Abstract
Chitin is a major component of fungal cell wall and is synthesized by chitin synthases (Chs). Plant pathogenic fungi normally have multiple chitin synthase genes. To determine their roles in development and pathogenesis, we functionally characterized all seven CHS genes in Magnaporthe oryzae. Three of them, CHS1, CHS6, and CHS7, were found to be important for plant infection. While the chs6 mutant was non-pathogenic, the chs1 and chs7 mutants were significantly reduced in virulence. CHS1 plays a specific role in conidiogenesis, an essential step for natural infection cycle. Most of chs1 conidia had no septum and spore tip mucilage. The chs6 mutant was reduced in hyphal growth and conidiation. It failed to penetrate and grow invasively in plant cells. The two MMD-containing chitin synthase genes, CHS5 and CHS6, have a similar expression pattern. Although deletion of CHS5 had no detectable phenotype, the chs5 chs6 double mutant had more severe defects than the chs6 mutant, indicating that they may have overlapping functions in maintaining polarized growth in vegetative and invasive hyphae. Unlike the other CHS genes, CHS7 has a unique function in appressorium formation. Although it was blocked in appressorium formation by germ tubes on artificial hydrophobic surfaces, the chs7 mutant still produced melanized appressoria by hyphal tips or on plant surfaces, indicating that chitin synthase genes have distinct impacts on appressorium formation by hyphal tip and germ tube. The chs7 mutant also was defective in appressorium penetration and invasive growth. Overall, our results indicate that individual CHS genes play diverse roles in hyphal growth, conidiogenesis, appressorium development, and pathogenesis in M. oryzae, and provided potential new leads in the control of this devastating pathogen by targeting specific chitin synthases.
Author Summary
Chitin is one of the major components of cell wall that plays vital roles in hyphal tip growth and fungal morphogenesis. Biosynthesis of chitin is catalyzed by chitin synthases, a well-known fungicide target. However, systematic characterization of chitin synthase genes has not yet been reported in plant pathogenic ascomycetes. To determine their roles in development and pathogenesis, in this study we functionally characterized all the seven chitin synthase genes (CHS1-CHS7) in Magnaporthe oryzae, a model for studying fungal development and pathogenesis. While CHS2, CHS3, CHS4, and CHS5 are dispensable for plant infection, CHS6 is essential for pathogenesis. The chs6 mutant failed to penetrate plant cells and develop infectious hyphae. Two other chitin synthase genes, CHS1 and CHS7, also are important for virulence. The chs1 and chs7 mutants caused only rare lesions on rice seedlings. Other than plant infection, CHS1 and CHS7 also play specific roles during conidiogenesis and appressorium formation, respectively. Interestingly, the chs7 mutant was blocked in appressorium formation by germ tubes on artificial hydrophobic surfaces but was normal by hyphal tips and on plant surfaces. Different chitin synthase genes may be involved during appressorium formation by different fungal tissues or on different surfaces in M. oryzae.
Introduction
Chitin, a microfibrillar β-1,4-linked homopolymer of N-acetyl-glucosamine (GlcNAc), is one of the major structural components of the fungal cell wall [1]. Chitin synthases are key enzymes catalyzing the polymerization of GlcNAc [1]. They are usually localized to cytoplasmic membrane and have attracted considerable attentions as targets for developing fungicides [2], [3]. Chitin synthase (CHS) genes from various fungi have been grouped into seven classes [4]. All chitin synthases have chitin synthase domains and transmembrane domains in common. In addition, the class V and class VI chitin synthases contain the myosin motor domain (MMD) at their N-terminal [5], [6]. Myosins are known as mechanoenzymes that convert chemical energy released by ATP hydrolysis into a mechanical force that is directed along actin filaments [7].
Fungi are different in chitin contents and in the composition of chitin synthases. In the budding yeast Saccharomyces cerevisiae, chitin constitutes 1–2% of the total dry weight, and is a minor cell wall component and mainly exists at the mother-daughter cell junction and septum [8]. S. cerevisiae has three CHS genes with distinct functions in cell wall expansion, septum formation, and budding [9]–[11]. Chs1 repairs the weakened cell wall of daughter cells after separation. Chs2 synthesizes chitin in primary septa. It is essential for both septum formation and cell division [12]. Chs3 chitin synthase is required for chitin ring formation at the base of emerging buds and chitin synthesis in the lateral cell. Chs3 is responsible for 90% of chitin synthesis while Chs1 and Chs2 are involved in only small amounts of chitin synthesis at extreme parts of cells [13]. Unlike the budding yeast, Schizosaccharomyces pombe has only two chitin synthase genes [14]. Ashbya gossybii and Candida albicans have three and four CHS genes, respectively [15].
In filamentous fungi, chitin content has been reported to reach up to 10–20% [16]. Filamentous ascomycetes generally have seven or eight CHS genes, which may reflect their greater complexity of growth and development and higher chitin content than the ascomycetous yeasts. A number of CHS genes have been characterized in the model filamentous ascomycetous fungi Neurospora crassa and Aspergillus nidulans. N. crassa has seven CHS genes, one for each class [17]. CHS1 (I) plays a major role in cell wall biogenesis [18]. The chs-1 RIP mutant had abnormal branching, swollen hyphal tips, reduced growth rate, and increased sensitivity to Nikkomycin Z [18]. Morphologically, the chs-2 RIP mutant was indistinguishable from the wild type. However, it had an increased sensitivity to the phosphatidylcholine biosynthesis inhibitor edifenphos [19]. The chs-4 RIP mutant did not show any reduction in chitin content under standard growth conditions. However, chitin enrichment in the cell wall induced by sorbose treatment was impaired in the chs-4 RIP [20]. In transformants expressing the CHS3-GFP or CHS6-GFP constructs, GFP signals were mainly detected in the hyphal tip, suggesting that they may be invovled in polarized growth [21]. A. nidulans has eight CHS genes [3]. One of the two class I CHS genes, chsB, functions in the formation of normal cell wall in vegetative hyphae, conidiophores, and developing conidia [22], [23]. The chsB mutant has similar defects in hyphal growth with the chs-1 RIP mutant [18]. The csmA and csmB genes encode class VI and V chitin synthases, respectively. The growth rate of the csmA mutant was similar to that of the wild type, but it was defective in cell wall integrity and tended to produce swollen hyphal tips and intrahyphal hyphae [24]. The csmB mutant had similar phenotypes with the csmA mutant and deletion of both csmA and csmB appeared to be lethal [6], suggesting that these two CHS genes had overlapping functions in A. nidulans. Although the chsA (II), chsC (III), and chsD (IV) mutants have no obvious phenotypes, the chsA chsD double mutant was reduced in conidiation and the chsA chsC double mutant was defective in hyphal growth and sexual/asexual development [25]–[27]. A. fumigatus also has eight chitin synthase genes. In general, the A. fumigatus CHS genes have similar functions as their orthologs in A. nidulans [3], [28].
The corn smut fungus Ustilago maydis, a basidiomycete, has two each for class II (CHS3, and CHS4), and class IV (CHS5 and CHS7), and one each for class I (CHS1), class III (CHS2), class V (CHS6), and class VI (MCS1) chitin synthase genes [5]. Deletion of the chs1 gene has no obvious effects on mating, virulence and dimorphic behavior, but chs1 cells are slightly shorter [5], [29]. The chs2 mutant has no obvious effects on mating, virulence, or dimorphism but the chs3 and chs4 mutants produced fewer hyphae on medium with synthetic pheromone [5], [29]. The CHS5, CHS6, CHS7, and MCS1 genes are all important for virulence [5], [30], [31]. While the chs5 mutant forms irregular yeast cells, the chs7 mutant has a cell separation defect. The chs6 and mcs1 mutants have thicker cell wall than the wild type. The mcs1 mutant has no morphological defects in vitro but forms large aggregates of spherical cells.
In plant pathogenic ascomycetes, to date there is no report on systematic characterization of CHS genes although a number of chitin synthase genes have been shown to be important for virulence in various plant pathogenic fungi, including BcCHS1 and BcCHSIIIa of Botrytis cinerea [32], [33], GzCHS5 and GzCHS7 of Fusarium graminearum [34], CHS5 and CHS7 of F. verticillioides [35], and CgCHSV of Colletotrichum graminicola [36]. In the vascular wilt pathogen F. oxysporum, four of its nine CHS genes have been characterized. The CHS1 (III) gene is not important for plant infection but the chs2 (II) mutant is reduced in virulence on tomato plants. Moreover, the stress conditions affected normal development in the chs2 mutant but not in the chs1 mutant [37]. The CHSV (V) and CHSVb (VI) are two MMD CHS genes. Deletion of the CHSV or CHSVb gene resulted in a significant reduction in virulence and production of swollen, balloon-like structures along the hyphae [38]. The chsV chsVb double mutant was viable but failed to infect and colonize tomato plants [39].
In the genome of the rice blast fungus Magnaporthe oryzae [40], there are seven predicted CHS genes, including CHS7 [41]. This study was initiated when the genome sequence of M. oryzae was first published [40]. We functionally characterized all seven CHS genes that were named after their N. crassa orthologs. Three of them, CHS1, CHS6, and CHS7, were important for plant infection. The chs1 mutant had altered conidium morphology and was defective in conidium germination. It was reduced in invasive growth and virulence. The chs6 mutant was non-pathogenic. Appressoria formed by the chs6 mutant were defective in penetration and invasive hyphae in plant cells. Germ tubes of the chs7 mutant failed to form appressoria on artificial hydrophobic surfaces. However, it was normal in appressorium formation on plant surfaces or by hyphal tips, indicating that defects of the chs7 mutant in appressorium formation are surface- and cell type-specific. Appressorium formation by hyphal tips and germ tubes may involve different chitin synthase genes in M. oryzae. In addition, we generated and characterized chs5 chs6 double mutants to determine the relationship among these two chitin synthase genes with similar structures and expression profiles. Results from this study indicate that individual CHS gene plays diverse roles in hyphal growth, conidiogenesis, appressorium development, and pathogenesis in M. oryzae, and provided potential new leads in the control of this devastating pathogen by targeting specific chitin synthases.
Results
Seven chitin synthase genes of M. oryzae are differentially expressed
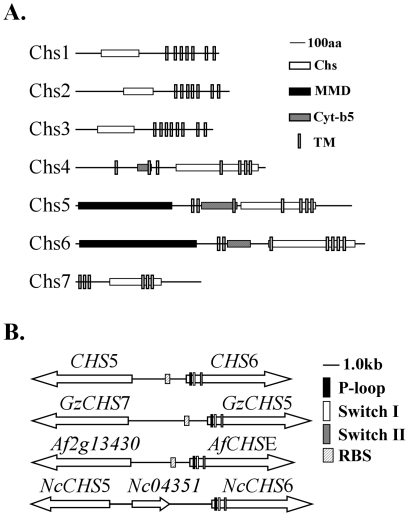
The M. oryzae genome contains seven predicted chitin synthase genes MGG_01802.6, MGG_04145.6, MGG_09551.6, MGG_09962.6, MGG_13014.6, MGG_13013.6, and MGG_06064.6 (www.broad.mit.edu/annotation/genome/magnaporthe_grisea). They are named as CHS1-CHS7 (Figure S1) according to their orthologs in N. crassa [17]. In addition to the core chitin synthase domain (Chs, IPR004835), all the M. oryzae chitin synthase genes have multiple transmembrane (TM) domains (Figure 1A). CHS4, CHS5, and CHS6 also have one cytochrome b5-like heme/steroid binding domain (Cyt-b5, IPR001199) upstream from the Chs domain. Structurally, CHS5 and CHS6 are more similar to each other than to other CHS genes in M. oryzae (Figure 1A). Both of them have an N-terminal myosin motor domain (MMD, IPR001609) and they are closely linked in the genome (Figure 1B).
Figure 1. The seven predicted CHS genes in M. oryzae.
(A) Domain structures of the seven chitin synthases in M. oryzae. Chs, chitin synthase domain; Cyt-b5, cytochrome b5-like heme/steroid binding domain; MMD, myosin motor domain; TM, transmembrane domain. (B) Directions and chromosomal positions of CHS5 and CHS6 in M. oryzae and their orthologs in F. graminearum, A. fumigatus, and N. crassa. RBS, Rlm1p-binding sequence.
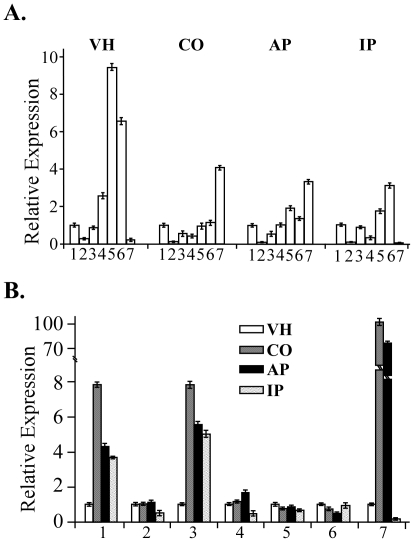
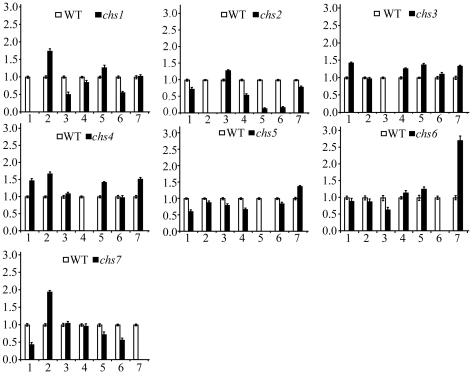
The expression levels of the CHS genes in vegetative hyphae, conidia, appressoria, and infected rice leaves were assayed by quantitative RT-PCR (qRT-PCR). In comparison with other CHS genes, the abundance of CHS2 transcripts was relatively low in all four fungal tissues (Figure 2A). The CHS7 gene had the highest expression level in conidia and appressoria but the lowest expression level in vegetative hyphae and infected rice leaves among all seven CHS genes (Figure 2A). In contrast, both CHS5 and CHS6 had higher expression levels in vegetative hyphae and infected rice leaves than other CHS genes (Figure 2A), suggesting that they played more important roles in the vegetative or invasive hyphal growth than other CHS genes.
Figure 2. Expression profiles of seven CHS genes assayed by qRT-PCR.
RNA samples of the wild-type strain P131 were isolated from vegetative hyphae grown in minimal medium (VH), conidia harvested from 10-day-old oatmeal agar cultures (CO), 24 h appressoria (AP), and infected plant leaves (IP). The relative expression level of individual CHS genes was analyzed with the 2−ΔΔCt method with the actin gene as the internal control for normalization. (A) Comparison of the transcript abundance of seven CHS genes in VH, CO, AP, and IP. The expression level of CHS1 was arbitrarily set to 1. Columns 1 to 7 represent the chitin synthase genes CHS1 to CHS7. (B) Comparison of the transcript abundance of individual CHS genes in four different fungal tissues. The expression level of each CHS gene in vegetative hyphae (VH) was arbitrarily set to 1. Mean and standard errors were determined with data from three independent replicates.
The CHS1 and CHS3 genes had similar expression profiles, with the lowest expression level in vegetative hyphae and the highest in conidia (Figure 2B). Their expression levels also were increased in appressoria and infected rice leaves in comparison with vegetative hyphae. CHS7 is known to be important for appressorium formation on artificial surfaces [41]. However, its expression level was higher in conidia than in appressoria (Figure 2B). The open reading frames (ORFs) of CHS5 and CHS6 are only 2,957-bp apart. Their orthologs are also located next to each other in F. oxysporum [39], A. nidulans [6], A. fumigatus, and F. graminearum (Figure 1B). Similar expression profiles of CHS5 and CHS6 indicate that they may have some common promoter elements due to their head-to-head arrangement on the chromosome (Figure 1B). As expected, one putative Rlm1p-binding sequence (RBS) CTAcgcaTAG was found at 917-bp upstream of the CHS6 ORF. Putative RBS also exists in the promoter regions of the CHS5 and CHS6 orthologs in F. graminearum and A. fumigatus (Figure 1B).
Generation of mutants deleted of individual CHS genes
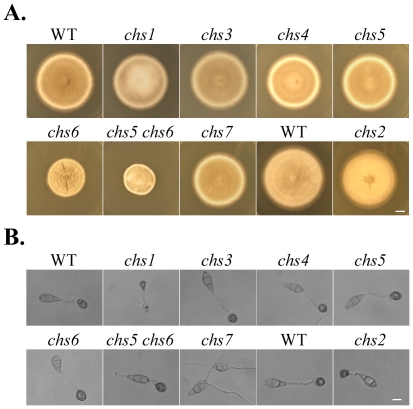
In order to systematically characterize their biological functions in M. oryzae, mutants deleted of individual CHS genes were generated by the conventional gene replacement approach (Table 1; Table S1). For each gene, at least two independent null mutants with identical phenotypes were generated (Table 1). All the chs mutants were identified by PCR and confirmed by Southern blot analyses (Figure S2). Although the growth rate or colony morphology was altered in some mutants, none of the CHS genes were essential for vegetative growth. While deletion of CHS2 and CHS6 resulted in a 20% and 31% reduction, respectively, in the growth rate, all the other chs mutants had no significant changes in vegetative growth (Table 2). In comparison with the wild type, all the chs mutants had normal colony surface hydrophobicity (Figure S3). However, unlike other chs mutant, the chs6 mutant formed colonies with short, compact aerial hyphae and wrinkled surface on CM plates (Figure 3A). In contrast, the chs1 mutant tended to produce fluffy colonies and was slightly reduced in the melanization of aerial hyphae (Figure 3A).
Table 1. Fungal strains used in this study.
| Strain | Brief description | Reference |
| P131 | Wild type, MAT1-1 | [55] |
| S1528 | Wild type, MAT1-2 | [53] |
| LA11 | chs1 mutant of P131 | this study |
| LA40 | chs1 mutant of P131 | this study |
| LA1 | chs3 mutant of P131 | this study |
| LA3 | chs4 mutant of P131 | this study |
| LA28 | chs4 mutant of P131 | this study |
| LA8 | chs5 mutant of P131 | this study |
| LA14 | chs6 mutant of P131 | this study |
| LA26 | chs6 mutant of P131 | this study |
| LA12 | chs7 mutant of P131 | this study |
| LA2 | chs2 mutant of S1528 | this study |
| LA6 | chs2 mutant of S1528 | this study |
| LA51 | chs3 mutant of S1528 | this study |
| LA53 | chs3 mutant of S1528 | this study |
| LA41 | chs5 mutant of S1528 | this study |
| LA43 | chs5 mutant of S1528 | this study |
| LA49 | chs5 chs6 double mutant of P131 | this study |
| LA66 | chs5 chs6 double mutant of P131 | this study |
| LA33 | Transformant of LA40 expressing CHS1-eGFP | this study |
| LA71 | Transformant of LA12 expressing CHS7-eGFP | this study |
Table 2. Phenotype characterization of chs mutants of M. oryzae.
| Strain | Growth rate (mm/day)a | Germination (%)b | Conidiation (105spores/plate)c | Appressoria formation (%)d | Disease indexe |
| P131 (WT) | 3.4±0.1α * | 95.5±1.9α | 76.9±3.8α | 96.2±0.1α | 62±4α |
| LA40 (chs1) | 3.1±0.1α | 64.3±1.9α | 14.4±0.9β | 61.0±4.1β | 2±1β |
| LA1 (chs3) | 3.2±0.1α | 98.4±1.3α | 73.6±0.7α | 97.4±0.3α | 53±9α |
| LA28 (chs4) | 3.1±0.1α | 98.1±2.9α | 74.4±7.1α | 95.4±1.9α | 60±5α |
| LA8 (chs5) | 3.2±0.1α | 94.8±4.5α | 74.9±3.6α | 96.2±2.3α | 61±3α |
| LA26 (chs6) | 2.1±0.1β | 93.9±1.3α | 5.6±0.2β | 91.5±0.5α | 0±0β |
| LA49 (chs5 chs6) | 1.7±0.1β | 90.4±1.2α | 0.5±0.1γ | 85.2±1.0α | 0±0β |
| LA12 (chs7) | 3.1±0.1α | 95.1±1.8α | 74.2±4.9α | 0.0±0.0β | 10±6β |
| S1528 (WT) | 3.5±0.1α | 95.6±1.9α | 25.3±2.1α | 97.6±2.0α | 56±10α |
| LA6 (chs2) | 2.9±0.1β | 98.1±1.5α | 4.3±0.3β | 90.6±1.7α | 61±2α |
Average daily extension in the diameter of complete medium (CM) cultures. Means and standard deviations were calculated from three independent experiments.
Percentage of conidia germinated after 1 h incubation on hydrophobic surfaces.
Conidia produced by 14-day-old oatmeal agar plates (Φ 9 cm).
Percentage of germ tubes formed appressoria after incubation for 20 h on hydrophobic surfaces.
The number of lesions formed on the 5 cm tip region of infected leaves.
*Data from three replicates were analyzed with two sample t-test. The same greek letter indicated that there was no significant difference with the wild-type or single mutant. Different letters were used to mark statically significant difference (P = 0.01).
Figure 3. Colony morphology and melanized appressoria of the chs deletion mutants.
(A) Colonies of the wild-type strains P131 and S1528 and the chs1-chs7 mutants formed on the complete medium (CM) at 25°C. Photos were taken at 7 days post-inoculation. Bar = 10 mm. (B) Appressorium formation assays with conidia from the same set of the wild-type and mutant strains. Representative images were taken after 24 h incubation on glass coverslips. Germ tubes of the chs7 mutant failed to form appressoria. Bar = 10 µm.
Except for the chs1 mutant, all the other chs mutants produced three-celled pyriform conidia with normal morphology. However, conidiation was reduced over 5-fold in the chs2 and chs6 mutants (Table 2). The chs1 mutant tended to form pear-shaped, single-celled conidia. It also was reduced in conidiation but to a less degree than the chs2 and chs6 mutants (Table 2). When tested with different stresses, we found that the chs6 mutant but not the other chs mutants had increased susceptibility to osmotic and cell wall stresses (Table 3). Its growth rate on CM with 0.7 M NaCl was reduced over 50% in comparison with that on regular CM (Table 3).
Table 3. Assay for inhibitory effects of various chemicals on vegetative growth.
| 0.7 M NaCl | 1.2 M Sorbitol | 5 mM H2O2 | 50 µg/ml CFW | 50 µg/ml CR | 5 mM NZ | |
| P131 (WT) | 37.9±2.0α | 34.6±0.8α | 8.0±1.1α | 6.4±1.2α | 7.2±2.0α | 3.2±1.2α |
| LA40 (chs1) | 40.9±0.7α | 35.2±1.1α | 13.1±1.1β | 4.1±2.5α | 5.7±1.2α | 4.9±2.1α |
| LA1 (chs3) | 40.6±1.3α | 33.3±1.2α | 6.5±1.2α | 8.1±1.2α | 20.3±1.2β | 5.6±1.2α |
| LA28 (chs4) | 38.9±0.7α | 38.9±0.7α | 11.8±1.2α | 14.4±1.2β | 6.7±2.5α | 7.6±2.1α |
| LA8 (chs5) | 38.2±1.2α | 33.3±1.2α | 5.6±1.2α | 4.8±0.0α | 6.5±1.2α | 1.6±1.2α |
| LA26 (chs6) | 52.3±3.5β | 41.5±1.0β | 6.1±2.2α | 32.3±0.7β | 36.9±3.4β | 23.0±3.6β |
| LA49 (chs5chs6) | 34.0±5.1α | 6.4±1.9γ | 13.5±5.8β | 31.9±4.2β | 36.3±3β | N/A |
| LA12 (chs7) | 37.5±0.0α | 34.1±1.2α | 7.5±0.0α | 9.1±1.2α | 7.5±0.0α | 7.5±0.0α |
| S1528 (WT) | 36.9±1.6α | 45.3±1.0α | 2.3±0.0α | 12.2±1.1α | 1.5±0.4α | 5.3±1.2α |
| LA6 (chs2) | 39.7±8.0α | 55.9±0.7β | 8.4±1.2α | 38.1±4.4β | 9.3±1.0α | 12.6±1.2β |
The inhibition extent (%) = ((the diameter of regular CM cultures-the diameter of cultures on CM with different chemical)/the diameter of regular CM cultures)×100; CFW, Calcofluor white; CR, Congo red; NZ, Nikkomycin Z. N/A, not assayed. Data from three replicates were analyzed with two sample t-test. The same greek letter indicated that there was no significant difference with the wild-type or single mutant. Different letters were used to mark statically significant difference (P = 0.01).
Chitin contents and chitin synthase activities in the chs mutants
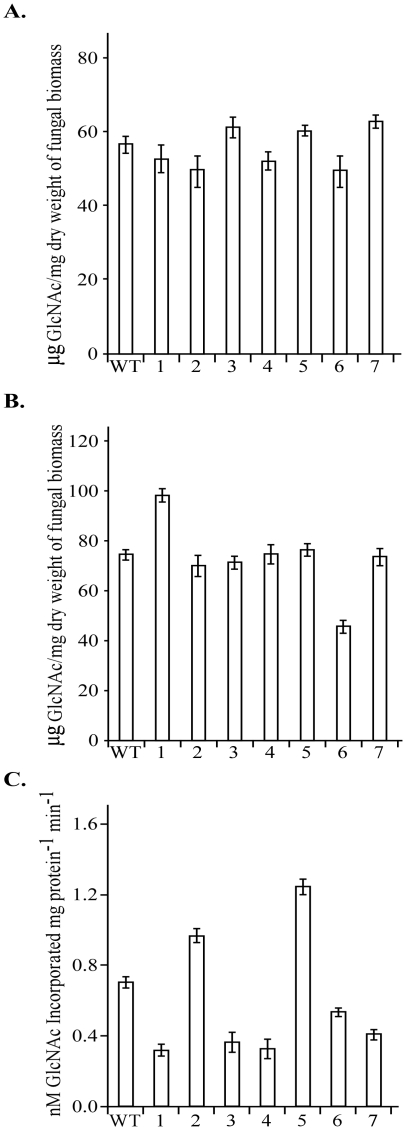
We then measured the chitin contents in vegetative hyphae and conidia of the chs mutants. In comparison with the wild type, none of the chs mutants (chs1-chs7) had significant changes in the chitin content in vegetative hyphae (Figure 4A). However, the chitin content was increased approximately 30% in the chs1 mutant but reduced approximately 40% in the chs6 mutant in conidia (Figure 4B). The other chs mutants had no obvious changes in the chitin content in conidia.
Figure 4. Chitin contents and chitin synthase activities in the wild-type and chs mutant strains.
The chitin content was assayed with vegetative hyphae harvested from 2-day-old CM cultures (A) or conidia harvested from 10-day-old OTA plates (B) of the wild type and chs mutants. The chitin content is expressed in µg of glucosamine hydrochloride per mg of dry-weight of fungal biomass. (C) Chitin synthase activities (nM UDP-N-acetyl-glucosamine incorporated into chitin per mg protein per minute) were assayed with microsomal fractions of proteins isolated from vegetative hyphae of the wild-type and chs mutant strains. Mean and standard deviation were calculated with results from three biological replicates.
Chitin synthase activities were assayed with the microsomal fraction extracts from vegetative hyphae of the wild type and chs mutants. In comparison with the wild type, the chs1, chs3, chs4, and chs7 mutants had approximately 2-fold reduction in chitin synthesis activity (Figure 4C). The chitin synthase activity also was reduced in the chs6 mutant but the reduction was less than 2-fold. In contrast, the chs2 and chs5 mutants had higher chitin synthase activity than the wild-type strain (Figure 4C). Nevertheless, their increase in chitin synthase activity was less than 2-fold in comparison with the wild type.
Expression levels of individual CHS genes in the chs mutants
The expression levels of individual CHS genes were assayed by qRT-PCR with RNA isolated from vegetative hyphae of the wild-type and chs mutant strains (Figure 5). In the chs1 mutant, CHS2 was increased 1.7-fold, and changes in expression levels in other CHS genes were less significantly than that of CHS2. In the chs2 mutant, the expression levels of CHS5 and CHS6 were reduced over 5-fold. Although CHS5 and CHS6 are closely linked, deletion of any one of these two genes had no significant effects on the expression level of the other one. Among all the CHS genes, only the expression level of CHS7 was increased 2.7-fold in the chs6 mutant. The chs7 mutant had 2.2-fold reduced expression of CHS1 and 1.9-fold increased expression of CHS2, respectively. These results indicated that deletion of one specific CHS gene had different impacts on the expression of other CHS genes during vegetative growth.
Figure 5. The expression levels of individual CHS genes in the wild type and chs mutants.
RNA samples isolated from vegetative hyphae harvested from 2-day-old CM cultures were used for qRT-PCR assays. The actin gene was used as the endogenous control for normalization. Relative expression levels were estimated with the 2−ΔΔCt method. The expression level of each CHS gene in the wild type was arbitrarily set to 1. Mean and standard errors were determined with data from three independent replicates.
The CHS1 and CHS7 genes play important roles in appressorium formation
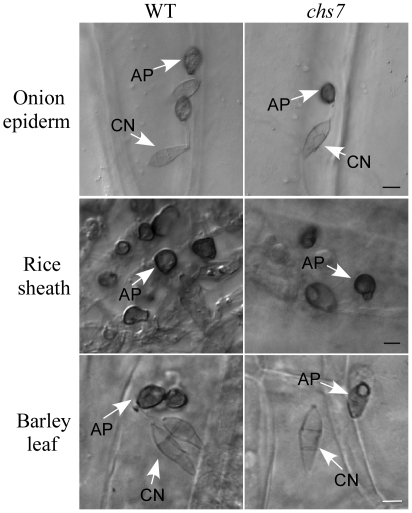
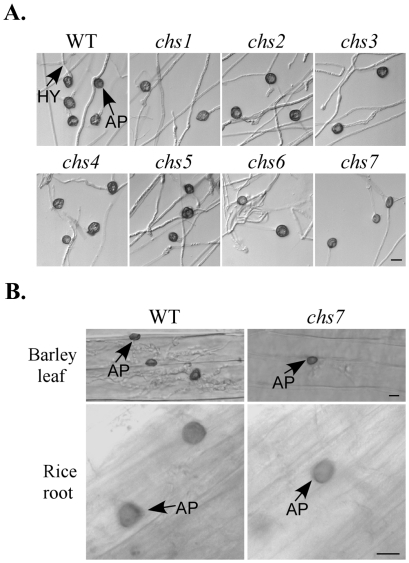
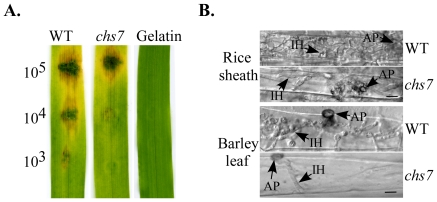
On artificial hydrophobic surfaces, all but the chs1 and chs7 mutants were normal in conidium germination and produced melanized appressoria (Table 2). The chs1 mutant was reduced 33% in conidium germination and only 61% of the germ tubes formed melanized appressoria. Appressoria formed by the chs1 mutant tended to be smaller than those of the wild type (Figure 3B). Germ tubes of the chs7 mutant failed to form melanized appressoria on artificial hydrophobic surfaces (Figure 3B), which was consistent with the previous report [41]. However, when the chs7 mutant was assayed for appressorium formation on plant surfaces, melanized appressoria were observed. Germ tubes of the chs7 mutant produced appressoria on onion epidermis, rice leaf sheaths, and barley leaves (Figure 6; Table S2), suggesting that its defects in appressorium formation is surface-dependent. Interestingly, like other chs mutants, hyphal tips of the chs7 mutant formed melanized appressoria on artificial hydrophobic surfaces (Figure 7A). Hyphal tips of the chs7 mutant also formed melanized appressoria on plant surfaces assayed (Figure 7B).
Figure 6. Appressorium formation assays with the chs7 mutant.
Melanized appressoria were formed by the germ tubes of the wild-type strain P131 and chs7 mutant on onion epidermis cells, rice leaf sheath, and barley leaves. AP, appressoria formed by germ tubes; CO, conidia. Bar = 10 µm.
Figure 7. Appressorium formation assays with hyphal tips.
(A) Appressoria formed by hyphal tips of the wild-type strain P131 and chs1-chs7 mutants on glass cover slips. Bar = 10 µm. (B) Appressoria formed by hyphal tips of P131 and chs7 mutant on barley leaf and rice root surfaces. AP, appressoria formed by hyphal tips; HY, hyphae. Bar = 10 µm.
We then assayed the effects of 1,16-hexadecanediol on appressorium formation on artificial surfaces. Germ tubes of the wild type and chs7 mutant had similar appressoria formation efficiency in the presence of the cutin monomer on hydrophobic surfaces (Table S2), which is consistent with the previous report [41]. Similar to the wild type, cutin monomer treatment had no significant effect on appressorium formation by hyphal tips in the chs7 mutant (Table S2). These data suggested that appressoria formation by germ tubes and hyphal tips were regulated differently in the chs7 mutant, and defects of the chs7 mutant in appressorium formation appeared to be specific to its germ tubes. It is possible that chitin synthase genes have distinct impacts on appressorium formation by germ tubes and hyphal tips.
The CHS1, CHS6, and CHS7 genes are important for plant infection
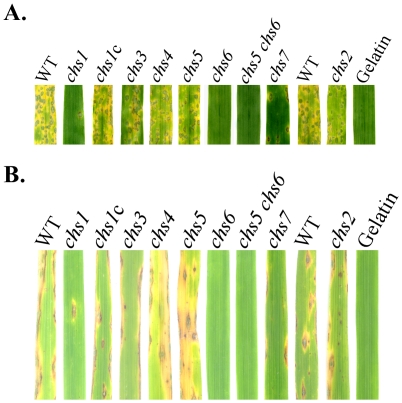
When conidia of the chitin synthase mutants were used to spray barley seedlings, the chs2, chs3, chs4, and chs5 mutants had no significant changes in virulence (Figure 8A; Table 2). The chs7 mutant was reduced in virulence, which is consistent with the earlier report [41]. However, the chs1 mutant was more significantly reduced in virulence than the chs7 mutant. On leaves sprayed with the chs1 mutant, only rare lesions were observed (Figure 8A). Unlike the other chs mutant, the chs6 mutant was non-pathogenic. On barley leaves sprayed with the chs6 mutant, no lesions were observed 5 days post inocultion (dpi) (Figure 8A; Table 2). Under the same conditions, numerous blast lesions were observed on leaves inoculated with the wild-type strain P131 (Figure 8A). Even on wounded barley leaves, the chs6 mutant failed to form any lesions.
Figure 8. Infection assays with rice and barley seedlings.
Conidia were harvested from 10-day-old oatmeal agar cultures of wild-type strains P131 and S1528, and the chs1-chs7 mutants. (A) Eight-day-old seedlings of barley cultivar Golden Promise were spray inoculated. Typical leaves were photographed 5 dpi. (B) Two-week-old seedlings of rice cultivar LTH were sprayed with conidium suspensions of the wild-type and chs mutant strains or 0.25% gelatin solution as the control. Photos were taken at 7 dpi.
We then repeated infection assays with rice seedlings. Similar results were obtained for all the chs mutants (Figure 8B). The CHS2, CHS3, CHS4, and CHS5 appeared to be dispensable for plant infection. In contract, the CHS6 gene is essential for pathogenesis in M. oryzae. The chs6 mutant failed to cause any lesion. On rice leaves sprayed with the chs1 and chs7 mutants, fewer blast lesions were observed than those sprayed with the wild type (Figure 8B).
CHS7 is important for appressorium penetration and invasive growth in penetration assays with rice leaf sheaths and barley leaves
To further confirm that CHS7 is important for virulence, conidium suspensions with different spore concentrations were drop-inoculated onto barley leaves (Figure 9A). In comparison with the wild type, the chs7 mutant failed to cause extensive necrosis and chlorosis at 104 conidia/ml or lower. Even at 105 conidia/ml, the chs7 mutant was less virulent (Figure 9A). In penetration assays with rice leaf sheaths and barley leaves, the wild type produced branching invasive hyphae in plant cells 48 hours post inoculation (hpi) (Figure 9B). Under the same conditions, the chs7 mutant had only limited invasive hyphae in underlying plant cells (Figure 9B), indicating that CHS7 is important for invasive growth in M. oryzae. Expression of the CHS7-eGFP suppressed the defects of the observed mutant defects, including appressorium formation and invasive growth.
Figure 9. The chs7 mutant was reduced in virulence.
(A) Eight-day-old barley leaves were drop-inoculated with conidium suspensions of the wild type and chs7 mutant. The concentration of each conidium suspensions was marked to the left. Inoculation with 0.25% gelatin was used as the control. Typical leaves were photographed 5 dpi. (B) Rice sheaths and barley leaves were drop-inoculated with conidia of the wild type and chs7 mutant. Penetration and invasive hyphae were examined 48 hpi. AP, appressoria; IH, infection hyphae. Bar = 10 µm.
CHS1 is important for infection-related morphogenesis
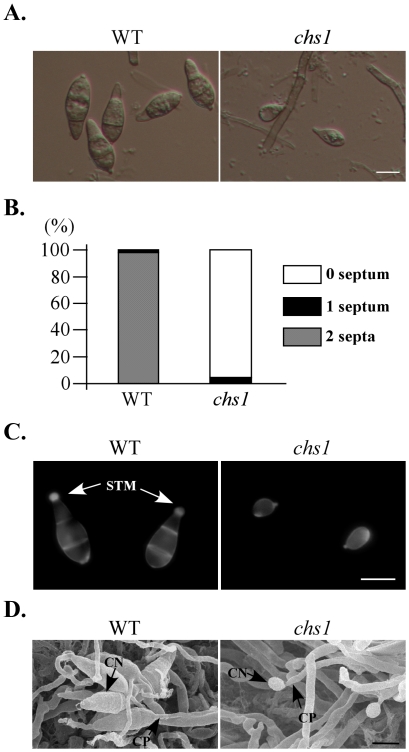
The chs1 mutant had severe defects in conidium morphology. More than 90% of chs1 conidia had no septum (Figure 10A; Figure 10B). No three-celled conidium was observed in the chs1 mutant. The shape of chs1 conidia also differed from that of the wild type (Figure 10A). One distinct defect of the chs1 mutant was the lack of spore tip and spore tip mucilage (STM) because the top of conidia were rounded (Figure 10A). When conidia were stained with Calcofluor white (CFW), the STM of the wild-type conidia had strong fluorescence signals. Conidia of the chs1 mutant lack a clear tip and STM staining (Figure 10C). When examined by scanning electron microscope (SEM), the chs1 mutant produced conidia sympodially on conidiophores (Figure 10D), indicating that CHS1 may be dispensable for conidiophore development and the initiation of conidiogenesis. However, later stages of conidiogenesis were blocked or defective in the chs1 mutant after the initial development of young conidia. CHS1 may be responsible for the synthesis of the apical landmark in the developing conidia.
Figure 10. CHS1 is important for conidiogenesis.
(A) Typical conidia of the wild-type strain P131 and chs1 mutant were examined under differential interference contrast (DIC) microscope. (B) The percentage of conidia with 0-, 1-, and 2-septa in the wild-type strain P131 and chs1 mutant. The chs1 mutant failed to produce normal, three-celled pyriform conidia. (C) Spore tip mucilage of the conidia of the wild type and chs1 mutant were stained with Calcofluor White (CFW) and examined by DIC and epifluorescence (UV) microscopy. Arrows pointed to STM. Bar = 10 µm. (D) Conidia and conidiophores of the wild-type P131 and chs1 mutant examined under scanning electron microscope (SEM). CO, conidium; CP, conidiophores; STM spore tip mucilage. Bar = 10 µm.
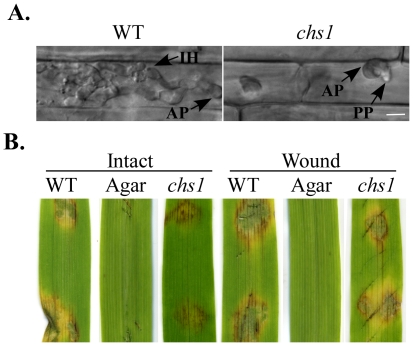
Although the chs1 mutant was reduced in conidium germination and appressorium formation, nuclear division and migration appeared to be normal during appressorium formation (Figure S4). Because appressoria formed by chs1 germ tubes were smaller than those of the wild-type appressoria, we conducted penetration assays with rice leaf sheaths (Figure 11A). As expected, the chs1 mutant formed melanized appressoria but failed to develop invasive hyphae at 48 h in the epidermal cells of rice leaf sheaths, although penetration pegs may be present. Under the same conditions, the wild type produced branching, bulbous invasive hyphae (Figure 11A).
Figure 11. The chs1 mutant was defective in appressorium penetration and plant infection.
(A) Appressorium penetration assays with rice leaf sheaths. Appressoria of the chs1 mutant failed to penetrate plant cells and develop infectious hyphae at 48 hpi. (B) Intact and wounded barley leaves were inoculated with culture blocks of the wild-type P131 and chs1 mutant. Lesions were visible 5 dpi after removing the inoculum. Inoculation with water agar blocks was the negative control. AP, appressorium; IH, invasive hyphae; PP, penetration peg.
We then assayed the virulence of the chs1 mutant on detached barley leaves. On intact leaves, culture blocks of the wild type cause extensive necrosis surrounding the inoculation site but the chs1 mutant only caused limited necrosis directly beneath culture blocks (Figure 11B). On wounded barley leaves, the wild type and chs1 mutant caused lesions of similar sizes at the wounding sites (Figure 11B). These results indicate that the reduction of the chs1 mutant in virulence may be directly related to its defect in appressorium penetration.
CHS1 is highly expressed in young, developing conidia
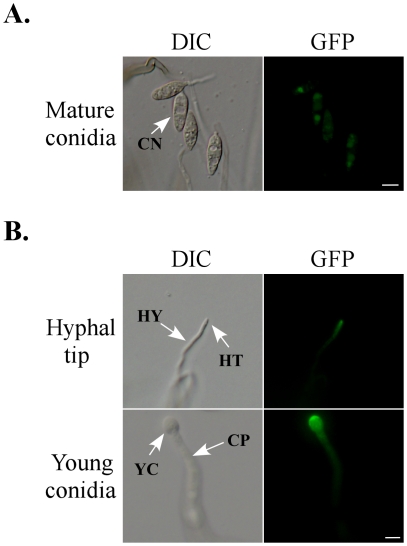
To determine the expression and localization of CHS1, a CHS1-eGFP fusion construct under the control of its native promoter was generated and transformed into the chs1 mutant LA40 (Table 1). Transformant LA33 (Table 1) was identified by PCR and confirmed by Southern blot analysis to contain a single copy of pLA8. It had no obvious defects in vegetative growth, appressorium formation, penetration, and plant infection. Moreover, the conidium morphology and conidiation defects of the chs1 mutant were rescued in transformant LA33 (Figure 12A). In infection assays, transformant LA33 was as virulent as the wild type (Figure 8), indicating that the GFP fusion had no effect on CHS1 function and the CHS1-eGFP fusion complemented the chs1 mutant in virulence.
Figure 12. Chs1-eGFP fusion proteins accumulate to the tips.
(A) Conidia produced by transformant LA33 expressing the CHS1-eGFP construct were observed by DIC and epifluorescence microscopy. (B) Hyphal tips and developing conidia of transformant LA33. The Chs1-eGFP fusion protein localized to the tip region in vegetative hyphae (the upper panel) and developing a young conidium (the bottom panel) at the tip of a conidiophore. CO, conidia; CP, conidiophores; HT, hyphal tips; HY, hyphae; YC, young conidia. Bar = 10 µm.
In transformant LA33 (Table 1), weak GFP signals were detected in the cytoplasm and vacuoles of vegetative hyphae, conidia (Figure 12A), and infectious hyphae (data not shown). Most likely, Chs1-eGFP fusion proteins that normally localize to the inner side of cytoplasm membrane were internalized and possibly degraded in these tissues. GFP signals were much stronger at the tip region of hyphae and conidiophores (Figure 12B). In developing conidia, the distal end region had the strongest GFP signals (Figure 12B).
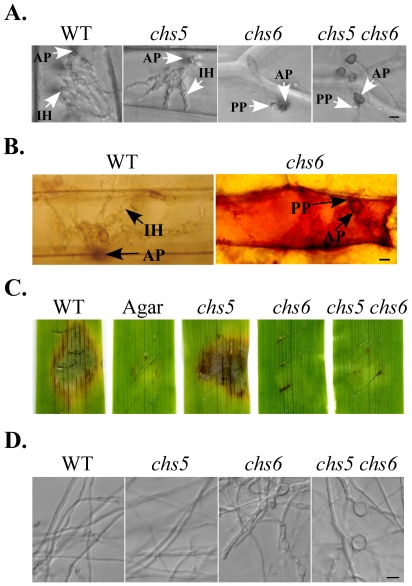
Deletion of CHS6 blocks appressorium penetration and invasive growth
Because the chs6 mutant still formed appressoria but was non-pathogenic (Figure 3; Figure 8), we then assayed its defects in penetration and invasive growth. Similar to what was observed on artificial surfaces, the chs6 mutant had no defects in appressorium formation on onion epidermal cells. However, appressoria formed by the mutant failed to successfully penetrate and develop invasive hyphae in plant cells 48 hpi (Figure 13A), which was consistent with spray infection results (Figure 8). Under the same conditions, extensive invasive hyphae were formed by the wild type. When stained with 3,3′-diaminobenzidine (DAB), strong ROS accumulation was observed only in plant cells penetrated by the chs6 mutant (Figure 13B). The chs6 mutant appeared to develop penetration pegs and elicited cytoplasm aggregation in underlying plant cells (Figure 13A), indicating that the chs6 penetration was blocked by plant defense responses [42].
Figure 13. The chs6 mutant and chs5 chs6 double mutant failed to penetrate the plant cell.
(A) Appressorium penetration assays with onion epidermal cells. Appressoria of the chs6 mutant and chs5 chs6 double mutant failed to penetrate plant cells and develop infectious hyphae at 48 hpi. Bar = 10 µm. (B) ROS accumulation in the infected barley leaves by the chs6 mutant stained with DAB. Strong ROS accumulations were observed in the leaves inoculated with the chs6 mutant. Bar = 10 µm. (C) Wounded barley leaves were inoculated with culture blocks of the wild-type P131, chs5, chs6, and chs5 ch6 double mutant. Lesions were visible 5 dpi after removing the inocula. Inoculation with water agar blocks was the negative control. (D) Vegetative hyphae of the wild-type P131 and the chs5, chs6, and chs5 chs6 mutants from 2-day-old 5×YEG cultures. Bar = 10 µm. AP, appressorium; IH, invasive hyphae; PP, penetration peg.
To further characterize the defects of the chs6 mutant in plant infection, we conducted wound-inoculation assays with barley leaves. On wounding sites inoculated with culture blocks of the wild type, extensive necrosis was observed at the wounding sites and in the surrounding area (Figure 13C). The chs6 mutant only caused limited darkening at the wounding site. No extensive, spreading zone of necrosis was observed (Figure 13C). Therefore, the chs6 mutant was defective in the differentiation and growth of invasive hyphae in planta. To confirm that these phenotypes were directly related to the deletion of CHS6, we conducted co-segregation analysis. Among 42 progeny isolated from the LA26 (chs6)×S1528 (WT) genetic cross (Table 1), 22 of them were resistant to hygromycin and had the same defects with the chs6 mutant. The rest 20 hygromycin-sensitive progeny were similar to the wild type.
The chs5 chs6 double mutant has more severe defects than the chs6 mutant
Because the CHS5 and CHS6 genes have similar structures and expression profiles, to further characterize their relationship, we generated the chs5 chs6 double mutant by deleting the 7.2-kb fragment containing the N-terminal portions of these two chitin synthase genes (Figure S2H). The double mutant LA49 (Table 1) was confirmed by Southern blot analysis (Figure S2H). Although the chs5 mutant had no obvious defects in growth or conidiation, the chs5 chs6 double mutant had more severe defects in vegetative growth (Figure 3A; Figure S5; Table S3) and conidiation (Table 2) than the chs6 mutant. Unlike colonies of the chs6 mutant with a wrinkled surface, colonies formed by the double mutant were smaller in size and had smooth appearance (Figure 3A). When examined under DIC microscope, the hyphal growth defects observed in the chs6 mutant also was enhanced in the double mutants (Figure 13D). The intercalary and apical hyphal swellings tended to be larger in the double mutant than the chs6 mutant (Figure 13D). These results indicate that CHS5 and CHS6 have overlapping functions in maintaining polarized growth in vegetative hyphae.
Same as the chs6 mutant, the chs5 chs6 double mutant formed melanized appressoria (Figure 3B). No obvious defects were observed in appressorium formation. In spray infection assays with both rice and barley leaves, the chs5 chs6 double mutant was non-pathogenic (Figure 8). In penetration assays with onion epidermal cells, the chs5 chs6 mutant, like the chs6 mutant failed to successfully penetrate underlying plant cells and develop invasive hyphae 48 hpi (Figure 13A). In wound-inoculation assays, the wild type and chs5 mutant caused extensive necrosis at the wounding sites and surrounding areas. The chs5 chs6 mutant caused limited darkening at the wounding sites but failed to cause extensive necrosis in the surrounding areas (Figure 13C). The expression levels of CHS3 and CHS4 were increased 2.6- and 2.4-fold, respectively, in the chs5 chs6 double mutant (Figure S6). Increased expression of these CHS genes may be able to partially compensate for the deletion of both CHS5 and CHS6 genes.
Discussion
Chitin and glucan are two major components of fungal cell wall. The F. graminearum and A. fumigatus genomes have eight CHS genes [1], [43]. Like N. crassa, M. oryzae has seven predicted chitin synthase genes. The CHS1 gene was highly expressed in conidia, especially in young conidia in M. oryzae (Figure 12). Deletion of CHS1 resulted in the production of conidia with a defective morphology. Majority of chs1 conidia were globular, single-celled (Figure 10). It appears that CHS1 plays a conidium-specific role in cytokinesis in developing conidia. CHS1 may be responsible for the synthesis of the landmark for the apical cell during conidiogenesis. In N. crassa, the chs-1 RIP mutant had no defects in conidiogenesis [18]. In A. nidulans, there are two class I chitin synthase genes, chsB and chsF [3]. Although ChsB functions in conidiophore and conidium development, the chsB mutant has normal conidium morphology [23]. In A. fumigatus, deletion of chsC has no detectable phenotype. The chsG and chsC chsG mutants were defective in hyphal growth and conidiation but produced normal conidia [44].
As the inocula for dispersal in the field, conidia play a critical in the initiation of plant infection and spreading. In this study, we found that three CHS genes, CHS1, CHS2 and CHS 6 are important in conidiation of M. oryzae. In particular, the chs1 mutant produced single cell conidia. Because conidia produced by N. crassa and Aspergilli are single-celled, defects of the chs1 mutant may be unique to cell division and cytokinesis in developing conidia. To test this hypothesis, we generated the Fgchs1 (FGSG_10116.3) deletion mutant in F. graminearum. Our preliminary data indicate that the Fgchs1 mutant were shorter and narrower than those of the wild type. While approximately 80% of the wild-type conidia had 3-4 septa, the majority of Fgchs1 conidia were three-celled (two septa), indicating that the CHS1 ortholog also is important for septation in conidia in F. graminearum. We would like to point out here that F. graminearum has two class I chitin synthase genes (Figure S1). Although FGSG_10116.3 shares higher similarity with the M. oryzae CHS1 gene than FGSG_03418.3, they may have overlapping functions in F. graminearum during conidiogenesis.
For many plant pathogenic fungi, conidium attachment and germination is the initial infection step. We noticed that many of the single-celled conidia were defective in germination in the chs1 mutant. Although the wild-type and chs1 mutant strains had similar chitin content in vegetative hyphae, the latter had higher chitin content in conidia (Figure 4). In consistent with this observation, conidia of the chs1 mutant had patches of strong CFW staining cell wall areas (Figure 10C). Higher chitin content and thicker cell wall may be responsible for reduced conidium germination in the chs1 mutant. Deletion of a chitin synthase gene resulted in an increased in overall chitin content has been reported in other fungi, such as the chs2 mutant of C. albicans [45] and the chsA chsC double mutant of A. nidulans [46].
Among the seven CHS genes in M. oryzae, CHS2 had the lowest expression level in conidia, appressoria, and infected leaves. Its expression level in vegetative hyphae was only slightly higher than that of CHS7 (Figure 2). In F. oxysporum, the chs2 mutant had a significant reduction in virulence [37]. However, the chs2 mutant of M. oryzae was as virulent as the wild type on the seedlings of rice and barley although it was reduced in vegetative growth and conidiation, indicating that CHS2 was not important for plant infection. Therefore, CHS2 must have distinct functions in invasive hyphae and vegetative hyphae. In U. maydis, there are three class II chitin synthase genes, CHS2, CHS3, and CHS4. The chs2 mutant has no obvious defects in mating, dimorphism, and plant infection, but deletion of either chs3 or chs4 resulted in a significant reduction in mating efficiency and virulence [5], [29]. In M. oryzae, the chs2 mutant had increased sensitivity to Calcofluor white, Congo red, and Nikkomycin Z (Table 3), indicating that CHS2 is important for cell wall integrity, which was similar to the Bcchs1 (II) mutant in B. cinerea [32].
The CHS3 and CHS1 genes had similar structures (Figure 1A) and similar expression patterns (Figure 2B) in M. oryzae. In the chs3 mutant, the CHS1 expression was slightly up-regulated in the chs3 mutant (Figure 5). Our preliminary data showed that the chs1 chs3 double mutant had increased sensitivity to hyperosmotic and oxidative stresses, indicating that the CHS1 and CHS3 genes may have compensatory functions in cell wall modifications in responses to stresses in M. oryzae.
In M. oryzae, deletion of the CHS3, CHS4, and CHS5 genes that encode classes III, IV, and V chitin synthases, respectively, had no significant changes in conidiation, appressorium formation, and plant infection although the growth rate might be slightly reduced. In A. nidulans, deletion of the chsC gene had no obvious effects on growth and reproduction but the chsA (II) chsC (III) double mutant was defective in septum formation and asexual or sexual development [47]. It is possible that CHS3 has overlapping functions with CHS2 or other CHS genes in M. oryzae. In Colletotrichum graminicola and F. oxysporum, the class III chitin synthase genes are dispensable for hyphal growth, conidiation, and plant infection [36], [37]. Deletion of CHS3 and CHS4 led to up-regulated expression of other CHS genes (Figure 5), suggesting that these two genes might function as negative regulators on expression of other CHS genes.
Both the CHS5 and CHS6 MMD-containing chitin synthase genes had the highest expression level in vegetative hyphae. Like their orthologs in several other filamentous ascomycetes, CHS5 and CHS6 are located next to each other in a head-to-head arrangement in M. oryzae (Figure 1B). Because their ORFs are only 2,957-bp apart, it is possible that CHS5 and CHS6 share some regulatory elements in their promoters. Putative Rlm1-binding sites are present in the promoter regions of the CHS5 and CHS6 orthologs in A. nidulans [6], and F. oxysporum [42]. In S. cerevisiae, transcription factor Rlm1 functions downstream of the PKC pathway for regulating cell wall integrity [48]. In M. oryzae, the MIG1 gene, an ortholog of RLM1, is required for the development and growth of invasive hyphae in plant cells [49]. The chs6 mutant had similar defects in plant infection (Figure 8) with the mig1 mutant [49]. It is possible that the expression of CHS6 may be under the control of Mig1 in M. oryzae.
In A. nidulans, the csmA and csmB single deletion mutants had no significant changes in the growth rate but the csmA csmB double mutant was not viable [6], suggesting an essential role for these CHS genes in hyphal tip growth [50], [51]. In F. oxysporum, the chsV chsVb double mutant is viable but both chsV and chsVb single mutants were non-pathogenic [39]. Similar results were reported in F. graminearum. Both GzCHS5 and GzCHS7 are important for hyphal growth and pathogenicity [34]. In M. oryzae, deletion of CHS5 had no effects on vegetative growth and plant infection but the chs6 mutant was non-pathogenic and defective in hyphal growth. The chs5 chs6 double mutant was more significantly reduced in vegetative growth and conidiation than the chs6 mutant, indicating that CHS5 may play a minor overlapping role with CHS6 in growth and differentiation. Hyphal swelling observed in the chs6 and chs5 chs6 mutants indicated a weakened cell wall. Chs5 is 92-amino-acid residues shorter than Chs6 in the MMD domain. It lacks the consensus motifs of myosins, such as the P-loop, Switch I, and Switch II, that are present in Chs6 (Figure 1B), suggesting that functional differences of these two genes may derive from their structural differences in their MMDs.
Although there was a previous report that appressorium formation by germ tubes was blocked in the chs7 mutant [41], we found that its hyphal tips had no defects in developing melanized appressoria on artificial hydrophobic surfaces, suggesting that the role of CHS7 in appressorium formation is cell type-specific. It is possible that germ tubes, but not hyphal tips, of the chs7 mutant are defective in surface recognition. Recently, it has been shown that the msb2 mutant was defective in appressorium formation on artificial surfaces but formed appressoria efficiently on plant surfaces [52]. We also found that germ tubes of the chs7 mutant formed melanized appressoria on plant surfaces (Figure 6). These results indicate that the CHS7 gene is not essential for appressorium formation and defects of the chs7 mutant in appressorium formation are surface-dependent and cell-type specific. Appressorium formation by the hyphal tips and germ tubes may require different chitin synthase genes in M. oryzae. Therefore, it will be important to further characterize the role of CHS7 in appressorium formation by germ tubes and hyphal tips. It is also critical to identify and characterize the CHS gene(s) with elevated expression levels in the germ tubes or hyphal tips of the chs7 mutant during appressorium formation on different surfaces.
To our knowledge, systemic characterization of chitin synthase genes in fungal pathogens has been carried out only in U. maydis, a basidiomycetous fungal pathogen. Our functional characterization of all the seven predicted chitin synthase genes in M. oryzae seems to be the first such study in an ascomycetous phytopathogenic fungus. Although this approach is not novel, we did observe unique phenotypes in the chs1, chs6, and chs7 mutants that are not reported in previous studies. CHS1 is the first fungal chitin synthase gene that specifically affects conidium morphology and germination. The fact that the chs1 mutant produced more conidia than the chs2 and chs6 mutants suggests that CHS1 may be dispensable for conidiophore development and play a unique role after the de-limitation of young conidia. Because the chitin content was increased in chs1 conidia, other CHS genes must be able to compensate the deletion of CHS1 in M. oryzae. The chs5 chs6 double mutant is viable, which also indicate that other CHS genes can support hyphal tip growth. Therefore, it will be important to further characterize the interactions and relationships among the CHS genes during different developmental and plant infection stages in M. oryzae. In conclusion, the chitin synthases important for hyphal growth, conidiogenesis, appressorium development, and pathogenesis could be provided as potential targets to develop new strategies or fungicides to control the rice blast disease.
Materials and Methods
Strains and culture conditions
The M. oryzae wild-type stains P131 and S1528, and various transformants generated (Table 1) in this study were cultured on oatmeal agar (OTA) at 25°C under bright light [53]–[55]. Fungal genomic DNA isolation, protoplast preparation, transformation, measurements of vegetative growth rate and conidiation were performed as described [52], [56], [57]. For testing sensitivities to various stresses, fungal growth was assayed after incubation at 25°C for 5 days on complete medium (CM) plates with 0.7 M NaCl, 1.2 M sorbitol (Amresco), 50 µg/ml Congo red (Sigma), 50 µg/ml Calcofluor white (Sigma), 5 mM Nikkomycin Z (Sigma) as described [5], [52], [58], [59]. 10 µM cutin monomer 1,16-hexadecanediol (Sigma) was used to examine the effect on appressoria formation by germ tubes or hyphal tips [52], [60]. Examination of GFP expression in young conidia and conidiophores were performed as described [61]. Genetic crosses were performed as described [53], [62], [63].
Generation of the CHS gene replacement constructs and mutants
For generating the CHS1 gene replacement construct pLA1, the 1.3-kb upstream and 1.7-kb downstream fragments of the CHS1 gene were amplified with primers 1P1/1P2 and 1P3/1P4 (Table S1), respectively. The resulting PCR products were cloned into the SpeI-EcoRI and SalI-KpnI sites of pKOV21 [53]. After linearization with NotI, pLA1 was transformed into protoplasts of the wild-type strain P131. Putative chs1 mutants were identified by PCR and further confirmed by Southern blot hybridizations [64]. Similar strategies were used to generate the gene replacement constructs for the other chitin synthase genes.
qRT-PCR analysis
Total RNAs were isolated from mycelia harvested from 2-day-old minimal media or complete media, conidia harvested from 10-day-old oatmeal agar cultures, 24 h appressoria, and infected rice leave harvested 5 dpi with the TRIzol reagent (Invitrogen) and purified with the DNA-free kit (Ambion). First strand cDNA was synthesized with the AccuScript High Fidelity RT-PCR System (Stratagene) and used as the template for qRT-PCR as described [65], [66]. The M. oryzae actin gene MGG_03982.6 was used as the endogenous control for normalization. Relative expression levels were estimated with the 2−ΔΔCt method [67]. Data from three biological replicates were used to calculate the mean and standard deviation. Primers used for qRT-PCR are listed in Table S1.
Measurement of the chitin content
The chitin content was determined by measuring the amount of glucosamine released by acid hydrolysis of fungal cell wall [68]–[70]. One gram of freshly harvested vegetative hyphae or conidia was grinded in liquid nitrogen and suspended in 5 ml of deionized water. After centrifugation at 13,000 g for 10 min at 4°C, the pellets were freeze-dried overnight (Labconco). For each 5 mg of the dried pellet, 1 ml of 6 M HCl was added. After hydrolyzed at 100°C for 4 h, the hydrolysate was adjusted with 10 N NaOH to pH 7.0. An aliquot (0.2 ml) of the resulting mixture was added to 0.25 ml of 4% acetylacetone in 1.25 M sodium carbonate and heated for 30 min at 100°C. The mixture was heated for 1 h at 60°C after adding 2 ml of ethanol and 0.25 ml of the Ehrlich reagent [71] and centrifugated at 13,000 g for 10 min. The supernatant was measured for absorbance at 530 nm (BioTek). The chitin content (µg of glucosamine/mg of dry weight of fungal biomass) was calculated in accordance with a standard curve established by measuring the absorbance of known amounts of glucosamine hydrochloride (Sigma).
Chitin synthase activity assay
Microsomal fractions of proteins were isolated with a similar procedure as described [32], [70], [72] with minor modifications. Homogenates of 1 g of mycelia grinded in liquid nitrogen were suspended in 20 ml of 50 mM Tris-HCl (pH 7.4) and 2.5 mM MgCl2. After centrifugation at 13,000 g for 10 min, the supernatant was centrifugated at 40,000 g for 1 h at 4°C. The resulting pellet was dissolved in 0.5 ml of 50 mM Tris-HCl (pH 7.4) with 30% glycerol (w/v) and assayed for protein contents with a NanoDrop (Thermo). Chitin synthase activities were assayed as described [18], [72] in a 50 µl of reaction mixture containing 10 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 1.0 mM UDP-N-acetyl-D-glucosamine, 667 nM (0.01 uCi) UDP-N-acetyl-D-[U-14C]glucosamine (300 mCi/mmol; American Radiolabled Chemicals), and 32 mM N-acetyl-glucosamine. After incubation at 30°C for 1 h, the reactions were terminated by adding 50 µl of acetic acid. The final mixture was loaded onto a glass fiber filter (Φ 2.5 cm, GF/C, Whatman) and washed three times with 1 ml of 30% acetic acid and 70% ethanol, and once with 1 ml of ethanol. The discs were dried and the radioactivity retained on the filters was measured by a liquid scintillation counter (PerkinElme). The chitin synthase activity was expressed as nM of GlcNAc incorporated into chitin per mg of protein per minute [32].
Generation of the CHS1-eGFP construct and transformant
A 4.5-kb fragment of CHS1 gene along with its 1.5-kb promoter region was amplified with primers DW1F/DW1R (Table S1) and cloned into pKNTG [53]. The resulting CHS1-eGFP fusion construct pLA8 was confirmed by sequencing analysis. After NotI digestion, pLA8 was transformed into protoplasts of the chs1 mutant LA40 (Table 1). Monoconidial cultures of neomycin-resistant transformants were screened for GFP expression and confirmed by Southern blot analysis. Similar strategies were used to generate the CHS7-eGFP construct and transformant.
Assay for appressorium formation and plant infection
Appressorium formation by germ tubes on artificial surface were assayed as described [52], [54], [73]. Appressorium formation assays with hyphal tips were performed as described [74]. Appressorial penetration and invasive hyphae development were assayed with rice leaf sheaths and onion epidermal cells [53], [64], [75]. For appressoria formation and penetration assay on plant surface, 20 µl of conidia suspension or fresh hyphal bocks were dropped on the 5-day-old barley leaf cultivar, Golden Promise, and incubated within a moist chamber for 24 h and 48 h, respectively. In order to observe the appressoria and invasive hyphae clearly, 96% ethanol (v/v) were used to remove the chlorophyll [76]. After shaking the sample in 96% ethanol (v/v) for 12 h, a bright microscope was used to observe the appressoria and invasive hyphae. For testing the appressoria formed by hyphal tips on rice root surface, 5-day-old root of rice cultivar, LTH was placed on the surface of MS (Calsson Lab) media with 1% agar [77], [78], and the fresh hyphal blocks were placed on the root surface. Appressoria formation was examined with a bright microscope at 48 hpi.
For spray infection assays, conidia were adjusted to 1×105 conidia/ml in 0.25% gelatin and used for inoculation with 14-day-old seedlings of rice cultivar, LTH and 8-day-old seedlings of barley cultivar, Golden Promise [53], [79]. Lesion formation was examined at 5–7 days post-inoculation. Mean number of lesions formed on 5-cm leaf tips was determined as described [63], [80]. Infection assays with mycelial blocks on intact or wounded barley leaves were preformed as described [54]. Infection assay with dilution-drop conidia suspension was performed as previously described [81].
Scanning electron microscope (SEM) examination
Culture blocks from 10-day-old OTA cultures were fixed in 4% glutaraldehyde at 4°C for 16 h. The samples were then dehydrated and coated with gold as described [52], [82]. Conidia and conidiophores were observed with a JSM-6360LV (Jeol Ltd., Tokyo) scanning electron microscope.
Calcofluor white, DAPI staining, and DAB assays
Conidia and hyphae were fixed in a fixation solution (3.75% formaldehryde, 50 mM PBS, pH 7.5, 0.2% Triton X-100) for 15 min and then stained with 10 µg/ml Calcoflour white and/or 1 µg/ml DAPI (Sigma) for 5 min [54], [83]. After washing twice with sterile distilled water, the chitin deposition in cell wall and septa, and neclear devision were examined with an Eclipse 800 epifluorescence microscope (Nikon). For staining with 3,3′-diaminobenzidine (DAB, Sigma), 5-day-old barley leaves were inoculated with the wild type and chs mutant. After incubation at room temperature for 48 h, samples were stained with 1 mg/ml DAB solution (pH 3.8) for 8 h and de-stained with ethanol/acetic acid (94/4, v/v) for 1 h before examination [65], [76], [84].
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JF912404 (CHS1), JF912405 (CHS2), JF912406 (CHS3), JF912407 (CHS4), JF912408 (CHS5), JF912409 (CHS6), and EU935590 (CHS7).
Supporting Information
Phylogenetic tree of fungal chitin synthase. Phylogenetic tree was generated by the Clustalx1.83. The scale bar indicates 0.1 distance units. Species abbreviation are Af (Aspergillus fumigatus), An (Aspergillus nidulans), Bc (Botrytis cinerea), Cg (Colletotrichum graminicola), Fo (Fusarium oxysporum), Gz (Gibberella zeae), and Nc (Neurospora crassa).
(TIF)
CHS gene deletion construct and confirmation. (A) CHS1 gene deletion strategy and confirmation. CHS1 deletion construct (the upper) was created by replacing the CHS1 gene with hygromycin phosphotransferase cassette (hph). The upstream and downstream flanking sequences were amplified with primers 1P1/1P2 and 1P3/1P4, and ligated with the hph cassette. The positions of primers 1P1, 1P2, 1P3, 1P4, 1out, and PF are indicated with small arrows. E, EcoRI; EV, EcoRV; K, KpnI; S, SalI; Sp, SpeI. This construct was introduced into the wild-type strain P131. The putative mutants were screened with primers 1out/PF, and the putative mutants LA11 and LA40 had 1.9-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRV-digested genomic DNA of wild-type P131 and chs1 mutant LA11 and LA40 hybridized with Probe 1, which was amplified with primers 1P1/1P2. The results show a single 5.0-kb band (lane1) for the wild-type P131 and a single 6.0-kb band (lane 2 and lane 3) for chs1 mutant LA11 and LA40 (right bottom). (B) CHS2 gene deletion strategy and confirmation. CHS2 deletion construct (the upper) was created by replacing the CHS2 gene with hph. The upstream and downstream flanking sequences were amplified with primers 2P1/2P2 and 2P3/2P4, and ligated with the hph cassette. The positions of primers 2P1, 2P2, 2P3, 2P4, 2out, and PF are indicated with small arrows. B, BamHI; E, EcoRI; H, HindIII; K, KpnI. This construct was introduced into the wild-type S1528. The deletion mutants were screened with primers 2out/PF, and the putative mutants LA2 and LA6 had 2.3-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of KpnI-digested genomic DNA of wild-type strain S1528 and chs2 mutant LA2 and LA6 hybridized with Probe 2, which was amplified with primers 2P1/1P2. The results show a single 3.0-kb band (lane 1) for the wild-type S1528 and a single 6.1-kb band (lane 2 and lane 3) for chs2 mutant LA2 and LA6 (right bottom). (C) CHS3 gene deletion strategy and confirmation. CHS3 deletion construct (the upper) was created by introducing the hph within the CHS3 gene. The fragment within CHS3 gene was amplified with primers 3P1/3P2. The hph gene fragment was amplified with primers 3P3/3P4. The hph gene fragment was inserted into the fragment within CHS3 by SalI restriction enzyme. The positions of primers 3P1, 3P2, 3P3, 3P4, 3out, and PF are indicated with small arrows. B, BamHI; E, EcoRI; H, HindIII; S, SalI; X, XhoI. This construct was introduced into the wild-type P131. The deletion mutants were screened with primers 3out/PF, and the putative mutant LA1 had 2.5-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI and HindIII-digested genomic DNA of the wild-type P131 and chs3 mutant (LA1) hybridized with Probe 3, which was amplified with primers 3P3/3P4. The results show a single 6.1-kb band (lane 1) for the chs3 mutant LA1 and no band (lane 2) for wild-type strain P131 when using EcoRI-digesting the genomic DNA; a single 4.7-kb band (lane 3) for the chs3 mutant LA1 and no band (lane 4) for the wild-type P131 when using HindIII-digesting the genomic DNA (right bottom). (D) CHS4 gene deletion strategy and confirmation. CHS4 deletion construct (the upper) was created by replacing the CHS4 gene with hph. The upstream and downstream flanking sequences were amplified with primers4P1/4P2 and 4P3/4P4, and ligated with the hph cassette. The positions of primers 4P1, 4P2, 4P3, 4P4, 4out, and PF are indicated with small arrows. A, ApaI; E, EcoRI; S, SalI; Sp, SpeI. This construct was introduced into the wild-type P131. The deletion mutants were screened with primers 4out/PF, the putative mutants LA3 and LA28 had 2.1-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of SalI-digested genomic DNA of the wild-type P131 and chs4 mutants LA3 and LA28 hybridized with Probe 4, which was amplified with primers 4P1/4P2. The results show a single 3.5-kb band (lane 1) for the wild-type P131 and a single 4.4-kb band (lane 2 and lane 3) for chs4 mutants LA3 and LA28 (right bottom). (E) CHS5 gene deletion strategy and confirmation. CHS5 deletion construct (the upper) was created by replacing the CHS5 gene with hph. The upstream and downstream flanking sequences were amplified with primers 5P1/5P2 and 5P3/5P4, and ligated with the hph cassette. The positions of primers 5P1, 5P2, 5P3, 5P4, 5out, and PF are indicated with small arrows. B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; K, KpnI; S, SalI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 5out/PF, and the putative mutant (LA8) had 2.0-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of BglII and HindIII-digested genomic DNA of wild-type P131 and CHS5 deletion mutant LA8 hybridized with Probe 5, which was amplified with primers 5P3/5P4. The results show a single 6.2-kb band (lane 1) for the CHS5 deletion mutant LA8 and no band (lane 2) for the wild-type P131 when using BglII-digesting the genomic DNA; a single 2.2-kb band (lane 3) for the chs5 mutant (LA8) and no band (lane4) for the wild-type P131 when using HindIII-digesting the genomic DNA (right bottom). (F) CHS6 gene deletion strategy and confirmation. CHS6 deletion construct (the upper) was created by replacing the fragment within CHS6 gene with hph. The upstream and downstream flanking sequences were amplified with primers 6P1/6P2 and 6P3/6P4, and ligated with the hph cassette. The positions of primers 6P1, 6P2, 6P3, 6P4, 6out, and PF are indicated with small arrows. E, EcoRI; H, HindIII; K, KpnI; S, SpeI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 6out/PF, and the putative mutants LA14 and LA26 had 2.0-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI-digested genomic DNA of wild-type strain P131 and chs6 mutants LA14 and LA26 hybridized with Probe 6, which was amplified with primers 6P1/6P2. The results show a single 3.5-kb band (lane 3) for the wild-type P131 anda single 4.4-kb band (lane 1 and lane 2) for chs6 mutants LA14 and LA26 (right bottom). (G) CHS7 gene deletion strategy and confirmation. CHS7 deletion construct (the upper) was created by replacing fragment within CHS7 gene with hph. The upstream and downstream flanking sequences were amplified with primers 7P1/7P2 and 7P3/7P4, and ligated with the hph cassette. The positions of primers 7P1, 7P2, 7P3, 7P4, 7out, and PR are indicated with small arrows. E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SpeI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 7out/PR, and the putative mutant LA12 had 1.7-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI and PstI-digested genomic DNA of the wild-type P131 and chs7 mutant LA12 hybridized with Probe 7, which was amplified with primers 7P3/7P4. The results show a single 6.2-kb band (lane 1) for the chs7 mutant LA12 and no band (lane 2) for wild-type P131 when using BglII-digesting the genomic DNA; a single 2.9-kb band (lane 3) for the chs7 mutant LA12 and no band (lane 4) for the wild-type P131 when using PstI-digesting the genomic DNA (right bottom). (H) CHS5 CHS6 gene deletion strategy and confirmation. The CHS5 CHS6 deletion construct was generated by replacing the N-terminal regions of both CHS5 and CHS6 with the hph cassette. The upstream and downstream flanking sequences were amplified with primers 8P1/8P2 and 8P3/8P4, and ligated with the hph cassette. B, BamHI; E, EcoRI; H, HindIII; K, KpnI. The putative double mutants LA49 and LA66 were identified by PCR with primers 8out/PF with a 1.8-kb specific band. A 0.2-kb band within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of PstI-digested genomic DNA of the wild-type P131 and chs5 chs6 double mutants LA49 and LA66 hybridized with Probe 8, which was amplified with primers 8P3/8P4. The results show a single 2.7-kb band for the wild-type P131 and a single 4.2-kb band for chs5 chs6 double mutants LA49 and LA66 (right bottom).
(TIF)
Hydrophobicity assay. Drops of 20 µl sterile distilled water were placed on the surface of vegetative colony from 7-day-old complement media agar cultures. The results show the surface of vegetative colony from the wild-type P131 and S1528, and seven CHS gene deletion mutants were hydrophobic.
(TIF)
Nuclear division and movement during appressorium formation in the chs1 mutant. Conidia of the wild type and chs1 mutant incubated on hydrophobic glass coverslips for 0, 1, 4, and 24 h were stained with CFW and DAPI and examined with an epifluorescence microscope. Bar = 10 mm.
(TIF)
The chs5 chs6 double mutant was more significantly reduced in growth rate than chs6 mutant. The wild-type P131, chs5, chs6, and chs5 chs6 double mutant were cultured on OTA plates under light at 25°C. Photographs were taken at 7 and 14 dpi, respectively. Bar = 10 mm.
(TIF)
qRT-assays with the expression levels of CHS genes in the wild-type P131 and chs5 chs6 mutants. The RNA was isolated from vegetative mycelia shaken in CM for two days. The actin gene was used as the endogenous control for normalization. Relative expression levels were estimated with the 2−ΔΔCt method. The expression level of each CHS gene in the wild type was arbitrarily set to 1. Mean and standard errors were determined with data from three independent replicates.
(TIF)
PCR primers used in this study.
(DOC)
The effects of cutin monomer treatment on appressorium formation by the wild type and chs7 mutant on different surfaces.
(DOC)
Defect of the chs 5 chs 6 mutants in vegetative growth.
(DOC)
Acknowledgments
We thank M.F. Xue for assistance with bioinformatic analyses and X.Y. Zhou for fruitful discussions. We also thank Dr. Larry Dunkle at Purdue University for critical reading of this manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the 973 program (Grant No. 2012CB114002) from the Ministry of Sciences and Technology, China, the Program for Changjiang Scholars and Innovative Research Team in University from the Ministry of Education, China, and a Special Fund from the State Key Laboratory of Agrobiotechnology to Y.L. Peng. L.A. Kong's stay in Purdue University was supported by the China Scholarship Council fellowship (2009635040). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lenardon MD, Munro CA, Gow NA. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 2010;13:416–423. doi: 10.1016/j.mib.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horiuchi H. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med Mycol. 2009;47:S47–52. doi: 10.1080/13693780802213332. [DOI] [PubMed] [Google Scholar]
- 4.Choquer M, Boccara M, Goncalves IR, Soulie MC, Vidal-Cros A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur J Biochem. 2004;271:2153–2164. doi: 10.1111/j.1432-1033.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 5.Weber I, Assmann D, Thines E, Steinberg G. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell. 2006;18:225–242. doi: 10.1105/tpc.105.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeshita N, Yamashita S, Ohta A, Horiuchi H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol Microbiol. 2006;59:1380–1394. doi: 10.1111/j.1365-2958.2006.05030.x. [DOI] [PubMed] [Google Scholar]
- 7.Hodge T, Cope MJ. A myosin family tree. J Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- 8.Cid VJ, Duran A, del Rey F, Snyder MP, Nombela C, et al. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford RA, Shaw JA, Cabib E. Yeast chitin synthases 1 and 2 consist of a non-homologous and dispensable N-terminal region and of a homologous moiety essential for function. Mol Gen Genet. 1996;252:420–428. doi: 10.1007/BF02173007. [DOI] [PubMed] [Google Scholar]
- 10.Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- 11.Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- 12.Roncero C, Sanchez Y. Cell separation and the maintenance of cell integrity during cytokinesis in yeast: the assembly of a septum. Yeast. 2010;27:521–530. doi: 10.1002/yea.1779. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo Y, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. Genetic analysis of chs1 + and chs2 + encoding chitin synthases from Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2004;68:1489–1499. doi: 10.1271/bbb.68.1489. [DOI] [PubMed] [Google Scholar]
- 15.Munro CA, Whitton RK, Hughes HB, Rella M, Selvaggini S, et al. CHS8-a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet Biol. 2003;40:146–158. doi: 10.1016/s1087-1845(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 16.Klis FM. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Leon E, Verdin J, Freitag M, Roberson RW, Bartnicki-Garcia S, et al. Traffic of chitin synthase 1 (CHS-1) to the Spitzenkorper and developing septa in hyphae of Neurospora crassa: actin dependence and evidence of distinct microvesicle populations. Eukaryot Cell. 2011;10:683–695. doi: 10.1128/EC.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yarden O, Yanofsky C. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 1991;5:2420–2430. doi: 10.1101/gad.5.12b.2420. [DOI] [PubMed] [Google Scholar]
- 19.Din AB, Yarden O. The Neurospora crassa chs-2 gene encodes a non-essential chitin synthase. Microbiol. 1994;140:2189–2197. doi: 10.1099/13500872-140-9-2189. [DOI] [PubMed] [Google Scholar]
- 20.Din AB, Specht CA, Robbins PW, Yarden O. chs-4, a class IV chitin synthase gene from Neurospora crassa. Mol Gen Genet. 1996;250:214–222. doi: 10.1007/BF02174181. [DOI] [PubMed] [Google Scholar]
- 21.Riquelme M, Bartnicki-Garcia S, Gonzalez-Prieto JM, Sanchez-Leon E, Verdin-Ramos JA, et al. Spitzenkorper localization and intracellular traffic of green fluorescent protein-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot Cell. 2007;6:1853–1864. doi: 10.1128/EC.00088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borgia PT, Iartchouk N, Riggle PJ, Winter KR, Koltin Y, et al. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet Biol. 1996;20:193–203. doi: 10.1006/fgbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda K, Yamada K, Deoka K, Yamashita S, Ohta A, et al. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot Cell. 2009;8:945–956. doi: 10.1128/EC.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiuchi H, Fujiwara M, Yamashita S, Ohta A, Takagi M. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J Bacteriol. 1999;181:3721–3729. doi: 10.1128/jb.181.12.3721-3729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motoyama T, Fujiwara M, Kojima N, Horiuchi H, Ohta A, et al. The Aspergillus nidulans genes chsA and chsD encode chitin synthases which have redundant functions in conidia formation. Mol Gen Genet. 1997;253:520–528. doi: 10.1007/BF02172373. [DOI] [PubMed] [Google Scholar]
- 26.Culp DW, Dodge CL, Miao Y, Li L, Sag-Ozkal D, et al. The chsA gene from Aspergillus nidulans is necessary for maximal conidiation. FEMS Microbiol Lett. 2000;182:349–353. doi: 10.1111/j.1574-6968.2000.tb08920.x. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara M, Ichinomiya M, Motoyama T, Horiuchi H, Ohta A, et al. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J Biochem. 2000;127:359–366. doi: 10.1093/oxfordjournals.jbchem.a022616. [DOI] [PubMed] [Google Scholar]
- 28.Latge JP, Mouyna I, Tekaia F, Beauvais A, Debeaupuis JP, et al. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med Mycol. 2005;43:S15–22. doi: 10.1080/13693780400029155. [DOI] [PubMed] [Google Scholar]
- 29.Xoconostle-Cazares B, Leon-Ramirez C, Ruiz-Herrera J. Two chitin synthase genes from Ustilago maydis. Microbiol. 1996;142:377–387. doi: 10.1099/13500872-142-2-377. [DOI] [PubMed] [Google Scholar]
- 30.Garcera-Teruel A, Xoconostle-Cazares B, Rosas-Quijano R, Ortiz L, Leon-Ramirez C, et al. Loss of virulence in Ustilago maydis by Umchs6 gene disruption. Res Microbiol. 2004;155:87–97. doi: 10.1016/j.resmic.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Treitschke S, Doehlemann G, Schuster M, Steinberg G. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell. 2010;22:2476–2494. doi: 10.1105/tpc.110.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soulie MC, Piffeteau A, Choquer M, Boccara M, Vidal-Cros A. Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet Biol. 2003;40:38–46. doi: 10.1016/s1087-1845(03)00065-3. [DOI] [PubMed] [Google Scholar]
- 33.Soulie MC, Perino C, Piffeteau A, Choquer M, Malfatti P, et al. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol. 2006;8:1310–1321. doi: 10.1111/j.1462-5822.2006.00711.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim JE, Lee HJ, Lee J, Kim KW, Yun SH, et al. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr Genet. 2009;55:449–459. doi: 10.1007/s00294-009-0258-6. [DOI] [PubMed] [Google Scholar]
- 35.Larson TM, Kendra DF, Busman M, Brown DW. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr Genet. 2011;57:177–189. doi: 10.1007/s00294-011-0334-6. [DOI] [PubMed] [Google Scholar]
- 36.Werner S, Sugui JA, Steinberg G, Deising HB. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Microbe Interact. 2007;20:1555–1567. doi: 10.1094/MPMI-20-12-1555. [DOI] [PubMed] [Google Scholar]
- 37.Martin-Udiroz M, Madrid MP, Roncero MI. Role of chitin synthase genes in Fusarium oxysporum. Microbiol. 2004;150:3175–3187. doi: 10.1099/mic.0.27236-0. [DOI] [PubMed] [Google Scholar]
- 38.Madrid MP, Di Pietro A, Roncero MI. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol Microbiol. 2003;47:257–266. doi: 10.1046/j.1365-2958.2003.03299.x. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Urdiroz M, Roncero MI, Gonzalez-Reyes JA, Ruiz-Roldan C. ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot Cell. 2008;7:112–121. doi: 10.1128/EC.00347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 41.Odenbach D, Thines E, Anke H, Foster AJ. The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Mol Plant Pathol. 2009;10:81–94. doi: 10.1111/j.1364-3703.2008.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaime YP, Urdiroz MM, Roncero MI, Reyes JA, Roldan MC. Chitin synthase-deficient mutant of Fusarium oxysporum elicits tomato plant defence response and protects against wild-type infection. Mol Plant Pathol. 2010;11:479–493. doi: 10.1111/j.1364-3703.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 44.Mellado E, Aufauvre-Brown A, Gow NA, Holden DW. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol Microbiol. 1996;20:667–679. doi: 10.1046/j.1365-2958.1996.5571084.x. [DOI] [PubMed] [Google Scholar]
- 45.Munro CA, Schofield DA, Gooday GW, Gow NA. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiol. 1998;144:391–401. doi: 10.1099/00221287-144-2-391. [DOI] [PubMed] [Google Scholar]
- 46.Ichinomiya M, Horiuchi H, Ohta A. Different functions of the class I and class II chitin synthase genes, chsC and chsA, are revealed by repression of chsB expression in Aspergillus nidulans. Curr Genet. 2002;42:51–58. doi: 10.1007/s00294-002-0329-4. [DOI] [PubMed] [Google Scholar]
- 47.Ichinomiya M, Yamada E, Yamashita S, Ohta A, Horiuchi H. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot Cell. 2005;4:1125–1136. doi: 10.1128/EC.4.6.1125-1136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung US, Sobering AK, Romeo MJ, Levin DE. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol. 2002;46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 49.Mehrabi R, Ding S, Xu JR. MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell. 2008;7:791–799. doi: 10.1128/EC.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruiz-Herrera J, Gonzalez-Prieto JM, Ruiz-Medrano R. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 2002;1:247–256. doi: 10.1111/j.1567-1364.2002.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 51.Cope MJ, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain: implications for structure and function. Structure. 1996;4:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 52.Liu W, Zhou X, Li G, Li L, Kong L, et al. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 2011;7:e1001261. doi: 10.1371/journal.ppat.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Zhao X, Sun J, Kang Z, Ding S, et al. A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant Microbe Interact. 2010;23:112–123. doi: 10.1094/MPMI-23-1-0112. [DOI] [PubMed] [Google Scholar]
- 54.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 55.Peng YL, Shishiyama J. Temporal sequence of cytological events in rice leaves infected with Pyricularia oryzae. Can J Bot. 1991;66:730–735. [Google Scholar]
- 56.Park G, Xue C, Zhao X, Kim Y, Orbach M, et al. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen XL, Yang J, Peng YL. Large-scale insertional mutagenesis in Magnaporthe oryzae by Agrobacterium tumefaciens-mediated transformation. Methods Mol Biol. 2011;722:213–224. doi: 10.1007/978-1-61779-040-9_16. [DOI] [PubMed] [Google Scholar]
- 58.Guo M, Chen Y, Du Y, Dong Y, Guo W, et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011;7:e1001302. doi: 10.1371/journal.ppat.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Liu W, Hou Z, Wang C, Zhou X, et al. A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol Plant Microbe Interact. 2011;24:118–128. doi: 10.1094/MPMI-06-10-0129. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Xue C, Kong L, Li G, Xu JR. A Pmk1-interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae. Eukaryot Cell. 2011;10:1062–1070. doi: 10.1128/EC.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell. 2004;3:1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue C, Park G, Choi W, Zheng L, Dean RA, et al. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chi MH, Park SY, Kim S, Lee YH. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009;5:e1000401. doi: 10.1371/journal.ppat.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding SL, Liu W, Iliuk A, Ribot C, Vallet J, et al. The Tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell. 2010;22:2495–2508. doi: 10.1105/tpc.110.074302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramanujam R, Naqvi NI. PdeH, a high-affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae. PLoS Pathog. 2010;6:e1000897. doi: 10.1371/journal.ppat.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68.Lee JI, Yu YM, Rho YM, Park BC, Choi JH, et al. Differential expression of the chsE gene encoding a chitin synthase of Aspergillus nidulans in response to developmental status and growth conditions. FEMS Microbiol Lett. 2005;249:121–129. doi: 10.1016/j.femsle.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Roy SK, Chiba Y, Takeuchi M, Jigami Y. Characterization of Yeast Yea4p, a uridine diphosphate-N-acetylglucosamine transporter localized in the endoplasmic reticulum and required for chitin synthesis. J Biol Chem. 2000;275:13580–13587. doi: 10.1074/jbc.275.18.13580. [DOI] [PubMed] [Google Scholar]
- 70.Guerriero G, Avino M, Zhou Q, Fugelstad J, Clergeot PH, et al. Chitin synthases from Saprolegnia are involved in tip growth and represent a potential target for anti-oomycete drugs. PLoS Pathog. 2010;6:e1001070. doi: 10.1371/journal.ppat.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elson LA, Morgan WT. A colorimetric method for the determination of glucosamine and chondrosamine. Biochem J. 1933;27:1824–1828. doi: 10.1042/bj0271824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi WJ, Sburlati A, Cabib E. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc Natl Acad Sci U S A. 1994;91:4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim S, Park SY, Kim KS, Rho HS, Chi MH, et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009;5:e1000757. doi: 10.1371/journal.pgen.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu W, Xie S, Zhao X, Chen X, Zheng W, et al. A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2010;23:366–375. doi: 10.1094/MPMI-23-4-0366. [DOI] [PubMed] [Google Scholar]
- 75.Yi M, Chi MH, Khang CH, Park SY, Kang S, et al. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell. 2009;21:681–695. doi: 10.1105/tpc.107.055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Christensen HT, Zhang ZG, Wei YD, Collige DB. Subcellar localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- 77.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 78.Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol. 2009;69:337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- 79.Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci U S A. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talbot NJ, Kershaw MJ, Wakley GE, De Vries O, Wessels J, et al. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li G, Zhou X, Kong L, Wang Y, Zhang H, et al. MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS One. 2011;6:e19951. doi: 10.1371/journal.pone.0019951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang Z, Sell I, H B. Infection of wheat spikes by Fusarium avenaceum and alterations of cell wall components in the infected tissue. Eur J Plant Pathol. 2005;111:19–28. [Google Scholar]
- 83.Xu JR, Urban M, Sweigard JA, JE H. The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol Plant Microbe Interact. 1997;10:187–194. [Google Scholar]
- 84.Huang K, Czymmek KJ, Caplan JL, Sweigard JA, Donofrio NM. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 2011;7:e1001335. doi: 10.1371/journal.ppat.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of fungal chitin synthase. Phylogenetic tree was generated by the Clustalx1.83. The scale bar indicates 0.1 distance units. Species abbreviation are Af (Aspergillus fumigatus), An (Aspergillus nidulans), Bc (Botrytis cinerea), Cg (Colletotrichum graminicola), Fo (Fusarium oxysporum), Gz (Gibberella zeae), and Nc (Neurospora crassa).
(TIF)
CHS gene deletion construct and confirmation. (A) CHS1 gene deletion strategy and confirmation. CHS1 deletion construct (the upper) was created by replacing the CHS1 gene with hygromycin phosphotransferase cassette (hph). The upstream and downstream flanking sequences were amplified with primers 1P1/1P2 and 1P3/1P4, and ligated with the hph cassette. The positions of primers 1P1, 1P2, 1P3, 1P4, 1out, and PF are indicated with small arrows. E, EcoRI; EV, EcoRV; K, KpnI; S, SalI; Sp, SpeI. This construct was introduced into the wild-type strain P131. The putative mutants were screened with primers 1out/PF, and the putative mutants LA11 and LA40 had 1.9-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRV-digested genomic DNA of wild-type P131 and chs1 mutant LA11 and LA40 hybridized with Probe 1, which was amplified with primers 1P1/1P2. The results show a single 5.0-kb band (lane1) for the wild-type P131 and a single 6.0-kb band (lane 2 and lane 3) for chs1 mutant LA11 and LA40 (right bottom). (B) CHS2 gene deletion strategy and confirmation. CHS2 deletion construct (the upper) was created by replacing the CHS2 gene with hph. The upstream and downstream flanking sequences were amplified with primers 2P1/2P2 and 2P3/2P4, and ligated with the hph cassette. The positions of primers 2P1, 2P2, 2P3, 2P4, 2out, and PF are indicated with small arrows. B, BamHI; E, EcoRI; H, HindIII; K, KpnI. This construct was introduced into the wild-type S1528. The deletion mutants were screened with primers 2out/PF, and the putative mutants LA2 and LA6 had 2.3-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of KpnI-digested genomic DNA of wild-type strain S1528 and chs2 mutant LA2 and LA6 hybridized with Probe 2, which was amplified with primers 2P1/1P2. The results show a single 3.0-kb band (lane 1) for the wild-type S1528 and a single 6.1-kb band (lane 2 and lane 3) for chs2 mutant LA2 and LA6 (right bottom). (C) CHS3 gene deletion strategy and confirmation. CHS3 deletion construct (the upper) was created by introducing the hph within the CHS3 gene. The fragment within CHS3 gene was amplified with primers 3P1/3P2. The hph gene fragment was amplified with primers 3P3/3P4. The hph gene fragment was inserted into the fragment within CHS3 by SalI restriction enzyme. The positions of primers 3P1, 3P2, 3P3, 3P4, 3out, and PF are indicated with small arrows. B, BamHI; E, EcoRI; H, HindIII; S, SalI; X, XhoI. This construct was introduced into the wild-type P131. The deletion mutants were screened with primers 3out/PF, and the putative mutant LA1 had 2.5-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI and HindIII-digested genomic DNA of the wild-type P131 and chs3 mutant (LA1) hybridized with Probe 3, which was amplified with primers 3P3/3P4. The results show a single 6.1-kb band (lane 1) for the chs3 mutant LA1 and no band (lane 2) for wild-type strain P131 when using EcoRI-digesting the genomic DNA; a single 4.7-kb band (lane 3) for the chs3 mutant LA1 and no band (lane 4) for the wild-type P131 when using HindIII-digesting the genomic DNA (right bottom). (D) CHS4 gene deletion strategy and confirmation. CHS4 deletion construct (the upper) was created by replacing the CHS4 gene with hph. The upstream and downstream flanking sequences were amplified with primers4P1/4P2 and 4P3/4P4, and ligated with the hph cassette. The positions of primers 4P1, 4P2, 4P3, 4P4, 4out, and PF are indicated with small arrows. A, ApaI; E, EcoRI; S, SalI; Sp, SpeI. This construct was introduced into the wild-type P131. The deletion mutants were screened with primers 4out/PF, the putative mutants LA3 and LA28 had 2.1-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of SalI-digested genomic DNA of the wild-type P131 and chs4 mutants LA3 and LA28 hybridized with Probe 4, which was amplified with primers 4P1/4P2. The results show a single 3.5-kb band (lane 1) for the wild-type P131 and a single 4.4-kb band (lane 2 and lane 3) for chs4 mutants LA3 and LA28 (right bottom). (E) CHS5 gene deletion strategy and confirmation. CHS5 deletion construct (the upper) was created by replacing the CHS5 gene with hph. The upstream and downstream flanking sequences were amplified with primers 5P1/5P2 and 5P3/5P4, and ligated with the hph cassette. The positions of primers 5P1, 5P2, 5P3, 5P4, 5out, and PF are indicated with small arrows. B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; K, KpnI; S, SalI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 5out/PF, and the putative mutant (LA8) had 2.0-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of BglII and HindIII-digested genomic DNA of wild-type P131 and CHS5 deletion mutant LA8 hybridized with Probe 5, which was amplified with primers 5P3/5P4. The results show a single 6.2-kb band (lane 1) for the CHS5 deletion mutant LA8 and no band (lane 2) for the wild-type P131 when using BglII-digesting the genomic DNA; a single 2.2-kb band (lane 3) for the chs5 mutant (LA8) and no band (lane4) for the wild-type P131 when using HindIII-digesting the genomic DNA (right bottom). (F) CHS6 gene deletion strategy and confirmation. CHS6 deletion construct (the upper) was created by replacing the fragment within CHS6 gene with hph. The upstream and downstream flanking sequences were amplified with primers 6P1/6P2 and 6P3/6P4, and ligated with the hph cassette. The positions of primers 6P1, 6P2, 6P3, 6P4, 6out, and PF are indicated with small arrows. E, EcoRI; H, HindIII; K, KpnI; S, SpeI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 6out/PF, and the putative mutants LA14 and LA26 had 2.0-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI-digested genomic DNA of wild-type strain P131 and chs6 mutants LA14 and LA26 hybridized with Probe 6, which was amplified with primers 6P1/6P2. The results show a single 3.5-kb band (lane 3) for the wild-type P131 anda single 4.4-kb band (lane 1 and lane 2) for chs6 mutants LA14 and LA26 (right bottom). (G) CHS7 gene deletion strategy and confirmation. CHS7 deletion construct (the upper) was created by replacing fragment within CHS7 gene with hph. The upstream and downstream flanking sequences were amplified with primers 7P1/7P2 and 7P3/7P4, and ligated with the hph cassette. The positions of primers 7P1, 7P2, 7P3, 7P4, 7out, and PR are indicated with small arrows. E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SpeI. This construct was introduced into the wild-type P131. The deletion mutant was screened with primers 7out/PR, and the putative mutant LA12 had 1.7-kb specific band. 0.2-kb fragment within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of EcoRI and PstI-digested genomic DNA of the wild-type P131 and chs7 mutant LA12 hybridized with Probe 7, which was amplified with primers 7P3/7P4. The results show a single 6.2-kb band (lane 1) for the chs7 mutant LA12 and no band (lane 2) for wild-type P131 when using BglII-digesting the genomic DNA; a single 2.9-kb band (lane 3) for the chs7 mutant LA12 and no band (lane 4) for the wild-type P131 when using PstI-digesting the genomic DNA (right bottom). (H) CHS5 CHS6 gene deletion strategy and confirmation. The CHS5 CHS6 deletion construct was generated by replacing the N-terminal regions of both CHS5 and CHS6 with the hph cassette. The upstream and downstream flanking sequences were amplified with primers 8P1/8P2 and 8P3/8P4, and ligated with the hph cassette. B, BamHI; E, EcoRI; H, HindIII; K, KpnI. The putative double mutants LA49 and LA66 were identified by PCR with primers 8out/PF with a 1.8-kb specific band. A 0.2-kb band within MoACT1 gene was amplified as the endogenous reference (left bottom). Southern blot of PstI-digested genomic DNA of the wild-type P131 and chs5 chs6 double mutants LA49 and LA66 hybridized with Probe 8, which was amplified with primers 8P3/8P4. The results show a single 2.7-kb band for the wild-type P131 and a single 4.2-kb band for chs5 chs6 double mutants LA49 and LA66 (right bottom).
(TIF)
Hydrophobicity assay. Drops of 20 µl sterile distilled water were placed on the surface of vegetative colony from 7-day-old complement media agar cultures. The results show the surface of vegetative colony from the wild-type P131 and S1528, and seven CHS gene deletion mutants were hydrophobic.
(TIF)
Nuclear division and movement during appressorium formation in the chs1 mutant. Conidia of the wild type and chs1 mutant incubated on hydrophobic glass coverslips for 0, 1, 4, and 24 h were stained with CFW and DAPI and examined with an epifluorescence microscope. Bar = 10 mm.
(TIF)
The chs5 chs6 double mutant was more significantly reduced in growth rate than chs6 mutant. The wild-type P131, chs5, chs6, and chs5 chs6 double mutant were cultured on OTA plates under light at 25°C. Photographs were taken at 7 and 14 dpi, respectively. Bar = 10 mm.
(TIF)
qRT-assays with the expression levels of CHS genes in the wild-type P131 and chs5 chs6 mutants. The RNA was isolated from vegetative mycelia shaken in CM for two days. The actin gene was used as the endogenous control for normalization. Relative expression levels were estimated with the 2−ΔΔCt method. The expression level of each CHS gene in the wild type was arbitrarily set to 1. Mean and standard errors were determined with data from three independent replicates.
(TIF)
PCR primers used in this study.
(DOC)
The effects of cutin monomer treatment on appressorium formation by the wild type and chs7 mutant on different surfaces.
(DOC)
Defect of the chs 5 chs 6 mutants in vegetative growth.
(DOC)