Abstract
Extensive mutagenesis via massive recoding of retrotransposon Ty1 produced a synthetic codon-optimized retrotransposon (CO-Ty1). CO-Ty1 is defective for retrotransposition, suggesting a sequence capable of down-regulating retrotransposition. We mapped this sequence to a critical ∼20-bp region within CO-Ty1 reverse transcriptase (RT) and confirmed that it reduced Ty1 transposition, protein, and RNA levels. Repression was not Ty1 specific; when introduced immediately downstream of the green fluorescent protein (GFP) stop codon, GFP expression was similarly reduced. Rap1p mediated this down-regulation, as shown by mutagenesis and chromatin immunoprecipitation. A regular threefold drop is observed in different contexts, suggesting utility for synthetic circuits. A large reduction of RNAP II occupancy on the CO-Ty1 construct was observed 3′ to the identified Rap1p site and a novel 3′ truncated RNA species was observed. We propose a novel mechanism of transcriptional regulation by Rap1p whereby it serves as a transcriptional roadblock when bound to transcription unit sequences.
SACCHAROMYCES cerevisiae Ty1 is the best-characterized and most abundant of five retrotransposon families present in the genome. Ty1 shares many similarities with retroviruses, such as HIV-1 and makes similar use of an RNA intermediate in its propagation. Unsurprisingly, this Ty1 RNA intermediate is essential for Ty1 retrotransposition. Not only does this “mRNA” code for the GAG and POL proteins required for retrotransposition, but it also serves a critical function as the genetic material for replication. Both Ty1 mRNA and its cDNA product contain cis-acting nucleotide determinants required for retrotransposition.
Mutagenesis of Ty1 and screening for cis-acting nucleotide determinants has been previously performed with various strategies, including in-frame linker insertion mutagenesis (Braiterman et al. 1994; Devine and Boeke 1994; Monokian et al. 1994), analysis of synthetic Ty1 DNA fragments to determine minimal requirements for the integration reaction (Eichinger and Boeke 1990; Braiterman et al. 1994; Devine and Boeke 1994; Sharon et al. 1994), deletion analysis and PCR-mediated mutagenesis of mini-Ty1 elements (Xu and Boeke 1990; Bolton et al. 2005), and comparative sequence analysis followed by targeted mutagenesis and/or generation of complementary mutations in RNA stem regions (Chapman et al. 1992; Heyman et al. 1995; Lauermann et al. 1995; Friant et al. 1998; Cristofari et al. 2002; Bolton et al. 2005). These studies provided insights into mechanisms underlying Ty1 retrotransposition and revealed several cis-acting sequences, including the Meti-tRNA:primer binding site (PBS) interaction (Chapman et al. 1992; Friant et al. 1998), LTRs (Eichinger and Boeke 1990; Braiterman et al. 1994; Devine and Boeke 1994; Sharon et al. 1994), polypurine tracts (PPTs) (Heyman et al. 1995; Lauermann et al. 1995), the GAG-POL frameshift region (Belcourt and Farabaugh 1990; Lawler et al. 2001), CYC5 and CYC3 (Cristofari et al. 2002), and an extensive 5′ RNA structure required for efficient initiation of reverse transcription (Bolton et al. 2005). Additional cis-sequences may remain undiscovered in Ty1. To discover these requires mutagenesis on a much larger scale. This motivated the present study, which in the end led in a different direction.
A near-comprehensive method of coding region mutagenesis is based on gene synthesis. Typically, synthetic genes encode the same product as the gene of interest; extensively recoding affects expression and/or base composition. This technique has been used with great success (Han and Boeke 2004; Neves et al. 2004; Tian et al. 2004; Patterson et al. 2005), often producing codon-optimized (CO) genes capable of elevated expression or expression in nonhost organisms. Furthermore, this strategy has also been utilized on other retroelements, such as HIV-1, to optimize expression and to remove negative cis-acting sequence elements such as the Rev response element (Kotsopoulou et al. 2000; Nguyen et al. 2004). Despite these successes, synthetic biology’s use of silent mutations also has the potential to introduce negative cis-acting sequence elements and to generate synthetic genes with reduced expression and/or decreased RNA stability (Kim and Lee 2006; Kudla et al. 2009; Welch et al. 2009).
In this study, we designed a synthetic element on the basis of Ty1-H3 with GeneDesign (Richardson et al. 2006) to recode Ty1-H3 with codons optimized for Saccharomyces cerevisiae expression, introducing 1209 base changes. The resulting synthetic codon-optimized Ty1 (CO-Ty1) was defective for Ty1 transposition and had reduced Ty1 protein and full-length RNA levels even though the known Ty1 cis-sequences had not been altered, suggesting identification of a novel cis-acting sequence capable of regulating Ty1 expression. Mapping and testing of this cis-acting sequence, however, revealed that the defect was the consequence of an inhibitory sequence in CO-Ty1 in the form of a Rap1p binding site.
Rap1p (Repressor Activator Protein) is a multifunctional protein with the binding site consensus ACACCCRYACAYM (Lieb et al. 2001) with transcriptionally opposing roles in the activation of transcription at promoters of ribosomal and glycolytic genes (Woudt et al. 1987; Tornow and Santangelo 1990) as well as in transcriptional silencing through the recruitment of silencing factors to the HM loci and telomeres (Shore and Nasmyth 1987; Buchman et al. 1988; Kyrion et al. 1992). The latter activity has been attributed to the nonessential Rap1-S domain, which is separate from the DNA binding domain (Sussel and Shore 1991; Kyrion et al. 1992). We describe here a new role for Rap1p in controlling expression via its ability to bind Rap1p sites inside transcription units (TUs) and thereby block and/or terminate transcriptional elongation. As Rap1p has been shown to ChIP within several TUs (Lieb et al. 2001), this finding may reveal a previously unexplored level of expression regulation and/or sequence constraints available within TUs.
Materials and Methods
Strains and media
All yeast strains used in this study, unless specified otherwise, were in the genetic background of GRF167 (JB740; MATα his3Δ200 leu2Δ1 ura3-167). Saccharomyces Genome Deletion strains BY4742 and BY4743 were used for analysis of rrp6Δ and rap1 variants, respectively. Media were prepared as described (Sherman et al. 1987).
Gene synthesis and plasmid construction
CO-Ty1 design was performed with GeneDesign (Richardson et al. 2006); construction of CO-Ty1 has been described (Yarrington 2009).
Gal-Ty1-mhis3AI plasmids pRY022, pRY091, and all chimeric CO-Ty1 variants were described previously (Curcio and Garfinkel 1991; Yarrington 2009). GFP expression vectors used the GFP reporter cassette from pFA6a-kanMX6-Gal1-GFP (Longtine et al. 1998) subcloned into pRS416 (Sikorski and Hieter 1989) via the EcoRI and SalI sites of the multicloning site. GFP70WT and GFP70CO, as well as all mutants and deletions, were constructed by subcloning the appropriate hybridized oligos with AscI nucleotide overhangs into the AscI site immediately downstream of the GFP stop codon.
Primers and oligos
All primer and oligo sequences are available in Supporting Information, Table S3, Table S4, and Table S5.
RNA isolation
Yeast cells harboring the pGal-Ty1-mhis3AI and CO-Ty1 elements were grown in 5 ml 0.2% glucose at 30° overnight for 12–16 hr. The following day, yeast culture A600 was measured and 2 OD of culture was used to reinoculate a 10-ml 2% galactose culture at 0.2 A600 grown at 22° for 20–24 hr to an A600 of ∼1.0. Total RNA was extracted by hot acid phenol (Collart and Oliviero 2001), purified by RNeasy minikit (Qiagen), and either fractionated on formaldehyde agarose gels or reverse transcribed into cDNA (SuperScript III First Strand Synthesis, Invitrogen) for quantitative real-time PCR (Q-PCR) analysis.
RNA Northern blot analysis
For Northern quantification of Ty1-mhis3AI RNAs, 10 μg total RNA was heat denatured in sample buffer (55% deionized formamide, MOPS buffer, pH 7.0, 5% formaldehyde, 8 mM EDTA, and 0.1% bromophenol blue) before electrophoresis on 1% agarose gels containing MOPS buffer, pH 7.0 (40 mM MOPS, 10 mM sodium acetate, and 1 mM EDTA) and 2% formaldehyde. RNA was transferred by capillary action and fixed by UV crosslinking to Gene Screen Plus filters as described by the manufacturer (NEN Life Science Products, Boston, MA). Membrane-bound RNAs were hybridized with either a HIS3-specific (0.5-kb fragment of pECB4B) or CO-Ty1–specific (0.1-kb fragment, 1 kb upstream of the Rap1p site) probe. DNA probes were internally labeled and purified over G25 Sephadex spin columns. Filters were exposed to a Molecular Dynamics phosphoimager screen.
Q-PCR
Q-PCR was performed as previously described (Livak and Schmittgen 2001). RNA levels were calculated by the 2(-ΔΔC(T)) method, using actin as reference, and normalized to the amount of native Ty1-H3 at the 0 min time point.
Transposition assay
Transformants containing the URA3-marked Ty1 GAL-Ty1-mhis3AI plasmid, pRY022, and synthetic variants were patched onto SC −Ura with 2% glucose (Curcio and Garfinkel 1991). After 2 days at 30°, yeast patches were replica plated to SC −Ura medium with 2% galactose and incubated at 22° for 2 days. After this period, plates were replica plated to YPD and incubated at 30° for 1 day and then replica plated again the following day to SC −His for an additional day at 30°. Transposition was recorded quantitatively by the ability of patches to grow on SC −His plates.
Immunoblot analysis
Yeast cells harboring the pGal-Ty1mhisAI and CO-Ty1 chimeric elements were grown in 5 ml 0.2% glucose at 30° overnight for ∼12–16 hr. The following day, yeast culture ODs were measured and 1 OD was used to reinoculate a 5-ml 2% galactose culture at 0.2 A600 and grown at 22° for 20–24 hr. Yeast cultures were then harvested and 2.5 A600 of cells were spun down for immunoblot analysis.
Protein extraction of cells was performed by mild alkaline treatment (Kushnirov 2000) by resuspending 2.5 A600 of cells in 200 μl of 0.2 M NaOH for 10 min, centrifugation and removal of supernatant, and boiling in 100 μl of SDS/PAGE sample buffer for 10 min. Ten microliters of each extract was run onto a 4–20% Tris-glycine polyacrylamide gel (Invitrogen). Proteins were transferred onto 0.45 μm PVDF membranes (Immunobilon-P from Millipore, Bedford, MA) in transfer buffer (25 mM Tris base, 192 mM glycine, and 20% methanol) at 30 V for 12–16 hr or 100 V for 2 hr. Membranes were washed three times (10 min each) with blocking buffer (Tris-buffered saline containing 1% nonfat milk and 0.1% Tween 20) and incubated for 1 hr with a 1/1000 dilution of Anti-IN 8B11 ascites to detect Ty1 integrase (IN) (Eichinger and Boeke 1990). Membranes were washed as indicated above and incubated for 1 hr with 1/2000 dilutions of ECL antimouse (to detect IN). Immunoblots were washed again as indicated above and visualized by ECL Plus chemifluorescence (Amersham).
Flow cytometry
Flow cytometry on yeast was performed as previously described (Starling et al. 2003). Briefly, yeast cells harboring the GFP70WT and GFP70CO variants were grown in 5 ml 0.2% glucose at 30° overnight for 12–16 hr. The following day, yeast culture A600 was measured and 1 OD was used to inoculate a 5-ml 2% galactose culture at 0.2 A600 grown at 22° for 20–24 hr to an A600 of ∼1.0. Yeast culture A600 was measured again, and 5 × 105 cells were added to 1 ml solution of PBS and sorted on a BD LSR II (BD Biosciences) FACS machine. GFP fluorescence was measured using the fluorescein isothiocyanate (FITC) channel of the BD LSRII and analyzed by the BD FaCSDIVA software package. Yeast cells were gated such that an empty vector control plasmid reported only 0.1% GFP+ cells.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as previously described (Meluh and Broach 1999). Antibodies to yeast Rap1p (Y-300, Santa Cruz) and RNAP II subunit Rpb3p (IY26, Neoclone) were used for Rap1p and RNAP II IP, respectively. Occupancy values and percent IP were calculated by dividing the amount of PCR product (derived from the Ct value in the best-fit regression real-time PCR standard curve for the specific primer set) in the IP by the amount of the PCR product in the total chromatin prep. IP values were normalized to a control (i.e., GFP70WT for Rap1p ChIP) and enrichment of occupancy or IP was calculated by dividing the experimental percent IP by that of its specific control.
Circularized rapid amplification of cDNA ends analysis
Circularized rapid amplification of cDNA ends was performed as previously described (Rissland and Norbury 2009). Briefly, the isolated total RNA was first dephosphorylated with antarctic phosphatase (NEB, M0289) according to the manufacturer’s instruction. The reaction was then purified by RNeasy Mini kit (Qiagen, 74104) and the 5′ cap was removed using 2.5 units of tobacco acid pyrophosphatase (Epicentre Biotechnologies, T81050). The reaction was incubated at 37° for 1 hr and then treated with another round of cleanup using the RNeasy Mini kit. Three micrograms of the decapped product was ligated using T4 RNA ligase (Epicentre Biotechnologies, LR5010) at 16° overnight in a total reaction volume of 40 μl. A separate ligation reaction was performed without the 5′ decapping treatment as a control. Ligation products were reverse transcribed with SuperScript III First-Strand Synthesis system (Invitrogen, 18080-051) using the supplied random hexamers or a 20-bp reverse primer (CTTAGAAGTAACCGAAGCAC) mapping ∼100 bp after the 5′ end of the Ty1 message. Five microliters of the resulting cDNA was used for PCR (30 cycles, 1-min extension) using the above reverse primer and a forward primer (GTTTGGGTGGTATTGGTGACTCTAA) mapping ∼400 bp upstream of the Rap1p site to amplify the circularized product. The resulting amplicons were cloned into the T-Easy vector (Promega, A1360) and sequenced.
Results
Construction of a synthetic codon-optimized Ty1 element
The CO-Ty1 element was designed using the various modules of GeneDesign (Richardson et al. 2006) and synthesized using established methods (Stemmer et al. 1995). The Codon Juggle module offers DNA sequence recoding with a variety of constraints. For CO-Ty1 codon optimization, we used the set of most favored codons in highly expressed genes in S. cerevisiae (Sharp et al. 1988). Because the 5′ and 3′ LTRs of Ty1 are essential for transposition (Eichinger and Boeke 1990), the sequence used for codon optimization was the coding sequence for GAG-POL, corresponding to the region spanning bases 294–5562 of the Ty1-H3 sequence. To fuse GAG and POL into one ORF, C1596 (Belcourt and Farabaugh 1990) was deleted prior to optimization and restored afterward. Furthermore, as many cis-acting sequences are already known to affect Ty1 retrotransposition, only sequences outside these regions were recoded. The resulting CO-Ty1 sequence is 77% identical to that of the native sequence. Further details of oligo design, PCR assembly, and cloning have been described previously (Yarrington 2009).
CO-Ty1 is defective for both Ty1 transposition and processed protein levels
To assay retrotransposition of CO-Ty1, the retrotransposition indicator/reporter mhis3AI (Curcio and Garfinkel 1991) was inserted downstream of the GAG-POL stop codon and the construct was subcloned behind the GAL1 promoter, creating pRY091, in which retrotransposition is measured by formation of His+ colonies after exposure to galactose (Figure 1A). Retrotransposition assays revealed a significant defect in CO-Ty1 retrotransposition relative to native Ty1-H3 (Figure 1B), suggesting sequence changes deleterious to Ty1 transposition. A quantitative assay revealed a 19.8-fold reduction in retrotransposition frequency (see Table S1).
Figure 1 .
Functional and biochemical characterization of CO-Ty1. (A) GAL-Ty1-mhis3AI reporter plasmid used for native Ty1 and CO-Ty1 analysis (Curcio and Garfinkel 1991). (B) Transposition assay of pRY022 native Ty1 (WT) and pRY091 CO-Ty1 (CO) grown on galactose and replica plated to SC −His for 2 days. Transposition activity was demonstrated by the appearance of colonies on SC −His plates (see Table S1). (C) Integrase protein (IN) immunoblot analysis of EV, empty vector; WT, pRY022; CO, pRY091. IN is indicated by an arrow and an internal loading control is indicated by *. (D) Above, IN immunoblot analysis of native and CO-Ty1 with and without a protease inactivating in-frame linker insertion (Monokian et al. 1994). EV, empty vector; WT, pRY022; CO, pRY091; WTp, pRY159; COp, pRY160. Protease activity is indicated by a plus or minus sign. Upper arrow indicates GAG-POL; lower arrow indicates IN. Below, RNA blot analysis of samples from above. EV, empty vector; WT, pRY022; CO, pRY091; WTp, pRY159; COp, pRY160. Ty1-HIS3 RNA is indicated by an arrow.
To further characterize CO-Ty1, we examined Ty1 IN and RT levels. Surprisingly, the synthetic CO-Ty1 displayed ∼3-fold diminished levels of integrase protein (Figure 1C), in spite of the fact that protein sequences were not altered; a similar reduction in RT was also observed (not shown). In both cases, a reduction of ∼2.97-fold was observed (see Table S1).
CO-Ty1 is defective for GAG-POL expression
The IN and RT proteins are proteolytically processed from a GAG-POL precursor protein. To determine whether the effect was on the precursor protein or restricted to the individual products of precursor processing, we introduced an inactivating in-frame linker insertion into the Ty1 protease (Monokian et al. 1994) of the native and CO-Ty1 plasmids. Immunoblot analyses revealed that GAG-POL protein levels of the CO-Ty1 were similarly reduced in full-length precursor, as was processed IN (Figure 1D), consistent with an overall deficit in CO-Ty1 gene expression.
CO-Ty1 has reduced Ty1 RNA
Diminished GAG-POL gene expression in CO-Ty1 presumably results directly from a full-length mRNA deficit. Indeed, RNA levels in CO-Ty1-mhis3AI displayed reduced full-length Ty1 RNA levels (2.98-fold) relative to native Ty1 (Figure 1D and Table S1).
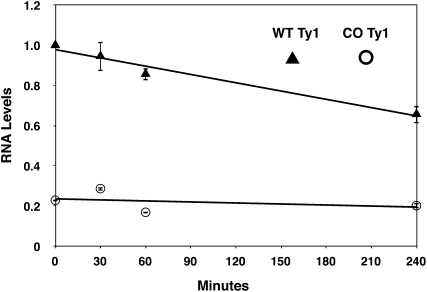
To determine whether the reduced CO-Ty1 RNA levels resulted from a defect in Ty1 transcription or in RNA stability, we assessed Ty1 RNA levels after adding glucose to shut off new transcription from the galactose-inducible Ty1 constructs (Figure 2). A 4.37-fold difference in RNA levels was observed before glucose was added, in close agreement with steady-state CO-Ty1 protein and RNA levels. The time course showed that CO-Ty1 did not have decreased RNA stability; rather it may have slightly increased RNA stability. The native Ty1 RNA level time course was consistent with a previous estimate of Ty1 RNA half-life (Nonet et al. 1987), validating the assay. These results suggest that RNA formation is defective in CO-Ty1.
Figure 2 .
Ty1 RNA stability. Native (WT) and CO-Ty1 (CO) Ty1-HIS3 RNA levels were measured by Q-PCR after addition of glucose to shut off new transcription at the indicated time points. Ty1 RNA levels were normalized to the native Ty1 0 min time point, and error bars are the standard deviation of two biological replicates. Native Ty1-HIS3 RNA is indicated by triangles and CO-Ty1-HIS3 RNA is indicated by circles.
Critical CO-Ty1 sequence maps to a 70-bp region in the N-terminal region of RT
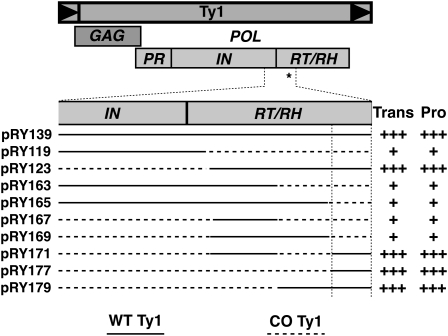
We used a systematic collection of native/synthetic chimeric clones, made possible by introduced common unique restriction sites, to map a critical cis-acting sequence capable of regulating Ty1 retrotransposition to the N-terminal region of the Ty1 RT gene (Figure 3) (Yarrington 2009). The critical CO-Ty1 sequence was mapped by examination of Ty1 transposition and IN levels in a series of chimeric CO-Ty1/native Ty1 clones.
Figure 3 .
Mapping and characterization of 70-bp retrotransposition critical sequence. Diagram of Ty1 with open reading frames as a mapping reference. Selected and expanded region of IN and RT represents the initially mapped EcoRI–AatII 556-bp region found to contain the critical cis-acting sequence (*). Solid line, native Ty1 sequence; dashed line, CO-Ty1 sequence. Transposition frequency (Trans) and protein levels (Pro) are indicated qualitatively by pluses. Vertical lines define inferred critical sequence affecting Ty1 retrotransposition.
Effects of critical CO-Ty1 sequence are transplantable
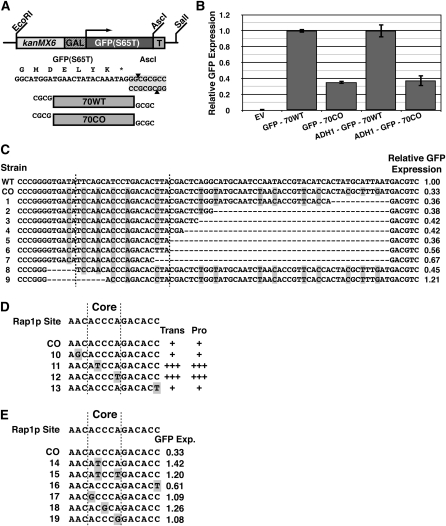
To determine whether or not the critical region represented a novel sequence requirement for Ty1 transposition, we determined its effects in a non-Ty1 context. The 70-bp critical region was cloned immediately after the stop codon of GFP in the pFA6a-kanMX6-Gal1-GFP construct (Figure 4A) (Longtine et al. 1998). The GFP reporter system was then further subcloned into pRS416 (Sikorski and Hieter 1989), creating GFP70CO. The corresponding native Ty1 sequence was similarly cloned into GFP in the same way, creating GFP70WT; this construct had a minimal impact on GFP expression (not shown).
Figure 4 .
Functional characterization and mapping of critical 70-bp sequence in GFP. (A) GFP reporter construct used for GFP-70WT and GFP-70CO analysis. Tested sequences were inserted into the reporter construct immediately downstream of the GFP stop codon. (B) GFP expression of indicated reporter constructs grown in galactose as measured by fluorescence-activated cell sorting. EV, empty vector; GFP-70WT, construct with 70-bp critical region from native Ty1 inserted immediately after GFP stop codon; GFP-70CO, construct with 70-bp critical region from CO-Ty1 inserted immediately after GFP stop codon; ADH1-GFP-70WT, same as GFP-70WT, but driven by ADH1 promoter; ADH1-GFP-70CO, same as GFP-70CO, but driven by ADH1 promoter (Table S2). (C) Deletion analysis of GFP-70CO reveals 20-bp critical sequence down-regulating GFP expression. GFP expression is normalized to GFP-70WT. Vertical lines enclose minimal critical sequence. Highlighted bases differ between GFP-70WT and GFP-70CO. WT, GFP-70WT; CO, GFP-70CO; 1–9, various mutations in GFP-70CO. (D) Mutation analysis of Rap1p site within CO-Ty1. Transposition frequency (Trans) and protein levels (Pro) are indicated qualitatively by pluses. Vertical lines represent the Rap1p core binding site. Highlighted bases differ between native Ty1 and CO-Ty1. CO, CO-Ty1; 1–4, various mutations in CO-Ty1. (E) Mutation analysis of the identified Rap1p site within GFP-70CO. Vertical lines represent the Rap1p core binding site. GFP expression is normalized to GFP-70WT. Highlighted bases differ between GFP-70WT and GFP-70CO. CO, GFP-70CO; 1–6, various mutations in GFP-70CO.
FACS analysis of the GFP70WT and GFP70CO GFP reporter constructs revealed that the critical CO-Ty1 sequence down-regulated GFP expression 2.84-fold, a value very similar to the reduced expression level in CO-Ty1 (Figure 4B and Table S2). Thus the CO-Ty1 sequence did not represent a novel Ty1 sequence requirement for retrotransposition but rather appeared to be a portable synthetic negative regulatory sequence inadvertently introduced by codon optimization. Repression via the synthetic sequence was independent of the GAL1 promoter of the CO-Ty1 and GFP70CO constructs as an ADH1-GFP70CO was similarly reduced in expression relative to ADH1-GFP70WT (Figure 4B). Furthermore, as this sequence was able to mediate its effects in the 3′-UTR of GFP and in multiple reading frames (not shown), the effect of this synthetic sequence was independent of protein translation. By monitoring FACS of various terminal deletion constructs of GFP70CO, the minimal required sequence for gene repression was mapped to a 20- to 25-bp region (Figure 4C).
Critical CO-Ty1 sequence contains a Rap1p binding site
As GFP and Ty1 repression were mediated by neither decreased RNA stability nor protein translation, the mechanism of repression was likely affecting RNA production. To that end, the identified critical CO-Ty1 sequence was entered into a matrix search for transcription factor binding sites program, Match (Kel et al. 2003), to determine whether the critical CO-Ty1 sequence matched a unique binding factor. Within both the 70-bp CO-Ty1 and the 20- to 25-bp subregion, Match identified a unique Rap1 binding site with a 1.00 core match and 0.979 matrix match.
Interestingly, the repressive Rap1p site identified here ACACCCAGACACC is nearly identical to Rap1p sites found at yeast telomeres (ACACCCAYACAYY in which the Ys are more likely to be Cs (Idrissi and Pina 1999). Mutating residue 12 to a T, a change more similar to a ribosomal protein promoter Rap1p site (Idrissi and Pina 1999), somewhat alleviated down-regulation of GFP (Figure 4C, compare strain 5 to 6). This result may indicate that not all Rap1p sites are equally able to down-regulate in this context.
Mutation of ACCCA core binding site of Rap1p rescues expression
Nearly all Rap1 binding sites contain a core ACCCA sequence; mutation of this site abrogates binding (Buchman et al. 1988; Lieb et al. 2001). Mutagenesis of the Rap1p binding site was used to confirm Rap1p specificity. Restoring either position 2 (C→T) or 5 (A→T) to the native Ty1 sequence in the ACCCA core was sufficient to entirely relieve Rap1p-mediated repression of CO-Ty1 transposition and protein levels (Figure 4D). No other bases, when restored to native Ty1 counterparts outside the Rap1p ACCCA core binding site could alleviate down-regulation in both Ty1 and GFP constructs (Figure 4E). Furthermore, mutation of the core at positions 1, 3, and 5 individually to G prevented down-regulation of GFP (Figure 4E). Interestingly, changes to the Rap1p ACCCA consensus at position 2 to T and/or position 5 to G are present in some ribosomal protein promoter Rap1p binding sites (Idrissi et al. 2001; Lieb et al. 2001). These specific changes in our system prevented down-regulation of GFP, thus not all Rap1p binding sites are equally capable of TU-specific down-regulation.
Down-regulation depends on orientation and number of sites
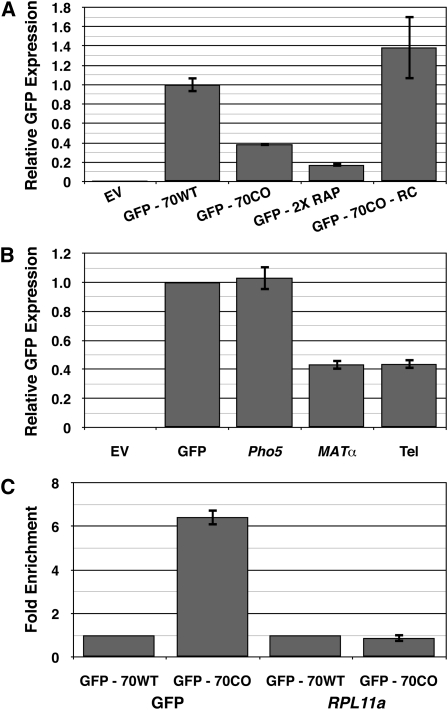
Rap1p sites are found in promoters of ribosomal and glycolytic genes in either orientation and, especially within ribosomal promoters, in multiple copies (Woudt et al. 1987; Tornow and Santangelo 1990). Surprisingly, the inverted Rap1p site had no effect on GFP expression (Figure 5A). This suggests that the Rap1p site must be presented in the same orientation as ongoing transcription to mediate down-regulation.
Figure 5 .
Rap1p binding sites are capable of down-regulating expression. (A) The orientation and number of Rap1p binding sites affect the ability of Rap1p to down-regulate expression. GFP expression was measured by fluorescence-activated cell sorting. EV, empty vector; GFP-70WT, construct with 70-bp critical region from native Ty1 inserted immediately after GFP stop codon; GFP-70CO, construct with 70-bp critical region from CO-Ty1 inserted immediately after GFP stop codon; GFP-2XRAP, same as GFP-70CO, but with additional Rap1p binding site in the forward orientation; GFP-70CO-RC, same as GFP-70CO, but with the insert inverted. (B) High-affinity Rap1p binding sites from the yeast genome are capable of down-regulating expression. GFP expression was measured by fluorescence-activated cell sorting. EV, empty vector; GFP, empty construct from Figure 4A; Pho5, same as GFP with a documented Rap1p binding site from the PHO5 ORF; MATα, same as GFP with a documented Rap1p binding site from the HMα locus; Tel, as GFP with a documented Rap1p binding site from yeast telomere-like sequence (Buchman et al. 1988). (C) Chromatin immunoprecipitation demonstrates that Rap1p binds the identified Rap1p site in GFP-70CO. DNA was sheared and bound fragments were pulled down by polyclonal antibody to C terminus of Rap1p. Enrichment was measured by Q-PCR and normalized to GFP-70WT. GFP, primers designed to amplify Rap1p binding site within the GFP reporter construct; RPL11a, primers designed to amplify a known Rap1p binding site in the RPL11a promoter (Lieb et al. 2001; Zhao et al. 2006). GFP-70WT, construct with 70-bp critical region from native Ty1 inserted immediately after stop codon of GFP; GFP-70CO, construct with 70-bp critical region from CO-Ty1 inserted immediately after stop codon of GFP.
Furthermore, inserting two Rap1p sites into GFP essentially doubled down-regulation, leading to an approximately sixfold drop in GFP expression of the GFP2XRAP construct compared to GFP70WT (Figure 5A). Introduction of additional Rap1p sites resulted in further, but diminishing, down-regulation of GFP (data not shown). The approximately twofold increase in GFP down-regulation observed with GFP2XRAP suggests that the Rap1p site can be modular for down-regulation within a TU. These results are less consistent with models based on internal transcriptional activation from the introduced Rap1p site or effects on histone repression as mechanisms for Rap1p-mediated down-regulation, as we did not see a synergistic effect from the addition of extra Rap1p sites, an effect that has been observed for both transcriptional activation and release from histone repression at the promoter of Rap1p bound genes (Woudt et al. 1987; Idrissi and Pina 1999; Idrissi et al. 2001).
Genomic Rap1 sites can down-regulate GFP
To further demonstrate that Rap1p mediated the observed transcriptional repression, we moved a series of genomic Rap1p sequences from a telomere-like sequence, MATα, and PHO5, which are known to bind Rap1p in vitro and activate transcription in vivo (Buchman et al. 1988) into GFP. With both telomere and MATα sites, we observed robust down-regulation when the respective Rap1p binding sites were subcloned into GFP (Figure 5B). We observed no down-regulation of GFP, however, when the PHO5 Rap1p binding site was inserted into GFP (Figure 5B). The latter lack of down-regulation likely reflects low affinity of the site, ∼40-fold less than that observed for the telomere sequence (Buchman et al. 1988). This result suggests that Rap1p-mediated down-regulation requires a certain affinity between Rap1p and its binding site for effective down-regulation, consistent with a transcriptional roadblock model for repression, and likely explains why not all Rap1p sites are capable of down-regulation.
Rap1p binds to critical CO-Ty1/Rap1 sequence
To demonstrate that Rap1p binds to the identified binding site, we employed ChIP. Rap1p was crosslinked to its binding sites by fixing exponentially growing cultures containing the GFP reporters with 1% formaldehyde and performing ChIP.
Q-PCR analysis revealed that DNA fragments surrounding the Rap1p site of GFP70CO were enriched >6.43-fold compared to amplification of the same region from GFP70WT (Figure 5C and Table S2). Q-PCR analysis of a known Rap1p site present in the promoter of RPL11A (Lieb et al. 2001; Zhao et al. 2006) revealed similar levels of RPL11A pull-down with the two GFP constructs, validating our ChIP results. This demonstrates that the site indeed binds Rap1p in vivo.
Rap1p-mediated down-regulation is not mediated via N- or C-terminal domains
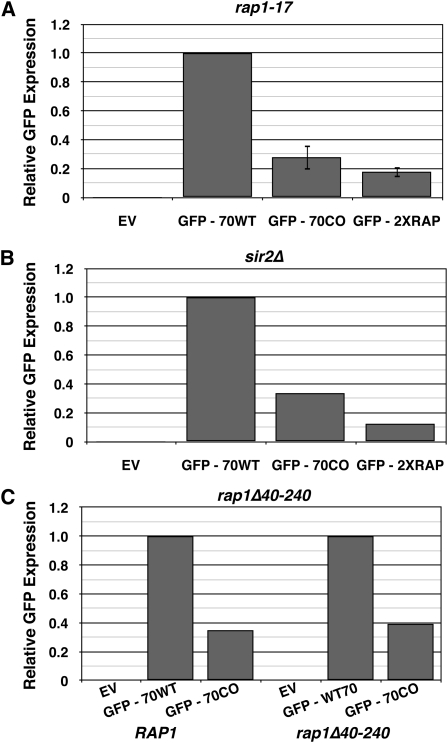
Because telomeric Rap1p binding sites, which consist of sequences very similar to the identified Rap1p binding site, silence transcription at and near telomeres via recruitment of Sir proteins (Shore and Nasmyth 1987; Buchman et al. 1988; Kyrion et al. 1992), gene silencing was a likely mechanism of down-regulation. To determine whether silencing was involved, a rap1-17 allele (Kyrion et al. 1992) was utilized. The rap1-17 allele lacks the C-terminal 165 amino acid residues and is defective for silencing at both HM loci and telomeres (Kyrion et al. 1992; Moretti et al. 1994). Yeast expressing rap1-17, however, did not rescue GFP expression from GFP70CO or GFP2XRAP (Figure 6A), indicating that neither the C-terminal domain of Rap1p nor silencing were responsible for Rap1p-mediated repression of GFP70CO.
Figure 6 .
Neither the N nor the C terminus of the Rap1p protein affect its down-regulation of GFP expression. (A) Constructs were transformed into a yeast strain with a nonsense mutation in Rap1p resulting in a 165-codon deletion of the C-terminal domain (Kyrion et al. 1992). GFP expression was measured by fluorescence-activated cell sorting. EV, empty vector; GFP-70WT, construct with 70 bp of critical region from native Ty1 inserted immediately after stop codon of GFP; GFP-70CO, construct with 70 bp of critical region from CO-Ty1 inserted immediately after stop codon of GFP; GFP-2XRAP, as GFP-70CO, but with additional Rap1p binding site in the forward orientation. (B) Constructs were transformed into a yeast strain with a sir2 deletion genotype. GFP expression was measured by fluorescence-activated cell sorting. EV, empty vector; GFP-70WT, construct with 70 bp of critical region from native Ty1 inserted immediately after stop codon of GFP; GFP-70CO, construct with 70 bp of critical region from CO-Ty1 inserted immediately after stop codon of GFP; GFP-2XRAP, as GFP-70CO, but with additional Rap1p binding site in the forward orientation. (C) Constructs were transformed into a yeast strain with a 200-amino-acid deletion within the N terminus of Rap1p. GRF167 was used as a wild-type control. GFP expression was measured by fluorescence-activated cell sorting. EV, empty vector; GFP-70WT, construct with 70 bp of critical region from native Ty1 inserted immediately after stop codon of GFP; GFP-70CO, construct with 70 bp of critical region from CO-Ty1 inserted immediately after stop codon of GFP.
The reporter constructs were also transformed into a sir2Δ strain incapable of supporting transcriptional silencing (Rine and Herskowitz 1987); no rescue of GFP expression was observed (Figure 6B). Thus the observed Rap1p-mediated down-regulation is not mediated by canonical silencing.
The N-terminal domain of Rap1p can be deleted without functional consequence to the cell, and is capable of causing >50° bends in DNA upon Rap1p binding (Muller et al. 1994). To determine whether this domain and/or DNA bending contributed to Rap1-mediated down-regulation, an N-terminal deletion (Δ40–240) of Rap1 was constructed. The rap1Δ40–240 was shuffled (Boeke et al. 1987) into a haploid rap1Δ strain, and the resulting strain was transformed with the GFP reporter constructs (Figure 6C); no rescue of GFP expression was observed. Thus neither N- nor C-terminal domains of Rap1p underlie Rap1p-mediated down-regulation.
Rap1p functions as a transcriptional roadblock or terminator
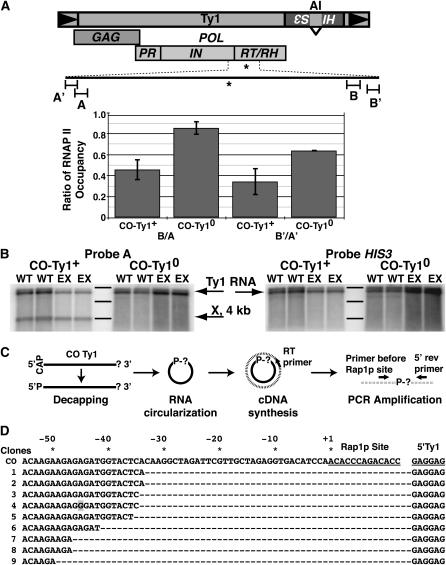
As Rap1p-mediated repression within TUs did not appear to be a function of the N- or C-terminal domains, it was formally possible that the Rap1p DNA-binding domain mediated repression via binding within our constructs, serving as a steric transcriptional block and/or terminator of elongation. To determine whether Rap1p inhibited expression in this way, RNA polymerase II (RNAP II) occupancy along CO-Ty1 was evaluated using ChIP/Q-PCR of sequences ∼1000-bp up- and downstream of the Rap1p site (Figure 7A). A CO-Ty1 from which the Rap1p site had been removed (CO-Ty10, pRY177) served as a control. DNA was sheared by sonication, and fragments occupied by RNAP II were selected using monoclonal antibody raised against Rpb3p, an RNAP II subunit.
Figure 7 .
Rap1p blocks elongation and produces a novel transcript. (A) Diagram of Ty1; Rap1p binding site in CO-Ty1 is indicated by *. Chromatin immunoprecipitation demonstrates RNAP II occupancy; DNA was sheared and bound fragments were pulled down by monoclonal antibody to RPB3, a core component of RNAP II. Occupancy was determined by Q-PCR and the ratio of RNAP II at the downstream primer in a pair to the upstream primer in a pair is shown for CO-Ty1 (CO-Ty1+) and for a variant in which the identified Rap1p binding site has been replaced with corresponding sequence from the native Ty1 (CO-Ty10). A, primers designed to amplify a 109-bp sequence 1086 bp 5′ of the identified Rap1p binding site; A′, primers designed to amplify a 103-bp sequence 1286 bp 5′ of the identified Rap1p binding site; B, primers designed to amplify a 115-bp sequence 1015 bp 3′ of the identified Rap1p binding site; B′, primers designed to amplify a 103-bp sequence 1191 bp 3′ of the identified Rap1p binding site. (B) RNA blot analysis of CO-Ty1+ and CO-Ty10 with probes amplified from a 109-bp sequence 1086 bp 5′ of the identified Rap1p binding site (A) and HIS3. Two different strains (WT, GRF167; EX, rrp6Δ) were used for analysis. Ty1-HIS3 RNA is indicated by an arrow and is the full-length RNA. X represents novel CO-Ty1–specific RNA species. Bands from the 0.5- to 10-kb ladder by Invitrogen are marked. From top to bottom, the sizes are 8, 6, and 4 kb. (C) cRT–PCR strategy used for mapping the 3′ end of the X RNA. Messenger RNA molecules are represented as solid black lines and cDNA as broken lines. The CAP and the “?” represent the 5′ cap and unknown 3′ end, respectively. The “P” indicates the 5′-phosphate group and “P-?” indicates the ligated 5′–3′ junction. (D) Sequence analysis of the 5′–3′ junction of representative X RNA molecules. The Rap1p site and the 5′ start of the Ty1 message are underlined. CO represents the sequence of the intact, nontruncated synthetic Ty1 element and the numbers 1–9 label the sequences of nine X RNA molecules after undergoing the cRT–PCR protocol. The sequence shown for the nine clones before the Rap1p site is immediately joined to the GAGGAG of the 5′ end of the Ty1 message. Dashes are used as a spacer to illustrate the distance the polymerase stalled before the Rap1p site. * and corresponding numbers indicate the distance before the start of the Rap1p site. The shaded box indicates a mutation observed in one of our clones. A few additional RNAs (not pictured) truncated further upstream. The differences in stalling position observed may be the result of polymerase backtracking.
Q-PCR with primers hybridizing 1000-bp 5′ and 1000-bp 3′ of the Rap1p site showed a large decrease in downstream RNAP II occupancy, which was reversed by mutating the binding site (Figure 7A). The decreased RNAP II occupancy downstream of the site agrees well with the observed decreases in RNA and protein expression levels in CO-Ty1, consistent with Rap1p’s mode of action in this context as a transcription block or elongation terminator.
To further probe the role of Rap1p as a block or terminator of transcription, we extracted total RNA from yeast cells and probed with CO-Ty1 sequences 1000 bp 5′ of the Rap1p binding site. A novel band (X) appears, which is consistent with RNAP II stalling or terminating transcription (Figure 7B). As a control we probed the same blot with sequence derived from the HIS3 gene 3′ of the CO-Ty1 coding sequence (Figure 7A) and no band was detected. The nuclear exosome complex does not affect the appearance of X, a finding consistent with the stalling model (Figure 8). To determine whether X was polyadenylated and thus a candidate to be degraded by the cytoplasmic exosome, the X RNA was circularized Rissland and Norbury(2009), converted to cDNA, and finally sequenced across the 5′–3′ junction (Figure 7C). Rather than polyadenylation of the X RNA message, we observed a direct ligation of CO-Ty1 sequence ∼34 bp upstream of the Rap1p site to the 5′ start of the Ty1 message; no poly(A) sequences were observed (Figure 7D). All findings are consistent with a polymerase stalling model.
Figure 8 .
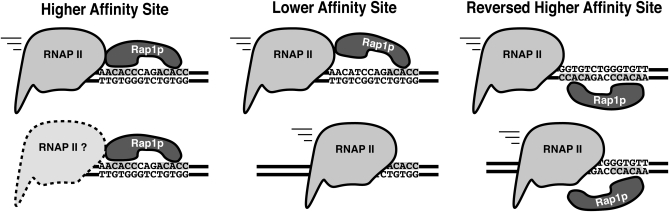
Model of Rap1p-mediated down-regulation of gene expression. (Left) High-affinity site in the sense orientation impedes RNAP II’s progress. (Middle) Lower-affinity site may not bind Rap1p avidly enough to prevent displacement by RNAP II. (Right) High-affinity site in inverted orientation may make Rap1p susceptible to displacement by RNAP II because the second Myb domain lacks the additional contacts provided by the C terminus of the Rap1p DNA binding domain.
Discussion
Rap1p is an essential DNA-binding factor found at the promoters of genes, especially those involved in ribosomal protein and glycolytic pathways, the HM locus, and at telomeres. At these sites, this multifunctional protein is capable of both transcriptional activation or transcriptional silencing, disparate functions dependent on adjoining sequence context, including the presence of other DNA binding factors, and the number and spacing of Rap1p sites (Grossi et al. 2001). We describe here a novel Rap1p transcriptional function independent of sequence context, provided the Rap1p site lies within a TU. The identified Rap1p site in this study was equally capable of regulating gene transcription within the Ty1 ORF or the 3′-UTR of GFP. Further, this repression was not specific to the isolated Rap1p site as the telomere and the HM locus Rap1p sites also regulated GFP expression. The major determinant for this down-regulatory capacity of Rap1p appears to be the binding affinity of Rap1p to its respective site. Interestingly, the Rap1p sites capable of transcriptional silencing at telomeres and the HM locus also have the highest affinities for Rap1p (Buchman et al. 1988; Pina et al. 2003).
Rap1p can bind very stably to the consensus ACACCCRYACAYM (Lieb et al. 2001) with koff rates of >60 min for some sequences (Woudt et al. 1987; Idrissi and Pina 1999). This very tight binding suggests a possible model for down-regulation by blocking elongating RNAP II during transcription (Figure 8). Requirements for Rap1 site orientation in regards to active transcription suggest that either Rap1p is easier to dislodge when presented in the reverse orientation or that the RNA polymerase must encounter a specific face of Rap1p. Evidence for the former comes from the Rap1p crystal structure. The two Myb DNA binding domains of Rap1p are similarly positioned in the major grooves of the binding site and can be superimposed upon each other by a translation of 8 bp. Despite these similarities, however, the C terminus of the Rap1p DNA binding domain folds back toward the first Myb DNA binding domain, making additional contacts with the first half of the Rap1p binding site, completely enveloping it. Furthermore, these additional contacts induce an opening of the major groove by 5–6 Å (Konig et al. 1996; Taylor et al. 2000). It is possible that the additional contacts with the C terminus of the Rap1p DNA binding domain and the distortion of the DNA at the 5′ half of the Rap1p site provide the mechanism for orientation specificity. Furthermore, this orientation specificity is also not without precedent. Reb1, the yeast terminator for RNAP1, shares the Myb-like DNA binding domain and functions as a terminator in only the forward orientation with regard to transcription (Reeder and Lang 1994). Consistent with this model, we observed a large decrease in RNAP II occupancy downstream of the Rap1p site found within our CO-Ty1 element that did not occur when the Rap1p site was replaced with native Ty1 sequence. A probe of a total yeast RNA blot with sequence upstream of the Rap1p site revealed a novel band consistent with a stalled or terminated transcript, further supporting this model. As native termination in yeast occurs either early within the first few hundred nucleotides of transcription or late with the cooperative actions of the polyadenylation machinery (Buratowski 2009; Perales and Bentley 2009), neither mechanism explains our results. Rather, our results are consistent with a pronounced transcriptional pause. Testing of CO-Ty1 in an rrp6 nuclear exosome mutant revealed that the nuclear exosome complex does not affect the appearance of the novel truncated band. Futhermore, the novel band is also unlikely to be affected by the cytoplasmic exosome as mapping of the 3′ end failed to reveal the presence of poly(A). Rather than termination per se, stalling is the likely mechanism of Rap1p-mediated down-regulation of transcription. Consistent with a stalling model, transcription and translation are similarly reduced, suggesting these stalled transcripts are not successfully translated, which would be expected if they are nuclear.
Such a mechanism is not unprecedented, although it has not been described for Rap1p. A related yeast protein, Reb1, has certain similarities to Rap1. Better known as the terminator for RNA polymerase I, the main role of Reb1p is to pause that polymerase at the release element, a role that can mostly be replaced by a heterologous DNA binding roadblock such as the lac repressor (Jeong et al. 1995). Interestingly, like Rap1p, the DNA binding domain of Reb1p is similar to that of the Myb oncoprotein and likewise is only capable of pausing transcription in an orientation-specific manner (Reeder and Lang 1994). In humans, the transcription factor MAZ shares similarities with Rap1p, and is found to bind to promoters as well as to pause RNAP II transcription (Ashfield et al. 1994; Yonaha and Proudfoot 1999). Interestingly, the authors speculate that the placement of MAZ in promoters of closely spaced genes might be required to prevent transcriptional interference. Lastly, another DNA binding roadblock, the Gln-111 mutant EcoRI, defective for cleavage but capable of high-affinity binding (Wright et al. 1989), can stably pause a bacteriophage polymerase in vitro (Pavco and Steege 1990). This protein was also found to be capable of pausing RNAP II ∼30 bp upstream of an EcoRI site (Yonaha and Proudfoot 1999), a distance very similar to that observed for Rap1p. However, unlike the ability of Rap1p to stall RNAP II driven by the GAL promoter, the Gln-111 roadblock could be rescued by multiple polymerases on the same template (Epshtein et al. 2003). The mechanism for this escape, and the likely rationale for the failure of Rap1p to completely repress expression, was recently elucidated by the Svejstrup lab. It was found that rear end collision between closely packed RNA polymerases could drive a stalled polymerase through a pause site (Saeki and Svejstrup 2009).
Our findings help explain the unusual distribution of Rap1p sites in the genome. Rap1p sites are found at much higher frequency within intergenic regions, and, furthermore, although Rap1p binds to the consensus ACACCCRYACAYM, it binds preferentially to intergenic sequences (ChIPs to 182 of 322 possible sites, 57%) than to ORF sequences (ChIPs to 23 of 163 possible sites, 14%) (Lieb et al. 2001). As Rap1p is capable of repressing transcription within ORFs, it is likely that the existing ORF sequences are either under additional transcription regulation by Rap1p or have evolved low affinity Rap1p sites, as in the case of PHO5. Our finding also explains the surprising spacing between divergent Rap1p-driven promoters in which Rap1p was shown to preferentially bind the −1 nucleosome (Koerber et al. 2009). While Rap1p was often found at divergent Rap1p-driven promoters that shared the same −1 nucleosome, Rap1p was never found when the −1 nucleosome of one gene also served as the +1 nucleosome of a divergently transcribed gene. These results indicate that both coding and intergenic sequences are evolutionarily constrained to prevent Rap1p sites within TU sequences. Lastly, high affinity Rap1p sites are most commonly found in regions of heterochromatin, namely the HM locus and telomeres. Given the abundance of Rap1p sites at these regions and the level of RNAP II stalling observed in this report, it is possible that the stalling effects of Rap1p may not be limited to just transcription but could also have a profound effects on both recombination and/or DNA replication (Selth et al. 2010). Interestingly, these regions are found to be sites of transient DNA replication fork pausing and require the aid of the Rrm3p helicase even in the absence of Sir proteins (Ivessa et al. 2003).
This study demonstrates that Rap1p can serve as a transcription terminator or roadblock within a TU with significant downfield effects on both RNA levels and protein abundance a novel function for Rap1p. Rap1p has been shown to ChIP within TUs of several genes (Lieb et al. 2001), indicating that such genes have either evolved low affinity Rap1p sites or may be under additional levels of transcriptional regulation via binding of Rap1p to sites inside TUs. It is of course possible that transcriptional repression within TUs via tight binding transcription factors may not be limited solely to Rap1p, allowing for more widespread repression within S. cerevisiae, and probably other species. Indeed, the DNA binding domain of Rap1p is similar to that of Engrailed, Matα2p, Antennapedia, POU, and Paired (Konig et al. 1996).
Lastly, this study serves as another important observation for the growing synthetic biology field that not all “silent” mutations are indeed silent and demonstrates that the recognition sequences of certain DNA binding factors inside coding regions should be important considerations in design of synthetic genes. However, the ability of this small portable element to “dial down” gene expression relatively predictably might represent an important new asset in designing and tweaking synthetic regulatory circuits.
Acknowledgments
We thank Jeffrey Corden for helpful discussions, Kathryn O’Donnell for invaluable help with Q-PCR and Northern blot analyses, and Pamela Meluh and Zheng Kuang for help with ChIP. S.M.R. was supported by Department of Energy grant DE-FG02097ER25308. This work was also supported in part by National Institutes of Health grant GM36481 to J.D.B.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Ashfield R., Patel A. J., Bossone S. A., Brown H., Campbell R. D., et al. , 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13: 5656–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J., 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62: 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R., 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Bolton E. C., Coombes C., Eby Y., Cardell M., Boeke J. D., 2005. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA 11: 308–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman L. T., Monokian G. M., Eichinger D. J., Merbs S. L., Gabriel A., et al. , 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene 139: 19–26 [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D., 1988. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol. Cell. Biol. 8: 5086–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K. B., Bystrom A. S., Boeke J. D., 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 89: 3236–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart, M. A., and S. Oliviero, 2001 Preparation of Yeast RNA, Chapter 13, Unit 13.12 in Current Protocols in Molecular Biology. John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Bampi C., Wilhelm M., Wilhelm F. X., Darlix J. L., 2002. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J 21: 4368–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine S. E., Boeke J. D., 1994. Efficient integration of artificial transposons into plasmid targets in vitro: a useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 22: 3765–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger D. J., Boeke J. D., 1990. A specific terminal structure is required for Ty1 transposition. Genes Dev. 4: 324–330 [DOI] [PubMed] [Google Scholar]
- Epshtein V., Toulmé F., Rahmouni A. R., Borukhov S., Nudler E., 2003. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 22: 4719–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S., Heyman T., Bystrom A. S., Wilhelm M., Wilhelm F. X., 1998. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol. 18: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi S., Bianchi A., Damay P., Shore D., 2001. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 21: 8117–8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. S., Boeke J. D., 2004. A highly active synthetic mammalian retrotransposon. Nature 429: 314–318 [DOI] [PubMed] [Google Scholar]
- Heyman T., Agoutin B., Friant S., Wilhelm F. X., Wilhelm M. L., 1995. Plus-strand DNA synthesis of the yeast retrotransposon Ty1 is initiated at two sites, PPT1 next to the 3′ LTR and PPT2 within the pol gene. PPT1 is sufficient for Ty1 transposition. J. Mol. Biol. 253: 291–303 [DOI] [PubMed] [Google Scholar]
- Idrissi F. Z., Pina B., 1999. Functional divergence between the half-sites of the DNA-binding sequence for the yeast transcriptional regulator Rap1p. Biochem. J. 341(Pt 3): 477–482 [PMC free article] [PubMed] [Google Scholar]
- Idrissi F. Z., Garcia-Reyero N., Fernandez-Larrea J. B., Pina B., 2001. Alternative mechanisms of transcriptional activation by Rap1p. J. Biol. Chem. 276: 26090–26098 [DOI] [PubMed] [Google Scholar]
- Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., et al. , 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Jeong S. W., Lang W. H., Reeder R. H., 1995. The release element of the yeast polymerase I transcription terminator can function independently of Reb1p. Mol. Cell. Biol. 15: 5929–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel A. E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O. V., et al. , 2003. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 31: 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee S. B., 2006. Rare codon clusters at 5′-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr. Purif. 50: 49–57 [DOI] [PubMed] [Google Scholar]
- Koerber R. T., Rhee H. S., Jiang C., Pugh B. F., 2009. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol. Cell 35: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P., Giraldo R., Chapman L., Rhodes D., 1996. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85: 125–136 [DOI] [PubMed] [Google Scholar]
- Kotsopoulou E., Kim V. N., Kingsman A. J., Kingsman S. M., Mitrophanous K. A., 2000. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74: 4839–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A. W., Tollervey D., Plotkin J. B., 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V. V., 2000. Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Kyrion G., Boakye K. A., Lustig A. J., 1992. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5159–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauermann V., Nam K., Trambley J., Boeke J. D., 1995. Plus-strand strong-stop DNA synthesis in retrotransposon Ty1. J. Virol. 69: 7845–7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J. F., Jr, Merkulov G. V., Boeke J. D., 2001. Frameshift signal transplantation and the unambiguous analysis of mutations in the yeast retrotransposon Ty1 Gag-Pol overlap region. J. Virol. 75: 6769–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb J. D., Liu X., Botstein D., Brown P. O., 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Broach J. R., 1999. Immunological analysis of yeast chromatin. Methods Enzymol. 304: 414–430 [DOI] [PubMed] [Google Scholar]
- Monokian G. M., Braiterman L. T., Boeke J. D., 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene 139: 9–18 [DOI] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D., 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Muller T., Gilson E., Schmidt R., Giraldo R., Sogo J., et al. , 1994. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J. Struct. Biol. 113: 1–12 [DOI] [PubMed] [Google Scholar]
- Neves F. O., Ho P. L., Raw I., Pereira C. A., Moreira C., et al. , 2004. Overexpression of a synthetic gene encoding human alpha interferon in Escherichia coli. Protein Expr. Purif. 35: 353–359 [DOI] [PubMed] [Google Scholar]
- Nguyen K. L., Ilano M., Akari H., Miyagi E., Poeschla E. M., et al. , 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319: 163–175 [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R., 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S. S., Dionisi H. M., Gupta R. K., Sayler G. S., 2005. Codon optimization of bacterial luciferase (lux) for expression in mammalian cells. J. Ind. Microbiol. Biotechnol. 32: 115–123 [DOI] [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A., 1990. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J. Biol. Chem. 265: 9960–9969 [PubMed] [Google Scholar]
- Perales R., Bentley D., 2009. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Fernandez-Larrea J., Garcia-Reyero N., Idrissi F. Z., 2003. The different (sur)faces of Rap1p. Mol. Genet. Genomics 268: 791–798 [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Lang W., 1994. The mechanism of transcription termination by RNA polymerase I. Mol. Microbiol. 12: 11–15 [DOI] [PubMed] [Google Scholar]
- Richardson S. M., Wheelan S. J., Yarrington R. M., Boeke J. D., 2006. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 16: 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Herskowitz I., 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland O. S., Norbury C. J., 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H., Svejstrup J. Q., 2009. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell 35: 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L. A., Sigurdsson S., Svejstrup J. Q., 2010. Transcript elongation by RNA Polymerase II. Annu. Rev. Biochem. 79: 271–293 [DOI] [PubMed] [Google Scholar]
- Sharon G., Burkett T. J., Garfinkel D. J., 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14: 6540–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., et al. , 1988. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 16: 8207–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Hicks J. B., Fink G. R., 1987. Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shore D., Nasmyth K., 1987. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell 51: 721–732 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starling A. L., Ortega J. M., Gollob K. J., Vicente E. J., Andrade-Nobrega G. M., et al. , 2003. Evaluation of alternative reporter genes for the yeast two-hybrid system. Genet. Mol. Res. 2: 124–135 [PubMed] [Google Scholar]
- Stemmer W. P., Crameri A., Ha K. D., Brennan T. M., Heyneker H. L., 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164: 49–53 [DOI] [PubMed] [Google Scholar]
- Sussel L., Shore D., 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88: 7749–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor H. O., O’Reilly M., Leslie A. G., Rhodes D., 2000. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol. 303: 693–707 [DOI] [PubMed] [Google Scholar]
- Tian J., Gong H., Sheng N., Zhou X., Gulari E., et al. , 2004. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Tornow J., Santangelo G. M., 1990. Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene 90: 79–85 [DOI] [PubMed] [Google Scholar]
- Welch M., Govindarajan S., Ness J. E., Villalobos A., Gurney A., et al. , 2009. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE 4: e7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Mager W. H., Nieuwint R. T., Wassenaar G. M., van der Kuyl A. C., et al. , 1987. Analysis of upstream activation sites of yeast ribosomal protein genes. Nucleic Acids Res. 15: 6037–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P., 1989. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem. 264: 11816–11821 [PubMed] [Google Scholar]
- Xu H., Boeke J. D., 1990. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell. Biol. 10: 2695–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrington, R. M., 2009 Synthetic and Biochemical Studies of Retrotransposon Ty1. Ph.D. Thesis, Johns Hopkins University School of Medicine, Baltimore.
- Yonaha M., Proudfoot N. J., 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3: 593–600 [DOI] [PubMed] [Google Scholar]
- Zhao Y., McIntosh K. B., Rudra D., Schawalder S., Shore D., et al. , 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 26: 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]