Abstract
Z-Guggulsterone is a major ingredient in the Indian traditional hypolipidemic remedy guggul. A study in mice has established that its hypolipidemic effect involves the farnesoid X receptor (FXR), presumably by acting as an antagonist of this receptor. It is generally assumed that the antagonism leads to induction of cytochrome P450 7A1 (CYP7A1), the rate-limiting enzyme converting free cholesterol to bile acids. In this study, we tested whether Z-guggulsterone indeed induces human CYP7A1. In addition, the expression of cholesteryl ester hydrolase CES1 and bile salt export pump (BSEP) was monitored. Contrary to the general assumption, Z-guggulsterone did not induce CYP7A1. Instead, this phytosterol significantly induced CES1 and BSEP through transactivation. Z-Guggulsterone underwent metabolism by CYP3A4, and the metabolites greatly increased the induction potency on BSEP but not on CES1. BSEP induction favors cholesterol elimination, whereas CES1 involves both elimination and retention (probably when excessively induced). Interestingly, clinical trials reported the hypolipidemic response rates from 18% to 80% and showed that higher dosages actually increased VLDL cholesterol. Our findings predict that better hypolipidemic outcomes likely occur in individuals who have a relatively higher capacity of metabolizing Z-guggulsterone with moderate CES1 induction, a scenario possibly achieved by lowering the dosing regimens.
Keywords: atherlosclerosis, bile acids, dyslipidemias, carboxylesterase, cytochrome P450 7A1, transactivation, antioxidant
Cardiovascular diseases remain the leading cause of death. The majority of these diseases involve a pathogenic process: atherosclerosis (1). Hyperlipidemia is the primary atherogenic risk factor (1, 2). In the plasma, lipids are encapsulated particles with apoproteins (1). High-density lipoproteins (HDL) transport lipids from peripheral tissues to the liver and, thus, are antiatherogenic (1). Low-density lipoproteins (LDL) are the major type of lipoproteins accumulated in the subendothelial matrix and, hence, are atherogenic. LDL is derived from very low-density lipoproteins (VLDL), which are assembled in and secreted by the liver (2). VLDL assembly starts when apolipoprotein B-100 (apo-B100) is translated and translocated into endoplasmic reticulum (ER). Maturation of VLDL particles undergoes multiple lipidation processes that traverse from the ER and post-ER/Golgi compartments (2). Lipids incorporated into VLDL particles undergo continuous hydrolysis/reesterification.
Carboxylesterase-1 (CES1) in humans and its functionally related mouse counterpart (Ces1d) in lipid catabolism are implicated in the hydrolysis/reesterification cycle (3, 4). Indeed, higher CES1 activity facilitates VLDL maturation (5), and transgenic expression of CES1 leads to increased secretion of apoB proteins (3). Conversely, hydrolysis of cholesterol esters by CES1 is linked to cholesterol elimination (6). In the classic pathway of bile acid synthesis, cholesterol is oxidized by cytochrome 7A1 (CYP7A1) (7), eventually converted into bile acids, and secreted by the bile acid export pump (BSEP). The secretion of bile acids represents the net elimination of excessive cholesterol (8).
Increased bile acid synthesis has long been explored for the development of drugs with cholesterol-lowering activity. The biliary secretion has a large capacity and is equally effective in eliminating both endogenous and dietary cholesterols. For quite some time, guggul has gained interest worldwide (9–11). Originally described in the Indian traditional medicine Ayurveda, guggul has been used for thousands of years to treat conditions such as atherosclerosis (9, 12). The hypolipidemic activity has been confirmed in multiple animal models (9). Human clinical trials, however, produced inconsistent results. Some clinical trials reported that guggul lowered plasma cholesterol by 20–30% and triglycerides by 10–20%, with the response rate being as high as 80% (9, 13, 14). Other trials, however, did not detect the same level of effectiveness (10, 11).
Nonetheless, Z-guggulsterone is recognized as the major active ingredient. Its hypolipidemic activity in mice requires the farnesoid X receptor (FXR) (15). This phytosterol is an antagonist of FXR, and such antagonism is assumed to accelerate bile acid synthesis by inducing CYP7A1. To directly test this hypothesis, human primary hepatocytes were treated with Z-guggulsterone, and the expression of CYP7A1 was determined. In addition, the expression of CES1 and BSEP was monitored. Contrary to the general assumption, Z-guggulsterone was an inducer of CES1 and BSEP but not of CYP7A1. In addition, Z-guggulsterone was metabolized, and the metabolites differentially altered CES1 and BSEP induction. Our findings predict that the hypolipidemic activity of Z-guggulsterone depends on two important interplays: CES1 induction versus CYP7A1 activity, and the metabolism of Z-guggulsterone versus the relative inducibility between CES1 and BSEP.
EXPERIMENTAL PROCEDURES
RT-qPCR
Human primary hepatocytes were obtained from the Liver Tissues Procurement and Distribution System (University of Minnesota) or CellzDirect (Pittsboro, NC). The use of the human samples was approved by the Institutional Review Board. Hepatocytes and hepatoma cells (Huh7) were cultured and treated with Z-guggulsterone, chenodeoxycholic acid (CDCA), or both as described previously (12). The mRNA levels were determined with TaqMan assays (16). The assay identification numbers were CES1, Hs00275607_m1; CYP7A1, Hs00167982_m1; BSEP, Hs00184824_m1; GAPDH, 4352934E; and RNA polymerase II, Hs00172187_m1. The CES1 probe recognized both CES1A1 and CES1A2, and both enzymes are identical, although encoded by distinct genes (17).
Reporter assays
The BSEP guggulsterone element reporter and the FXR expression construct were described elsewhere (12). The CES1A1 reporters containing the promoter and its upstream sequence at varying lengths were prepared by inserting the corresponding genomic fragment into the pGL3 basic luciferase vector at the Mlu I and Xho I sites. All genomic fragments were generated by PCR with high-fidelity Platinum Taq DNA polymerase. The primer sequences are listed in Table 1. Initially, human genomic DNA from the placenta was used as the template for the amplification of the genomic fragment from -7714 to -21 (from the initial translation codon) with primers CES1A1-21XhoIa and 7714MluIs. This fragment was inserted into the pGL3 basic luciferase vector to prepare reporter CES1A1-7714Luc. This construct was then used as the template to prepare reporters: CES1A1-6981Luc, CES1A1-3582Luc, CES1A1-3432Luc, and CES1A1-2932Luc with the same antisense primer (CES1A1-21XhoIa) but different sense primer (Table 1). The reporters CES1A1-9622Luc and CES1A1-9332Luc contained additional upstream sequences. To prepare these constructs, human genomic DNA was used as the template with primers CES1A1-4777a and CES1A1-9622Mlus or CES1A1-9333MluIs. The respective fragments were digested with Mlu I and Spe I (an internal site in the fragments). The digested fragments were then ligated to the reporter CES1A1-7714Luc pretreated with the same endonucleases. The CES1A1 guggulsterone element reporter was prepared by inserting the oligonucleotides (5′-CACAATCTGCAGAGTCATCATGAAG-3′) into the pGL3 promoter vector at the Nhe I and Xho I sites. All constructs were confirmed by sequencing. The reporter assays were performed as described previously (12).
TABLE 1.
Sequences of primers for reporter constructs
| Primer | Sequence | Reporter |
| CES1A1-21XhoIa | 5′-tcggggcctgcgaggtctcttgcagttca-3′ | All promoter reporters |
| CES1A1-7713MluIs | 5′-taaggtggatggattacctgaggtcaggagt-3′ | CES1A1-7714Luc |
| CES1A1-6980MluIs | 5′-ttgcatgtcaaagtgtcagaatggagaaat-3′ | CES1A1-6981Luc |
| CES1A1-3582MluIs | 5′-ttattgcttacagctgaagtg-3′ | CES1A1-3582Luc |
| CES1A1-3432MluIs | 5′-tcaggcaaaacctaggagtgg-3′ | CES1A1-3432Luc |
| CES1A1-2931MluIs | 5′-tccagcctgcacgtggctaga-3′ | CES1A1-2932Luc |
| CES1A1-4777a | 5′-catagctagccaccctcaatggcgcaaaca-3′ | 1A19622 and 9332Luc |
| CES1A1-9622MluIs | 5′-tgacacgcgtaccaactcagtccagattcccccagggcag-3′ | CES1A1-9622Luc |
| CES1A1-9332MluIs | 5′-tgacacgcgtaagagctcgctgtgctccatgaatggggtg-3′ | CES1A1-9332Luc |
Metabolism of Z-guggulsterone
Metabolism was conducted with recombinant CYPs (1 pmol) and pooled human liver microsomes (20 μg) in a total volume of 100 μl (18, 19). Preliminary studies were performed to determine conditions supporting the linear rate. Z-Guggulsterone was dissolved in acetonitrile, and the final concentration of the solvent was 0.1%. After 10 min preincubation at 37°C, reactions were initiated by adding the NADPH-regenerating system (NRS) and then were incubated for 40 min. The reactions were terminated by two volumes of ice-cold acetonitrile containing the internal standard spironolactone (10 μg/ml), followed by centrifugation. For the inhibition assay, the prereaction mixtures contained a CYP inhibitor at previously reported concentrations (20). The metabolism was monitored for the disappearance of Z-guggulsterone by LC-MS/MS (API 3200) (19). Detection of the analytes was performed in positive ion mode using the mass transitions of m/z 313.2→97.0 for Z-guggulsterone and m/z 416.6→341.2 for IS. Injection analysis was performed at a flow rate of 200 μl/min to obtain optimum source parameters. The assay was linear from 1.04 to 416.27 ng/ml for Z-guggulsterone.
Purification of metabolites
Reactions were set up in a total volume of 1 ml with CYP3A4. The supernatants were separated on a Chromolith SpeedROD column RP-18e (Merck, Germany) by a gradient mobile phase made of 0.1% aqueous formic acid (A) and acetonitrile (B). The mobile phase started with 10% acetonitrile for 2 min, 15% for 5 min, 70% for 3 min, 30% for 4 min, and 10% for 1 min with the flow rate of 2 ml/min. The metabolites were detected by a diode array detector at 240 nm. The concentrated metabolites were analyzed by HPLC, and the concentrations were estimated according to the curve generated with Z-guggulsterone at various concentrations. The metabolites were analyzed for the mass-to-charge ratios by a Q-Star Elite time-of-flight mass spectrometer (Applied Biosystems/MDS SCIEX). The LC conditions were the same as described above but with a reduced flow rate (0.5 ml/min). Analytic parameters were as follows: injection volume, 20 μl; column temperature, 30°C; DAD range, 210–400 nm with 240 as the detection wavelength; ionization mode, ESI+; scan range, 220–500 amu; and scan rate, 1 scan/s.
Mouse hepatocyte culture and treatment
Primary mouse hepatocytes were isolated from 9-week-old CD-1 mice (male) by a modified two-step collagenase digestion method, essentially as described previously (21). Hepatocytes were dispersed from the digested liver in Williams’ Medium E (Sigma-Aldrich, St. Louis, MO) without collagenase and washed by low-speed centrifugation (100–150 g, 5 min) three times. Hepatocytes were suspended in Williams’ Medium E containing 35% percoll. The resulting cell pellet was then suspended in Williams’ Medium E containing 10% fetal bovine serum, insulin-transferrin-sodium selenite (ITS) supplement, and dexamethasone (100 nM). Cell viability was determined by trypan blue exclusion. Hepatocytes were then plated onto collagen-coated 12-well culture plates. The cells were allowed to attach for 4 h at 37°C in a humidified chamber with 95%/5% air/CO2. Culture plates were then gently swirled, and the medium containing unattached cells was aspirated. Fresh medium (free serum) was added to each well, and the cultures were returned to the humidified chamber. Hepatocytes were cultured in the same medium for 48 h, but the medium was changed at 24 h. Thereafter, hepatocytes were cultured in DMEM containing treatment chemicals at appropriate concentrations for 48 h; the treatment medium was replaced once at 24 h. Treated hepatocytes were collected in RIPA buffer for Western blotting. Otherwise, hepatocytes were collected in Tris-HCl (50 mM, pH 7.4), and lysates were prepared by sonication as described previously (21). All mice were allowed free access to Purina Rodent Chow 5001 and water, and the use of animals was approved by the Institutional Animal Care and Use Committee.
Electrophoresis-coupled activity determination
Nondenaturing gel electrophoresis for staining esterase activity was conducted as described previously (22). Cell lysates (5 or 20 μg) was solubilized with 0.2% Lubrol and subjected to electrophoresis with a 3% acrylamide stacking gel and a 7.5% acrylamide separating gel. After electrophoresis, the gels were washed for 1 h in 100 mM potassium phosphate buffer (pH 6.5), followed by incubating in the same buffer containing 1-naphthylacetate (5 mM) and 4-benzolamino-2,5-dimethoxybenzenediazonium chloride hemi (zinc chloride) salt, usually termed Fast Blue RR (0.4 mg/ml). The staining of esterases by this method is based on the formation of a black, insoluble complex between 1-naphthol hydrolyzed from 1-naphthylacetate and Fast Blue RR.
Other analyses
Electrophoretic mobility shift assay (EMSA) was performed as described previously (21, 23). Western blotting was detailed elsewhere (19, 24). Preparation of CES1 antibody against a peptide was described previously (25). The antibody (PC-064) against BSEP (mouse and human) was purchased from Kamiya Biomedical Co. (Seattle, WA), the antibody against human CYP7A1 (ab79847) from Abcam, Inc. (Cambridge, MA), and the antibody against mouse cyp7a1 from Cosmo Bio Co. (Western Carlsbad, CA). Significant differences were determined by one-way ANOVA followed by a Duncan's multiple comparison test (P < 0.05).
RESULTS
Z-Guggulsterone induces CES1 and BSEP but not CYP7A1
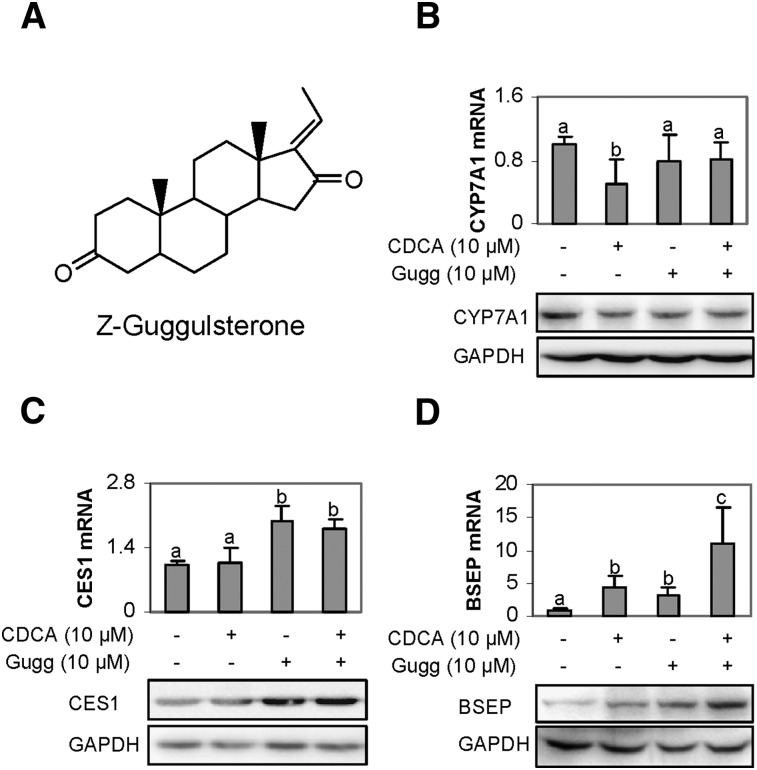
It is generally assumed that Z-guggulsterone exerts hypolipidemic activity based on a feed-back mechanism (7) by increasing bile acid synthesis through inducing CYP7A1. On the other hand, rats fed with guggul herb extract did not show increased expression of CYP7A1 (26). Nevertheless, we made an effort to determine whether Z-guggulsterone induces human CYP7A1. Human primary hepatocytes were treated with Z-guggulsterone, and the expression was determined. CDCA, a known suppressor of CYP7A1 (27), was included in this study as a negative control. Contrary to the general assumption, Z-guggulsterone caused a 20% decrease of CYP7A1 mRNA (Fig. 1B), although the decrease did not reach statistical significance. As expected, CDCA significantly decreased CYP7A1 mRNA (Fig. 1B). The decrease was partially reversed by Z-guggulsterone (Fig. 1B). The level of CYP7A1 protein showed a pattern of changes similar to the level of CYP7A1 mRNA. In contrast to the suppression of CYP7A1, Z-guggulsterone significantly induced both CES1 and BSEP mRNA with BSEP (Fig. 1C, D). The fold of induction of BSEP was higher than that of CES1 in the primary hepatocytes (P < 0.05). Cotreatment with CDCA synergistically increased BSEP induction but slightly decreased CES1 induction (Fig. 1C, D). As seen with CYP7A1, the levels of CES1 and BSEP proteins exhibited patterns of changes similar to those of the respective mRNA levels.
Fig. 1.
Effect of Z-guggulsterone on the expression of CES1, CYP7A1, and BSEP. (A) Chemical structure of Z-guggulsterone. (B–D) Regulated expression in human primary hepatocytes. Hepatocytes (n = 4–8) were treated with Z-guggulsterone (10 μM), CDCA (10 μM), or both for 24 h. DMSO (0.1%) was used as the control. The mRNA levels of CES1, CYP7A1, BSEP, GAPDH, or polymerase II by Taqman probes. Columns labeled with a different letter were statistically significant (P < 0.05). To determine the changes in the protein level, lysates (0.5 µg for CES1 and 20 µg for CYP7A1 or BSEP) from pooled samples (n = 4) were resolved by 7.5% SDS-PAGE and transferred electrophoretically to nitrocellulose membranes. The blots were incubated with an antibody against CYP7A1, CES1, or BSEP and then developed with chemiluminescent substrate and reprobed by GAPDH antibody. The signal was captured by Carestream 2200 PRO Imager.
Activation of the CES1A1 promoter
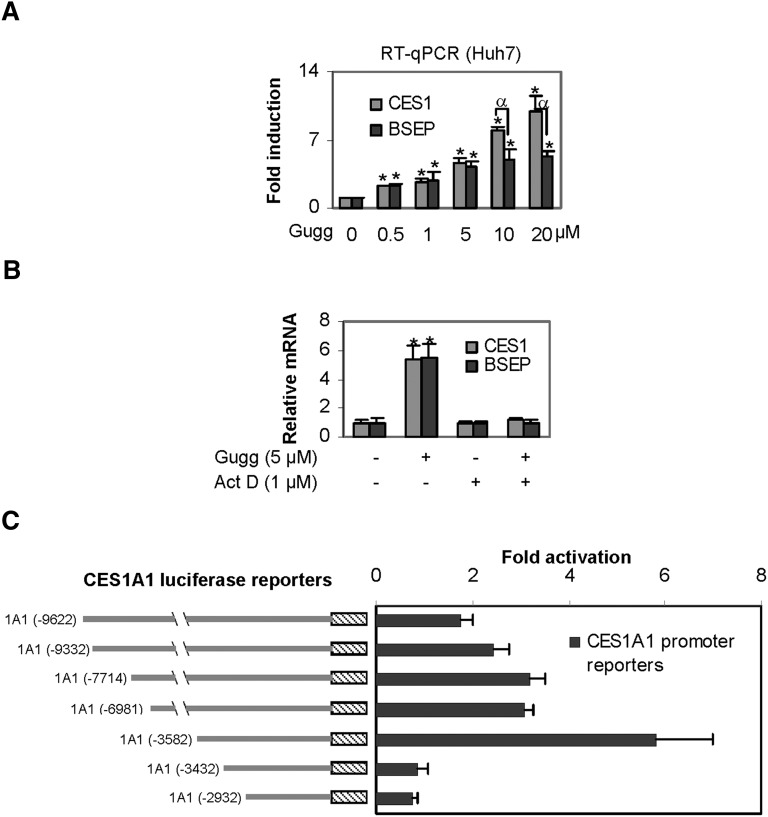
We have shown that Z-guggulsterone induces BSEP through transactivation (12). Next, we tested whether transactivation is also involved in CES1 induction. Huh7 cells (a human hepatic line) were used for this study. Initially, concentration-dependent induction was determined in this line. As shown in Fig. 2A, Z-guggulsterone induced CES1 and BSEP mRNA in a concentration-dependent manner; however, the magnitude of induction varied markedly. Both genes were induced comparably at lower concentrations, but CES1 mRNA was induced to a significantly greater extent at 10 and 20 µM. All data points on CES1 induction significantly differed from each other (P < 0.05), except those between 0.5 and 1 µM. Likewise, all concentrations caused statistically significant induction of BSEP. However, the magnitudes of the inductions between 0.5 and 1 µM did not differ significantly, and neither did those among 5, 10, and 20 µM. Next, we tested whether actinomycin D, a transcriptional inhibitor, abolishes the induction of both CES1 and BSEP. Huh7 cells were treated with Z-guggulsterone in the presence or absence of actinomycin D. As shown in Fig. 2B, cotreatment with actinomycin D completely abolished the induction of both CES1 and BSEP, suggesting that Z-guggulsterone induces CES1, like BSEP (12), through transactivation. We next tested whether Z-guggulsterone stimulates the promoter of CES1A1, the predominant form of the CES1 genes (17). As shown in Fig. 2C, all reporters were activated by Z-guggulsterone, except reporters CES1A1-3432luc and CES1A1-2932Luc. The maximum activation was 6-fold and occurred with the reporter CES1A1-3582Luc (Fig. 2C). CES1A1-3432luc, the immediate reporter shorter than CES1A1-3582Luc, no longer responded to Z-guggulsterone, suggesting that this 150-bp genomic fragment (-3582 to -3432) supports the transactivation.
Fig. 2.
Stimulation of the CES1A1 promoter. (A) Concentration-dependent induction of CES1 and BSEP Huh7 cells were treated with Z-guggulsterone (Gugg) at various concentrations (0–20 µM) for 24 h, and then the levels of CES1 and BSEP mRNA were determined as described above. An asterisk indicates statistical significance of a treatment over solvent control, and an alpha indicates statistical significance between two columns linked by a line (P < 0.05). (B) Effect of actinomycin D (Act D) on induction. Huh7 cells were treated with Z-guggulsterone (5 μM) for 24 h in the absence or presence of actinomycin D (1 μM), and the levels of CES1 and BSEP mRNA were determined. An asterisk indicates statistical significance of a treatment over solvent control (P < 0.05). (C) Activation of CES1A1 reporters. Huh7 cells were seeded in 48-well plates. After an overnight incubation, the cells were transfected with a reporter (50 ng) along with 5 ng of the null-Renilla luciferase plasmid. The transfected cells were treated with Z-guggulsterone (10 μM) or the same volume of DMSO for 24 h. Luciferase activities were determined with a dual-luciferase reporter assay. Data in this figure are from three separate experiments.
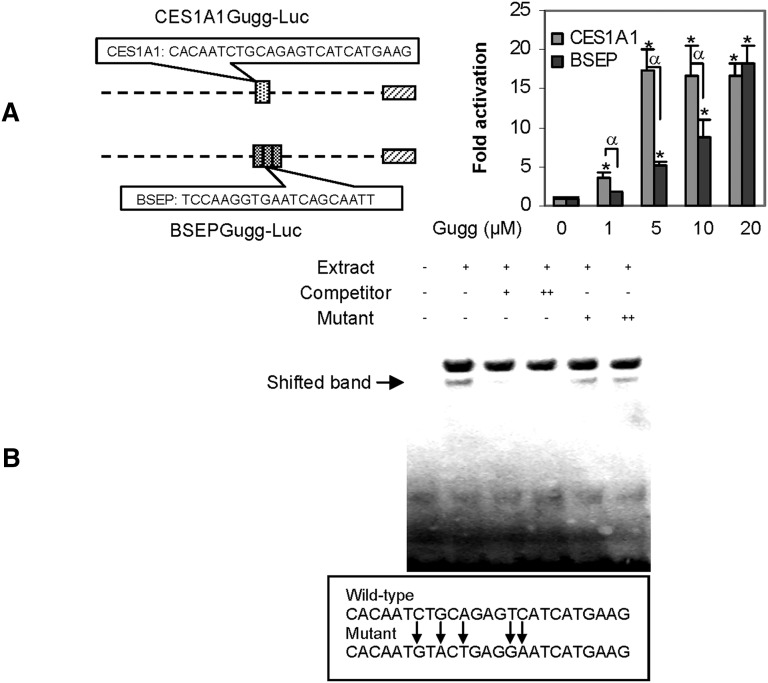
To precisely specify the sequence for the transactivation, element reporters (20–25 bases each) were prepared to span the entire region with overlapping sequence. Among the element reporters, only one reporter, designated CES1A1Gugg-Luc, responded to Z-guggulsterone (Fig. 3A). This and the BSEPGugg-Luc reporter were tested for the activation as a function of Z-guggulsterone. As shown in Fig. 3A, the CES1A1 element reporter was activated much higher, except at the 20 µM data point (Fig. 3A). Note that the CES1 reporter contained only a single copy of the element, whereas the BSEP reporter contained three copies. Nevertheless, EMSA with nuclear extract from Z-guggulsterone treated Huh7 cells detected a shifted band with the CES1A1 element. This binding was competed effectively by the corresponding nonlabeled element (Fig. 3B). In contrast, the mutant element (5′-CACAATGTACTGAGGAATCATGAAG-3) had no competitive effect (Fig. 3B).
Fig. 3.
Characterization of CES1A1 guggulsterone element. (A) Stimulation of the CES1A1 and BSEP element reporters. Reporter assays were performed as described above but with element reporters. An asterisk indicates statistical significance of a treatment over solvent control, and an alpha indicates statistical significance between two columns linked by a line (P < 0.05). (B) EMSA analysis. Nuclear extracts (5 μg) from Huh7 cells treated with Z-guggulsterone (10 μM) were incubated with a biotinylated probe (0.01 pmol) for 20 min. In the competition assay, nuclear extracts were preincubated with the unlabeled probe or mutant at 50× (+) or 100× (++) excess for 20 min, and then incubated with the biotinylated probe. The protein-DNA complexes were electrophoretically resolved and detected with streptavidin-conjugated horseradish peroxidase and chemiluminescent substrate.
Metabolism by CYP3A4
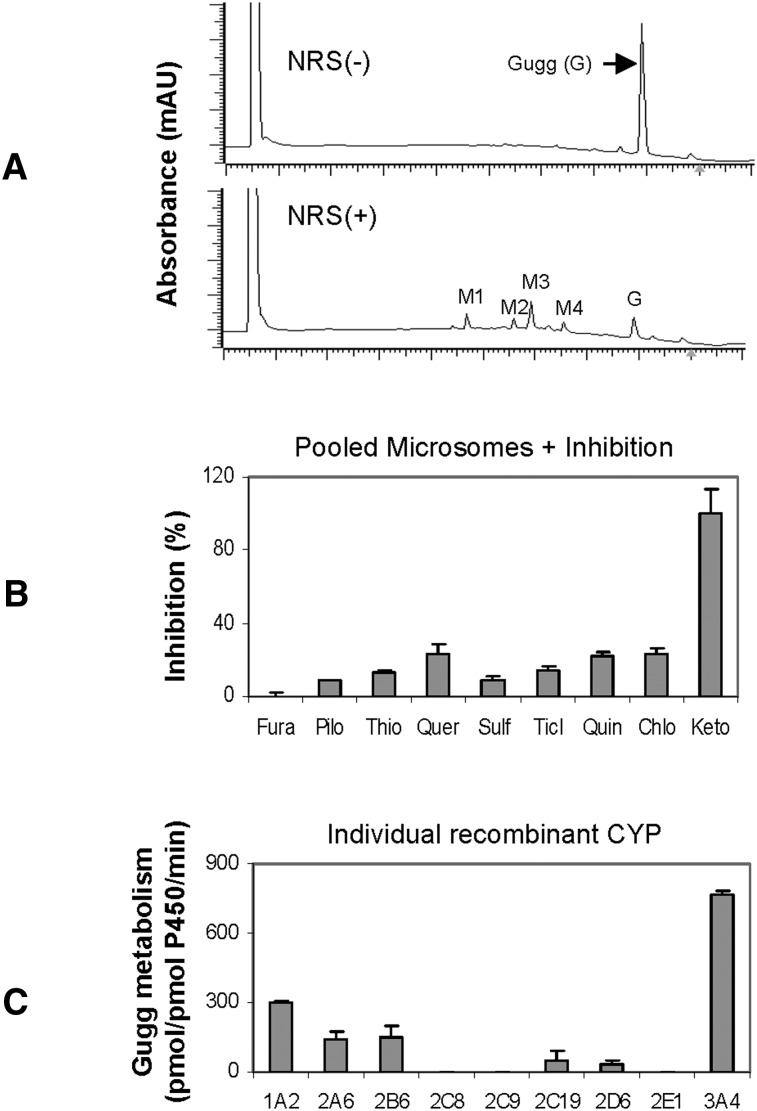
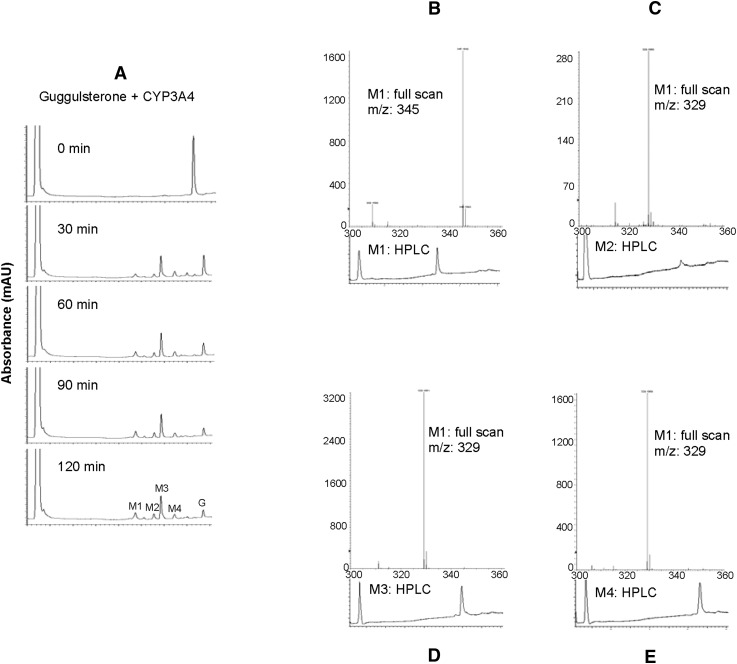
As described in Figs. 1 and 2, BSEP mRNA was induced higher in primary hepatocytes, whereas CES1 mRNA was induced comparably or higher in Huh7 cells, depending on the concentrations. One explanation is that primary hepatocytes express higher basal levels of CES1 and, thus, the inducibility is lower. Another explanation is that primary hepatocytes, but not Huh7 (28), effectively metabolize Z-guggulsterone. To test whether Z-guggulsterone actually undergoes metabolism, incubations were performed with pooled human liver microsomes in the presence or absence of the NADPH-regenerating system. This system is required for reactions catalyzed by CYPs and flavin monooxygenases (28). As shown in Fig. 4A, the reaction incubated without NRS, like zero-minute incubation, yielded a peak with a retention time of 7.92 min (i.e., Z-guggulsterone). This peak, however, disappeared by ∼90% when the incubation was performed in the presence of NRS (Fig. 4A). At the same time, several metabolite peaks appeared with a retention time of 4.69 min (M1), 5.60 (M2), 5.92 (M3), and 6.56 (M4), respectively. M3 was the most abundant (Fig. 4A).
Fig. 4.
Metabolism of Z-guggulsterone. (A) Chromatograms generated by pooled human liver microsomes. The incubations were performed at 37°C in a total volume of 100 μl containing pooled human liver microsomes (20 μg) and Z-guggulsterone (10 μM) in the presence of NRS. The incubations lasted for 90 min. The elution trace was generated at 240 nm by injecting 20 μl supernatants. (B) Inhibited metabolism of Z-guggulsterone. Z-guggulsterone (10 μM) was metabolized by microsomes (20 μg) in the presence or absence of a CYP inhibitor: 50 μM furafylline, 100 μM pilocarpine, 75 μM thio tepa A, 10 μM quercetin, 20 μM sulfaphenazole, 1 μM ticlopidine, 10 μM quinidine, 100 μM chlormethiazole, or 1 μM ketoconazole. The reactions lasted for 40 min. The disappearance of Z-guggulsterone was monitored. The percentage of inhibition was calculated. (C) Metabolism by recombinant CYPs. The incubations were performed with a recombinant CYP (1 pmol) and Z-guggulsterone (10 μl) for 40 min. All experiments were conducted twice with each in triplicate.
The majority of oxidations are catalyzed by the CYP system (28). We next tested whether one or more CYPs metabolize Z-guggulsterone. Incubations were conducted with pooled microsomes but in the presence of a CYP inhibitor. A total of nine inhibitors were used, including furafylline (CYP1A2), pilocarpine (CYP2A6), thio-tepa (CYP2B6), quercetin (CYP2C8), sulfaphenazole (CYP2C9), ticlopidine (CYP2C19), quinidine (CYP2D6), chlormethiazole (CYP2E1), and ketoconazole (CYP3A4/5). With the exceptions of furafylline (Fura) and ketoconazole (Keto), all inhibitors caused a 10–20% decrease in the metabolism (Fig. 4B). Ketoconazole caused complete inhibition, whereas the CYP1A2 inhibitor furafylline caused no inhibition. Isoform-specific substrates were used to confirm the activity of each inhibitor.
The inhibition study demonstrated that multiple CYPs metabolized Z-guggulsterone, with the CYP3A4 being the most active. To directly test this possibility, recombinant CYPs were tested for metabolism. Consistent with the inhibition study, CYP3A4 had the highest activity (Fig. 4C). Moderate metabolism was detected with CYP1A2, 2A6, and 2B6; slight metabolism was detected with CYP2C19 and 2D6; and no metabolism was detected with CYP2C8, 2C9, and 2E1 (Fig. 4C). Interestingly, CYP1A2 was the second most active toward Z-guggulsterone, although the corresponding inhibitor showed no activity (Fig. 4B). Specific substrates for each CYP were included to verify the catalytic activity of each recombinant CYP.
Purification of major metabolites
To determine whether the metabolism alters the induction potency, we purified several major metabolites. Initially, we performed a time-course study to determine the kinetics of the metabolism by recombinant CYP3A4. As shown in Fig. 5A, the parent compound was disappeared by ∼70% within 30 min of incubation. Prolonged incubation further increased the metabolism. The metabolite profile generated by CYP3A4 was very similar to that generated by liver microsomes (Figs. 4A and 5A), confirming that this CYP is the primary enzyme for the metabolism. Interestingly, the 30 min incubation produced similar amounts of M1 and M4; however, prolonged incubation produced more M1 than M4 (Fig. 5A). To this end, we purified M1, M2, M3, and M4 to homogeneity. As shown in Fig. 5B–E, all purified metabolites produced a single peak with a predicated retention time. Mass spectrometry revealed that M1 was at m/z 345 and the other three metabolites at 329. Z-Guggulsterone had a mass-to-charge ratio of 313. Therefore, M1 represented an addition of two hydroxyl groups, whereas the others resulted from an addition of a single hydroxyl group.
Fig. 5.
Analyses of metabolites by HPLC and LC-MS. (A) Time-course metabolism by CYP3A4. The incubations were performed at 37°C with 1 pmol CYP3A4 and 10 μM Z-guggulsterone for 0–120 min. The HPLC elution trace was generated at 240 nm. (B–E) HPLC chromatogram and full scan of metabolites M1, M2, M3, and M4, respectively.
Induction potency of major metabolites
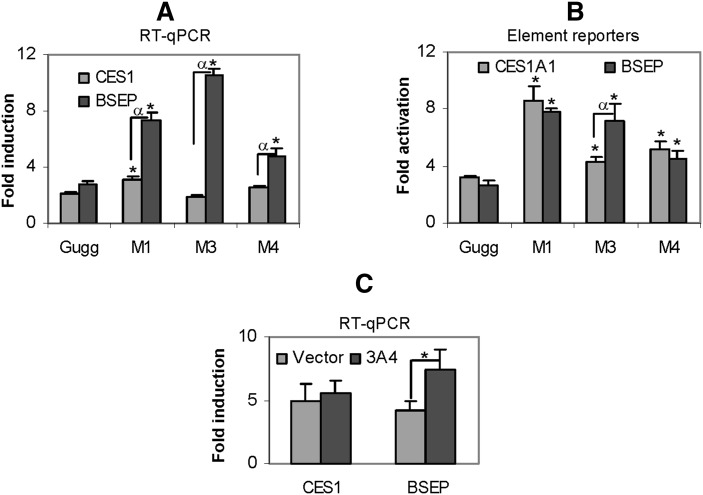
To determine whether the metabolites and Z-guggulsterone differ in the induction potency, RT-qPCR and reporter assays were performed. The concentrations of the metabolites were estimated based on the area under the curve. This study included all major metabolites except M2, which was not sufficient to generate the standardized concentration (2 µM). Compared with Z-guggulsterone, all metabolites caused significant increases in the level of BSEP mRNA (Fig. 6A), with M3 causing the highest increase (> 3-fold). In contrast, the magnitude of changes in CES1 mRNA was much less. Compared with Z-guggulsterone, M1 caused a 47% increase, M3 caused a 12% decrease, and M4 caused a 22% increase in CES1 induction (Fig. 6A). All metabolites tested showed significantly higher induction of BSEP than CES1 mRNA (Fig. 6A).
Fig. 6.
Functional characterization of the metabolites. (A) Differential induction of CES1 and BSEP Huh7 cells were treated with Z-guggulsterone (2 μM) or a metabolite (2 μM) for 24 h. The mRNA levels of CES1 and BSEP were determined. An asterisk indicates statistical significance of a metabolite over Z-guggulsterone, and an alpha indicates statistical significance between two columns linked by a line (P < 0.05). (B) Differential activation of CES1 and BSEP element reporters. Huh7 cells were seeded in 48-well plates. After overnight incubation, the cells were transfected with a reporter (50 ng) and 5 ng of the null-Renilla luciferase plasmid. The transfected cells were treated with Z-guggulsterone (2 μM), a metabolite (2 μM), or the same volume of DMSO for 24 h. The reporter luciferase activities (firefly) were normalized according to the null-Renilla luciferase activities. An asterisk indicates statistical significance of a metabolite over Z-guggulsterone, and an alpha indicates statistical significance between two columns linked by a line (P < 0.05). (C) Effect of CYP3A4 on the induction of CES1 and BSEP. Huh7 cells were seeded in 24-well plates at a density of 1.5 × 105 and cultured overnight. The cells were transfected with a CYP3A4 expression construct or the corresponding vector (0.5 µg). The transfected cells were cultured for 24 h and then treated with Z-guggulsterone at 5 μM for 24 h. Total RNA was isolated and analyzed for the levels of CES1 and BSEP mRNA. The data were collected from three separate experiments. *P < 0.05.
We next tested the activation of CES1 and BSEP element reporters by various metabolites. The results are summarized in Fig. 6B. The changes in element reporter activation, compared with those in the induction of CES1 and BSEP mRNA, had several important differences. First, all metabolites showed increases in the activation of the element reporters, which was not true in mRNA induction (Fig. 6A, B). For example, metabolite M3 actually caused a slight decrease in the induction of CES1 mRNA. Second, between CES1A1 and BSEP, metabolites generally caused less difference in the activation of the element reporters than in induction of the mRNA (Fig. 6A, B). For example, metabolite M1 caused much higher increases in the induction of BSEP mRNA than CES1 mRNA, whereas this metabolite caused similar increases in the activation of both element reporters (Fig. 6A, B). And third, the overall changes in the CES1 element reporter activation were greater than those in the induction of CES1 mRNA. For example, compared with Z-guggulsterone, the maximal change in the induction by M1 was 47% increase (Fig. 6A), whereas the maximal change in the activation by M1 was ∼2-fold (Fig. 6B). To further determine whether CYP3A4-based metabolism of Z-guggulsterone differentially alters BSEP and CES1 induction, Huh7 cells were transfected with CYP3A4 or the vector, treated with Z-guggulsterone (5 µM), and then analyzed for the induction of CES1 and BSEP mRNA. As shown in Fig. 6C, cotransfection of CYP3A4, compared with the vector, significantly increased BSEP but not CES1 induction.
Effect of Z-guggulsterone on the expression of mouse bsep, cyp7a1, and carboxylesterases (ces)
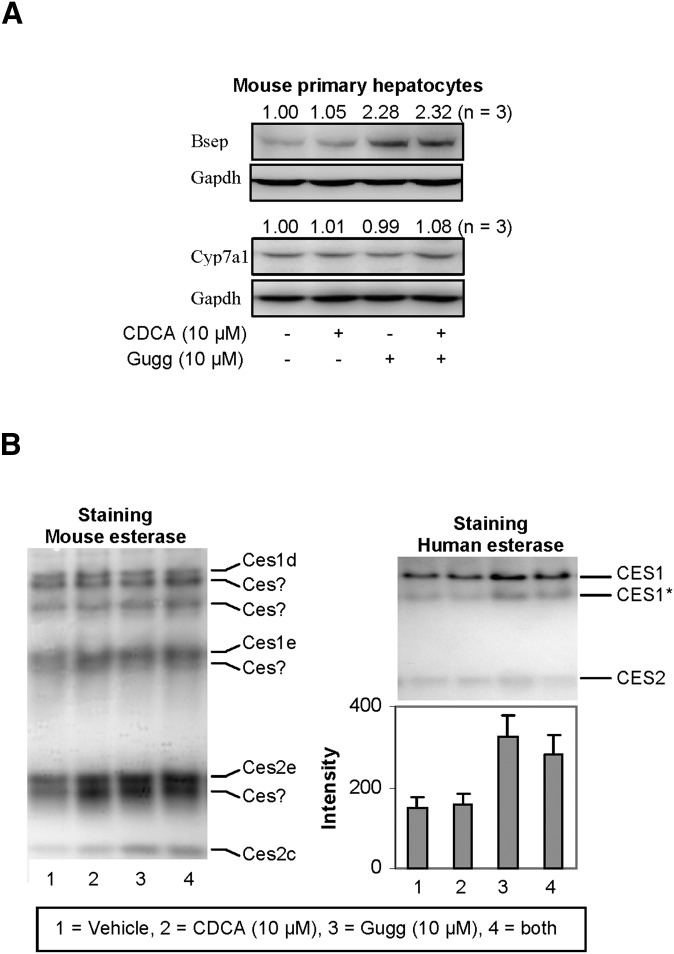
To determine whether Z-guggulsterone modulates the expression of the corresponding mouse genes, mouse primary hepatocytes were treated with Z-guggulsterone, CDCA, or both. As shown in Fig. 7A, Z-guggulsterone significantly induced bsep but not cyp7a1, and inductions were not affected by CDCA cotreatment. These responses were consistent with the responses on the expression of BSEP and CYP7A1 in rats fed with guggul extract (26). As expected, CDCA, a weak activator of mouse fxr (26), caused little changes in the expression of bsep or cyp7a1 (Fig. 7A). Next we tested whether these treatments alter the expression of mouse carboxylesterases. In terms of the number of carboxylesterase genes, mice have almost three times as many as humans (29). The relative level of carboxylesterases can be indirectly determined by nondenaturing electrophoresis, followed by staining for hydrolytic activity with 1-naphythalacetate (22). This standard substrate can be hydrolyzed by many carboxylesterases. As shown in Fig. 7B (left), both CDCA and Z-guggulsterone markedly induced several major mouse carboxylesterases, such as ces2e and ces2c, but not many others, such as ces1d (the mouse counterpart of human CES1). The identity of some of the electrophoretically distinct mouse carboxylesterases remains to be established. As expected, Z-guggulsterone significantly increased hydrolysis by CES1 but much less so by CES2 (Fig. 7B, right). Cotreatment with CDCA slightly decreased the increased hydrolysis of CES1 by Z-guggulsterone.
Fig. 7.
Effect of Z-guggulsterone, CDCA, and both on the expression of mouse bsep, mouse cyp7a1, and esterase activity staining. (A) Expression of mouse bsep and cyp7a1. Mouse primary hepatocytes (n = 3) were treated with Z-guggulsterone (10 μM), CDCA (10 μM), or both for 24 h. DMSO (0.1%) was used as the control. Lysates (25 µg) were resolved by 7.5% SDS-PAGE and transferred electrophoretically to nitrocellulose membranes. The blots were incubated with an antibody against bsep or cyp7a1, developed with chemiluminescent substrate, and reprobed by GAPDH antibody. The signal was captured by Carestream 2200 PRO Imager. (B) Nondenaturing electrophoresis stained for hydrolytic activity. Mouse primary hepatocytes and human HepG2 cells were treated as described above. Lysates from mouse hepatocytes (5 µg) or HepG2 cells (20 µg) were subjected to native gel electrophoresis and stained for esterase activity with 1-naphythalacetate as described in Experimental Procedures. The staining intensity was captured by Carestream 2200 PRO Imager. Note the glycosylation variants of CES.
DISCUSSION
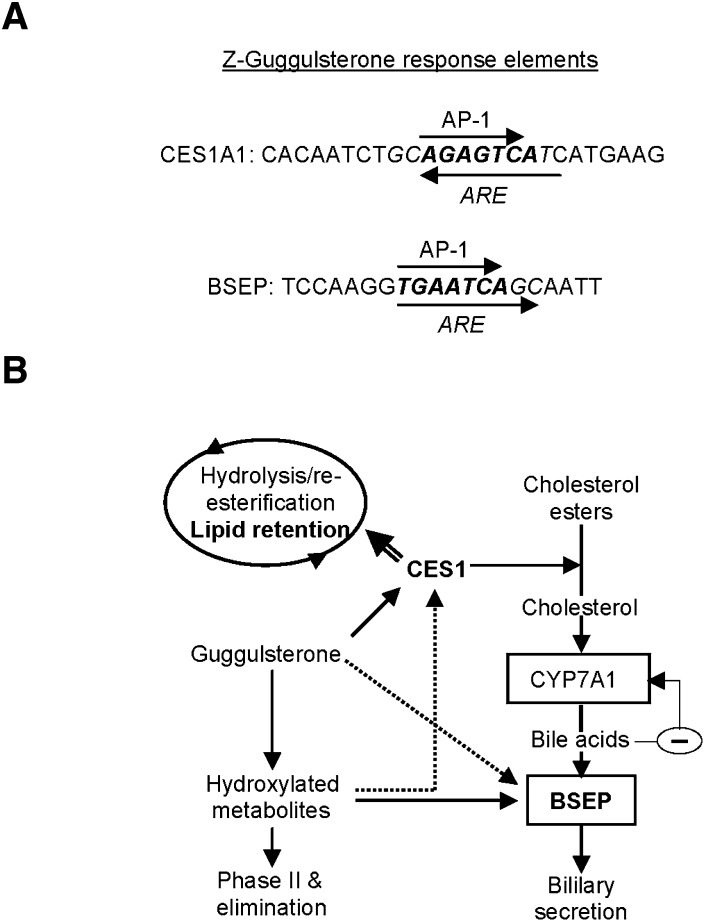
This study has shown that Z-guggulsterone significantly induced CES1 and BSEP. The induction of CES1 by Z-guggulsterone, like the induction of BSEP, was achieved by transactivation (Fig. 2). The transactivation depended on a particular genomic sequence designated as the CES1A1 guggulsterone element (Fig. 3A). Coincidently, both CES1A1 and BSEP guggulsterone response elements contain an a ntioxidant r esponse e lement (ARE) and an AP-1 site (Fig. 8A). However, the CES1A1 ARE is orientated in an opposite direction to that of BSEP (Fig. 8A). Also, the CES1A1 AP-1 site (AGAGTCA) slightly differs from that of BSEP (TGAATCA) (Fig. 8A). More importantly, these two elements differentially responded to Z-guggulsterone and its metabolites, particularly metabolite M3 (Fig. 6B). Compared with Z-guggulsterone, M3 increased the activation of the BSEP reporter from 2.6- to 7.3-fold, whereas the increase was much less in the activation of the CES1A1 reporter from 3.2- to 4.3-fold (Fig. 6B). On the other hand, similar responses were detected between the reporter activation and mRNA induction of BSEP (Fig. 6A, B), whereas the reporter activation of CES1 was much higher than the corresponding mRNA induction (Fig. 6A, B). One explanation is that the BSEP element provides the primary mechanism for BSEP induction by Z-guggulsterone, whereas the CES1A1 element represents one of the mechanisms for CES1 induction.
Fig. 8.
Response elements and interplays among CES1, BSEP, and CYP7A1. (A) CES1A1 and BSEP guggulsterone response elements. The antioxidant response elements (ARE) are italicized, whereas the activator protein-1 sites (AP-1) are bolded. (B) Multiple interplays among metabolism, CES1/BSEP induction, lipid retention, and bile acid synthesis The arrows point to the targets that an enzyme or compound acts on or is converted into. When two arrows points to the same target, the solid line denotes a major effect and the dotted line denotes a minor effect. The double-lined arrow denotes an increased role when the induction of CES1 exceeds the capacity of bile acid synthesis.
Clearly, the metabolism of Z-guggulsterone is determined largely by CYP3A4. The metabolite profile generated by human microsomes was similar to that by CYP3A4 (Figs. 4A and 5A). On the other hand, there were several interesting observations, particularly regarding the metabolism by CYP1A2 (Fig. 4B, C). Furafylline, an inhibitor of CYP1A2, showed little inhibition of Z-guggulsterone metabolism by liver microsomes (Fig. 4B), yet recombinant CYP1A2 significantly metabolized this phytosterol (Fig. 4C). One explanation is that Z-guggulsterone is a substrate of both CYP3A4 and CYP1A2, but CYP3A4 has higher affinity and more efficiently metabolizes the phytosterol. In addition, CYP3A4 is two to three times more abundant than CYP1A2 in the liver (28). Therefore, CYP1A2 normally contributes less, if any, to the hepatic metabolism of Z-guggulsterone. Another interesting observation was that CYP3A4 produced various metabolites (Fig. 5A). Most of them resulted from addition of a single hydroxyl group (Fig. 5C–E). In contrast, the formation of M1 resulted from addition of two hydroxyl groups (Fig. 5B). Prolonged incubation increased the production of M1 but proportionally decreased M4 (Fig. 5A), suggesting that M4 is a substrate of CYP3A4 to produce M1 (Fig. 5A).
Although the purified metabolites generally increased the induction of both CES1 and BSEP, the magnitude was much greater on BSEP (Fig. 6). Secretion of bile acids represents net elimination of cholesterol (8); therefore, induction of BSEP favors lowering cholesterol. In contrast, induction of CES1 may favor the elimination or recycling of cholesterol, probably depending on the level of CYP7A1 (Fig. 8B). Given that the action of CES1 leads to increased free cholesterol, high levels of CYP7A1 in individuals would facilitate the conversion of cholesterol into bile acids, whereas low levels of CYP7A1 in others would favor the pathway of VLDL assembling (double-lined arrow in Fig. 8B). Therefore, individuals with a high level of BSEP induction relative to CES1 induction (probably with high capacity of metabolizing guggulsterones) would benefit more in lowering cholesterol. Accordingly, the purified metabolites likely deliver more favorable hypolipidemic profiles than Z-guggulsterone. The dosage, on the other hand, is another factor that may influence the hypolipidemic efficacy. The standard dosage (3 × 1,000 mg guggul extract/daily) in a random trial increased the VLDL-cholesterol by 6.4%, whereas the high dosage (3 × 2,000 mg/daily) increased it by 9.8%, pointing to the excessive induction of CES1 (14). We have shown that CYP3A4 efficiently metabolized Z-guggulsterone (Figs. 4B, C and 5A) and that cotransfection of CYP3A4 favors the induction of BSEP but not CES1 (Fig. 6C). A previous study identified Z-guggulsterone as a potent activator of the pregnane X receptor (PXR) (30), a nuclear receptor supporting CYP3A4 induction. Therefore, dosages even lower than the standard dosage likely produce sufficient metabolites with high potency on BSEP induction and, thus, produce better plasma lipid profiles. Note that Z-guggulsterone also induced several mouse carboxylesterases (Fig. 7B, left); however, it remains to be determined whether these carboxylesterases favor lipid retention or elimination.
The metabolism-based alteration of the biological activity may contribute significantly to the observed individual variations in guggul hypolipidemic efficacy. In Indian populations, the positive rate in lowering LDL cholesterol is 60–80% (13, 14), whereas the rate in American populations is only 18% (10). The precise mechanism on the large variation in the positive rates remains to be determined. In addition to a much lower dosing regimen in the India trials, differences in metabolism likely contribute to the variations (31). Unfortunately, none of the clinical trials reported the blood concentrations of guggulsterones (10, 14). We and other investigators have shown that Z-guggulsterone is a substrate (this study) and an inducer of CYP3A4 (27). Therefore, the involvement of CYP3A4 in the metabolism presents a major variable for guggul therapy due to its variants, diverse substrates, and altered expression by pathological conditions and coadministered drugs (21, 32, 33). People normally consume guggul supplement but not purified guggulsterones. As a result, other components in the extracts (34) may modulate the hypolipidemic activity of guggulsterones by acting on the genes relevant to the hypolipidemic action or the metabolism of guggulsterones.
Z-Guggulsterone, an FXR antagonist, is assumed to downregulate CYP7A1 through the bile acid–FXR feedback mechanism. It was surprising that this phytosterol alone downregulated CYP7A1 by ∼20% (Fig. 1B). It has been reported that primary hepatocytes produce CDCA, the most potent bile acid in triggering the feedback mechanism (27), at the level (∼1 µM) in medium comparable to that in normal human plasma (35, 36), suggesting that the feedback mechanism is operational in primary hepatocytes. The lack of the induction of CYP7A1 by Z-guggulsterone points to a pathway that overrides the feedback mechanism. While Z-guggulsterone has been shown to modulate the activities of many nuclear receptors, activation of PXR led to the repression of the CYP7A1 promoter (36, 37). On the other hand, treatment with CDCA (10 µM) caused greater CYP7A1 suppression than Z-guggulsterone (Fig. 1B); and interestingly, the suppression was partially reversed by Z-guggulsterone. These findings suggest that Z-guggulsterone downregulates CYP7A1 through both PXR and FXR, with FXR requiring higher presence of bile acids. In addition, Z-guggulsterone and CDCA synergistically induced BSEP (Fig. 1D), further weakening the feedback mechanism, although it remains to be ascertained whether and to which extent the feed-back mechanism contributes to the hypolipidemic activity of Z-guggulsterone.
In summary, our study presents two interplays that likely determine the overall hypolipidemic activity of guggul (Fig. 7). The first interplay occurs between CES1 induction and CYP7A1 activity. CES1 induction within the capacity of CYP7A1 favors the synthesis of bile acids and elimination of cholesterol. Excessive induction of CES1 beyond the capacity of CYP7A1 likely increases the engagement of this enzyme in the hydrolysis/reesterification cycle, favoring VLDL assembly. The second interplay occurs between the metabolism of guggulsterones and the induction of BSEP/CES1. Increased metabolism enhances BSEP induction and probably decreases the induction of CES1. It is, therefore, expected that individuals with a higher capacity of metabolizing guggulsterones (probably lower dosing) will show lower induction of CES1 and have better hypolipidemic outcomes.
Acknowledgments
The authors appreciate the use of the Core Facility supported by National Center for Research Resources, National Institutes of Health Grant P20 RR-016457.
Footnotes
Abbreviations:
- BSEP
- bile salt export pump
- CDCA
- chenodeoxycholic acid
- CES1
- carboxylesterase-1
- CYP7A1
- cytochrome P450 7A1
- EMSA
- electrophoretic mobility shift assay
- ER
- endoplasmic reticulum
- FXR
- farnesoid X receptor
- NRS
- NADPH-regenerating system
- PXR
- pregnane X receptor
This work was supported by National Institutes of Health Grants R01 GM-61988, R01 ES-07965, R01 DK-087755, and F05 AT-003019. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Buemi M., Aloisi C., Fulvio F., Caccamo C., Cavallaro E., Crascì E., Criseo M., Corica F., Frisina N. 2005. Cardiorenal consequences of atherosclerosis and statins therapy: from the past to the future. Curr. Pharm. Des. 11: 3973–3984. [DOI] [PubMed] [Google Scholar]
- 2.Sundaram M., Yao Z. 2010. Recent progress in understanding protein and lipid factors affecting hepatic VLDL assembly and secretion. Nutr. Metab. (Lond). 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei E., Alam M., Sun F., Agellon L. B., Vance D. E., Lehner R. 2007. Apolipoprotein B and triacylglycerol secretion in human triacylglycerol hydrolase transgenic mice. J. Lipid Res. 48: 2597–2606. [DOI] [PubMed] [Google Scholar]
- 4.Wei E., Ben Ali Y., Lyon J., Wang H., Nelson R., Dolinsky V. W., Dyck J. R., Mitchell G., Korbutt G. S., Lehner R. 2010. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 11: 183–193. [DOI] [PubMed] [Google Scholar]
- 5.Blais D. R., Lyn R. K., Joyce M. A., Rouleau Y., Steenbergen R., Barsby N., Zhu L. F., Pegoraro A. F., Stolow A., Tyrrell D. L., et al. 2010. Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication. J. Biol. Chem. 285: 25602–25612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B., Natarajan R., Ghosh S. 2005. Human liver cholesteryl ester hydrolase: cloning, molecular characterization, and role in cellular cholesterol homeostasis. Physiol. Genomics. 23: 304–310. [DOI] [PubMed] [Google Scholar]
- 7.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson P. A., Lan T., Rao A. 2009. Bile acid transporters. J. Lipid Res. 50: 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urizar N. L., Moore D. D. 2003. GUGULIPID: a natural cholesterol-lowering agent. Annu. Rev. Nutr. 23: 303–313. [DOI] [PubMed] [Google Scholar]
- 10.Szapary P. O., Wolfe M. L., Bloedon L. T., Cucchiara A. J., DerMarderosian A. H., Cirigliano M. D., Rader D. J. 2003. Guggulipid for the treatment of hypercholesterolemia: a randomized controlled trial. JAMA. 290: 765–772. [DOI] [PubMed] [Google Scholar]
- 11.Nohr L. A., Rasmussen L. B., Straand J. 2009. Resin from the mukul myrrh tree, guggul, can it be used for treating hypercholesterolemia? A randomized, controlled study. Complement. Ther. Med. 17: 16–22. [DOI] [PubMed] [Google Scholar]
- 12.Deng R., Yang D., Radke A., Yang J., Yan B. 2007. The hypolipidemic agent guggulsterone regulates the expression of human bile salt export pump: dominance of transactivation over farsenoid X receptor-mediated antagonism. J. Pharmacol. Exp. Ther. 320: 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal K., Saran R. K., Nityanand S., Gupta P. P., Hasan M., Das S. K., Sinha N., Agarwal S. S. 1986. Clinical trial of ethyl acetate extract of gum gugulu (gugulipid) in primary hyperlipidemia. J. Assoc. Physicians India. 34: 249–251. [PubMed] [Google Scholar]
- 14.Nityanand S., Srivastava J. S., Asthana O. P. 1989. Clinical trials with gugulipid. A new hypolipidaemic agent. J. Assoc. Physicians India. 37: 323–328. [PubMed] [Google Scholar]
- 15.Urizar N. L., Liverman A. B., Dodds D. T., Silva F. V., Ordentlich P., Yan Y., Gonzalez F. J., Heyman R. A., Mangelsdorf D. J., Moore D. D. 2002. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 296: 1703–1706. [DOI] [PubMed] [Google Scholar]
- 16.Yang D., Pearce R. E., Wang X., Gaedigk R., Wan Y. J. Y., Yan B. 2009. Human carboxylesterases HCE1 and HCE2: ontogenic expression, inter-individual variability and differential hydrolysis of oseltamivir, aspirin, deltamethrin and permethrin. Biochem. Pharmacol. 77: 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukami T., Nakajima M., Maruichi T., Takahashi S., Takamiya M., Aoki Y., McLeod H. L., Yokoi T. 2008. Structure and characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes. Pharmacogenet. Genomics. 18: 911–920. [DOI] [PubMed] [Google Scholar]
- 18.Shi D., Yang J., Yang D., LeCluyse E. L., Black C., You L., Akhlaghi F., Yan B. 2006. Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the activation is inhibited by antiplatelet agent clopidogrel. J. Pharmacol. Exp. Ther. 319: 1477–1484. [DOI] [PubMed] [Google Scholar]
- 19.Shi D., Yang D., Prinssen E. P., Davies B. E., Yan B. 2011. Surge in expression of carboxylesterase 1 during the post-neonatal stage enables a rapid gain of the capacity to activate the anti-influenza prodrug oseltamivir. J. Infect. Dis. 203: 937–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perloff E. S., Mason A. K., Dehal S. S., Blanchard A. P., Morgan L., Ho T., Dandeneau A., Crocker R. M., Chandler C. M., Boily N., et al. 2009. Validation of cytochrome P450 time-dependent inhibition assays: a two-time point IC50 shift approach facilitates kinact assay design. Xenobiotica. 39: 99–112. [DOI] [PubMed] [Google Scholar]
- 21.Shi D., Yang D., Yan B. 2010. Dexamethasone transcriptionally increases the expression of the pregnane X receptor and synergistically enhances pyrethroid esfenvalerate in the induction of cytochrome P450 3A23. Biochem. Pharmacol. 80: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D., Li Y., Yuan X., Matoney L., Yan B. 2001. Regulation of rat carboxylesterase expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): a dose-dependent decrease in mRNA levels but a biphasic change in protein levels and activity. Toxicol. Sci. 64: 20–27. [DOI] [PubMed] [Google Scholar]
- 23.Liu F., Song X., Yang D., Deng R., Yan B. 2008. The far and distal enhancers in the CYP3A4 gene co-ordinate the proximal promoter in responding similarly to the pregnane X receptor but differentially to hepatocyte nuclear factor-4alpha. Biochem. J. 409: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D., Yang J., Shi D., Deng R., Yan B. 2011. Scoparone potentiates transactivation of the bile salt export pump gene and this effect is enhanced by cytochrome P450 metabolism but abolished by a PKC inhibitor. Br. J. Pharmacol. 164: 1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W., Song L., Zhang H., Matoney L., LeCluyse E., Yan B. 2000. Dexamethasone differentially regulates expression of carboxylesterase genes in humans and rats. Drug Metab. Dispos. 28: 186–191. [PubMed] [Google Scholar]
- 26.Cui J., Huang L., Zhao A., Lew J. L., Yu J., Sahoo S., Meinke P. T., Royo I., Pelaez F., Wright S. D. 2003. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J. Biol. Chem. 278: 10214–10220. [DOI] [PubMed] [Google Scholar]
- 27.Ellis E., Axelson M., Abrahamsson A., Eggertsen G., Thörne A., Nowak G., Ericzon B. G., Björkhem I., Einarsson C. 2003. Feedback regulation of bile acid synthesis in primary human hepatocytes: evidence that CDCA is the strongest inhibitor. Hepatology. 38: 930–938. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson A. 2001. Biotransformation of zenobiotics. In Casarett and Doull's Toxicology: The Basic Science of Poisons. 6th edition. C. D. Klaassen, editor. McGraw-Hill, New York. 139–162.
- 29.Holmes R. S., Wright M. W., Laulederkind S. J., Cox L. A., Hosokawa M., Imai T., Ishibashi S., Lehner R., Miyazaki M., Perkins E. J., et al. 2010. Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm. Genome. 21: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding X., Staudinger J. L. 2005. The ratio of constitutive androstane receptor to pregnane X receptor determines the activity of guggulsterone against the Cyp2b10 promoter. J. Pharmacol. Exp. Ther. 314: 120–127. [DOI] [PubMed] [Google Scholar]
- 31.Ingelman-Sundberg M., Sim S. C., Gomez A., Rodriguez-Antona C. 2007. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 116: 496–526. [DOI] [PubMed] [Google Scholar]
- 32.Keshava C., McCanlies E. C., Weston A. 2004. CYP3A4 polymorphisms--potential risk factors for breast and prostate cancer: a HuGE review. Am. J. Epidemiol. 160: 825–841. [DOI] [PubMed] [Google Scholar]
- 33.Martínez-Jiménez C. P., Jover R., Donato M. T., Castell J. V., Gómez-Lechón M. J. 2007. Transcriptional regulation and expression of CYP3A4 in hepatocytes. Curr. Drug Metab. 8: 185–194. [DOI] [PubMed] [Google Scholar]
- 34.Yu B. Z., Kaimal R., Bai S., El Sayed K. A., Tatulian S. A., Apitz R. J., Jain M. K., Deng R., Berg O. G. 2009. Effect of guggulsterone and cembranoids of Commiphora mukul on pancreatic phospholipase A(2): role in hypocholesterolemia. J. Nat. Prod. 72: 24–28. [DOI] [PubMed] [Google Scholar]
- 35.Ellis E. C. S. 2006. Suppression of bile acid synthesis by thyroid hormone in primary human hepatocytes. World J. Gastroenterol. 12: 4640–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang X., Han Y., Neuvonen M., Laitila J., Neuvonen P. J., Niemi M. 2010. High performance liquid chromatography-tandem mass spectrometry for the determination of bile acid concentrations in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 51–60. [DOI] [PubMed] [Google Scholar]
- 37.Owsley E., Chiang J. Y. 2003. Guggulsterone antagonizes farnesoid X receptor induction of bile salt export pump but activates pregnane X receptor to inhibit cholesterol 7alpha-hydroxylase gene. Biochem. Biophys. Res. Commun. 304: 191–195. [DOI] [PubMed] [Google Scholar]