Abstract
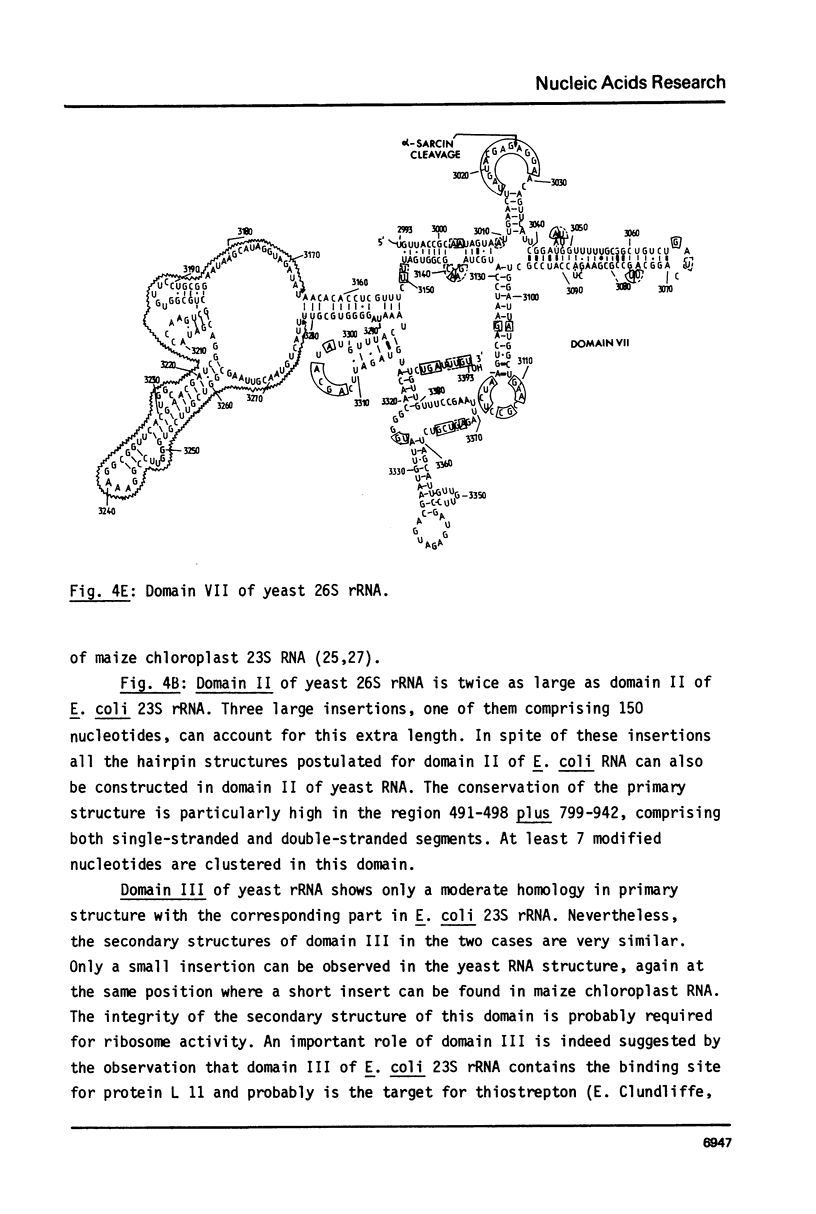
We present the sequence of the 26S rRNA of the yeast Saccharomyces carlsbergensis as inferred from the gene sequence. The molecule is 3393 nucleotides long and consists of 48% G+C; 30 of the 43 methyl groups can be located in the sequence. Starting from the recently proposed structure of E. coli 23S rRNA (see ref. 25) we constructed a secondary structure model for yeast 26S rRNA. This structure is composed of 7 domains closed by long-range base pairings as n the bacterial counterpart. Most domains show considerable conservation of the overall structure; unpaired regions show extended sequence homology and the base-paired regions contain many compensating base pair changes. The extra length of the yeast molecule is due to a number of insertions in most of the domains, particularly in domain II. Domain VI, which is extremely conserved, is probably part of the ribosomal A site. alpha-Sarcin, which apparently inhibits the EF-1 dependent binding of aminoacyl-tRNA, causes a cleavage between position 3025 and 3026 in a conserved loop structure, just outside domain VI. Nearly all of the located methyl groups, like in E. coli, are present in domain II, V and VI and clustered to a certain extent mainly in regions with a strongly conserved primary structure. The only three methyl groups of 26S rRNA which are introduced relatively late during the processing are found in single stranded loops in domain VI very close to positions which have been shown in E. coli 23S rRNA to be at the interface of the ribosome.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Rochaix J. D. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell. 1979 Sep;18(1):55–60. doi: 10.1016/0092-8674(79)90353-2. [DOI] [PubMed] [Google Scholar]
- Baer R. J., Dubin D. T. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981 Jan 24;9(2):323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R. C., Gerbi S. A. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1497–1511. doi: 10.1093/nar/7.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R. C., Klootwijk J., Sibum C. P., Planta R. J. Pseudouridylation of yeast ribosomal precursor RNA. Nucleic Acids Res. 1979 Sep 11;7(1):121–134. doi: 10.1093/nar/7.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R. C., Klootwijk J., Van Steenbergen T. J., De Kok A. J., Planta R. J. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977 May 2;75(1):311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Planta R. J. The molecular weights of yeast ribosomal precursor RNAs. Mol Biol Rep. 1975 Dec;2(4):321–325. doi: 10.1007/BF00357019. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Kelly J. M. Mature 23 SrRNA of prokaryotes appears homologous with the precursor of 25--28 rRNA of eukaryotes: comments on the evolution of 23--28 rRNA. FEBS Lett. 1981 Jul 20;130(1):1–6. doi: 10.1016/0014-5793(81)80652-7. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Planta R. J. Analysis of the methylation sites in yeast ribosomal RNA. Eur J Biochem. 1973 Nov 15;39(2):325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- Maden B. E. Methylation map of Xenopus laevis ribosomal RNA. Nature. 1980 Nov 20;288(5788):293–296. doi: 10.1038/288293a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. Ribosomal precursor RNA in Saccharomyces carlsbergensis. Eur J Biochem. 1967 Dec;3(2):248–258. doi: 10.1111/j.1432-1033.1967.tb19524.x. [DOI] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Schindler D. G., Davies J. E. Specific cleavage of ribosomal RNA caused by alpha sarcin. Nucleic Acids Res. 1977 Apr;4(4):1097–1110. doi: 10.1093/nar/4.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Zuker M., Ebel J. P., Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981 May 11;9(9):2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M. A., Sommer R. Sequence of a ribosomal RNA gene intron from Tetrahymena. Nature. 1980 Feb 14;283(5748):693–694. doi: 10.1038/283693a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]



