Abstract
Many essential aspects of genome function, including gene expression and chromosome segregation, are mediated throughout development and differentiation by changes in the chromatin state. Along with genomic signals encoded in the DNA, epigenetic processes regulate heritable gene expression patterns. Genomic signals such as enhancers, silencers, and repetitive DNA, while required for the establishment of alternative chromatin states, have an unclear role in epigenetic processes that underlie the persistence of chromatin states throughout development. Here, we demonstrate in fission yeast that the maintenance and inheritance of ectopic heterochromatin domains are independent of the genomic sequences necessary for their de novo establishment. We find that both structural heterochromatin and gene silencing can be stably maintained over an ∼10-kb domain for up to hundreds of cell divisions in the absence of genomic sequences required for heterochromatin establishment, demonstrating the long-term persistence and stability of this chromatin state. The de novo heterochromatin, despite the absence of nucleation sequences, is also stably inherited through meiosis. Together, these studies provide evidence for chromatin-dependent, epigenetic control of gene silencing that is heritable, stable, and self-sustaining, even in the absence of the originating genomic signals.
THE establishment and maintenance of alternative chromatin states over the course of multiple cell divisions requires the complex integration of both genomic and nongenomic signals (reviewed in Straub and Becker 2008). Such signals work in concert throughout development to guide both cell specialization and adaptation to environmental changes in vivo (Blasco 2007; Feinberg 2007; Surani et al. 2007). Much of our current understanding of alternate patterns of gene expression comes from experiments performed in model organisms, including Drosophila melanogaster (reviewed in Pirrotta and Gross 2005; Girton and Johansen 2008), Saccharomyces cerevisiae (reviewed in Buhler and Gasser 2009), and Schizosaccharomyces pombe (reviewed in Grewal and Elgin 2002). These studies have demonstrated that repositioning of a euchromatic gene to a genomic location adjacent to transcriptionally silent heterochromatin results in variegated patterns of gene expression, a phenomenon called position-effect variegation. In addition to establishing a functional link between chromatin structure and gene expression state, these studies demonstrated that genetically identical cells can achieve alternate gene expression states that are stably maintained through cell division, thereby supporting an epigenetic mechanism of inheritance.
In different organisms, the maintenance of alternative structural and functional chromatin states is regulated in part by chromatin modifications that are both physically associated with and inherited with the chromosome on which they act, including DNA methylation, histone modifications and substitutions, nonhistone chromatin proteins, and noncoding RNAs (Bonasio et al. 2010). However, while specific sequences necessary for the nucleation of heterochromatin have been identified in various organisms, an ongoing role of such sequences in the inheritance of the heterochromatic state following DNA replication and cell division has been demonstrated in some circumstances and organisms, but not others. For example, maintenance of gene repression at silent loci in both budding yeast (Holmes and Broach 1996; Cheng and Gartenberg 2000) and Drosophila (Busturia et al. 1997; Sengupta et al. 2004)—organisms that, to date, appear to lack DNA methylation (Lyko et al. 2000; Schaefer et al. 2010)—requires the continued persistence of the genomic nucleating elements necessary for the establishment of the repressive chromatin state. In these examples, the inheritance of alternative chromatin states cannot be uncoupled from the DNA sequences that direct establishment (or reassembly) of chromatin structure following each successive cell division. In contrast, however, the genomic nucleating sequences that direct inactivation of the X chromosome in female mammals are dispensable for continued maintenance of the silent chromatin state throughout development (Brown and Willard 1994). In the absence of nucleation sequences, DNA methylation (in addition to other epigenetic marks) serves as a molecular signal that guides reestablishment of the repressive chromatin structure following cell division. Importantly, methyl groups can remain stably associated with DNA throughout replication (reviewed in Goll and Bestor 2005). Thus, once selected for inactivation, the silent chromatin state is self-sustaining, and the chromosome remains both transcriptionally repressed and architecturally condensed throughout subsequent mitoses.
In fission yeast, reporter genes placed within or adjacent to the native mating-type loci (Grewal and Klar 1996), centromeres (Allshire et al. 1994, 1995), and telomeres (Nimmo et al. 1994) are subject to position-effect variegation. Likewise, the repositioning of specific genomic heterochromatin nucleation sequences from a native locus to an ectopic euchromatic locus results in transcriptional silencing of adjacent genes (Ayoub et al. 2000; Partridge et al. 2002; Wheeler et al. 2009). Alternative chromatin states are inherited clonally through both mitosis and meiosis via an epigenetic and DNA methylation-independent mechanism (Wilkinson et al. 1995). Previous attempts to address the role of genomic nucleating sequences in the inheritance of alternative chromatin states in fission yeast have been complicated by the presence of multiple genomic sites that direct nucleation of transcriptionally silent chromatin (Grewal and Klar 1996; Hall et al. 2002) as well as parallel, redundant pathways for heterochromatin assembly at the native mating-type loci (Jia et al. 2004). Thus, it remains an open question whether the stability, maintenance, and transmission of the heterochromatic state in fission yeast occur through a mechanism that depends on the persistence of the nucleating sequence.
Materials and Methods
Plasmids
To construct the L5flox-ade6+ targeting plasmid, oligonucleotides containing the LoxP sequence were cloned in the same orientation into SpeI and ClaI/BglII sites flanking the L5 element (Partridge et al. 2002) in a plasmid, BW7, that contains the L5-ade6+ reporter construct flanked by ura4+ homology at both the 3′ and 5′ ends (Wheeler et al. 2009). The resulting plasmid, BW38, (L5flox-ade6+ with ura4+ homology at both the 5′ and 3′ ends), was sequenced to ensure that no errors had been introduced during cloning.
Fission yeast strain construction
The genotypes for strains used in this study are listed in Supporting Information, Table S1. Experimental strains were constructed by transforming plasmid BW38 into two loci, the endogenous ura4+ locus (strain KFY 501; Chr3) and at an ectopic ura4+ locus located upstream of the trp1+ gene (KFY1481 and KFY 1482; Chr2; construction described below) and selecting for growth on pombe glutamate medium (PMG) lacking adenine (Moreno et al. 1991). Appropriate integration of the construct at each experimental locus was confirmed phenotypically by growth on media containing 2 g/liter of 5-fluoroorotic acid (FOA) (MP Biomedicals), as well as by Southern blot. All other strains in this study were generated through standard genetic crosses with strains carrying the L5flox-ade6+ allele.
To construct strains KFY 1481 and KFY 1482, primers BWP262F/R and BWP263F/R were used to amplify trp1+ homology regions. Following purification, 75 ng of each PCR product was ligated with 50 ng of the 1.7-kb HindIII fragment containing the ura4+ gene. Following ligation, the resulting sample was amplified using HiFi reagents (Invitrogen) purified, sequenced, and used to transform strain KFY 450. Following selection for growth on PMG −uracil media, strains were restreaked to PMG −uracil media and then streaked onto media containing 2 g/liter of FOA. Integration at the appropriate locus was confirmed by Southern blot. Following two rounds of backcrossing, strains KFY 1481 and KFY 1482 were isolated.
Excision of L5 using Cre recombinase
To induce excision of L5, L5flox-ade6+ strains were transformed with the pREP41-Cre plasmid (a gift from K. Takegawa) (Iwaki and Takegawa 2004) via electroporation (1.5 kV, 200 Ω, 25 μF) on a BioRad Gene Pulser II. Unexcised control strains were electroporated in the absence of pREP41-Cre plasmid. To allow for excision to occur prior to plating, strains were grown for 24 hr in liquid PMG media in the absence of leucine (pREP41-Cre) or in complete PMG (no DNA control). pREP41-Cre transformants were then plated twice on PMG −leucine and PMG complete plates, respectively. Control strains were plated directly onto PMG 1/10th adenine plates. Excision of L5 was confirmed in pREP41-Cre transformants using a PCR strategy (BWP33F, BWP33R, and BWP246F), followed by Southern blot analysis. Strains in which L5 was excised, L5ex-ade6+, were streaked onto PMG complete plates to allow for loss of the pREP41-Cre plasmid. Finally, individual colonies from which the pREP41-Cre plasmid was lost were streaked onto PMG 1/10th adenine. Phenotypically red colonies observed at this stage are referred to in the text as generation zero (Gen0) colonies.
Time course of ade6+ expression
All liquid cultures were grown in rich yeast extract with supplements (YES) media (Moreno et al. 1991), unless otherwise indicated. ade6+ and ade6− strains grow equivalently in rich media (Figure S1). For resolving ade6+ expression phenotypes, cells were plated on PMG 1/10th adenine plates and incubated at 32° for 4 days, shifted to 4° for one night, and then counted under a Leica MZ7.5 microscope. Evidence of silencing included phenotypically red, pink, and variegated colonies.
YES cultures were inoculated with a single colony of appropriate genotype and ade6+ phenotype, isolated from a Gen0 PMG 1/10th adenine plate. Subsequent time points derive from propagation of a single clone. Cultures were allowed to double for ∼10 generations, as determined by counting cell density with a hemocytometer. When appropriate density was achieved, a subset of the culture was plated on PMG 1/10th adenine plates. These plates were then used to calculate the proportion of colonies that exhibited silencing. In addition to plating, 2000 cells from the culture were used to inoculate a new culture that was allowed to double for ∼10 generations before plating. Using this scheme, cells were maintained in logarithmic growth throughout the time course. Figure 1C illustrates data from a single expressed clone and a single silenced clone. Figure S3 contains data from several independently derived strains of each phenotype (biological replicates). Figure 2 contains data from several independently derived strains following L5 excision. Data were graphed and mitotic stability was calculated and fit to a nonlinear curve using GraphPad Prism software.
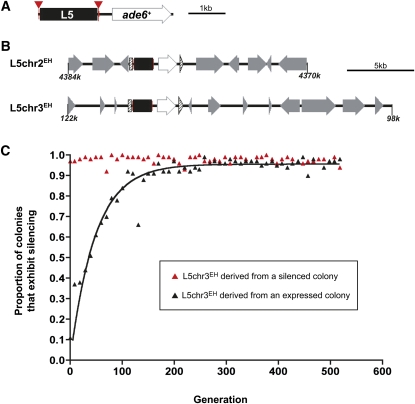
Figure 1 .
Heterochromatic silencing within a de novo heterochromatin domain exhibits parental state bias. (A) Schematic of the reporter gene construct. L5 (black box) is flanked by LoxP sites (inverted red triangles) upstream of the ade6+ reporter gene. (B) Experimental loci L5chr2EH and L5chr3EH; loci coordinates correspond to the S. pombe genome browser (http://old.genedb.org/gbrowse-bin/gbrowse/S.pombe/). The reporter gene construct was integrated at the ura4+ locus (hatched arrow), resulting in interruption of the gene into 5′ and 3′ fragments. Surrounding genes are designated by gray arrows in the direction of transcription. More detailed information about the experimental loci is provided in Figure S2. (C) The proportion of the culture, derived from a single colony, which exhibited silencing as determined by counting the number of colonies that had any phenotypic evidence of silencing. The red-derived culture is shown as red triangles and the white-derived culture is shown as black triangles with the corresponding exponential association curve y = ymax(1 − exp(−0.02 × x )), R2 = 0.9245. Additional time-course profiles for independently isolated colonies are shown in Figure S3).
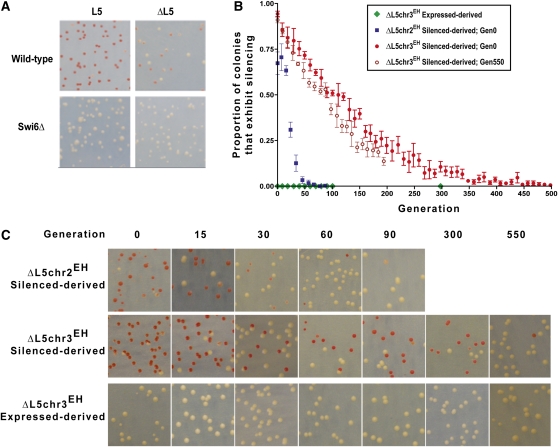
Figure 2 .
Heterochromatin is maintained and inherited in the absence of L5. (A) Wild-type and swi6Δ strains before and after transformation with the Cre plasmid. (B) The proportion of the culture that exhibited silencing as determined by counting the number of colonies that had any phenotypic evidence of silencing. The expressed-derived ΔL5chr3EH G0 culture is shown as green circles; the silenced-derived ΔL5chr2EH G0 culture is shown as blue squares; the silenced-derived ΔL5chr3EH G0 culture as red circles and silenced-derived ΔL5chr3EH G550 culture as dark red open circles. Error bars represent the SEM among at least three biological replicates. (C) Representative phenotypic images of the indicated cultures.
Chromatin immunoprecipitation
H3K9me2 ChIP was performed using a protocol modified from Wheeler et al. (2009). Single phenotypically red or white colonies from at least three independently derived biological replicates were selected and grown overnight to a density of between 2.0 × 106 and 7.0 × 106 cells/ml. Approximately 2.5 × 108 cells were fixed for 15 min in 1% paraformaldehyde. Cells were lysed by two rounds of bead beating for 30 sec in lysis buffer containing protease inhibitors. Chromatin was next sheared to an average DNA fragment size of 600 bp, precleared, and divided into input and IP samples. The immunoprecipitated (IP) sample was incubated overnight with 5 μl of anti-H3K9me2 antibody (Active Motif, 39239). Protein-A beads were added to the IP samples and incubated for 2 hr at 4° before washing. DNA was isolated from IP and input samples and the enrichment relative to an act1+ control locus was quantified using real-time PCR. Real-time PCR was performed in the presence of SYBR Green on a Bio-Rad iCycler. A standard curve was generated using DNA isolated from the appropriate parental strain. Standard curves had an R2 value of at least 0.990 and a PCR efficiency between 90 and 110%. Data were analyzed using iCycler iQ System software as the ratio of query locus/act1+ for IP relative to input samples and graphed using GraphPad Prism. All primers are listed in Table S2.
Meiotic crosses
Phenotypically red colonies from opposite mating types were crossed on malt extract plates and phenotypes analyzed by random spore analysis. At least three independent crosses were preformed among three biological replicate strains. Approximately 600 viable progeny from each cross were analyzed.
Four different crosses were performed between biological replicates for tetrad analysis. A total of 43 tetrads (172 spores) were analyzed.
Results
To address whether fission yeast can support heritable gene expression states in the absence of genomic nucleating sequences, we have capitalized on a previously described ectopic silencing assay in fission yeast (Wheeler et al. 2009). De novo domains of heterochromatin can be nucleated by a single copy of the L5 repetitive element (Figure 1A), a 1.6-kb AT-rich sequence isolated from the fission yeast centromeric repeat and present in multiple copies at each of the S. pombe endogenous centromeres (Partridge et al. 2002). Ectopic heterochromatin domains are characterized by transcriptional repression of surrounding genes and by the accumulation of heterochromatin markers, including H3K9me2-modified nucleosomes and the HP1 homolog, Swi6 (Partridge et al. 2002; Wheeler et al. 2009).
Genomic and epigenetic signals regulate inheritance of alternative gene expression states at ectopic loci
The L5 element was integrated at two genomic loci (Figure 1B and Figure S2); both loci, referred to throughout as L5chr2EH (chromosome 2 at the trp1+ locus; EH, ectopic heterochromatin) and L5chr3EH (chromosome 3 at the ura4+ locus) have previously been described as genomic locations that support the de novo formation of heterochromatin (Iida et al. 2008; Wheeler et al. 2009). At both L5chr2EH and L5chr3EH, the assembly of heterochromatin can be detected phenotypically; adjacent to the ectopic L5 element is an ade6+ reporter gene, the repression of which can be visualized as the presence of red pigment within a colony (Allshire et al. 1994), due to the accumulation of a red byproduct in the adenine biosynthetic pathway. Thus, L5-mediated silencing is monitored as a phenotypic change from white colonies to red colonies and loss of silencing as a change from red to white. L5-mediated silencing is subject to position-effect variegation, and colonies with expressed (white), silenced (red), and intermediate (pink or sectored) phenotypes are observed.
Alternative expression states resulting from position-effect variegation in fission yeast are mitotically metastable (Allshire et al. 1994). When the experimental reporter gene is positioned at the mating-type locus, <2% of cells change phenotype following <50 generations of growth on nonselective media (Grewal and Klar 1996). To characterize the stability of the expressed and silenced states associated with ectopic de novo heterochromatin domains further, single colonies were selected, and the phenotypes of the resulting mitotic progeny were analyzed at regular intervals over a period of ∼600 cell divisions (∼2 months) in nonselective media. At both ectopic loci, the parental expression state was maintained in the progeny during the initial generations of growth (<25 doublings); that is, a culture derived from a silenced colony remained nearly entirely silenced, while a culture derived from an expressing colony continued to express the ade6+ reporter gene (Figure 1C and Figure S3). Thus, consistent with earlier observations (Allshire et al. 1994; Grewal and Klar 1996), the parental state can influence the transcriptional state of the progeny through an epigenetic mechanism. Following additional rounds of cell division, the silenced culture remains stably silenced, with an average of 98% of the colonies exhibiting silencing throughout the duration of the time course, extending by >10-fold the observation of epigenetic persistence of a heterochromatic state (Grewal and Klar 1996). Most of the remaining colonies exhibited a variegated phenotype, indicative of a phenotypic switch between expressed and silent chromatin states as a single cell develops into a colony. The dynamic, reversible position effects observed here reflect the overlapping and competing activities between genome-directed activities (transcriptional activation of ade6+ vs. (re)establishment of heterochromatin via the L5 sequence) and epigenetic inheritance pathways (Cheutin et al. 2003, 2004).
In contrast, the expressed, active gene pattern was unstable over time. Approximately 2% of cells with an expressed gene pattern convert to the silent state following each cell division (half-life of the expressed state is 33 generations; Figure 1C and Figure S3). The instability observed over an extended period of 600 cell divisions reflects the ongoing influence of the L5 genomic nucleating element and its role in reestablishment of heterochromatin following each cell cycle. Thus, even when derived from a colony expressing ade6+, the genomic influence of the L5 element to direct heterochromatin assembly is favored in some cells over the epigenetic maintenance of the expressed state.
Gene silencing at de novo heterochromatin domains is maintained through cell division in the absence of the L5 nucleating element
Both epigenetic and genomic processes underlie the establishment and maintenance of transcriptionally active states within two de novo heterochromatin domains. To determine whether these linked processes can be experimentally uncoupled from one another at L5 ectopic loci, we engineered strains in which the L5 could be deleted in vivo following the establishment of the de novo heterochromatin domain (Holmes and Broach 1996; Cheng and Gartenberg 2000). The L5 element was flanked by LoxP site-specific recombination sites (Iwaki and Takegawa 2004), and transient expression of Cre recombinase in these strains resulted in the efficient excision of L5 (Figure S4). Following the removal of L5 (ΔL5), cultures derived from colonies expressing ade6+ retained the expressing state in 100% of cells (Figure 2, B and C). Thus, in contrast to colonies expressing ade6+ in the presence of L5, strains lacking L5 failed to reestablish silencing, even after extended cell divisions (Figure 2A). These strains also lack detectable levels of the heterochromatic histone modification, H3K9me2, at either ΔL5chr2EH or ΔL5chr3EH (Figure S5). Therefore, once silencing is lost, it can only be reestablished in the presence of the L5 genomic nucleating element, and gene expression is no longer subject to epigenetic silencing.
In contrast, however, at both ΔL5chr2EH and ΔL5chr3EH, cultures derived from cells repressing the ade6+ reporter gene initially maintained silencing despite removal of the L5 heterochromatin-nucleating sequence (Figure 2, B and C; Figure S6 and File S1). Maintenance of the transcriptionally silent state was dependent on the presence of a functional heterochromatin pathway (Figure 2A). Together, these data demonstrate that once established (a step that is, as shown above, dependent on the presence of the heterochromatin nucleating sequence), the maintenance of the transcriptionally silent state can be uncoupled from the genomic heterochromatin nucleating sequence. Thus, in the absence of the genomic signal that directs heterochromatin (re)assembly, epigenetic signals are sufficient to maintain the transcriptionally silent chromatin state at two distinct ectopic loci in fission yeast.
Notably, the proportion of transcriptionally silent colonies decreases throughout the time course, and differences in the stability of heterochromatin are observed between the two ectopic loci tested here (Figure 2, B and C). At ΔL5chr2EH, the silenced phenotype had a half-life of 18 generations, whereas at ΔL5chr3EH, the switch between epigenetic states occurs more slowly, with a half-life of ∼100 generations, reflecting an estimated loss rate of 0.7% per generation. Accompanying this loss, ade6+ transcript levels increased over time (Figure S5A) and enrichment of H3K9me2 heterochromatin mark at the ade6+ locus in ΔL5chr3EH strains decreased with time (Figure S5B). Thus, while the maintenance of the heterochromatic state through ∼35 cell divisions is independent of genomic nucleating sequences at both ectopic loci, the epigenetic stability of the domain varies between loci (Wheeler et al. 2009 and see Discussion).
To ascertain whether the stability of the epigenetically determined state at ΔL5chr3EH might change during the extended time course, we compared the stability of transcriptional silencing in colonies derived from ΔL5chr3EH at generation zero (Gen0; following confirmation of excision, see Materials and Methods) to the stability of Gen550 colonies by which time silencing is relatively rare. Over the course of 200 generations, the mitotic stability of heterochromatin in cells derived from generation 550 was largely unchanged, relative to cells derived from earlier in the time course (half-life of ∼85 generations) (Figure 2B). Thus, regardless of when in the time course a silent colony is selected, the mitotic stability of the transcriptionally silent state remains constant.
Structural heterochromatin is maintained at ectopic loci in the absence of the L5 nucleating element
In addition to silencing genes adjacent to the ade6+ reporter gene, the L5 element recruits structural heterochromatin components, which spread into neighboring endogenous sequences, creating a de novo heterochromatin domain (Wheeler et al. 2009). To further explore the chromatin structure associated with the ectopic heterochromatin domains established at L5chr2EH and L5chr3EH loci, enrichment levels of H3K9me2 throughout the domains were determined by chromatin immunoprecipitation. Importantly, both genomic insertion sites lack detectable heterochromatin marks in a wild-type strain, prior to the insertion of L5 (Cam et al. 2005). In the presence of the L5 element, adjacent sequences are highly enriched in H3K9me2. Thus, together with the observation that heterochromatin shows comparable mitotic stability at L5chr2EH and L5chr3EH, the similar chromatin structure near L5 suggests that establishment of de novo heterochromatin domains is similar at both loci. In contrast, local genomic features likely influence H3K9me2 enrichment at the boundaries of the domains (spanning ∼9 kb at L5chr2EH and 12 kb at L5chr3EH) that result from the bidirectional spread of heterochromatin from the nucleating L5 element (Wheeler et al. 2009 and Discussion).
To determine whether maintenance of the ade6+ transcriptionally silent state correlates with the continued presence of structural heterochromatin following excision of L5, cultures were analyzed for enrichment of H3K9me2 throughout the de novo domain at Gen0. Importantly, both ΔL5chr2EH and ΔL5chr3EH de novo domains remained enriched in H3K9me2 with similar levels of enrichment, though the levels are moderately reduced relative to the parental, L5-containing strains (Figure 3A). Thus, while the initial establishment of heterochromatin is dependent on a nucleating element, the ongoing maintenance of structural heterochromatin, as well as the persistence of the gene expression state, is independent of the nucleating sequence and is not a peculiarity of the local genomic environment. As expected, no detectable H3K9me2 enrichment was present at ectopic loci in cultures derived from colonies that had lost silencing at the ade6+ reporter gene at Gen0 (Figure S7), confirming that once the transcriptionally silent chromatin state converts to a transcriptionally expressed chromatin state, the silent state can only be reestablished in the presence of the genomic L5 nucleating element.
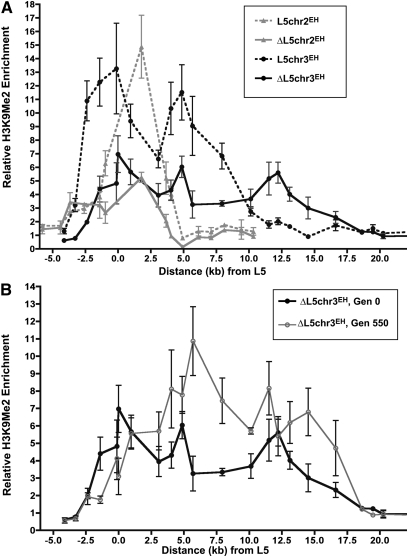
Figure 3 .
Heterochromatin is maintained throughout the de novo heterochromatin domain. (A) The relative enrichment of H3K9me2 throughout the de novo heterochromatin domains relative to the distance from the L5 element (centered at 0) or the remaining LoxP site following L5 excision (centered at 0). Error bars represent the SEM among biological replicates. (B) The level of H3K9me2 enrichment in ΔL5chr3EH cultures at Gen0 and Gen550.
Interestingly, distal to the site of L5 excision at the ΔL5chr3EH locus, the level of H3K9me2 enrichment actually exceeds the level of enrichment in L5-containing strains. This unexpected pattern of heterochromatin distribution is even more readily apparent over time, as assessed by the distribution of H3K9me2 in the progeny of transcriptionally silenced ΔL5chr3EH colonies selected at Gen550 (Figure 3B). Despite similar rates of stability between these two cultures (Figure 2B), the relative enrichment of H3K9me2 in cultures derived from colonies at Gen550 is increased at least twofold over levels seen in colonies derived from Gen0. Thus, while H3K9me2 marks are redistributed over time, redistribution has no evident effect on the stability of the chromatin domain.
Heterochromatin is maintained through meiosis in the absence of L5
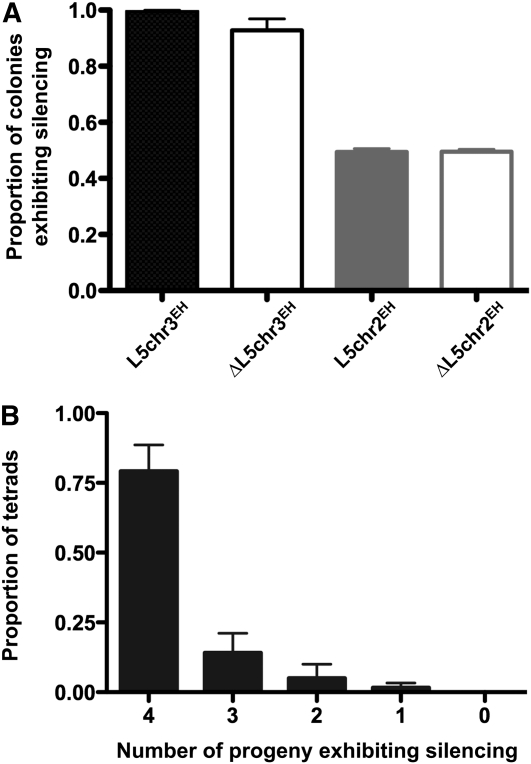
Our data demonstrate uncoupling of genomic and epigenetic signals needed for the establishment and maintenance of chromatin states in mitosis. However, the extent to which and the manner by which heterochromatin can be inherited through meiosis by an epigenetic pathway has only been examined in the presence of genomic nucleating elements (Grewal and Klar 1996). To determine whether the transcriptionally silenced state can be inherited through meiosis in the absence of L5, two strains, each carrying a phenotypically silenced locus, were mated, followed by random spore analysis of progeny. Remarkably, 49.6% of ΔL5chr2EH progeny and 94.2% of ΔL5chr3EH progeny exhibited silencing after meiosis (Figure 4A). To determine whether the loss of the transcriptionally silent state among meiotic products was random or nonrandom, phenotypes were observed for each of the four meiotic products from independent asci derived from crosses of ΔL5chr3EH strains. Figure 4B illustrates that the transcriptional status of the ade6+ gene in a given progeny is independent of the remaining three meiotic products and suggests that the loss of silencing occurs randomly during meiosis. Thus, these data point to a nucleation-sequence–independent epigenetic pathway of inheritance of transcriptional states through meiosis.
Figure 4 .
The silenced phenotype is heritable through meiosis. (A) The number of meiotic progeny that exhibit silencing per meiosis was determined for the indicated crosses. (B) The number of progeny that exhibited silencing following meiosis was scored.
Discussion
We demonstrate a nucleation-sequence–independent pathway of epigenetic inheritance of chromatin states in fission yeast, as measured by both gene expression and presence of chromatin modifications associated with transcriptionally silent heterochromatin. Although evidence for epigenetic inheritance in fission yeast has been well documented previously (Allshire et al. 1994; Grewal and Klar 1996; reviewed in Grewal and Jia 2007), this study resolves prior uncertainty about the role of nucleation sequences in the epigenetic inheritance and persistence of alternative chromatin states in this organism. In sharp contrast to epigenetic inheritance pathways in both budding yeast (Holmes and Broach 1996; Cheng and Gartenberg 2000) and flies (Busturia et al. 1997; Sengupta et al. 2004), maintenance and inheritance of chromatin states in fission yeast may be uncoupled from the genomic DNA sequences that direct initial establishment of heterochromatin following cell division. The nucleation-sequence–independent de novo heterochromatin domain is self-sustaining through as many as 600 cell divisions (Figures 2 and 3), as well as through meiosis (Figure 4), distinguishing it from previous studies that demonstrated only transient (Gullerova and Proudfoot 2008) and/or short-lived (Allshire et al. 1994; Grewal and Klar 1996) maintenance of chromatin states associated with gene silencing. Similar nucleation-sequence–independent, or DNA binding-component–independent inheritance of epigenetic chromatin states has also been observed in maize (Chandler 2007) and mammalian cell culture (Brown and Willard 1994; Ayyanathan et al. 2003). These studies extend current views of the inheritance of chromatin states and provide a framework (Figure 5) for exploring the intricate relationship among genetics, epigenetics, and genome function in the context of heterochromatin.
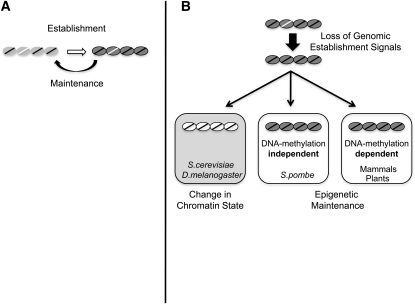
Figure 5 .
Model of the transmission of chromatin states. (A) Chromatin states are established (open arrow) by DNA sequences present in the genome (white line), which direct assembly of transcriptionally silent chromatin. Examples include the E and I silencers in S. cerevisiae (Holmes and Broach 1996; Cheng and Gartenberg 2000), polycomb response elements in D. melanogaster (Busturia et al. 1997; Sengupta et al. 2004), the L5 element described here, and the XIC in mammals (Brown and Willard 1994). The chromatin state is maintained through cell division by both epigenetic processes and sequence-dependent reassembly. (B) Following removal of genomic sequences that direct establishment, the silent chromatin state is lost in both S. cerevisiae and D. melanogaster. In these organisms, available evidence suggests that genomic elements are required at each cell division to reestablish repressive chromatin states (Holmes and Broach 1996; Busturia et al. 1997; Cheng and Gartenberg 2000; Sengupta et al. 2004). In contrast, despite the lack of genomic nucleation sequences, the chromatin state is heritable through cell division in both fission yeast and mammals. Mammals utilize DNA methylation as a molecular signal to faithfully transmit repressive chromatin states throughout development (Feng et al. 2006), whereas epigenetic information is passed through cell division in fission yeast in a DNA methylation-independent pathway (Wilkinson et al. 1995). These findings have important implications for additional mechanisms that can mediate inheritance of chromatin states.
While persistence of the chromatin state is independent of the nucleating genomic element, features of the genomic locus do, nonetheless, influence both the level of enrichment of H3K9me2 throughout the larger de novo domain, as well as the endpoints of the de novo domains (Figure 3) (Wheeler et al. 2009). In addition, although the maintenance of the heterochromatic state through cell division is independent of nucleating sequences at experimental loci, the dynamics of maintenance (and thus the preservation of the domain) varies between the two genomic loci. Importantly, both loci lack detectable H3K9me2 and Swi6p in their native state prior to L5 insertion (Cam et al. 2005); thus it is unlikely that the stability observed at ΔL5chr3EH is a consequence of chromatin marks present at the ectopic insertion site. Further, the ectopic heterochromatin is easily detected in asynchronous cells; it is therefore distinct from previously described examples of transient heterochromatin that assemble only during G1-S (Gullerova and Proudfoot 2008; Gullerova et al. 2011). Intriguingly, at L5chr3 EH the recently identified tam14+ gene (Rhind et al. 2011) is convergent with ura4+ and may contribute to the stability of the de novo domain at this locus (Gullerova and Proudfoot 2008). Moreover, a Tf2 retrotransposon, as well as two LTR fragments, are present near the L5chr3EH insertion site (Figure S2). Although these loci are not enriched in H3K9Me or Swi6 in a wild-type cell (Cam et al. 2005), histone deacetylases Clr3 and Clr6 are present at Tf2 elements (Cam et al. 2008) and may therefore provide a chromatin environment or serve as a protosilencer (Fourel et al. 2002) that may enhance the stability of de novo domains of heterochromatin. Stability may also occur through an H3K9me-independent pathway (Hansen et al. 2011). This is unlikely, however, because the transcriptionally silenced state is abrogated upon crossing strains into clr4− or ago1− background (Wheeler et al. 2009) (Table S3). These observations invite approaches to identify different sequence elements that can sustain (at ΔL5chr3EH) or antagonize (at ΔL5chr2EH) inheritance of heterochromatin genome-wide and to thus provide further insight into the nature of genomic code(s) that underlie different chromatin states (Wheeler et al. 2009).
Epigenetic principles underlie many fundamental and/or pathological processes in biology, such as maintenance of genome integrity (Ekwall 2007; Martienssen et al. 2008), cellular differentiation (Bartova et al. 2008), stem cell pluripotency (Surani et al. 2007), tumorigenesis (Mathews et al. 2009; Nise et al. 2009), and regeneration and aging (Blasco 2007; Vaiserman 2008). Despite an appreciation of the various histone and DNA modification patterns associated with the epigenetic regulation of gene expression, a detailed understanding of the molecular processes that underlie the transfer of epigenetic information through cell division remains limited to DNA methylation (Goll and Bestor 2005; Feng et al. 2006). Propagation mechanisms have been proposed for histone H3 modifications associated with transcriptionally silent chromatin (Kouzarides 2007; Probst et al. 2009), although experimental evidence is lacking. The ability to experimentally uncouple the genomic and epigenetic influences on the inheritance of chromatin states will permit further elucidation of the underlying genetic and molecular mechanisms that act on the chromatin fiber, studies that can be initially approached in this system using genetic methods. These alternative pathways may act in concert with DNA methylation and genomic nucleating sequences in multicellular eukaryotes to generate stable chromatin states that are also reversible or may regulate biological phenomena that result, at least in part, from nucleation-independent epigenetic events (Figure 5). Furthermore, alternative pathways may be critical carriers of epigenetic information following fertilization, when most parental methylation patterns are erased (Morgan et al. 2005).
Acknowledgments
We thank K. Takegawa for the gift of the Cre expression plasmid; Terilyn Gaither for technical assistance; and Beth Sullivan, Laura Rusche, and members of the Scott, Willard, and Rusche laboratories for critical discussion.
Footnotes
Communicating editor: F. Winston
Literature Cited
- Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G., 1994. Position effect variegation at fission yeast centromeres. Cell 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G., 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Ayoub N., Goldshmidt I., Lyakhovetsky R., Cohen A., 2000. A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics 156: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M. S., Bell P., Maul G. G., Schultz D. C., et al. , 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17: 1855–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartova E., Galiova G., Krejci J., Harnicarova A., Strasak L., et al. , 2008. Epigenome and chromatin structure in human embryonic stem cells undergoing differentiation. Dev. Dyn. 237: 3690–3702 [DOI] [PubMed] [Google Scholar]
- Blasco M. A., 2007. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D., 2010. Molecular signals of epigenetic states. Science 330: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. J., Willard H. F., 1994. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368: 154–156 [DOI] [PubMed] [Google Scholar]
- Buhler M., Gasser S. M., 2009. Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 28: 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A., Wightman C. D., Sakonju S., 1997. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 124: 4343–4350 [DOI] [PubMed] [Google Scholar]
- Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., et al. , 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Cam H. P., Noma K., Ebina H., Levin H. L., Grewal S. I., 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436 [DOI] [PubMed] [Google Scholar]
- Chandler V. L., 2007. Paramutation: from maize to mice. Cell 128: 641–645 [DOI] [PubMed] [Google Scholar]
- Cheng T. H., Gartenberg M. R., 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14: 452–463 [PMC free article] [PubMed] [Google Scholar]
- Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., et al. , 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Cheutin T., Gorski S. A., May K. M., Singh P. B., Misteli T., 2004. In vivo dynamics of Swi6 in yeast: evidence for a stochastic model of heterochromatin. Mol. Cell. Biol. 24: 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., 2007. Epigenetic control of centromere behavior. Annu. Rev. Genet. 41: 63–81 [DOI] [PubMed] [Google Scholar]
- Feinberg A., 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433–440 [DOI] [PubMed] [Google Scholar]
- Feng Y. Q., Desprat R., Fu H., Olivier E., Lin C. M., et al. , 2006. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G., Lebrun E., Gilson E., 2002. Protosilencers as building blocks for heterochromatin. Bioessays 24: 828–835 [DOI] [PubMed] [Google Scholar]
- Girton J. R., Johansen K. M., 2008. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv. Genet. 61: 1–43 [DOI] [PubMed] [Google Scholar]
- Goll M. G., Bestor T. H., 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Klar A. J., 1996. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86: 95–101 [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Elgin S. C., 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12: 178–187 [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gullerova M., Proudfoot N. J., 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Gullerova M., Moazed D., Proudfoot N. J., 2011. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 25: 556–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., et al. , 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Hansen K. R., Hazan I., Shanker S., Watt S., Verhein-Hansen J., et al. , 2011. H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases. PLoS Genet. 7: e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S. G., Broach J. R., 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Iida T., Nakayama J., Moazed D., 2008. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 31: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T., Takegawa K., 2004. A set of loxP marker cassettes for Cre-mediated multiple gene disruption in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68: 545–550 [DOI] [PubMed] [Google Scholar]
- Jia S., Noma K., Grewal S. I., 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Kouzarides T., 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lyko F., Ramsahoye B. H., Jaenisch R., 2000. DNA methylation in Drosophila melanogaster. Nature 408: 538–540 [DOI] [PubMed] [Google Scholar]
- Martienssen R. A., Kloc A., Slotkin R. K., Tanurdzic M., 2008. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb. Symp. Quant. Biol. 73: 265–271 [DOI] [PubMed] [Google Scholar]
- Mathews L. A., Crea F., Farrar W. L., 2009. Epigenetic gene regulation in stem cells and correlation to cancer. Differentiation 78: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–832 [DOI] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W., Reik W., 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14(Spec No 1): R47–R58 [DOI] [PubMed] [Google Scholar]
- Nimmo E. R., Cranston G., Allshire R. C., 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 13: 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nise, M. S., P. Falaturi and T. C. Erren, 2009 Epigenetics: origins and implications for cancer epidemiology. Med Hypotheses 74: 377–382. [DOI] [PubMed]
- Partridge J. F., Scott K. S., Bannister A. J., Kouzarides T., Allshire R. C., 2002. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Gross D. S., 2005. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell 18: 395–398 [DOI] [PubMed] [Google Scholar]
- Probst A. V., Dunleavy E., Almouzni G., 2009. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Rhind N., Chen Z., Yassour M., Thompson D. A., Haas B. J., et al. , 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., et al. , 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A. K., Kuhrs A., Müller J., 2004. General transcriptional silencing by a Polycomb response element in Drosophila. Development 131: 1959–1965 [DOI] [PubMed] [Google Scholar]
- Straub T., Becker P. B., 2008. DNA sequence and the organization of chromosomal domains. Curr. Opin. Genet. Dev. 18: 175–180 [DOI] [PubMed] [Google Scholar]
- Surani M. A., Hayashi K., Hajkova P., 2007. Genetic and epigenetic regulators of pluripotency. Cell 128: 747–762 [DOI] [PubMed] [Google Scholar]
- Vaiserman A. M., 2008. Epigenetic engineering and its possible role in anti-aging intervention. Rejuvenation Res. 11: 39–42 [DOI] [PubMed] [Google Scholar]
- Wheeler B. S., Blau J. A., Willard H., Scott K. C., 2009. The impact of local genome sequence on defining heterochromatin domains. PLoS Genet. 5: e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. R., Bartlett R., Nurse P., Bird A. P., 1995. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 23: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]