the synthesis of VLC-PUFA by ELOVL4 protein.
Keywords: elongase of very long chain fatty acids, very long chain polyunsaturated fatty acids, chylomicrons, fatty acids/biosynthesis, juvenile autosomal dominant Stargardt-like macular dystrophy type 3 disease, arachidonic acid, fish oil
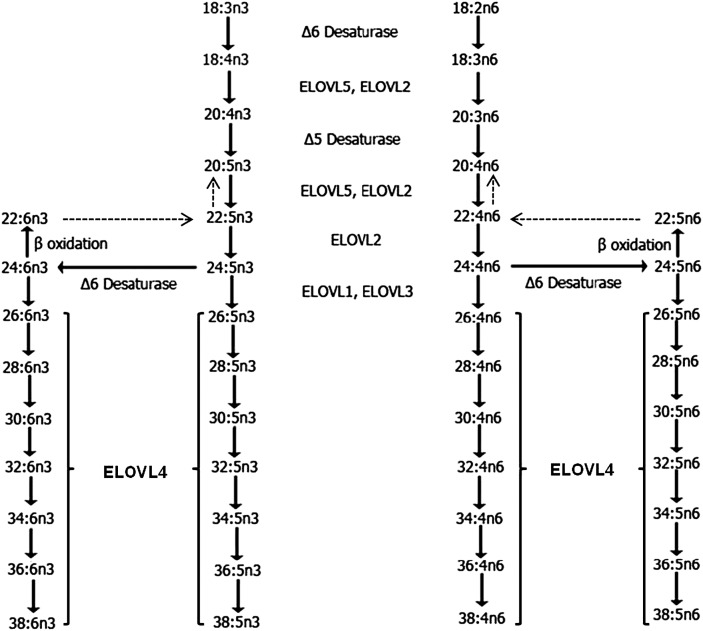
Very long chain PUFAs (VLC-PUFAs) are FAs of the n3 and n6 series with greater than C26 carbons. These unique groups of FAs are found mostly in the retina, brain, testis, and spermatozoa (1–5). Their extremely long carbon chains with poly methylene-interrupted cis double bonds that are prone to oxidative damage, coupled with their low occurrence in tissues in which they are found, make them difficult to analyze. Hence, little progress has been made in understanding the roles of VLC-PUFA in tissues (6). We now know that the biosynthesis of VLC-PUFA is catalyzed by the ELOngase of very long chain FA-4 (ELOVL4) from C26 precursors that are derived through a series of elongations and desaturations of shorter chain essential PUFAs. Common essential PUFAs include 18:2n6 (linoleic acid, LA), 18:3n3 (α-linolenic acid, α-LA) (7–9), eicosapentaenoic acid (EPA, 20:5n3), docosapentaenoic acid (22:5n3), docosahexaenoic acid (DHA, 22:6n3), and arachidonic acid (AA, 20:4n6), from which C26 PUFA precursors are generated utilizing seven different elongase and FA desaturase enzymes (Fig. 1) (10–13).
Fig. 1.
Schematic in vivo biosynthetic pathway from 18:3n3 and 18:2n6 mediated by ELOVL4 and other ELOVL family proteins. Desaturase and elongation steps are consecutively performed by fatty acid desaturase-1 (FADS1 or Δ5 desaturase), fatty acid desaturase-2 (FADS2 or Δ6 desaturase), and ELOVL1-5. Although some elongases are specific for a single step, others are nonspecific or multifunctional and act at several steps (e.g., human ELOVL5 and murine ELOVL2) (59).
No VLC-PUFAs are found in the liver or plasma because the liver does not express the ELOVL4 protein, suggesting that VLC-PUFAs are synthesized in situ from available C18-C26 PUFA precursors in ELOVL4-expressing tissues (14). When present, the VLC-PUFAs are generally found in amide bond linkages with ceramides and sphingomyelin in testis and sperm, or esterified to the sn-1 position of phosphatidylcholine (PC) in the retina (15–19).
EPA and DHA, which are precursors for the biosynthesis of VLC-PUFA, are major dietary n3 long-chain PUFAs (LC-PUFAs) of important physiological significance in tissues. The highest body concentration of 22:6n3 per unit weight is found in the phospholipids of retinal photoreceptor outer segments (20, 21). EPA is found in cholesteryl esters, triglycerides, and phospholipids of plasma, but is not found in tissues in substantial amounts; it has always been assumed to be efficiently used in 22:6n3 or eicosanoid biosynthesis (20, 22, 23). AA, which is mostly acquired from the diet or synthesized through elongation and desaturation of 18:2n6, is the major n6 LC-PUFA of the neural and vascular tissues of the retina and brain (22). Over the decades, much has been done showing that these LC-PUFAs are necessary for normal neural development and function (24–32). They also play significant roles in maintaining cell structure and physiological function by modulating cell differentiation and normal growth through signal transduction and cellular metabolism (33–35). Epidemiological studies found that the dietary intake of LC-PUFA decreases the progression and risk for age-related macular degeneration (AMD) (36–41).
The normal ELOVL4 protein is responsible for elongation of LC-PUFA to VLC-PUFA found in retinal photoreceptor cells and in several other tissues (15, 18, 19, 42). Reduced levels of C32-36 acyl PC have been found in the retina of genetic mouse models of autosomal dominant Stargardt-like macular dystrophy (STGD3) (43). These STGD3 animal models, just as humans with STGD3, eventually develop retinal degeneration (43–46) The reduction in VLC-PUFA in these animals is accompanied by reduced retinal function as measured by electroretinography (ERG) and build-up of toxic lipofuscin products in the retina and retinal pigment epithelium (RPE).
Recent and ongoing studies have reported reduced levels of VLC-PUFA in aging retinas and in retinas of donor eyes of patients with history of AMD (42). Hubbard et al. reported that FAs from red blood cell membranes of a family of STGD3 patients showed a significant inverse relationship between the degree of retinal phenotype and n3 LC-PUFA levels. While patients with severe retinal degeneration had average 20:5n3 and 22:6n3 levels, patients with higher levels of 20:5n3 and 22:6n3 had less-severe retinal phenotypes (47). These studies collectively suggest that different levels of 20:5n3 and 22:6n3 could influence the biosynthesis of VLC-PUFA in the retina, which could in turn lead to the variable phenotypic expression of STGD3 secondary to ELOVL4 mutation. This reasoning is supported by an in vivo study in which [3H]20:5n3 or [3H]22:6n3 was intravitreally injected into rat eyes. While the [3H]20:5n3 was shown to be actively elongated into VLC-PUFA up to 34 carbons, 90% of [3H]22:6n3 was esterified into retinal phospholipids without further elongation. It was concluded that 20:5n3 is the preferred substrate for the synthesis of n3 VLC-PUFA (48). However, at that time, the elongase responsible for converting the 20:5n3 to the VLC-PUFA was not known. Studies from our laboratory confirmed that ELOVL4 is the enzyme necessary for elongating C26 PUFA, generated from elongation of the 20:5n3 and 22:5n3, to VLC-PUFA (10). Recent studies have supported these findings and revealed that Zebra fish possess two genes encoding putative ELOVL4a and ELOVL4b proteins, with the latter having activity toward 20:5n3 and 22:5n3 but not 22:6n3 (13). Also, the Atlantic salmon ELOVL4 protein has been shown to effectively elongate C24-C26 polyunsaturated FAs to polyenoic products up to C36 (49). All of these findings collectively suggest that VLC-PUFAs play important but yet to be determined physiological and/or structural roles in tissues where they are found.
We are interested in understanding the biosynthesis and possible physiological, molecular, and structural role of VLC-PUFAs in the tissues in which they are found, since inheritance of mutations in the ELOVL4 gene leads to juvenile onset of retinal degeneration in STGD3 patients. We hypothesized that reduced levels of VLC-PUFA may be one of the underlying causes of retinal degeneration seen in STGD3 patients and sought to determine the efficiency of elongation of n3 and n6 C20-C22 PUFA by the ELOVL4 protein. We over-expressed recombinant ELOVL4 protein in pheochromocytoma cells (PC12s) and treated them with the precursors needed for the biosynthesis of VLC-PUFA. Using improved analytical and quantitative methods, we determined the efficiency of elongation of LC-PUFA to VLC-PUFA by the ELOVL4 protein and confirmed that 20:5n3 is indeed preferred over 22:6n3 and 20:4n6 as the substrate of VLC-PUFA biosynthesis. Understanding the mechanisms that favor the incorporation of one precursor over another and what roles the VLC-PUFAs play in the retina and other tissues will contribute to our efforts in finding therapeutic treatments for STGD3 patients and the more-prevalent AMD.
EXPERIMENTAL PROCEDURES
Materials
PC12 cells were purchased from American Type Culture Collection (Manassas, VA). Plasmids were isolated with NucleoBond plasmid isolation kit (Clontech Laboratories, Inc.; Mountain View, CA). Adenovirus shuttle and acceptor vectors (plp Adeno X Expression Systems 2 with Creator technology) were purchased from Clontech. Ad5-GFP under control of the cytomegalovirus promoter was kindly provided by Dr. Daniel J. J. Carr (University of Oklahoma Health Sciences Center; Oklahoma City, OK). Rabbit polyclonal green fluorescent protein (GFP) antibody was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) and the actin antibody was from Novus Biologicals (Littleton, CO). The sodium salts of 20:5n3 and 20:4n6 were >99% pure (as determined by gas liquid chromatography on the methyl ester) and were purchased from Nu-Chek Prep, Inc. (Elysian, MN). The sodium salt of 22:6n3 at ≥95% purity was purchased from Sigma Chemical Co. (St. Louis, MO). Complete mini, EDTA-free protease inhibitor tablets were purchased from Roche Applied Sciences (Indianapolis, IN). All tissue culture reagents were from Invitrogen Corporation (Carlsbad, CA). All other reagents including those for FA extraction and derivatization were of high quality, available from Sigma.
Construction of recombinant adenovirus carrying mouse ELOVL4
Recombinant adenovirus carrying the mouse Elovl4 gene was constructed using AdenoX Expression Systems 2 with the Creator technology (Clontech) as previously described (10). The recombinant viruses were prepared as high-titer stocks through the propagation in HEK293 cells by double cesium chloride purification, and dialyzed against a 10 mM Tris buffer (pH 8.0) that contained 80 mM NaCl, 2 mM MgCl2, and 10% glycerol (50). Infectious adenovirus titer was determined in triplicate by plaque-forming assay and expressed as plaque-forming units (pfu) per ml.
Cell culture, viral transductions, and FA treatments
PC12 cells were cultured using standard tissue culture procedures. The cells were dissociated and plated at 2 × 106 cells in 10 cm tissue culture plates in DMEM supplemented with 10% (v/v) heat-inactivated horse serum, 5% (v/v) heat-inactivated FBS, 100 U/ml penicillin, and 100 U/ml streptomycin. Cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2. The following day, the cells were transduced with either the Ad5-GFP (control) or Ad5-Elovl4 by adding viral particles at a concentration of 1 × 104 to 2 × 104 pfu/(ml) to the culture medium containing 2% (v/v) FBS (51, 52). After 24 h of incubation, the infection medium was replaced with normal culture medium containing 15% (v/v) serum. FAs were supplemented as sodium salts of the respective FAs conjugated to BSA fraction V. To the cells, 30–40 μg/ml of individual FA and combinations at 15–20 μg/ml for each FA were added. After incubation with FAs for 48 or 72 h, cells were washed in PBS containing 50 μM FA-free BSA fraction V and then washed two times with PBS only. The cells were then scraped and stored as pellets at −80°C until used.
RNA isolation and cDNA synthesis
RNA was isolated and purified from PC12 adenovirus-transduced cells using the PureLink Micro to Midi Total RNA Purification System from Invitrogen Life Sciences (Carlsbad, CA), following the manufacturer's protocol. Equal quantities (1.0 μg) of total RNA from cells were converted to first-strand cDNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) for quantitative real-time PCR (qRT-PCR). The first-strand cDNA was used for quantitative and normal RT-PCR. Degenerate primers for Elovl1-5, delta-5-desaturase (Δ5D or fatty acids desaturase 1; FADS1), and delta-6-desaturase (Δ6D or fatty acids desaturase 2; FADS2) were used. We used the housekeeping gene RPL19 that was designed to amplify human, mouse, and rat cDNAs for RT-PCR. Primers were designed in such a way that they spanned at least one intron so as to eliminate the chance of amplifying residual genomic DNA contaminations. Quantitative PCR and melt-curve analyses were performed using iQ SYBR Green Supermix (Bio-Rad; Hercules, CA) and an iCycler machine. Expression data were calculated from three independent samples, each with three qRT-PCRs, and are presented relative to the expression of RPL19 (mean ± SD).
Western blot analysis
PC12 cells transduced with Ad5-Elovl4, Ad5-GFP, or noninfected controls were collected and lysed in 200 μl of lysis buffer containing 20 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, complete protease inhibitor, and 1% Triton X-100. The lysates were briefly sonicated on ice, incubated with gentle rocking at 4°C for 1 h, and centrifuged at 27,000 g for 30 min at 4°C. Supernatants were collected, and protein concentrations determined by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific; Rockford, IL). Equal amounts of protein (30 μg) were separated by SDS/PAGE on 12% polyacrylamide gels, followed by electro-transfer to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk and incubated with custom-made rabbit polyclonal ELOVL4 antibodies (Bethyl Laboratories, Inc.; Montgomery, TX) at 1:1,000 dilution. Membranes were stripped and reprobed with a polyclonal antibody against GFP (Santa Cruz Biotechnology, CA) at 1:1,000 and then with the monoclonal antibody against β-actin (Sigma-Aldrich; St Louis, MO). Immunoreaction of horseradish peroxidase-conjugated donkey anti-rabbit or goat anti-mouse IgG secondary antibodies was detected by using Super-Signal West Dura Extended Duration Substrate (Pierce; Rockford, IL).
FA extraction
Total lipids were extracted from about 2.0 mg of protein following the method of Bligh and Dyer (53) with minor modification (54). The purified lipid extract was stored under nitrogen until used. To the purified lipid extracts, 50 nmol of 15:0, 17:0, 23:0 each and 4 nmol of 30:3n6 were added as internal standards. One milliliter of 16.6% concentrated HCl in methanol was then added and the tubes were sealed under N2 with Teflon-lined caps and heated at 100°C overnight. The tubes were cooled on ice and FA methyl esters (FAMEs) were extracted and processed as previously reported (10).
Lipid analysis by GC-MS
FAMEs were identified using an Agilent Technologies 7890A gas chromatograph (GC) with a 5975C inert XL mass spectrometer (MS) detector (Agilent Technologies; Santa Clara, CA). The GC-MS was operated in the electron impact total-ion and single-ion monitoring (SIM) modes. The injection volume was 1 μl and the inlet, held at 280°C, was set to pulsed splitless mode. An Agilent Technologies DB-23 column (60 m × 0.32 mm × 0.25 μm) was used with a helium carrier gas flow rate of 1.9 ml/min. The oven temperature began at 130°C for 1.0 min, was ramped to 170°C at 6.8°C/min, and then ramped to 215°C at 2.9°C/min. After holding at 215°C for 11.4 min, the oven was ramped to 230°C at 42°C/min and held for 9.6 min. The oven was then ramped to 290°C at 10°C/min and held for 14.4 min. The MS transfer line, ion source, and quadrupole temperatures were 290°C, 230°C, and 150°C, respectively. The PUFAs were identified by using the m/z ratios 79.1, 108.1, and 150.1 in SIM mode and the full scan mass spectra in total-ion mode.
FAMEs were quantified using an Agilent Technologies 6890N GC with flame ionization detector (FID). Sample concentrations were determined by comparison to internal standards 15:0, 17:0, 23:0, and 30:3n6. The injection volume was 1 μl and the inlet, held at 280°C, was set to pulsed split mode (10:1 ratio). An Agilent Technologies DB-23 column (60 m × 0.32 mm × 0.25 μm) was used with a hydrogen carrier gas constant pressure of 13.1 psi. The oven temperature began at 130°C for 0.8 min, was ramped to 170°C at 8.2°C/min then ramped to 215°C at 3.5°C/min. After holding at 215°C for 9.5 min, the oven was ramped to 230°C at 50°C/min and held for 8 min. The oven was then ramped to 290°C at 12.0°C/min and held for 12 min. The FID was held at 290°C.
We also tried analyzing pentylfluorobenzyl derivatives of VLC-PUFA utilizing negative-ion chemical ionization GC-MS, which would have shown intact (no fragmentation) molecular ions and would provide for a better means of quantification compared with flame ionization detection (55). However, with our current analytical methods, it seems that adding such a relatively large (compared with a methyl group) functional group to already heavy, nonvolatile VLC-PUFA molecules made it unsuitable to go through the GC system under reasonable analytical conditions. We are currently exploring methods and means that will allow us to quantify intact VLC-PUFA molecules by using electrospray ionization MS-MS or liquid chromatography MS-MS.
Statistical analysis
Statistical analyses were done using StatSoft Inc., Statistica 2000 and GraphPad Prism 5 software. Results are expressed as the mean ± SD. Differences were assessed by multivariate ANOVA followed by posthoc Scheffe test. All relative mol%s (±SD, n = 3) of n3 and n6 LC-PUFA and VLC-PUFA are from the total lipids extracted from sample homogenates equivalent to 2.0 mg of protein and were converted to FAMEs and quantified using GC-FID. Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, whereas # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3).
RESULTS
Expression profile of FA elongase and desaturase enzymes in PC12 cells
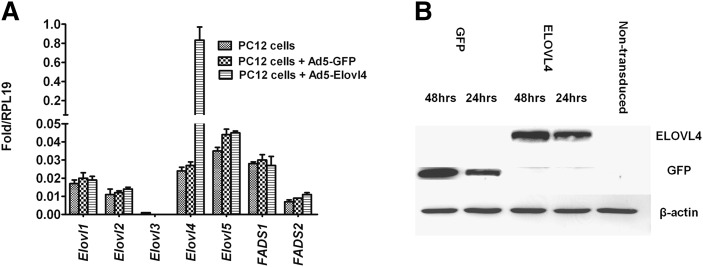
Initial studies were done in four different cell lines, namely immortalized hepatocytes (Hep2G), PC12 cells, mouse retinal cone photoreceptor-derived 661W cells, and the retinoblastoma cell line Y-79. Although the retinal cell lines have the machinery for VLC-PUFA biosynthesis, we excluded them because they endogenously express relatively higher levels of the ELOVL4 protein, which could affect the interpretation of our results. Because the major pathological phenotype seen in humans with mutations in the ELOVL4 gene is restricted to retinal neurons (photoreceptors), we chose PC12 cells, which are easily differentiated to neuronal cell lineage, can be easily transduced with viral particles, and have been extensively employed as a model system for neuronal differentiation and neuronal FA metabolism (56, 57). The expression profile of PC12 cell FA elongase and FA desaturase genes involved in VLC-PUFA biosynthesis was determined by using qRT-PCR (Fig. 2A). Expression levels of endogenous Elovl1, -2, and -4 were almost the same in all treatment groups. The endogenous level of Elovl5 was higher than that of endogenous Elovl1-4; however, Elovl3 expression levels were very low. FA desaturase 1 (Fads1) levels were higher than those of Fads2. However, when the PC12 cells were transduced with Ad5-Elovl4, the expression levels of ELOVL4 protein in the cells were increased by 40-fold when compared with expression levels in the control cells (nontransduced and Ad5-GFP-transduced cells) (Fig. 2A). The previously described affinity-purified rabbit polyclonal ELOVL4 antibodies (10) that recognize both mouse and rat ELOVL4 protein were used to confirm the ELOVL4 protein expression by using Western blot analysis (Fig. 2B). Although qRT-PCR results suggest that the PC12 cells express Elovl4 message, we did not have any detectible ELOVL4 protein in these cells (Fig. 2).
Fig. 2.
Gene expression profile of Elovl1-5, fatty acid desaturase-1 (FADS1 or D5 desaturase), and fatty acid desaturase-2 (FADS2 or D6 desaturase) in PC12 cells using qRT-PCR and Western blots. A: Comparison of quantitative expression of different genes involved in the n3 and n6 FA elongation pathway in PC12 cells presented relative to the expression of the housekeeping gene RPL19. The values represent the mean ± SD (n = 3) after normalizing with RPL19 calculated by the comparative threshold cycle method. Significant expression of ELOVL4 was observed in transduced ELOVL4-expressing PC12 cells. B: Western blots of the expression of ELOVL4, GFP, and β-actin in PC12 cells showed abundant ELOVL4 protein expression.
ELOVL4 protein elongates 20:5n3 and 22:6n3 to n3 VLC-PUFA
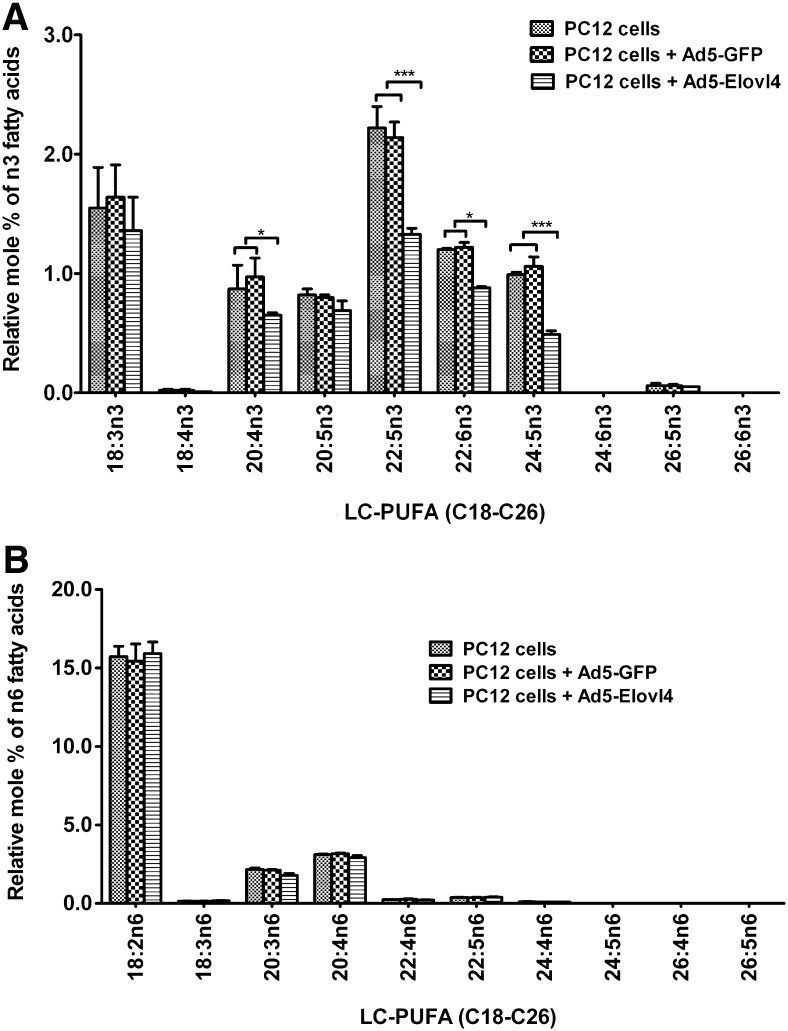
We have previously shown that the ELOVL4 protein expressed in cultured neonatal rat cardiomyocytes and human ARPE-19 cells is capable of elongating 20:5n3 and 22:5n3 into a series of n3 VLC-PUFAs (10). However, at that time, we could not accurately quantify the amount of VLC-PUFA synthesized because of lack of appropriate VLC-PUFA internal standards. In this study, we used chemically synthesized 30:3n6 (58), which is not present in any mammalian tissue, as an internal standard to quantify VLC-PUFA in PC12 cells supplemented with 20:5n3, 22:6n3, and 20:4n6. In the absence of FA treatment, ELOVL4-transduced PC12 cells had low levels of elongation products of n3 VLC-PUFA, whereas the control cells (nontransduced and Ad5-GFP-transduced cells) did not have any detectible VLC-PUFA products. The relative mol%s of the major elongation products include 0.18% 32:5n3, 0.13% 34:5n3, and 0.14% 36:5n3 (see supplementary Fig. IA). These products were derived either from elongation of VLC-PUFA precursors in the cells or from the FAs present in the serum that were part of the culture medium. Formation of VLC-PUFA in the ELOVL4 protein-expressing cells, but not in control cells, was accompanied by reductions in 20:4n3, 22:5n3, 22:6n3, and 24:5n3 (Fig. 3A). Also, 34:6n3 and 36:6n3, which might be from elongation of 22:6n3 or through activity of FADS2 protein on 34:5n3 and 36:5n3, respectively, were detected in the ELOVL4-transduced cells (see supplementary Fig. IA). No significant presence of n6 VLC-PUFA was detected in the ELOVL4-transduced cells, although the precursor, 20:4n6 (∼3%), was present in both control and ELOVL4-transduced cells (Fig. 3B).
Fig. 3.
FA composition of PC12 cells expressing ELOVL4 protein without FA treatment. Relative mol%s (±SD, n = 3) of n3 and n6 PUFA from total lipids extracted from sample homogenate equivalent to 2.0 mg of protein were converted to FAME and analyzed by GC-FID. A: Significant reductions in 20:4n3, 22:5n3, 22:6n3, and 24:5n3 were present in the ELOVL4-expressing cells but not in the controls. B: There were no significant changes in n6 LC-PUFA composition in the ELOVL4-expressing cells, and no n6 VLC-PUFAs were detected.Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, whereas # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3).
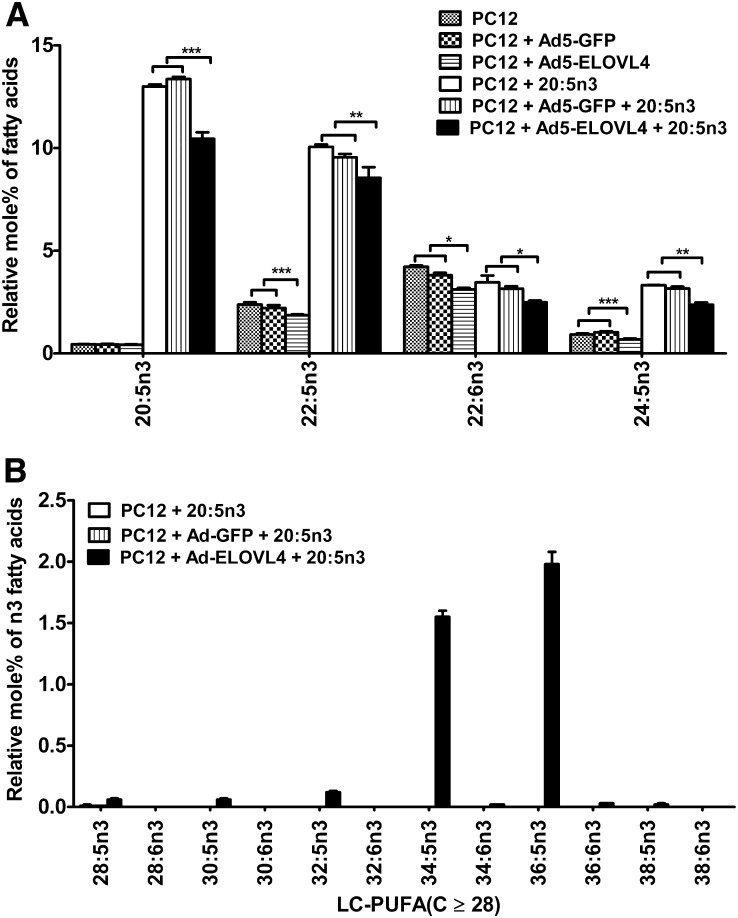
When the PC12 cells were treated with the sodium salt of 20:5n3 conjugated to BSA fraction V, the ELOVL4-transduced cells and controls took up the FA in almost the same amounts (Fig. 4A). There was 13% 20:5n3 in both PC12 controls and GFP-expressing cells but 11% in ELOVL4-expressing cells (Fig. 4A). The 20:5n3 precursor was elongated independently of the ELOVL4 protein to 22:5n3, which accounts for 10% (relative mol% of the total FAs) in PC12 cells, 10% in GFP-expressing cells, and 9% in ELOVL4-expressing cells. This elongation is probably catalyzed by the endogenously expressed elongases ELOVL5 or ELOVL2 (12, 59, 60). There was no significant difference in the levels of 20:5n3 and 22:5n3 between the two control groups, which supports our earlier findings that the mouse ELOVL4 protein does not appear to elongate C20-C22 LC-PUFA when 20:5n3 is used as the precursor (61). Also, 24:5n3 was synthesized in both ELOVL4 and control cells (Fig. 4A). Significant decreases in the levels of 20:5n3 (P < 0.001) and 22:5n3 (P < 0.01) in ELOVL4-transduced cells were accompanied by increases of 0.06% 28:5n3, 0.06% 30:5n3, 0.12% 32:5n3, 1.55% 34:5n3, and 1.98% 36:5n3 VLC-PUFA (Fig. 4B). The control cells treated with 20:5n3 did not have these elongation products (Fig. 4B). There were some 34:6n3 and 36:6n3 FA products, which were probably products from either FADS2 desaturase activity on 34:5n3 and 36:5n3 or endogenous 22:6n3 elongation.
Fig. 4.
PC12 cells expressing ELOVL4 protein efficiently elongate exogenous supplemented sodium salt of 20:5n3 to n3 VLC-PUFAs. A: Sodium salt of 20:5n3 conjugated to BSA fraction V were efficiently taken up at almost the same amounts in the Ad5 Elovl4-transduced PC12 cells and in control cells. Both 20:5n3 and 22:5n3 were drastically increased in response to 20:5n3 treatment. The elongation of 20:5n3 to 22:5n3, irrespective of ELOVL4 protein overexpression, is probably catalyzed by endogenously expressed elongases like Elovl5 or Elovl2 (60). There were no significant differences in the levels of 20:5n3, 22:5n3, or 24:5n3 between the ELOVL4-expressing and the two control groups. However, the levels of 20:5n3, 22:5n3, and 24:5n3 in the ELOVL4-transduced group were significantly lower than those in the two controls. B: Biosynthesis of n3 VLC-PUFA (C ≥ 28) elongation products occurred in the ELOVL4 protein-expressing PC12 cells treated with 20:5n3, but not in the controls. The major products were 34:5n3 and 36:5n3, which account for 1.5 and 1.98 relative mol% of total FAs, respectively. Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, whereas # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3)..
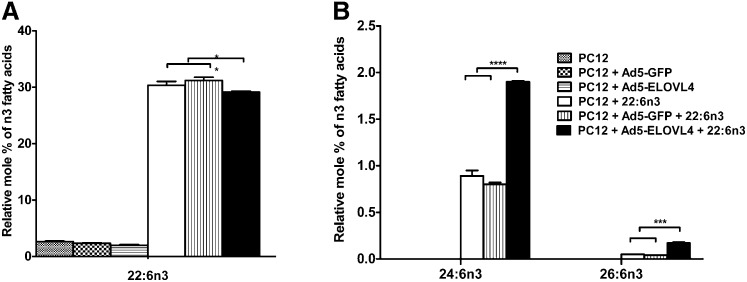
Considering the fact that 22:6n3 is the most abundant polyunsaturated FA in retinal phospholipids, one might expect that its elongation products would be more abundant in the retinal PC species. However, this is not the case, suggesting that 22:6n3 is either an end-product of LC-PUFA biosynthesis and not a preferred substrate for further elongation to VLC-PUFA (48, 62) or it is efficiently esterified to phospholipids, which makes it unavailable for further elongation. To determine the relative efficiency of elongation of 22:6n3 to VLC-PUFA, we treated ELOVL4-expressing cells with the BSA-conjugated sodium salt of 22:6n3. As shown in (Fig. 5A), ELOVL4-transduced PC12 cells and controls efficiently internalized 22:6n3, which accounted for 29 relative mol% in the ELOVL4-expressing cells and 30% and 31% in the nontransduced and Ad5-GFP-transduced cells, respectively. While there were no detectable levels of 22:5n3 in the PC12 cells treated with 22:6n3, the levels of 20:5n3, a retro-conversion product of 22:6n3, were slightly greater in the PC12 cells treated with 22:6n3 than in PC12 cells without 22:6n3 treatment (data not shown). The levels of 24:6n3 and 26:6n3 synthesized from 22:6n3 were increased in the ELOVL4-expressing cells compared to controls (Fig. 5B), however, we did not observe a corresponding increase in levels of 24:5n3 and 26:5n3 in 20:5n3 treated cells.
Fig. 5.
Quantification of 22:6n3 VLC-PUFA elongation products in PC12 cells expressing ELOVL4 protein and treated with the sodium salt of 22:6n3. A: Elovl4-transduced PC12 cells and control cells internalized the supplemented 22:6n3 in almost the same amounts. The level of 22:6n3 was slightly lower in PC12 cells expressing the ELOVL4 protein than in the controls. B: Increased biosynthesis of 24:6n3 and 26:6n3 in ELOVL4-expressing cells. Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, while # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3).
Also, increased levels of 0.12% 32:6n3, 0.27% 34:6n3, and 0.18% 36:6n3 VLC-PUFA were found in the ELOVL4-expressing cells but not in the controls (see supplementary Fig. IB). These values are much lower than those seen after 20:5n3 supplementation (Fig. 4B). Low amounts of 34:5n3 (0.07%) and 36:5n3 (0.08%) were also present in the ELOVL4-expressing cells but not in controls. These are from either 20:5n3 retro-converted from 22:6n3 or from serum FAs present in the culture medium. Similar results were obtained in Hep2G-expressing cells (data not shown).
20:4n6 is efficiently elongated to n6 VLC-PUFA by the ELOVL4 protein
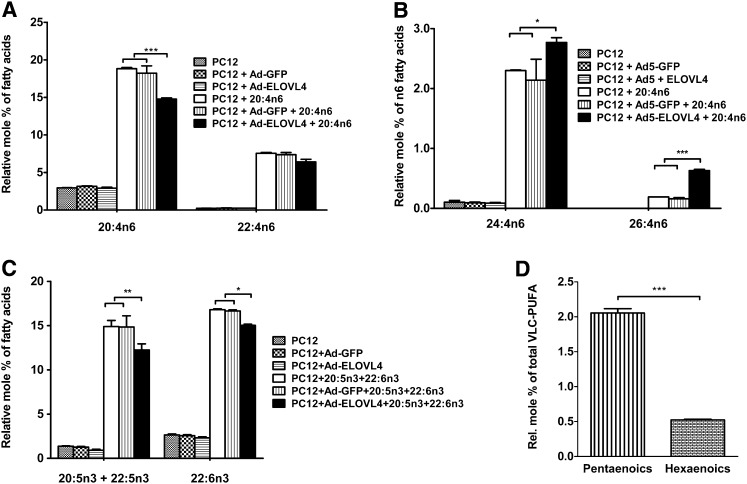
We next determined the efficiency of the ELOVL4 protein in elongating the second most abundant retinal LC-PUFA, 20:4n6, to n6 VLC-PUFA. The cells efficiently took up the supplemented 20:4n6 and elongated it to 22:4n6 independent of ELOVL4 expression (Fig. 6A). Relatively higher levels of 24:4n6 and 26:4n6 were synthesized in the ELOVL4-transduced cells than in the controls (Fig. 6B). The main VLC-PUFA products from 20:4n6 elongation in the ELOVL4-transduced cells included 0.04% 28:4n6, 0.04% 30:4n6, 0.13% 32:4n6, 2.26% 34:4n6, 1.38% 36:4n6, and 0.41% 38:4n6 (see supplementary Fig. IC). The desaturase activity of FADS2 on 20:4n6 and its elongation products were not evident, since there were no n6 VLC-PUFAs with 5-double bonds. The major elongation products were 34:4n6 and 36:4n6, representing 2.3 and 1.4 mol%, respectively.
Fig. 6.
PC12 cells expressing ELOVL4 protein are capable of elongating 20:4n6 to a series of n6 VLC-PUFAs. Also, combined supplementation of 20:5n3 and 22:6n3 in the PC12 cells expressing the ELOVL4 protein results in preferential elongation of 20:5n3 over 22:6n3. A: Elovl4-transduced PC12 cells and control cells efficiently internalized the supplemented sodium salt of 20:4n6 at almost the same amounts. Both 20:4n6 and 22:4n6 were substantially increased in all three groups, indicating that the synthesis of 22:4n6 was independent of ELOVL4 protein. There was no significant difference in the level of 20:4n6 between the two control groups, whereas the level of 20:4n6 in the ELOVL4 protein-expressing group was lower than in the two controls. B: Increased biosynthesis of 24:4n6 and 26:4n6 occurred in ELOVL4-expressing cells but not in controls. C: Supplemented 20:5n3 and 22:6n3 were efficiently taken up at almost the same amounts in the PC12 cells expressing ELOVL4 and in control cells. Elongated 22:5n3 from 20:5n3 was added to the 20:5n3 values. D: Relative mol% of pentaenoics synthesized from 20:5n3 was almost four times that of hexaenoics synthesized from 22:6n3. Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, whereas # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3).
Elovl4 preferentially elongates 20:5n3 over 20:4n6 and 22:6n3
To determine the elongation efficiency of the ELOVL4 protein for 20:5n3, 20:4n6, and 22:6n3 when supplemented in combinations as found in diets and dietary FA supplements, we treated ELOVL4-expressing cells and the controls with equal amounts of either 20:5n3 and 22:6n3 or 20:5n3 and 20:4n6.
When supplemented together, 20:5n3 and 22:6n3 were efficiently taken up at almost the same amounts (15–17%) in the PC12 cells regardless of ELOVL4 expression (Fig. 6C). Because there was an active elongation of 20:5n3 to 22:5n3, values of each were added for the histogram. The ELOVL4-expressing cells elongated both 20:5n3 and 22:6n3 to a series of n3 VLC-PUFAs. From 20:5n3, 34:5n3 and 36:5n3 account for 0.85% and 1.11%, respectively (see supplementary Fig. ID). On the other hand, 34:6n3 and 36:6n3 were only 0.16% and 0.29%, respectively (see supplementary Fig. ID). The total relative mol% of VLC-PUFA synthesized from 20:5n3 was almost four times that of those synthesized from 22:6n3 (Fig. 6D).
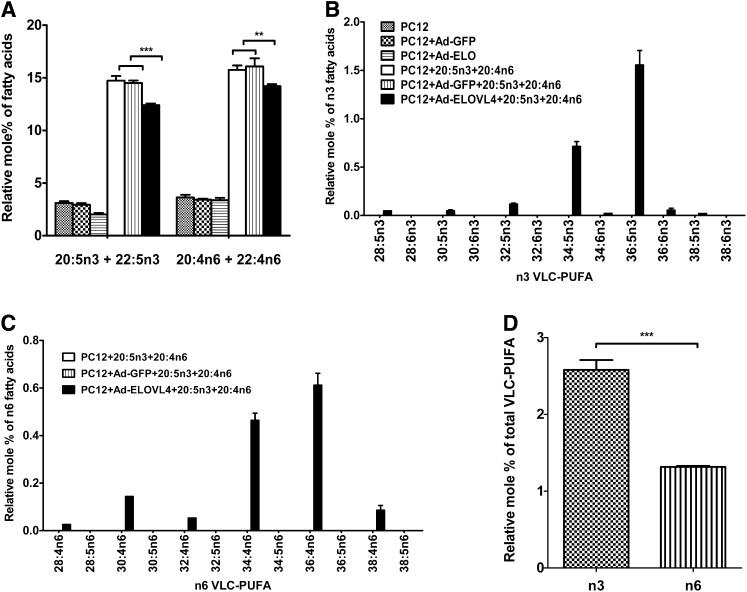
Similarly, when we supplemented the PC12 cells with equal amounts of 20:5n3 and 20:4n6, both FAs were taken up with approximately the same efficiency. There was 12% 20:5n3 plus 22:5n3 in ELOVL4-expressing cells and 15% 20:5n3 plus 22:5n3 in PC12 and GFP-expressing controls (Fig. 7A). Similarly, 14% 20:4n6 plus 22:4n6 was found in ELOVL4-expressing cells, whereas 15–16% was in the control cells. In ELOVL4-expressing cells, 0.71% 34:5n3 and 1.6% 36:5n3 were synthesized from 20:5n3 but 0.46% 34:4n6 and 0.61% 36:4n6 from 20:4n6 (Fig. 7B, C). Total amounts of n3 VLC-PUFA synthesized from 20:5n3 were twice the amount of n6 VLC-PUFA synthesized from 20:4n6 (Fig. 7D).
Fig. 7.
Comparison of the elongation efficiency of 20:4n6 in the presence of 20:5n3 in ELOVL4-expressing PC12 cells treated with equal amounts of 20:5n3 and 20:4n6. A: 20:5n3 and 20:4n6 were efficiently taken up at almost the same amounts in the PC12 cells expressing the ELOVL4 and in control cells. The C22 elongation products of both FAs were added to their respective precursors. B, C: N3 and n6 VLC-PUFAs were only present in ELOVL4-transduced cells. Similar to 20:5n3 and 22:6n3 cotreatment, C34 and C36 PUFAs were the major VLC-PUFA products from both 20:5n3 and 20:4n6 cotreatment. Comparatively, more n3 VLC-PUFA products from 20:5n3 were generated than n6 VLC-PUFA products from 20:4n6. D: Relative mol% of n3 VLC-PUFA synthesized from 20:5n3 was twice that of n6 VLC-PUFA synthesized from 20:4n6. Statistically significant differences are indicated as (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, whereas # indicates no significant difference (p > 0.05). Error bars represent mean ± SD (n = 3).
DISCUSSION
There are seven elongase enzymes (ELOVL1–7) that share sequence homologies with the Saccharomyces cerevisiae ELO group of enzymes (63, 64). The first and rate limiting condensation step in a four-step cycling process of FA elongation is catalyzed in a substrate-specific manner by the ELOVL enzymes (14, 60, 65). In humans, mutations in the ELOVL4 gene cause STGD3, which is characterized by progressive macular degeneration and eventual loss of vision. The ELOVL4 protein is the only member of the mammalian ELOVL family of proteins found thus far to be associated with a human disease. It is also the only elongase that has been shown to mediate biosynthesis of both saturated and polyunsaturated very long chain FAs (C28–C40) in different species (10–13, 49, 66). The presence of VLC-PUFA has been demonstrated in human donor retinas as well (19, 42).
Transgenic and knock-in animal models, expressing the mutant ELOVL4 protein in the retina, have reduced levels of retinal VLC-PUFA and develop retinal degeneration (43–46). Liu et al. (42) found VLC-PUFA in the human retina and RPE/choroid and reported that DHA and VLC-PUFA concentrations in AMD retinas were significantly lower than those in age-matched normal donor retinas. Because the retina of genetic mouse models of the human STGD3 has been found to exhibit reduced retinal function and to accumulate toxic lipid products, we hypothesize that VLC-PUFAs play uniquely important roles in retinal structure and function and that absence or reduction in retinal VLC-PUFA due to mutations in the ELOVL4 gene may contribute to the development of retinal degeneration in STGD3 patients. If our hypothesis can be confirmed, then dietary supplementation of VLC-PUFA may provide both STGD3 and possibly AMD patients with some therapeutic benefits (10, 43). However, there are no readily/commercially available sources of VLC-PUFA for such studies to be conducted. The question also remains as to whether VLC-PUFA provided through the diet can be absorbed by the enterocytes in the small intestine and packaged into chylomicrons for subsequent delivery and incorporation into target tissues such as the retina, since these FAs are not normally found in the liver and the blood. We do not have answers to these questions at the moment. However, we explored the possibility of finding the most suitable n3 or n6 dietary precursors of VLC-PUFA that could help increase the levels of VLC-PUFA in the retina of STGD3 patients with the hope that increasing retinal levels of VLC-PUFA will help protect or attenuate the rate of retinal degeneration in these patients.
We focused our attention on n3 LC-PUFAs, especially 20:5n3 and 22:6n3, as well as the n6 LC-PUFA 20:4n6, inasmuch as some of these FAs have been examined in clinical trials involving patients with AMD or retinitis pigmentosa with promising results (67–71). Also, in animal models, n3 PUFA supplementation has been shown to enhance (72) or prevent the reduction in ERG a- and b-wave amplitudes after transient ischemia (73, 74). Dietary n3 PUFAs have also been shown to be effective in suppressing inflammation and in reducing the incidence of arteriosclerosis and heart failure by lowering plasma triglycerides and blood pressure (75, 76). Thus, we evaluated which of the LC-PUFAs are metabolically active precursors for the synthesis of VLC-PUFA using cell cultures as a first step to eventually determine whether dietary supplementation of these FAs could provide beneficial effects to STGD3 patients, as has been shown for AMD patients in some of the Age-Related Eye Disease Study (AREDS) studies (37, 39).
We found that 20:5n3, 22:6n3, and 20:4n6 can be metabolically converted to VLC-PUFA when provided individually to PC12 cells expressing Elovl4. The supplemented C20-C22 FAs were efficiently elongated to C26 VLC-PUFA by endogenously expressed ELOVL1, 2, 5 proteins, but possibly very little by ELOVL3 protein, because low levels of Elovl3 were detected in the PC12 cells (60). It was interesting, however, that although the endogenous levels of Elovl4 were not very different from the other Elovls, it did not demonstrate any elongation activity on the supplemented FAs in the control cells. This suggests that the catalytic ability of the ELOVL4 protein to synthesize VLC-PUFA might be concentration dependent, as demonstrated in the biosynthesis of 24:5n3, 26:5n3, 24:4n6, and 26:4n6 (Figs. 5B and 6B).
As we have demonstrated, the ELOVL4 protein is capable of efficiently elongating individually supplemented FAs to VLC-PUFA. The major point of our study, however, is that when the FAs are supplemented in combinations, the ELOVL4 protein preferentially elongates C26 elongation products of 20:5n3 > 20:4n6 > 22:6n3. These findings are consistent with the finding that Atlantic salmon (Salmo salar) Elovl4 synthesized greater amounts of VLC-PUFA from 20:5n3 than from 20:4n6 and 22:6n3 (49). Whether supplemented individually or in combination with 20:5n3, most of the 22:6n3 seems to be more efficiently esterified into phospholipids and hence may not be available for elongation, whereas 20:5n3 and 20:4n6 are more readily converted to VLC-PUFA. If loss or reduction in VLC-PUFA plays a role in STGD3 progression, our findings may help explain why increased levels of 22:6n3 in the retina of transgenic mice carrying the human ELOVL4 5-base pair deletion mutation did not rescue the rate of retinal degeneration in these mice (77). Similarly, our results also shed light on the findings that when Elovl2 knockout mice were fed diets enriched in 22:6n3, significant amounts of the 22:6n3 were found in the liver and plasma, but little 22:6n3 was elongated to VLC-PUFA in the testis, and thus was unable to rescue infertility associated with loss of VLC-PUFA (78). An in vivo study also presented evidence that 20:5n3 is the preferred substrate for the synthesis of retinal VLC-PUFA (48). These findings have led us to conclude that although supplementation of 22:6n3 may be beneficial, as shown in some of the AREDS studies, 22:6n3 supplementation alone in STGD3 patients might not be a potential therapy. We suggest that if absence of VLC-PUFA contributes to the rate of retinal degeneration in STGD3 patients, and VLC-PUFA synthesis is much more efficiently achieved through elongation of 20:5n3 and 20:4n6, then early dietary supplementation of 20:5n3 and/or 20:4n6 in STGD3 patients may help slow the onset of retinal degeneration. One major caveat to this suggestion, which needs to be critically evaluated, is that we are driving the expression of the Elovl4 gene in the PC12 cells, a cell type that has no or low levels of ELOVL4 protein expression. As a result, the natural transcriptional, translational, and FA metabolic control mechanisms may be subverted in these cells and may not hold true in vivo in Elovl4-expressing cells such as the retina, skin, and testis. Also, the presence of the mutant protein in STGD3 patients could have an effect on the biosynthetic activity of the wild-type protein in vivo. However, our results provide an important indication on the selection of possible FAs for dietary supplementations in patients with ELOVL4 mutations, inasmuch as some substrates are preferentially elongated over others by this enzyme. We are currently evaluating this possibility in animal models of STGD3 to determine whether indeed in vivo supplementation of 20:5n3 and 20:4n6 will offer some therapeutic benefits to STGD3 patients.
Supplementary Material
Footnotes
Abbreviations:
- AA
- arachidonic acid
- AMD
- age-related macular degeneration
- AREDS
- Age-Related Eye Disease Study
- DHA
- docosahexaenoic acid
- EPA
- eicosapentaenoic acid
- ELOVL4
- ELOngase of very long chain fatty acid-4
- FAME
- fatty acid methyl ester
- FID
- flame ionization detector
- GC
- gas chromatograph
- LA
- linoleic acid
- LC-PUFA
- long-chain PUFA
- PC
- phosphatidylcholine
- PC12
- pheochromocytoma cell
- pfu
- plaque-forming units
- qRT-PCR
- quantitative real-time PCR
- RPE
- retinal pigment epithelium
- SIM
- single-ion monitoring
- STGD3
- autosomal dominant Stargardt-like macular dystrophy
- VLC-FA
- very long chain saturated or monounsaturated fatty acid
- VLC-PUFA
- very long chain PUFA
This work was supported by National Institutes of Health, National Eye Institute, Grants EY-04149, EY-00871, and EY-12190 (R.E.A.); National Center for Research Resources Grant RR-17703 (R.E.A.); grants from Research to Prevent Blindness, Inc. and the Foundation Fighting Blindness (R.E.A.); and from the Hope for Vision and Knights Templar Eye Foundation, Inc. (M.P.A.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Aveldaño M. I., Robinson B. S., Johnson D. W., Poulos A. 1993. Long and very long chain polyunsaturated fatty acids of the n-6 series in rat seminiferous tubules. Active desaturation of 24:4n-6 to 24:5n-6 and concomitant formation of odd and even chain tetraenoic and pentaenoic fatty acids up to C32. J. Biol. Chem. 268: 11663–11669. [PubMed] [Google Scholar]
- 2.Furland N. E., Oresti G. M., Antollini S. S., Venturino A., Maldonado E. N., Aveldano M. I. 2007. Very long-chain polyunsaturated fatty acids are the major acyl groups of sphingomyelins and ceramides in the head of mammalian spermatozoa. J. Biol. Chem. 282: 18151–18161. [DOI] [PubMed] [Google Scholar]
- 3.Furland N. E., Zanetti S. R., Oresti G. M., Maldonado E. N., Aveldano M. I. 2007. Ceramides and sphingomyelins with high proportions of very long-chain polyunsaturated fatty acids in mammalian germ cells. J. Biol. Chem. 282: 18141–18150. [DOI] [PubMed] [Google Scholar]
- 4.Poulos A., Sharp P., Johnson D., Easton C. 1988. The occurrence of polyenoic very long chain fatty acids with greater than 32 carbon atoms in molecular species of phosphatidylcholine in normal and peroxisome-deficient (Zellweger's syndrome) brain. Biochem. J. 253: 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rezanka T. 1989. Very-long-chain fatty acids from the animal and plant kingdoms. Prog. Lipid Res. 28: 147–187. [DOI] [PubMed] [Google Scholar]
- 6.Rezanka T., Sigler K. 2009. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 48: 206–238. [DOI] [PubMed] [Google Scholar]
- 7.Bjerve K. S., Thoresen L., Mostad I. L., Alme K. 1987. Alpha-linolenic acid deficiency in man: effect of essential fatty acids on fatty acid composition. Adv. Prostaglandin Thromboxane Leukot. Res. 17B: 862–865. [PubMed] [Google Scholar]
- 8.Burr G. O., Burr M. M. 1973. Nutrition classics from The Journal of Biological Chemistry 82:345–67, 1929. A new deficiency disease produced by the rigid exclusion of fat from the diet. Nutr. Rev. 31: 248–249. [DOI] [PubMed] [Google Scholar]
- 9.Connor W. E., Neuringer M., Reisbick S. 1992. Essential fatty acids: the importance of n-3 fatty acids in the retina and brain. Nutr. Rev. 50: 21–29. [DOI] [PubMed] [Google Scholar]
- 10.Agbaga M. P., Brush R. S., Mandal M. N., Henry K., Elliott M. H., Anderson R. E. 2008. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. USA. 105: 12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedell M., Harkewicz R., Wang X., Zhang K. 2010. Focus on molecules: ELOVL4. Exp. Eye Res. 90: 476–477. [DOI] [PubMed] [Google Scholar]
- 12.Okuda A., Naganuma T., Ohno Y., Abe K., Yamagata M., Igarashi Y., Kihara A. 2010. Hetero-oligomeric interactions of an ELOVL4 mutant protein: implications in the molecular mechanism of Stargardt-3 macular dystrophy. Mol. Vis. 16: 2438–2445. [PMC free article] [PubMed] [Google Scholar]
- 13.Monroig O., Rotllant J., Cerda-Reverter J. M., Dick J. R., Figueras A., Tocher D. R. 2010. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim. Biophys. Acta. 1801: 1145–1154. [DOI] [PubMed] [Google Scholar]
- 14.Agbaga M. P., Mandal M. N., Anderson R. E. 2010. Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J. Lipid Res. 51: 1624–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aveldaño M. I. 1987. A novel group of very long chain polyenoic fatty acids in dipolyunsaturated phosphatidylcholines from vertebrate retina. J. Biol. Chem. 262: 1172–1179. [PubMed] [Google Scholar]
- 16.Aveldaño M. I. 1992. Long and very long polyunsaturated fatty acids of retina and spermatozoa: the whole complement of polyenoic fatty acid series. Adv. Exp. Med. Biol. 318: 231–242. [DOI] [PubMed] [Google Scholar]
- 17.Aveldaño M. I. 1988. Phospholipid species containing long and very long polyenoic fatty acids remain with rhodopsin after hexane extraction of photoreceptor membranes. Biochemistry. 27: 1229–1239. [DOI] [PubMed] [Google Scholar]
- 18.Brush R. S., Tran J. T., Henry K. R., McClellan M. E., Elliott M. H., Mandal M. N. 2010. Retinal sphingolipids and their very-long-chain fatty acid-containing species. Invest. Ophthalmol. Vis. Sci. 51: 4422–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berdeaux O., Juaneda P., Martine L., Cabaret S., Bretillon L., Acar N. 2010. Identification and quantification of phosphatidylcholines containing very-long-chain polyunsaturated fatty acid in bovine and human retina using liquid chromatography/tandem mass spectrometry. J. Chromatogr. A. 1217: 7738–7748. [DOI] [PubMed] [Google Scholar]
- 20.Fliesler S. J., Anderson R. E. 1983. Chemistry and metabolism of lipids in the vertebrate retina. Prog. Lipid Res. 22: 79–131. [DOI] [PubMed] [Google Scholar]
- 21.Anderson R. E., O'Brien P. J., Wiegand R. D., Koutz C. A., Stinson A. M. 1992. Conservation of docosahexaenoic acid in the retina. Adv. Exp. Med. Biol. 318: 285–294. [DOI] [PubMed] [Google Scholar]
- 22.SanGiovanni J. P., Chew E. Y. 2005. The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog. Retin. Eye Res. 24: 87–138. [DOI] [PubMed] [Google Scholar]
- 23.Delton-Vandenbroucke I., Grammas P., Anderson R. E. 1997. Polyunsaturated fatty acid metabolism in retinal and cerebral microvascular endothelial cells. J. Lipid Res. 38: 147–159. [PubMed] [Google Scholar]
- 24.Catalan J., Moriguchi T., Slotnick B., Murthy M., Greiner R. S., Salem N., Jr 2002. Cognitive deficits in docosahexaenoic acid-deficient rats. Behav. Neurosci. 116: 1022–1031. [DOI] [PubMed] [Google Scholar]
- 25.Connor K. M., SanGiovanni J. P., Lofqvist C., Aderman C. M., Chen J., Higuchi A., Hong S., Pravda E. A., Majchrzak S., Carper D., et al. 2007. Increased dietary intake of omega-3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat. Med. 13: 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeMar J. C., Jr, DiMartino C., Baca A. W., Lefkowitz W., Salem N., Jr 2008. Effect of dietary docosahexaenoic acid on biosynthesis of docosahexaenoic acid from alpha-linolenic acid in young rats. J. Lipid Res. 49: 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffrey B. G., Mitchell D. C., Hibbeln J. R., Gibson R. A., Chedester A. L., Salem N., Jr 2002. Visual acuity and retinal function in infant monkeys fed long-chain PUFA. Lipids. 37: 839–848. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y. H., Salem N., Jr 2005. In vivo conversion of 18- and 20-C essential fatty acids in rats using the multiple simultaneous stable isotope method. J. Lipid Res. 46: 1962–1973. [DOI] [PubMed] [Google Scholar]
- 29.Niu S. L., Mitchell D. C., Lim S. Y., Wen Z. M., Kim H. Y., Salem N., Jr, Litman B. J. 2004. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J. Biol. Chem. 279: 31098–31104. [DOI] [PubMed] [Google Scholar]
- 30.Weisinger H. S., Armitage J. A., Jeffrey B. G., Mitchell D. C., Moriguchi T., Sinclair A. J., Weisinger R. S., Salem N., Jr 2002. Retinal sensitivity loss in third-generation n-3 PUFA-deficient rats. Lipids. 37: 759–765. [DOI] [PubMed] [Google Scholar]
- 31.Benolken R. M., Anderson R. E., Wheeler T. G. 1973. Membrane fatty acids associated with the electrical response in visual excitation. Science. 182: 1253–1254. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler T. G., Benolken R. M., Anderson R. E. 1975. Visual membranes: specificity of fatty acid precursors for the electrical response to illumination. Science. 188: 1312–1314. [DOI] [PubMed] [Google Scholar]
- 33.Greiner R. S., Moriguchi T., Hutton A., Slotnick B. M., Salem N., Jr 1999. Rats with low levels of brain docosahexaenoic acid show impaired performance in olfactory-based and spatial learning tasks. Lipids. 34(Suppl): 239–243. [DOI] [PubMed] [Google Scholar]
- 34.Garelli A., Rotstein N. P., Politi L. E. 2006. Docosahexaenoic acid promotes photoreceptor differentiation without altering Crx expression. Invest. Ophthalmol. Vis. Sci. 47: 3017–3027. [DOI] [PubMed] [Google Scholar]
- 35.German O. L., Insua M. F., Gentili C., Rotstein N. P., Politi L. E. 2006. Docosahexaenoic acid prevents apoptosis of retina photoreceptors by activating the ERK/MAPK pathway. J. Neurochem. 98: 1507–1520. [DOI] [PubMed] [Google Scholar]
- 36.Seddon J. M., Cote J., Rosner B. 2003. Progression of age-related macular degeneration: association with dietary fat, transunsaturated fat, nuts, and fish intake. Arch. Ophthalmol. 121: 1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.SanGiovanni J. P., Chew E. Y., Clemons T. E., Davis M. D., Ferris F. L., III, Gensler G. R., Kurinij N., Lindblad A. S., Milton R. C., Seddon J. M., et al. 2007. The relationship of dietary lipid intake and age-related macular degeneration in a case-control study: AREDS Report No. 20. Arch. Ophthalmol. 125: 671–679. [DOI] [PubMed] [Google Scholar]
- 38.Cho E., Hung S., Willett W. C., Spiegelman D., Rimm E. B., Seddon J. M., Colditz G. A., Hankinson S. E. 2001. Prospective study of dietary fat and the risk of age-related macular degeneration. Am. J. Clin. Nutr. 73: 209–218. [DOI] [PubMed] [Google Scholar]
- 39.SanGiovanni J. P., Chew E. Y., Agron E., Clemons T. E., Ferris F. L., III, Gensler G., Lindblad A. S., Milton R. C., Seddon J. M., Klein R., et al. 2008. The relationship of dietary omega-3 long-chain polyunsaturated fatty acid intake with incident age-related macular degeneration: AREDS report no. 23. Arch. Ophthalmol. 126: 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seddon J. M., Rosner B., Sperduto R. D., Yannuzzi L., Haller J. A., Blair N. P., Willett W. 2001. Dietary fat and risk for advanced age-related macular degeneration. Arch. Ophthalmol. 119: 1191–1199. [DOI] [PubMed] [Google Scholar]
- 41.Seddon J. M., George S., Rosner B. 2006. Cigarette smoking, fish consumption, omega-3 fatty acid intake, and associations with age-related macular degeneration: the US Twin Study of Age-Related Macular Degeneration. Arch. Ophthalmol. 124: 995–1001. [DOI] [PubMed] [Google Scholar]
- 42.Liu A., Chang J., Lin Y., Shen Z., Bernstein P. S. 2010. Long-chain and very long-chain polyunsaturated fatty acids in ocular aging and age-related macular degeneration. J. Lipid Res. 51: 3217–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon A., Jackson S. N., Woods A. S., Kedzierski W. 2007. A Stargardt disease-3 mutation in the mouse Elovl4 gene causes retinal deficiency of C32-C36 acyl phosphatidylcholines. FEBS Lett. 581: 5459–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMahon A., Butovich I. A., Mata N. L., Klein M., Ritter R., III, Richardson J., Birch D. G., Edwards A. O., Kedzierski W. 2007. Retinal pathology and skin barrier defect in mice carrying a Stargardt disease-3 mutation in elongase of very long chain fatty acids-4. Mol. Vis. 13: 258–272. [PMC free article] [PubMed] [Google Scholar]
- 45.Vasireddy V., Jablonski M. M., Khan N. W., Wang X. F., Sahu P., Sparrow J. R., Ayyagari R. 2009. Elovl4 5-bp deletion knock-in mouse model for Stargardt-like macular degeneration demonstrates accumulation of ELOVL4 and lipofuscin. Exp. Eye Res. 89: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasireddy V., Jablonski M. M., Mandal M. N., Raz-Prag D., Wang X. F., Nizol L., Iannaccone A., Musch D. C., Bush R. A., Salem N., Jr, et al. 2006. Elovl4 5-bp-deletion knock-in mice develop progressive photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 47: 4558–4568. [DOI] [PubMed] [Google Scholar]
- 47.Hubbard A. F., Askew E. W., Singh N., Leppert M., Bernstein P. S. 2006. Association of adipose and red blood cell lipids with severity of dominant Stargardt macular dystrophy (STGD3) secondary to an ELOVL4 mutation. Arch. Ophthalmol. 124: 257–263. [DOI] [PubMed] [Google Scholar]
- 48.Suh M., Clandinin M. T. 2005. 20:5n-3 but not 22:6n-3 is a preferred substrate for synthesis of n-3 very-long-chain fatty acids (C24-C36) in retina. Curr. Eye Res. 30: 959–968. [DOI] [PubMed] [Google Scholar]
- 49.Carmona-Antoñanzas G., Monroig O., Dick J. R., Davie A., Tocher D. R. 2011. Biosynthesis of very long-chain fatty acids (C>24) in Atlantic salmon: cloning, functional characterisation, and tissue distribution of an Elovl4 elongase. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 159: 122–129. [DOI] [PubMed] [Google Scholar]
- 50.Xiao J., Chodosh J. 2005. JNK regulates MCP-1 expression in adenovirus type 19-infected human corneal fibroblasts. Invest. Ophthalmol. Vis. Sci. 46: 3777–3782. [DOI] [PubMed] [Google Scholar]
- 51.Kang Z. B., Ge Y., Chen Z., Cluette-Brown J., Laposata M., Leaf A., Kang J. X. 2001. Adenoviral gene transfer of Caenorhabditis elegans n-3 fatty acid desaturase optimizes fatty acid composition in mammalian cells. Proc. Natl. Acad. Sci. USA. 98: 4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge Y., Wang X., Chen Z., Landman N., Lo E. H., Kang J. X. 2002. Gene transfer of the Caenorhabditis elegans n-3 fatty acid desaturase inhibits neuronal apoptosis. J. Neurochem. 82: 1360–1366. [DOI] [PubMed] [Google Scholar]
- 53.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 54.Martin R. E., Elliott M. H., Brush R. S., Anderson R. E. 2005. Detailed characterization of the lipid composition of detergent-resistant membranes from photoreceptor rod outer segment membranes. Invest. Ophthalmol. Vis. Sci. 46: 1147–1154. [DOI] [PubMed] [Google Scholar]
- 55.Pawlosky R. J., Sprecher H. W., Salem N., Jr 1992. High sensitivity negative ion GC-MS method for detection of desaturated and chain-elongated products of deuterated linoleic and linolenic acids. J. Lipid Res. 33: 1711–1717. [PubMed] [Google Scholar]
- 56.Richardson U. I., Wurtman R. J. 2007. Polyunsaturated fatty acids stimulate phosphatidylcholine synthesis in PC12 cells. Biochim. Biophys. Acta. 1771: 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin R. E., Wickham J. Q., Om A. S., Sanders J., Ceballos N. 2000. Uptake and incorporation of docosahexaenoic acid (DHA) into neuronal cell body and neurite/nerve growth cone lipids: evidence of compartmental DHA metabolism in nerve growth factor-differentiated PC12 cells. Neurochem. Res. 25: 715–723. [DOI] [PubMed] [Google Scholar]
- 58.Maharvi G. M., Edwards A. O., Fauq A. H. 2010. Chemical synthesis of deuterium-labeled and unlabeled very long chain polyunsaturated fatty acids. Tetrahedron Lett. 51: 6426–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer A., Kirsch H., Domergue F., Abbadi A., Sperling P., Bauer J., Cirpus P., Zank T. K., Moreau H., Roscoe T. J., et al. 2004. Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J. Lipid Res. 45: 1899–1909. [DOI] [PubMed] [Google Scholar]
- 60.Ohno Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y., Sassa T., Kihara A. 2010. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. USA. 107: 18439–18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Agbaga M. P., Brush R. S., Mandal M. N., Elliott M. H., Al-Ubaidi M. R., Anderson R. E. 2010. Role of Elovl4 protein in the biosynthesis of docosahexaenoic acid. Adv. Exp. Med. Biol. 664: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rotstein N. P., Pennacchiotti G. L., Sprecher H., Aveldano M. I. 1996. Active synthesis of C24:5, n-3 fatty acid in retina. Biochem. J. 316: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakobsson A., Westerberg R., Jacobsson A. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45: 237–249. [DOI] [PubMed] [Google Scholar]
- 64.Leonard A. E., Pereira S. L., Sprecher H., Huang Y. S. 2004. Elongation of long-chain fatty acids. Prog. Lipid Res. 43: 36–54. [DOI] [PubMed] [Google Scholar]
- 65.Ofman R., Dijkstra I. M., van Roermund C. W., Burger N., Turkenburg M., van Cruchten A., van Engen C. E., Wanders R. J., Kemp S. 2010. The role of ELOVL1 in very long-chain fatty acid homeostasis and X-linked adrenoleukodystrophy. EMBO Mol Med. 2: 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMahon A., Butovich I. A., Kedzierski W. 2011. Epidermal expression of an Elovl4 transgene rescues neonatal lethality of homozygous Stargardt disease-3 mice. J. Lipid Res. 52: 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodge W. G., Barnes D., Schachter H. M., Pan Y. I., Lowcock E. C., Zhang L., Sampson M., Morrison A., Tran K., Miguelez M., et al. 2006. The evidence for efficacy of omega-3 fatty acids in preventing or slowing the progression of retinitis pigmentosa: a systematic review. Can. J. Ophthalmol. 41: 481–490. [DOI] [PubMed] [Google Scholar]
- 68.Augood C., Chakravarthy U., Young I., Vioque J., de Jong P. T., Bentham G., Rahu M., Seland J., Soubrane G., Tomazzoli L., et al. 2008. Oily fish consumption, dietary docosahexaenoic acid and eicosapentaenoic acid intakes, and associations with neovascular age-related macular degeneration. Am. J. Clin. Nutr. 88: 398–406. [DOI] [PubMed] [Google Scholar]
- 69.Berson E. L., Rosner B., Sandberg M. A., Weigel-DiFranco C., Moser A., Brockhurst R. J., Hayes K. C., Johnson C. A., Anderson E. J., Gaudio A. R., et al. 2004. Further evaluation of docosahexaenoic acid in patients with retinitis pigmentosa receiving vitamin A treatment: subgroup analyses. Arch. Ophthalmol. 122: 1306–1314. [DOI] [PubMed] [Google Scholar]
- 70.Hoffman D. R., Birch D. G. 1995. Docosahexaenoic acid in red blood cells of patients with X-linked retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 36: 1009–1018. [PubMed] [Google Scholar]
- 71.Hodge W. G., Schachter H. M., Barnes D., Pan Y., Lowcock E. C., Zhang L., Sampson M., Morrison A., Tran K., Miguelez M., Lewin G. 2006. Efficacy of omega-3 fatty acids in preventing age-related macular degeneration: a systematic review. Ophthalmology.113: 1165–1172. [PMC free article] [PubMed]
- 72.Suh M., Sauve Y., Merrells K. J., Kang J. X., Ma D. W. 2009. Supranormal electroretinogram in fat-1 mice with retinas enriched in docosahexaenoic acid and n-3 very long chain fatty acids (C24-C36). Invest. Ophthalmol. Vis. Sci. 50: 4394–4401. [DOI] [PubMed] [Google Scholar]
- 73.Miyauchi O., Mizota A., Adachi-Usami E., Nishikawa M. 2001. Protective effect of docosahexaenoic acid against retinal ischemic injury: an electroretinographic study. Ophthalmic Res. 33: 191–195. [DOI] [PubMed] [Google Scholar]
- 74.Murayama K., Yoneya S., Miyauchi O., Adachi-Usami E., Nishikawa M. 2002. Fish oil (polyunsaturated fatty acid) prevents ischemic-induced injury in the mammalian retina. Exp. Eye Res. 74: 671–676. [DOI] [PubMed] [Google Scholar]
- 75.Jung U. J., Torrejon C., Tighe A. P., Deckelbaum R. J. 2008. n-3 Fatty acids and cardiovascular disease: mechanisms underlying beneficial effects. Am. J. Clin. Nutr. 87: 2003–2009. [DOI] [PubMed] [Google Scholar]
- 76.Torrejon C., Jung U. J., Deckelbaum R. J. 2007. n-3 Fatty acids and cardiovascular disease: actions and molecular mechanisms. Prostaglandins Leukot. Essent. Fatty Acids. 77: 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li F., Marchette L. D., Brush R. S., Elliott M. H., Le Y. Z., Henry K. A., Anderson A. G., Zhao C., Sun X., Zhang K., et al. 2009. DHA does not protect ELOVL4 transgenic mice from retinal degeneration. Mol. Vis. 15: 1185–1193. [PMC free article] [PubMed] [Google Scholar]
- 78.Guillou H., Zadravec D., Martin P. G., Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog. Lipid Res. 49: 186–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.