Abstract
Due to its genetic tractability and increasing wealth of accessible data, the yeast Saccharomyces cerevisiae is a model system of choice for the study of the genetics, biochemistry, and cell biology of eukaryotic lipid metabolism. Glycerolipids (e.g., phospholipids and triacylglycerol) and their precursors are synthesized and metabolized by enzymes associated with the cytosol and membranous organelles, including endoplasmic reticulum, mitochondria, and lipid droplets. Genetic and biochemical analyses have revealed that glycerolipids play important roles in cell signaling, membrane trafficking, and anchoring of membrane proteins in addition to membrane structure. The expression of glycerolipid enzymes is controlled by a variety of conditions including growth stage and nutrient availability. Much of this regulation occurs at the transcriptional level and involves the Ino2–Ino4 activation complex and the Opi1 repressor, which interacts with Ino2 to attenuate transcriptional activation of UASINO-containing glycerolipid biosynthetic genes. Cellular levels of phosphatidic acid, precursor to all membrane phospholipids and the storage lipid triacylglycerol, regulates transcription of UASINO-containing genes by tethering Opi1 to the nuclear/endoplasmic reticulum membrane and controlling its translocation into the nucleus, a mechanism largely controlled by inositol availability. The transcriptional activator Zap1 controls the expression of some phospholipid synthesis genes in response to zinc availability. Regulatory mechanisms also include control of catalytic activity of glycerolipid enzymes by water-soluble precursors, products and lipids, and covalent modification of phosphorylation, while in vivo function of some enzymes is governed by their subcellular location. Genome-wide genetic analysis indicates coordinate regulation between glycerolipid metabolism and a broad spectrum of metabolic pathways.
THE yeast, Saccharomyces cerevisiae, has emerged as a powerful model system for the elucidation of the metabolism, cell biology, and regulation of eukaryotic lipids. Due to the strong homology of yeast proteins, pathways, and regulatory networks with those in higher eukaryotes, yeast has provided numerous insights into the genetics and biochemistry of lipid-related diseases. As a system for the study of eukaryotic lipid metabolism, the advantages of yeast include its vast, well-curated, and electronically accessible archives of genetic data, including those detailing gene–enzyme relationships in the pathways for lipid synthesis and turnover. Another major advantage is the rapidly increasing understanding of the regulation and localization of enzymes and the movement of lipids within and among cellular membranes and compartments in yeast.
This article presents an overview of progress in elucidating gene–enzyme relationships, cellular localization, and regulatory mechanisms governing glycerolipid metabolism in yeast. The metabolism covered in this YeastBook chapter includes the regulation, synthesis, and turnover of phospholipids and triacylglycerol (TAG) and their precursors in the context of changing growth conditions and nutrient availability. All glycerophospholipids in yeast, including the major phospholipids, phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylethanolamine (PE), and phosphatidylcholine (PC), are derived from the precursor lipid, phosphatidic acid (PA) (Figures 1 and 2). A major topic of this review article is the tremendous recent progress in elucidating the complex regulatory mechanisms that control the connected and coordinated pathways involved in the synthesis of glycerophospholipids and TAG, which is also derived from PA (Figures 2, 3, and 4). The regulation of glycerolipid metabolism occurs at many levels and is a major topic discussed in this article. Adding complexity to analysis of the regulatory networks connected to lipid metabolism is the fact that critical signals controlling this regulation arise during the ongoing biosynthesis and turnover of the lipids themselves and involve precursors and metabolites embedded in the metabolism.
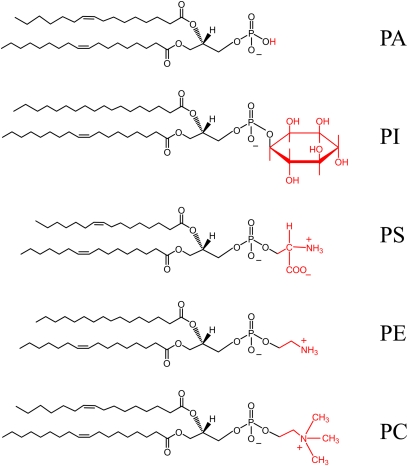
Figure 1 .
Phospholipid structures. The diagram shows the structures of the phospholipid PA and the major phospholipids PI, PS, PE, and PC that are derived from PA. The hydrophilic head groups (H, inositol, serine, ethanolamine, and choline) that are attached to the basic phospholipid structure are shown in red. The four most abundant fatty acids esterified to the glycerol-3-phosphate backbone of the phospholipids are palmitic acid, palmitoleic acid, steric acid, and oleic acid. The type and position of the fatty acyl moieties in the phospholipids are arbitrarily drawn. The relative amounts of the phospholipids as well as their fatty acyl compositions vary depending on strain (e.g., mutation) and growth condition.
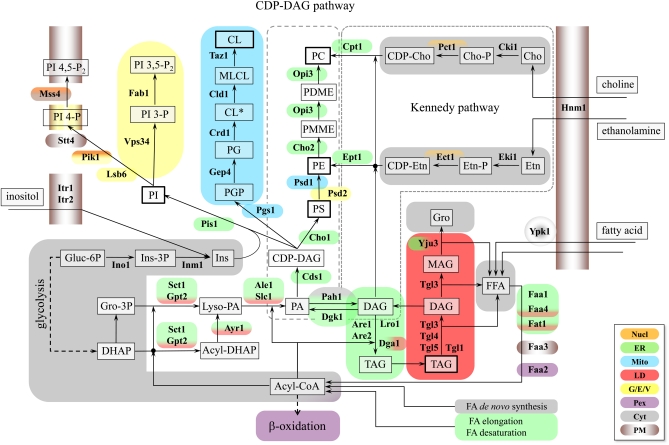
Figure 2 .
Pathways for the synthesis of glycerolipids and their subcellular localization. Phospholipids and TAG share DAG and PA as common precursors. In the de novo synthesis of phospholipids, PA serves as the immediate precursor of CDP-DAG, precursor to PI, PG, and PS. PS is decarboxylated to form PE, which undergoes three sequential methylations resulting in PC. PA also serves as a precursor for PGP, PG, and ultimately CL, which undergoes acyl-chain remodeling to the mature lipid. Alternatively, PA is dephosphorylated, producing DAG, which serves as the precursor of PE and PC in the Kennedy pathway. DAG also serves as the precursor for TAG and can be phosphorylated, regenerating PA. The names of the enzymes that are discussed in detail in this YeastBook chapter are shown adjacent to the arrows of the metabolic conversions in which they are involved and the gene–enzyme relationships are shown in Tables 1–3. Lipids and intermediates are boxed, with the most abundant lipid classes boxed in boldface type. Enzyme names are indicated in boldface type. The abbreviations used are: TAG, triacylglycerols; PI, phosphatidylinositol; PA, phosphatidic acid; CDP-DAG, CDP-diacylglycerol; DAG, diacylglycerol; MAG, monoacylglycerol; Gro, glycerol; DHAP, dihydroxyacetone phosphate, PS, phosphatidylserine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PGP phosphatidylglycerol phosphate; CL* precursor cardiolipin; MLCL, monolyso-cardiolipin; CL, mature cardiolipin; PMME, phosphatidylmonomethylethanolamine; PDME, phosphatidyl-dimethylethanolamine; PC, phosphatidylcholine; FFA, free fatty acids; Cho, choline, Etn, ethanolamine, Ins, inositol; Cho-P, choline phosphate; CDP-Cho, CDP-choline; Etn-P, ethanolamine phosphate; CDP-Etn, CDP-ethanolamine; PI 3-P, phosphatidylinositol 3-phosphate; PI 4-P, phosphatidylinositol 4-phosphate; PI 4,5-P2, phosphatidylinositol 4,5-bisphosphate; PI 3,5-P2, phosphatidylinositol 3,5-bisphosphate. Nucl, nucleus; ER, endoplasmic reticulum; Mito, mitochondria; LD, lipid droplets; G/E/V, Golgi, endosomes, vacuole; Pex, peroxisomes; Cyt, cytoplasma; PM, plasma membrane. CL* indicates a precursor of cardiolipin (CL) with saturated acyl-chain that undergoes deacylation/reacylation to mature CL. See text for details.
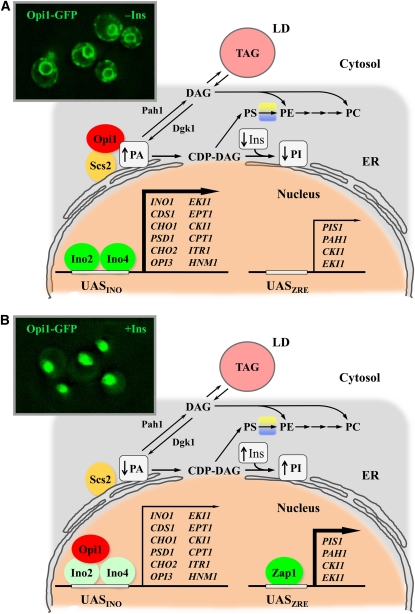
Figure 3 .
Model for PA-mediated regulation of phospholipid synthesis genes. (A) Growth conditions (e.g., exponential phase, inositol depletion, or zinc supplementation) under which the levels of PA are relatively high, the Opi1 repressor is tethered to the nuclear/ER membrane, and UASINO-containing genes are maximally expressed (boldface arrow) by the Ino2-Ino4 activator complex. (Inset) Localization of Opi1, fused with GFP at its C-terminal end and integrated into the chromosome, being expressed under its own promoter in live cells growing logarithmically in synthetic complete medium lacking inositol (−Ins) and analyzed by fluorescence microscopy. (B) Growth conditions (e.g., stationary phase, inositol supplementation, or zinc depletion) under which the levels of PA are reduced, Opi1 dissociates from the nuclear/ER membrane, and enters into the nucleus where it binds to Ino2 and attenuates (thin arrow) transcriptional activation by the Ino2–Ino4 complex. (Inset) Localization of Opi1, as described in A, except that the cells are growing logarithmically in medium containing 75 μM inositol. PA level decreases by the stimulation of PI synthesis in response to inositol (Ins) supplementation and by Zap1-mediated induction of PIS1, that results in an increase in PI synthesis in response to zinc depletion. The regulation in response to zinc depletion and stationary phase occurs without inositol supplementation. Pah1 and Dgk1 play important roles in controlling PA content and transcriptional regulation of UASINO-containing genes. The synthesis of TAG (which is stored in lipid droplets, LD) and phospholipids (with the exception of PE, which occurs in the mitochondria and Golgi) occurs in the ER. Fluorescence microscopy images of Opi1 localization courtesy of Yu-Fang Chang, Henry Laboratory, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY.
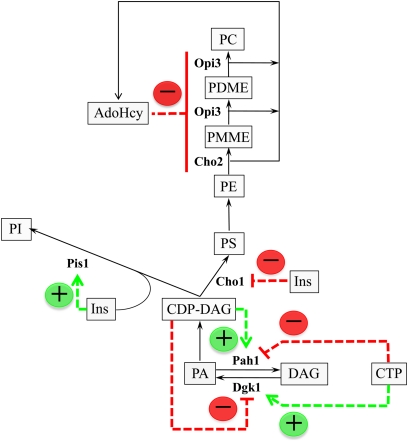
Figure 4 .
Regulation of phospholipid synthesis by soluble lipid precursors and metabolites. The diagram shows the major steps in the synthesis of phospholipids. The enzymes that are biochemically regulated by phospholipid precursors and products are shown. The green arrow designates the stimulation of enzyme activity, whereas the red line designates the inhibition of enzyme activity. Details on the biochemical regulation of these enzymes are discussed in the section Water soluble precursors of phospholipids, metabolism, and regulatory roles and in Carman and Han (2009a). See Figure 2 for abbreviations.
For example, PA plays a number of signaling roles vital to the regulation of lipid metabolism in yeast (Figure 3), in addition to its function as precursor to all phospholipids and TAG (Figure 2). PI synthesis is regulated in response to its precursor, inositol, on several levels (Figures 3 and 4) and PI also serves as precursor to both phosphatidylinositolphosphates and inositol-containing sphingolipids, both of which are implicated in a wide range of signaling and regulatory activities (Strahl and Thorner 2007; Dickson 2008, 2010), topics that will not be dealt with in detail in this YeastBook chapter. In addition, the enzymes controlling the metabolism of gycerolipids are localized to specific cellular compartments (Figure 2; Tables 1–3), while the lipids themselves are, for the most part, distributed to a much wider range of cellular compartments. Moreover, the regulation of TAG metabolism plays a major role in lipid droplet (LD) formation and depletion (Murphy and Vance 1999; Rajakumari et al. 2008; Kurat et al. 2009; Kohlwein 2010a), a topic that will also be addressed in detail in the chapter on Lipid Droplets and Peroxisomes. Thus, detailed knowledge of pathways, gene–enzyme relationships, and subcellular localization of enzymes and pools of lipids and metabolites involved in the synthesis and turnover of glycerolipids (Figures 2–4, Tables 1–3) is essential to the elucidation of the complex mechanisms responsible for their regulation.
Table 1 . Glycerolipid synthesis enzymes.
| Gene | Ino− or Opi− phenotype | Enzyme | Mol mass (kDa) | Isoelectric point | Molecules per cellb | Locationc | Transmembrane domains | Phosphorylation sitesd |
|---|---|---|---|---|---|---|---|---|
| SCT1 (GAT2) | Ino− | Glycerol-3-P /dihydroxyacetone-P acyltransferase | 85.7 | 7.27 | 1050 | ER | 4 | Few |
| GPT2 (GAT1) | — | Glycerol-3-P /dihydroxyacetone-P acyltransferase | 83.6 | 10.3 | 3100 | ER, lipid droplets | 4 | Several |
| AYR1 | — | Acyl DHAP reductase | 32.8 | 9.92 | 3670 | ER, lipid droplets | None | None |
| SLC1 | — | LysoPA/Acylglycerol-3-P acyltransferase | 33.8 | 10.41 | ND | ER, lipid droplets | 1 | None |
| ALE1 (SLC4,LPT1,LCA1) | — | LysoPA/Acylglycerol-3-P acyltransferase | 72.2 | 10.3 | ND | ER | 7 | Several |
| PSI1 (CST26) | — | LysoPI acyltransferase | 45.5 | 10.15 | 2010 | ER | 4 | None |
| TAZ1 | — | LysoPC acyltransferase/ monolysoCL acyltransferase | 44.2 | 9.38 | 1340 | Mitochondria | None | None |
| CDS1 (CDG1)a | Opi− | CDP-DAG synthase | 51.8 | 8.64 | ND | ER, mitochondria | 6 | Few |
| CHO1 (PSS1)a | Opi− | PS synthase | 30.8 | 6.23 | ND | ER | 2 | Several |
| PSD1a | Opi− | PS decarboxylase | 56.6 | 9.84 | 1080 | Mitochondria | None | None |
| PSD2 | — | PS decarboxylase | 130 | 7.85 | ND | Vacuole, endomembranes | None | Few |
| CHO2 (PEM1)a | Opi− | PE methyltransferase | 101.2 | 8.56 | 1810 | ER | 8 | Few |
| OPI3 (PEM2)a | Opi− | Phospholipid methyltransferase | 23.1 | 9.6 | 5890 | ER, mitochondria | None | None |
| PAH1 (SMP2) | — | PA phosphatase | 95 | 4.68 | 3910 | Cytoplasm, ER | None | Several |
| DGK1 (HSD1) | — | DAG kinase | 32.8 | 9.48 | 784 | ER | 4 | Few |
| EKI1a | — | Ethanolamine kinase | 61.6 | 5.69 | 3420 | Cytoplasm | None | Few |
| ECT1 (MUQ1) | — | Ethanolamine-P cytidylyltransferase | 36.8 | 6.44 | 4700 | Cytoplasm | None | None |
| EPT1a | — | Ethanolamine/choline phosphotransferase | 44.5 | 6.5 | ND | ER | 7 | None |
| CKI1a | — | Choline kinase | 66.3 | 5.43 | 3930 | Cytoplasm | None | Several |
| PCT1 (CCT1,BSR2)a | Opi− | Choline-P cytidylyltransferase | 49.4 | 9.26 | 3050 | Cytoplasm, nucleus | None | Several |
| CPT1a | — | Choline phosphotransferase | 44.8 | 6.57 | 981 | Membrane | 8 | None |
| PGS1 (PEL1) | — | PGP synthase | 59.3 | 10.5 | ND | Mitochondria | None | None |
| GEP4 | — | PGP phosphatase | 20.9 | 9.18 | ND | Mitochondria | None | None |
| CRD1 (CLS1) | — | CL synthase | 32 | 10.55 | 876 | Mitochondria | 3 | None |
| PIS1 | Ino− | PI synthase | 24.8 | 8.92 | 3810 | ER | 3 | None |
| LSB6 | — | PI 4-kinase | 70.2 | 6.68 | 57 | Plasma membrane, vacuole membrane | None | None |
| STT4 | Ino− | PI 4-kinase | 214.6 | 7.44 | 846 | Plasma membrane | None | Few |
| PIK1 | — | PI 4-kinase | 119.9 | 6.46 | 1600 | Plasma membrane, nucleus, Golgi | None | Several |
| VPS34 (END12,PEP15,VPL7,VPT29,STT8,VPS7) | Opi− | PI 3-kinase | 100.9 | 7.79 | 1080 | Vacuole | None | None |
| MSS4 | Ino− | PI 4-P 5-kinase | 89.3 | 10.13 | ND | Cytoplasm | None | Several |
| FAB1 (SVL7) | Ino− | PI 3-P 5-kinase | 257.4 | 8.45 | 149 | Vacuole | None | Several |
| DGA1a | — | Acyl-CoA diacylglycerol acyltransferase | 47.7 | 10.39 | 907 | ER, lipid droplets | 1 | Few |
| LRO1a | — | Phospholipid diacylglycerol acyltransferase | 75.3 | 6.67 | ND | ER | 1 | Few |
| ARE1 (SAT2)a | — | Acyl-CoA sterol acyltransferase | 71.6 | 8.27 | ND | ER | 9 | Several |
| ARE2 (SAT1)a | — | Acyl-CoA sterol acyltransferase | 74.0 | 7.71 | 279 | ER | 9 | Several |
| Phospholipid synthesis regulatory proteins | ||||||||
| INO2 (DIE1, SCS1)a | Ino− | Transcriptional activator | 34.2 | 6.23 | 784 | Nucleus | None | None |
| INO4 | Ino− | Transcriptional activator | 17.4 | 10.21 | 521 | Nucleus, cytoplasm | None | None |
| OPI1 | Opi− | Transcriptional repressor | 46 | 4.87 | 1280 | Nuclear/ER membrane | None | Few |
Much of the information in the table may be found in the Saccharomyces Genome Database. Ino−, inositol auxotrophy; Opi−, inositol excretion; ND, not determined.
Genes containing the UASINO element and regulated by the Ino2-Ino4-Opi1 circuit. The names in parentheses are aliases.
Table 3 . Glycerolipid turnover enzymes.
| Gene | Ino− or Opi−phenotype | Enzyme | Mol mass (kDa) | Isoelectric point | Molecules per cella | Locationbb | Transmembrane domains | Phosphorylation sitesc |
|---|---|---|---|---|---|---|---|---|
| CLD1 | — | CL specific phospholipase A2 | 52 | 10.3 | ND | Mitochondria | None | None |
| NTE1 | Ino− | PC specific phospholipase B | 187.1 | 8.19 | 521 | ER | 3 | Several |
| PLB1 | — | PC/PE specific phospholipase B | 71.6 | 4.36 | ND | Plasma membrane, ER, vesicles, extracellular | None | None |
| PLB2 | — | Nonspecific phospholipase B | 75.4 | 4.35 | 623 | Plasma membrane, ER, vesicles | None | None |
| PLB3 | — | Nonspecific phospholipase B | 75 | 4.7 | ND | Cytoplasm, vacuole, vesicles | None | None |
| PLC1 | — | PI 4,5-P2 specific phospholipase C | 110.5 | 9.84 | ND | Cytoplasm | None | Few |
| PGC1 | — | PG specific phospholipase C | 37 | 8.93 | 3270 | Mitochondria | None | None |
| SPO14 (PLD1) | — | PC specific phospholipase D | 195.2 | 7.58 | 49 | Cytoplasm | None | Several |
| SAC1 (RSD1) | Ino− | Nonspecific polyphosphoinositide phosphatase | 71.1 | 7.75 | 48,000 | ER, Golgi, vacuole | 2 | None |
| INP51 (SJL1) | — | PI 4,5-P2 phosphatase | 108.4 | 6.7 | 98 | Cytoplasm | None | None |
| INP52 (SJL2) | — | Nonspecific polyphosphoinositide phosphatase | 133.3 | 8.97 | ND | Actin | None | Several |
| INP53 (SJL3, SOP2) | — | Nonspecific polyphosphoinositide phosphatase | 124.6 | 7.18 | 1520 | Actin | None | Several |
| INP54 | — | PI 4,5-P2 phosphatase | 43.8 | 7.6 | 1200 | ER | None | None |
| YRM1 | — | PI 3-P phosphatase | 91 | 7.19 | 125 | Cytoplasm | None | Few |
| FIG4 | — | PI 3,5-P2 phosphatase | 101.7 | 5.52 | 339 | Vacuole | None | None |
| DPP1 (ZRG1) | — | DGPP phosphatase/nonspecific lipid phosphate phosphatase | 33.5 | 6.42 | 3040 | Vacuole | 5 | Few |
| LPP1 | — | Nonspecific lipid phosphate phosphatase | 31.5 | 8.25 | 300 | Golgi | 4 | None |
| PHM8 | — | LysoPA phosphatase | 37.7 | 5.14 | 195 | Cytoplasm, nucleus | None | None |
| TGL1 | — | Triacylglycerol lipase, sterylester hydrolase | 63.0 | 6.83 | 1470 | ER, lipid droplets | None | Several |
| TGL2 | — | Acylglycerol lipase | 37.5 | 8.41 | ND | Mitochondria | None | None |
| TGL3 | — | Triacylglycerol lipase, lysoPA acyltransferase | 73.6 | 8.50 | 3210 | Lipid droplets | 1 | Few |
| TGL4 | — | Triacylglycerol lipase, Ca++ dependent phospholipase A2, lysoPA acyltransferase | 102.7 | 8.05 | 195 | Lipid droplets | None | Several |
| TGL5 | — | Triacylglycerol lipase, lysoPA acyltransferase | 84.7 | 9.84 | 358 | Lipid droplets | 1 | Several |
| YJU3 | — | Monoacylglycerol lipase | 35.6 | 8.5 | 2140 | Lipid droplet, ER | None | None |
Much of the information in the table may be found in the Saccharomyces Genome Database. The names in parentheses are aliases. Ino−, inositol auxotrophy; ND, not determined.
Notably, in eukaryotes phospholipids play many vital roles in the biology of the cell that extend beyond lipid metabolism itself. These include roles in membrane trafficking and membrane identity (Vicinanza et al. 2008) and anchoring of membrane proteins (Roth et al. 2006; Pittet and Conzelmann 2007; Fujita and Jigami 2008), complex topics in their own right, which will be discussed only in brief in this YeastBook chapter. Phospholipids also serve as signaling molecules and as precursors of signaling molecules (Strahl and Thorner 2007). Thus, the advancements in yeast glycerolipid metabolism discussed in this review article also have enormous potential to contribute critical insights into these vital roles of lipids and lipid-mediated signaling in eukaryotic cells.
Pathways of glycerolipid metabolism
Major glycerolipids of S. cerevisiae include the phospholipids PC, PE, PI, PS (Figure 1), phosphatidylglycerol (PG), and cardiolipin (CL) (Rattray et al. 1975; Henry 1982; Carman and Henry 1989; Paltauf et al. 1992; Guan and Wenk 2006; Ejsing et al. 2009). Minor phospholipids include intermediates such as PA, CDP-diacylglycerol (CDP-DAG), phosphatidylmonomethylethanolamine (PMME), phosphatidyldimethylethanolamine (PDME), the D-3, D-4, and D-5 polyphosphoinositides, and lysophospholipids (Rattray et al. 1975; Oshiro et al. 2003; Strahl and Thorner 2007). TAG and diacylglycerol (DAG) are the major neutral glycerolipids. The fatty acids that are commonly esterified to the glycerophosphate backbone of yeast glycerolipids include palmitic acid (C16:0), palmitoleic acid (C16:1), stearic acid (C18:0), and oleic acid (C18:1) (Rattray et al. 1975; Henry 1982; Bossie and Martin 1989; McDonough et al. 1992; Martin et al. 2007). The pathways for the synthesis of phospholipids and TAG are shown in Figure 2. The enzymes and transporters of glycerolipid metabolism and the genes that encode them are listed in Tables 1–3. The gene–protein relationships shown in the tables have been confirmed by the analysis of gene mutations and/or by the biochemical characterization of the enzymes and transporters (Carman and Henry 1989; Greenberg and Lopes 1996; Henry and Patton-Vogt 1998; Carman and Henry 1999; Black and Dirusso 2007; Tehlivets et al. 2007; Kohlwein 2010b; Carman and Han 2011).
Synthesis and turnover of phospholipids
In the de novo pathways (Figure 2, Table 1), all membrane phospholipids are synthesized from PA, which is derived from glycerol-3-P via lysoPA by two fatty acyl CoA-dependent reactions that are catalyzed in the endoplasmic reticulum (ER) by the SCT1- and GPT2-encoded glycerol-3-P acyltransferases and the SLC1- and ALE1-encoded lysoPA/lysophospholipid acyltransferases, respectively (Athenstaedt and Daum 1997; Athenstaedt et al. 1999b; Zheng and Zou 2001; Benghezal et al. 2007; Chen et al. 2007b; Jain et al. 2007; Riekhof et al. 2007b). The glycerol-3-P acyltransferase enzymes also utilize dihydroxyacetone-P as a substrate, and the product acyl dihydroxyacetone-P is converted to lysoPA by the lipid droplet (LD) and ER-associated AYR1-encoded reductase (Athenstaedt and Daum 2000). At this point in the pathway, PA is partitioned to CDP-DAG, catalyzed by CDS1-encoded CDP-DAG synthase (Carter and Kennedy 1966; Kelley and Carman 1987; Shen et al. 1996) or to DAG, catalyzed by PAH1-encoded PA phosphatase (Han et al. 2006) (Figure 1). CDP-DAG synthase activity has been detected in the ER and in mitochondria (Kuchler et al. 1986), whereas PA phosphatase is a cytosolic enzyme that must associate with membranes to catalyze the dephosphorylation of PA to produce DAG (Han et al. 2006; Carman and Han 2009a). CDP-DAG and DAG are used to synthesize PE and PC by two alternative routes, namely, the CDP-DAG and Kennedy pathways (Figure 2). In the CDP-DAG pathway, CDP-DAG is converted to PS by the ER localized CHO1-encoded PS synthase (Atkinson et al. 1980; Letts et al. 1983; Bae-Lee and Carman 1984; Kiyono et al. 1987; Nikawa et al. 1987b). Yeast has two PS decarboxylases encoded, by the PSD1 and PSD2 genes. Psd1, localized to the inner mitochondrial membrane, accounts for the majority of the enzymatic activity in yeast, while the minor activity, Psd2, associates with Golgi/vacuole (Clancey et al. 1993; Trotter et al. 1993, 1995; Voelker 2003). PE then undergoes three sequential methylation reactions in the ER (Gaynor and Carman 1990), the first of which is catalyzed by the CHO2-encoded PE methyltransferase, while the final two methylations are catalyzed by the OPI3-encoded phospholipid methyltransferase (Kodaki and Yamashita 1987; Summers et al. 1988; Kodaki and Yamashita 1989; McGraw and Henry 1989). The CDP-DAG pathway is the major route for synthesis of PE and PC when wild-type cells are grown in the absence of ethanolamine and choline, and cho1, psd1psd2, and cho2opi3 mutants defective in this pathway have choline/ethanolamine auxotrophy phenotypes (Atkinson et al. 1980a Summers et al. 1988; McGraw and Henry 1989; Trotter and Voelker 1995; Trotter et al. 1995). PE and PC synthesis in mutants defective in the CDP-DAG pathway can also be supported by exogenously supplied lysoPE, lysoPC, or PC with short acyl chains, which are transported into the cell. LysoPE and lysoPC are acylated by the ALE1-encoded lysophospholipid acyltransferase (Jain et al. 2007; Tamaki et al. 2007; Riekhof et al. 2007a,b). Short chain PE and PC are remodeled with C16 and C18 acyl chains prior to incorporation into the membrane (Tanaka et al. 2008; Deng et al. 2010).
In the Kennedy pathway (Hjelmstad and Bell 1990), PE and PC are synthesized, respectively, from ethanolamine and choline (Figure 2, Table1). Exogenous ethanolamine and choline are both transported into the cell by the HNM1-encoded choline/ethanolamine transporter (Nikawa et al. 1986). The EKI1-encoded ethanolamine kinase (Kim et al. 1999) and the CKI1-encoded choline kinase (Hosaka et al. 1989) are both cytosolic enzymes, which, respectively, phosphorylate ethanolamine and choline with ATP to form ethanolamine-P and choline-P. Ethanolamine-P may also be derived from sphingolipids by dihydrosphingosine-1-P lyase, encoded by the DPL1 gene (Saba et al. 1997; Schuiki et al. 2010). These intermediates are then activated with CTP to form CDP-ethanolamine and CDP-choline, respectively, by the ECT1-encoded ethanolamine-P cytidylyltransferase (Min-Seok et al. 1996) and the PCT1-encoded choline-P cytidylyltransferase (Tsukagoshi et al. 1987), which are associated with the nuclear/ER membrane (Huh et al. 2003; Natter et al. 2005). Finally, the EPT1-encoded ethanolamine phosphotransferase (Hjelmstad and Bell 1988, 1991) and the CPT1-encoded choline phosphotransferase (Hjelmstad and Bell 1987, 1990), respectively, catalyze the reactions of CDP-ethanolamine and CDP-choline with DAG provided by the PAH1-encoded PA phosphatase to form PE and PC. Ept1 will also catalyze the CDP-choline–dependent reaction (Hjelmstad and Bell 1988). Cpt1 and Ept1 have somewhat ambiguous patterns of localization to vesicular structures, but have also been described to localize to mitochondria or ER (Huh et al. 2003; Natter et al. 2005).
The CDP-DAG and Kennedy pathways are both used by wild-type cells, regardless of whether or not ethanolamine and choline are present in the growth medium (Patton-Vogt et al. 1997; Henry and Patton-Vogt 1998; Kim et al. 1999). In the absence of exogenous choline, this Kennedy pathway precursor may be derived from hydrolysis of PC synthesized by the CDP-DAG pathway and subsequently hydrolyzed by phospholipase D (Patton-Vogt et al. 1997; Xie et al. 1998) encoded by the SPO14 gene (Rose et al. 1995; Waksman et al. 1996). Choline for PC synthesis can also be derived from Nte1-catalyzed deacylation (Zaccheo et al. 2004) followed by hydrolysis by Gde1 to produce choline and glycerophosphate (Fernandez-Murray and McMaster 2005a; Fisher et al. 2005; Patton-Vogt 2007). The activity of a Ca++-dependent phospholipase D with preference for PS and PE has been detected (Mayr et al. 1996), but the corresponding gene encoding a PE-specific phospholipase D has yet to be identified. Thus, proof that ethanolamine derived from PE is recycled for PE synthesis via the Kennedy pathway is still lacking. Kennedy pathway mutants (e.g., cki1eki1 and cpt1ept1) defective in both the CDP-choline and CDP-ethanolamine branches can synthesize PC only by the CDP-DAG pathway (McMaster and Bell 1994; Morash et al. 1994; Kim et al. 1999). However, unlike the CDP-DAG pathway mutants, the Kennedy pathway mutants do not exhibit any auxotrophic requirements and have an essentially normal complement of phospholipids (Morash et al. 1994; Kim et al. 1999).
In the synthesis of the inositol-containing phospholipids (Figure 2, Table 1), CDP-DAG donates its phosphatidyl moiety to inositol to form PI (Paulus and Kennedy 1960; Fischl and Carman 1983) in a reaction that competes in the ER with PS synthase for their common substrate, CDP-DAG (Kelley et al. 1988). While PIS1-encoded PI synthase is essential, a strain expressing a mutant form of Pis1 with a lower affinity for inositol has been isolated as an inositol auxotroph (Nikawa and Yamashita 1984; Nikawa et al. 1987a). The inositol used in PI synthesis is either synthesized de novo (discussed below) or obtained from the growth medium via the ITR1- and ITR2-encoded inositol transporters (Table 2) (Nikawa et al. 1991). Once formed, PI may be converted to PI 3-P by the VPS34-encoded PI 3 kinase (Herman and Emr 1990; Schu et al. 1993) or to PI 4-P by the PI 4 kinases encoded by LSB6 (Han G-S et al. 2002; Shelton et al. 2003), STT4 (Yoshida et al. 1994a), and PIK1 (Flanagan et al. 1993; Garcia-Bustos et al. 1994). PI 4-P may be further phosphorylated to PI 4,5-P2 by the MSS4-encoded PI 4-P 5 kinase (Yoshida et al. 1994b), whereas PI 3-P may be phosphorylated to PI 3,5-P2 by the FAB1-encoded PI 3-P 5 kinase (Yamamoto et al. 1995). The specific localization of these kinases and the lipids they produce play important roles in signaling, membrane identity, and membrane trafficking (Strahl and Thorner 2007). PI also serves as a precursor in the synthesis of the complex sphingolipids in yeast, which also play essential roles in signaling and membrane function (Dickson and Lester 2002; Dickson 2008), topics that are beyond the scope of this YeastBook chapter.
Table 2 . Glycerolipid precursor enzymes and transporters.
| Gene | Ino− or Opi− phenotype | Enzyme | Mol mass (kDa) | Isoelectric point | Molecules per cellb | Locationc | Transmembrane domains | Phosphorylation sitesd |
|---|---|---|---|---|---|---|---|---|
| ACC1 (ABP2,FAS3,MTR7)a | — | Acetyl CoA carboxylase | 250.4 | 6.22 | 20,200 | Cytoplasm | None | Several |
| HFA1 | — | Acetyl CoA carboxylase | 241.8 | 8.05 | 396 | Mitochondria | None | Few |
| FAS1a | — | Fatty acid synthase (β subunit) | 228.7 | 5.79 | 91,800 | Cytoplasm | None | Several |
| FAS2a | — | Fatty acid synthase (α subunit) | 206.9 | 5.21 | 17,000 | Cytoplasm | None | Several |
| ETR1(MRF’) | Ino− | 2-Enoyl thioester reductase | 42 | 9.78 | 1560 | Mitochondria | None | None |
| HTD2 (RMD12) | — | 3-Hydroxyacyl-thioester dehydratase | 33 | 9 | 799 | Mitochondria | None | None |
| MCT1 | — | Malonyl-CoA:ACP transferase | 40.7 | 6.9 | 1360 | Mitochondria | None | None |
| OAR1 | — | 3-Oxoacyl-ACP reductase | 31.2 | 9.3 | 1760 | Mitochondria | None | None |
| ACP1 | — | Acyl carrier protein | 13.9 | 4.64 | 60,500 | Mitochondria | None | Few |
| PPT2 | — | Phosphopantetheine:protein transferase | 19.9 | 8.46 | 486 | Mitochondria | None | None |
| CEM1 | — | β-keto-acyl synthase | 47.6 | 8.26 | 1660 | Mitochondria | None | None |
| FAA1 | — | Fatty acyl CoA synthetase | 77.8 | 7.58 | 7470 | ER, lipid droplets | None | None |
| FAA2 (FAM1) | — | Fatty acyl CoA synthetase | 83.4 | 7.7 | 358 | Peroxisomes | None | None |
| FAA3 | — | Fatty acyl CoA synthetase | 77.9 | 9.7 | 6440 | Plasma membrane | None | None |
| FAA4 | — | Fatty acyl CoA synthetase | 77.2 | 6.52 | 31,200 | ER, lipid droplets | None | None |
| FAT1 | — | Fatty acid transporter and fatty acyl CoA synthetase | 77.1 | 8.47 | 16,900 | ER, lipid droplets | 3 | None |
| OLE1 (MDM2) | — | Δ9 Fatty acid desaturase | 58.4 | 9.71 | ND | ER | 4 | None |
| ELO1 | — | FA elongase, condensing enzyme | 36.2 | 10.2 | 937 | ER | 5 | None |
| FEN1 (ELO2) | — | FA elongase, condensing enzyme | 40.0 | 10.35 | 3510 | ER | 7 | Several |
| SUR4 (ELO3) | — | FA elongase, condensing enzyme | 39.5 | 10.13 | ND | ER | 6 | None |
| IFA38 (YBR159w) | — | β-keto acyl-CoA reductase | 38.7 | 10.28 | 41900 | ER | 1 | None |
| PHS1 | — | 3-Hydroxy acyl-CoA dehydratase | 24.5 | 10.84 | ND | ER | 6 | None |
| TSC13 | — | Enoyl-CoA reductase | 36.8 | 10.38 | 23600 | ER | 4 | None |
| INO1a (APR1) | Ino− | Inositol 3-P synthase | 59.6 | 5.77 | ND | Cytoplasm | None | None |
| INM1 | — | Inositol 3-P phosphatase | 32.8 | 5 | 2440 | Cytoplasm, nucleus | None | None |
| URA7 | Ino− | CTP synthetase | 64.7 | 5.93 | 57,600 | Cytoplasm | None | Several |
| URA8 | — | CTP synthetase | 63 | 6.02 | 5370 | Cytoplasm | None | None |
| ITR1a | Opi− | Inositol transporter | 63.5 | 6.51 | ND | Plasma membrane | 12 | Several |
| ITR2 (HRB612) | — | Inositol transporter | 66.7 | 8.25 | 468 | Plasma membrane | 12 | None |
| HNM1 (CTR1)a | — | Choline transporter | 62 | 6.83 | ND | Plasma membrane | 12 | Few |
| GIT1 | — | GroPIns/GroPCho transporter | 57.3 | 8.64 | ND | Plasma membrane | 11 | None |
Much of the information in the table may be found in the Saccharomyces Genome Database. Ino−, inositol auxotrophy; Opi−, inositol excretion; ND, not determined.
Genes containing the UASINO element and are regulated by the Ino2-Ino4-Opi1 circuit, The names in parentheses are aliases.
In the synthesis of mitochondrial-specific phospholipids (Figure 2, Table1), CDP-DAG donates its phosphatidyl moiety to glycerol-3-P to form phosphatidylglycerophosphate (PGP) in the reaction catalyzed by the PGS1-encoded PGP synthase (Janitor and Subik 1993; Chang et al. 1998a). PGP is then dephosphorylated to PG by the GEP4-encoded PGP phosphatase (Osman et al. 2010). The CRD1-encoded CL synthase (Jiang et al. 1997; Chang et al. 1998b; Tuller et al. 1998) catalyzes the reaction between PG and another molecule of CDP-DAG to generate CL. The CLD1-encoded cardiolipin-specific phospholipase and the TAZ1-encoded monolyso cardiolipin acyltransferase, involved in establishing specific unsaturated cardiolipin species, are also specifically associated with the mitochondria (Beranek et al. 2009; Brandner et al. 2005).
The enzymes that catalyze the turnover of phospholipids include both phospholipases and lipid phosphatases (Table 3). NTE1-encoded phospholipase B (Zaccheo et al. 2004; Fernandez-Murray and McMaster 2005b, 2007) is an integral ER membrane protein and removes both fatty acids from PC to produce glycerophosphocholine (GroPCho). GroPCho may be re-acylated by an uncharacterized acyltransferase to PC (Ståhlberg et al. 2008). The phospholipase B enzymes encoded by PLB1, PLB2, and PLB3 catalyze the same type of reaction, but they are not specific and their localization is ambiguous. Plb1 localizes to the ER, vesicles, the plasma membrane, and the extracellular space and primarily utilizes PC and PE as substrates, whereas Plb3 is found in vesicles, vacuoles, as well as the cytosol and primarily uses PI as a substrate (Lee et al. 1994; Fyrst et al. 1999; Merkel et al. 1999, 2005). The GroPCho and glycerophosphoinositol (GroPIns) produced by the phospholipase B enzymes may be excreted into the growth medium and then transported back into the cell by the GIT1-encoded GroPCho/GroPIns transporter localized in the plasma membrane (Patton-Vogt and Henry 1998; Fisher et al. 2005). In turn, these molecules are hydrolyzed by a phosphodiesterase to produce the phospholipid precursor molecules choline and inositol, respectively (Patton-Vogt 2007). PLC1-encoded phospholipase C is cytosolic and specific for PI 4,5-P2 and produces DAG and inositol 1,4,5-trisphosphate (Flick and Thorner 1993; Yoko-O et al. 1993), and the PGC1-encoded phospholipase C localizes to lipid droplets and mitochondria and is specific for PG and produces DAG and glycerol 3-P (Simockova et al. 2008). The SPO14-encoded cytosolic phospholipase D is specific for PC and produces PA and choline (Rose et al. 1995; Waksman et al. 1996). Most phospholipids undergo rapid turnover and acyl-chain remodeling, which yields the typical complex pattern of lipid molecular species found in yeast (Schneiter et al. 1999; Guan and Wenk 2006; Ejsing et al. 2009). This remodeling is governed by specific acyltransferases, such as the PSI1-encoded acyltransferase in the ER that is involved in stearoyl-acylation of PI (Le Guedard et al. 2009), or the CLD1-encoded cardiolipin-specific phospholipase A in mitochondria (Beranek et al. 2009) and the TAZ1-encoded acyltransferase (yeast tafazzin ortholog; Gu et al. 2004; Testet et al. 2005). Moreover, several enzymes of TAG synthesis and degradation have additional activities, suggesting that they also may play a role in phospholipid acyl-chain remodeling (Rajakumari et al. 2008; Kohlwein 2010b; Rajakumari and Daum 2010a,b).
There are several phosphatase enzymes that catalyze the dephosphorylation of the polyphosphoinositides. Some of these enzymes are specific and some have broad substrate specificity. The YMR1-encoded (Taylor et al. 2000) and FIG4-encoded (Gary et al. 2002) phosphatases are specific for PI 3-P and PI 3,5-P2, respectively, whereas INP51-encoded (Stolz et al. 1998b) and INP54-encoded (Wiradjaja et al. 2001) phosphatases are specific for PI 4,5-P2. The phosphatase encoded by SAC1 will utilize PI 3-P, PI 4-P, and PI 3,5-P2 (Guo et al. 1999), whereas the phosphatases encoded by INP52 and INP53 will utilize any polyphosphoinositide as a substrate (Stolz et al. 1998; Guo et al. 1999).
The DPP1- and LPP1-encoded lipid phosphate phosphatase enzymes dephosphorylate a broad spectrum of substrates that include DAG pyrophosphate (DGPP), PA, lysoPA, sphingoid-base phosphates, and isoprenoid phosphates (Toke et al. 1998, 1999; Faulkner et al. 1999). While these enzymes may utilize PA as a substrate, they are not involved in the de novo synthesis of phospholipids and TAG; the function of which is ascribed to the PAH1-encoded PA phosphatase (Han et al. 2006; Carman and Han 2009a). The PHM8 gene encodes a lipid phosphatase that is specific for lysoPA and yields monoacylglycerol (MAG) and Pi (Reddy et al. 2008). N-acyl PE is a minor phospholipid species implicated in signaling processes (Merkel et al. 2005) and is degraded by the FMP30-encoded phospholipase D to N-acylethanolamide (NAE), which is related to endocannabinoids (Muccioli et al. 2009). NAE may be catabolized by Yju3, the major MAG lipase in yeast (see below).
Synthesis and turnover of TAG
The pathways for the synthesis of TAG and phospholipids share the same initial steps (Figure 2; Table 1; Kohlwein 2010b). Indeed, TAG is derived from the phospholipid, PA. The PAH1-encoded PA phosphatase provides DAG, which is acylated by the DGA1- and LRO1-encoded acyl-CoA–dependent and phospholipid-dependent diacylglycerol acyltransferases, respectively, to TAG (Oelkers et al. 2000; Oelkers et al. 2002; Sorger and Daum 2002; Kohlwein 2010b). The ARE1- and ARE2-encoded sterol acyltransferases utilize acyl-CoA but contribute only marginally to TAG synthesis from DAG (Yang, H. et al. 1996). Notably, deletion of both DGA1 and LRO1 genes does not result in a readily detectable growth phenotype in wild-type cells, indicating that TAG synthesis is not essential (Oelkers et al. 2002; Sorger and Daum 2002). Similarly, simultaneous deletion of the ARE1 and ARE2 genes does not impair cell growth (Yang et al. 1996). Even the dga1Δlro1Δare1Δare2Δ quadruple mutant that lacks both TAG and steryl esters, and has no lipid droplets (LD), exhibits only a slight extension of the lag phase after recovery from quiescence (Petschnigg et al. 2009). The dga1Δlro1Δare1Δare2Δ quadruple mutant, however, does exhibit a defect in sterol synthesis (Sorger et al. 2004) and is highly sensitive to unsaturated fatty acid (FA) supplementation (Garbarino et al. 2009; Petschnigg et al. 2009). Strikingly, provision of exogenous unsaturated FA in the absence of TAG synthesis leads to respiration-dependent cell necrosis (Rockenfeller et al. 2010), challenging the dogma that lipid-induced cell death is exclusively apoptotic. The dga1Δlro1Δare1Δare2Δ quadruple mutant also displays an Ino– phenotype at elevated growth temperature, indicative of altered INO1 expression and/or defective PI synthesis (Gaspar et al. 2011), ER stress induced by tunicamycin, which inhibits protein glycosylation and stimulates LD formation in wild-type cells. However, the dga1Δlro1Δare1Δare2Δ quadruple mutant is no more sensitive to tunicamycin than wild type, indicating that LD formation is not protective against this form of stress (Fei et al. 2009). Dga1 and Lro1 acyltransferase-dependent TAG synthesis, on the other hand, is required for growth at semipermissive temperatures in the absence of inositol in the sec13ts-1 mutant, defective in COPII vesicle budding from the ER. When shifted to higher growth temperature, the sec13ts mutant channels PA and DAG precursors from phospholipid into TAG, which apparently provides a degree of protection from the secretory stress caused by a block in membrane trafficking (Gaspar et al. 2008).
TAG hydrolysis (Figure 2, Table 3) clearly contributes lipid precursors that are essential to the resumption of growth upon exit from stationary phase (Kurat et al. 2009; Kohlwein 2010b). TAG degradation provides substrates for the synthesis of phospholipids and sphingolipids (Rajakumari et al. 2010), which are required for efficient cell cycle progression upon exit from quiescence (Kurat et al. 2009). Degradation of TAG is catalyzed by TGL1-, TGL3-, TGL4-, and TGL5-encoded TAG lipases, all of which are localized to LD (Athenstaedt and Daum 2003, 2005; Jandrositz et al. 2005; Kurat et al. 2006; Rajakumari et al. 2008; Kurat et al. 2009; Kohlwein, 2010b). Tgl1 harbors the canonical lipase catalytic triad, consisting of serine201, aspartate369 and histidine396 (Jandrositz et al. 2005). In contrast, Tgl3, Tgl4, and Tgl5 (and also Nte1, see above) are members of the patatin domain-containing family of (phospho) lipases, characterized by a catalytic diad of serine and aspartate (Kurat et al. 2006). The substrate specificities of the Tgl3, Tgl4, and Tgl5 TAG lipases differ. Tgl5 preferentially hydrolyses TAG molecular species harboring very long chain fatty acids (VLCFA) (Athenstaedt and Daum 2005), while Tgl3 also accepts DAG as a substrate in addition to TAG (Kurat et al. 2006). Tgl3, Tgl4, and Tgl5 also possess lysoPA acyltransferase activities, and Tgl4 additionally functions as a Ca++-dependent phospholipase A2 and steryl ester hydrolase (Rajakumari and Daum 2010a,b). Although Tgl1 also contributes to TAG hydrolysis, its major activity is as a steryl ester hydrolase, in conjunction with the YEH1- and YEH2-encoded enzymes (Jandrositz et al. 2005; Koffel et al. 2005). TGL2 encodes an acylglycerol lipase localized to mitochondria, but its role in TAG homeostasis has not been clarified yet (Ham et al. 2010). YJU3 encodes the major monoacylglycerol (MAG) lipase in yeast (Heier et al. 2010), but yju3Δ mutants do not display any detectable growth phenotype when tested under multiple conditions (Heier et al. 2010).
Stationary phase cells shifted into fresh media rapidly initiate TAG and steryl ester breakdown, which leads to almost full depletion of cellular LD pools within 4–6 hr in lag phase. This initial phase of TAG breakdown is governed by the activity of TGL3- and TGL4-encoded lipases on LD and the tgl3Δtgl4Δ mutant strain exhibits a delay in entering vegetative growth during exit from stationary phase (Kurat et al. 2006). Tgl4 is constitutively present on LD and activation of Tgl4 by Cdc28p-dependent phosphorylation is involved in TAG lipolysis that contributes to bud formation exiting from stasis (Kurat et al. 2009). Resumption of growth following stasis is also dependent on the DGK1-encoded DAG kinase. In comparison to wild-type cells, stationary phase dgk1Δ cells fail to initiate growth when de novo FA synthesis is impaired. The dgk1Δ mutant also fails to mobilize TAG under these conditions and accumulates TAG, phenotypes that are partially suppressed by the pah1Δ mutation or by channeling DAG into PC synthesis when choline is present (Fakas et al. 2011).
TAG synthesis and breakdown are also coordinated with phospholipid metabolism during logarithmic growth. Mutants with defects in TAG hydrolysis exhibit decreased synthesis of inositol containing sphingolipids and decreased PC and PI content during active growth (Rajakumari et al. 2010). Furthermore, mutants defective in synthesis or hydrolysis of TAG exhibit reduced capacity to restore cellular levels of PI when exogenous inositol is resupplied following an interval of inositol starvation during logarithmic growth. Under these conditions, alterations in the synthesis of inositol-containing sphingolipids are also observed in the dga1Δlro1Δare1Δare2Δ strain (Gaspar et al. 2011).
Glycerolipid precursors
Fatty acid synthesis and regulation:
The FA that are esterified to phospholipids and TAG are derived from de novo synthesis, the growth medium, and from lipid turnover (Tehlivets et al. 2007). The spectrum of FA in yeast consists mainly of C16 and C18 FA that are either saturated or monounsaturated, harboring a single double bond between carbon atoms 9 and 10 (Δ9 desaturation). Whereas de novo FA synthesis mostly takes place in the cytosol, all the enzymes involved in FA desaturation and elongation are associated with the ER membrane (Table 2) (Tehlivets et al. 2007).
Minor FA species are C12, C14, and very long chain FA, up to C26. However, FA compositions vary substantially between strains and are also dependent on cultivation conditions (Martin et al. 2007). Two different FA synthesis pathways exist in yeast, as in mammalian cells (Tehlivets et al. 2007): the major cytosolic pathway, which resembles the “eukaryotic” type I pathway and is responsible for the bulk synthesis of all major FA, and the mitochondrial (type II pathway), which is organized similarly to the bacterial FA biosynthetic pathway (Hiltunen et al. 2010). The latter is believed to synthesize FA up to C8 carbons, which serve as precursors for the synthesis of lipoic acid. Cytosolic FA synthesis is initiated by the ACC1-encoded acetyl-CoA carboxylase, which synthesizes malonyl-CoA from acetyl-CoA (Al-Feel et al. 1992; Hasslacher et al. 1993). This reaction requires a covalently bound biotin prosthetic group, which is attached to lysine735 in the biotin-carrier domain of Acc1 by the BPL1-encoded biotin:apoprotein ligase. The FA synthase complex is composed of two subunits, encoded by FAS1 (Fas1, β-subunit) and FAS2 (Fas2, α-subunit) and is organized as an α6/β6 heterooligomeric complex (Kuziora et al. 1983; Schweizer et al. 1986; Chirala et al. 1987). Fas2 carries a pantetheine prostetic group on the acyl-carrier protein (ACP) domain, which is a central element in a cycling series of reactions. In a first condensation step, malonyl-CoA is condensed with pantetheine-bound acetyl-CoA to form 3-keto-acyl-ACP, which is reduced to 3-hydroxyacyl-ACP, dehydrated to 2,3-trans-enoyl-ACP, and further reduced to acyl(C+2)-ACP. Both reduction steps require NADPH and, as a result, FA synthesis is a major consumer of this dinucleotide. This multistep process results in the addition of two carbon units to the growing FA chain and cycles up to seven times, resulting in acyl-chains typically of 16 carbon atoms. The newly formed FA is transferred to coenzyme A, to yield cytosolic long chain acyl-CoA (Tehlivets et al. 2007). Acyl-CoA molecules are precursors for all acylation reactions involved in the synthesis of phospholipids, TAG, long chain bases, ceramide, and steryl esters, and also serve as precursors in protein acylation. The acyl-CoAs, whether derived from FA de novo synthesis or lipid recycling, are subject to elongation and desaturation, yielding the typical pattern of saturated and unsaturated FA species (see below). Yeast also contains an ACB1-encoded acyl-CoA binding protein, which plays an important regulatory function in delivering acyl-CoA into various pathways (Schjerling et al. 1996; Gaigg et al. 2001; Faergeman et al. 2004; Rijken et al. 2009). Mitochondrial FA synthesis involves enzymatic steps similar to those catalyzed by the cytosolic FAS complex. However, in contrast to cytosolic FA synthesis, but resembling archae- and eubacterial type II fatty acid synthase, the reactions of FA synthesis in mitochondria are catalyzed by individual polypeptides, encoded by separate genes (Hiltunen et al. 2010).
Defects in the FAS1 or FAS2 genes encoding cytosolic FA synthase lead to FA auxotrophy and can typically be rescued by the addition of exogenous C14 or C16 FAs. However, defects in ACC1 (Hasslacher et al. 1993) and BPL2 are lethal and cannot be suppressed by the addition of long chain FA. The essential role of these genes, when exogenous C14 and C16 FAs are present, is most likely the requirement for malonyl-CoA in the synthesis of essential VLCFA, which are components of glycerophosphoinositol (GPI) anchors and sphingolipids (Pittet and Conzelmann 2007; Dickson 2008; Dickson 2010). The synthesis of VLCFAs is accomplished by an ER membrane-localized elongase complex that acts on long chain acyl-CoA, consisting of the condensing enzymes encoded by ELO1, ELO2, and ELO3 (Oh et al. 1997), the β-ketoacyl-CoA reductase encoded by gene YBR159w (Han et al. 2002), the dehydratase encoded by PHS1 (Denic and Weissman 2007; Kihara et al. 2008), and the enoyl-CoA reductase, encoded by TSC13 (Kohlwein et al. 2001). Tsc13 accumulates within a specialized region of the ER in juxtaposition to the vacuole, termed the nucleus-vacuole junction (Pan et al. 2000) through interaction with Osh1p and Nvj1p (Kvam et al. 2005). The physiological role for a localization of just one component of the FA elongation complex to this subregion of the ER is currently unclear, but indicates a role for VLCFA in the formation of microautophagic vesicles involved in piecemeal autophagy of the nucleus (Kvam et al. 2005). Triple mutations in all three condensing enzyme genes as well as mutations in PHS1 or TSC13 are lethal, further supporting the notion that VLCFAs are essential in yeast. Yeast also harbors a single FA Δ9-desaturase encoded by OLE1, which localizes to the ER (Stukey et al. 1989) and mutants defective in Ole1 require exogenous C16:1 or C18:1 FA for growth. Since FA desaturation is an oxygen-dependent process (see below), growth of yeast under anaerobic conditions also requires the supplementation of these unsaturated FA.
In the absence of de novo FA synthesis or desaturation, as in fas1, fas2, or ole1 mutants, and in wild-type cells under anaerobic conditions, cellular growth is dependent on an exogenous supply of FA. Under these conditions, the activity of acyl-CoA synthetases (Table 2), encoded by FAA1, FAA2, FAA3, FAA4, and FAT1 (Duronio et al. 1992; Harington et al. 1994; Johnson et al. 1994a,b; Watkins et al. 1998; Choi and Martin 1999; Black and Dirusso 2007) is also required. The acyl-CoA synthetases activate free FA with coenzyme A in an ATP-dependent reaction, and are also believed to be required for FA uptake into yeast cells (Faergeman et al. 2001; Black and Dirusso 2007; Obermeyer et al. 2007). Acyl-CoA synthetases differ in their substrate specificity and subcellular localization and are present in the ER membrane, plasma membrane, peroxisome, and LD (Natter et al. 2005; Black and Dirusso 2007). The acyl-CoA synthetase Faa2 and the enzymes for FA β-oxidation, a cyclic series of reactions breaking down FA into acetyl-CoA and generating FADH2 and NADH, are localized to the peroxisomes (planned YeastBook chapter “Lipid particles and peroxisomes” by Kohlwein and van der Klei). FA uptake into yeast cells is also mediated by an endocytotic mechanism that requires the activity of Ypk1, the yeast ortholog of the human serum- and glucocorticoid-induced kinase (Jacquier and Schneiter 2010). In the absence of acyl-CoA synthetases, FA released by lipid turnover (see above) cannot be activated and utilized for lipid synthesis. For this reason, mutants lacking these activities excrete FA (Scharnewski et al. 2008).
FA synthesis is regulated at multiple levels (Tehlivets et al. 2007). ACC1, FAS1, and FAS2 expression is under control of an UASINO element and coregulated with genes involved in phospholipid synthesis (Chirala 1992; Schuller et al. 1992) (see below). ACC1 expression is also regulated by the SAGA protein complex and TFIID (Huisinga and Pugh 2004) and may therefore depend on histone acetylation. ACC1 expression was also found to fluctuate during the cell division cycle, with a peak of expression in early G1 phase (Cho et al. 1998). Compensatory changes in ACC1 expression in mutants defective in Acc1 activity indicate an autoregulatory loop (Shirra et al. 2001). Expression of the FAS1 and FAS2 genes is regulated by the transcription factors Gcr1, Reb1, Rap1, and Abf1 (Schuller et al. 1994; Greenberg and Lopes 1996) and repressed by long chain FA (Chirala 1992). FAS1 is also regulated by the SAGA complex and TFIID, and both genes show cell cycle-dependent expression, with a peak level at M/G1 (Spellman et al. 1998; Huisinga and Pugh 2004).
Acc1 enzyme activity is greatly elevated in the snf1Δ mutant and the data are consistent with Acc1 being a substrate of AMP-activated protein kinase, encoded by the SNF1 (Woods et al. 1994; Shirra et al. 2001). Snf1 also regulates chromatin by phosphorylating histones among other proteins and, therefore, has multiple effects on expression of genes, including ACC1, FAS1, and FAS2. Notably, FAS1 and FAS2 promoter sequences differ, and the stoichiometry of the FAS complex is established both by the level of FAS1 gene expression and Fas1 protein abundance (Schuller et al. 1992; Wenz et al. 2001; Tehlivets et al. 2007). Excess of either protein is rapidly eliminated by degradation via vacuolar proteases (Fas1) or ubiquitination and proteasomal digestion (Fas2), respectively (Peng et al. 2003).
Knowledge about the regulation of FA elongation leading to VLCFA synthesis is limited. Microarray experiments have shown that ELO1 expression is upregulated in the presence of myristic acid (C14:0), consistent with the preference of the Elo1 protein for this FA (Toke and Martin 1996). Expression of ELO1, ELO2, and ELO3 genes also responds to stationary phase, nitrogen limitation, glycosylation defects, or α-factor treatment, indicating a regulatory cross-talk between nutritional state and cell proliferation to VLCFA synthesis (Gasch et al. 2000).
Ole1 is a nonheme iron-containing integral ER membrane protein (Table 2), which harbors an intrinsic cytochrome b5 domain as an electron carrier (Martin et al. 2007). To subtract two electrons and two protons from the saturated acyl-chain and transfer them to oxygen, Ole1 activity is complemented by the activity of an NADH cytochrome b5 reductase (dehydrogenase) that is also localized to the ER. Desaturation of FA by Ole1 is a highly regulated, oxygen-requiring process (Martin et al. 2007), which is discussed in detail in the section below on cell biology of lipids.
Water soluble precursors of phospholipids, metabolism, and regulatory roles:
A number of water-soluble molecules used in phospholipid synthesis, including inositol, choline, ethanolamine, serine, CTP, and S-adenosyl methionine (AdoMet) and the enzymes involved in these reactions are largely cytosolic (Table 2). However, choline and ethanolamine are produced in yeast in the context of ongoing synthesis and turnover of PE and PC in the membranes as discussed above. Considerable attention has been paid to inositol and CTP, which have major regulatory roles in phospholipid metabolism. The effects of inositol on transcriptional regulation UASINO-containing phospholipid biosynthetic genes controlled by the Opi1 repressor are shown in Figure 3 and will be discussed in a subsequent section. The regulatory effects of the soluble metabolites inositol, CTP, and S-adenosyl homocysteine (AdoHcy) on enzyme activity and metabolic flux in the CDP-DAG pathway (Figure 4) are discussed here.
Inositol is the precursor to PI (Figure 2), which is essential to the synthesis of polyphosphoinositides (Strahl and Thorner 2007), sphingolipids (Cowart and Obeid 2007; Dickson 2008, 2010), and GPI anchors (Pittet and Conzelmann 2007). Inositol is derived from glucose-6-P via the reactions catalyzed by the cytosolic INO1-encoded inositol-3-P synthase (Donahue and Henry 1981; Klig and Henry 1984; Dean-Johnson and Henry 1989) and the INM1-encoded inositol-3-P phosphatase (Murray and Greenberg 2000). Exogenously supplied inositol stimulates PI synthase and inhibits PS synthase activity, alleviating the competition with PI synthase for their common substrate, CDP-DAG (Figure 4). The presence of exogenous inositol leads to increased PI synthesis and reduced levels of both CDP-DAG and its precursor PA in wild-type cells (Kelley et al. 1988; Loewen et al. 2004). TAG, which is derived from PA by the action of Pah1 (Figure 2), is decreased in the presence of inositol and increases in its absence (Gaspar et al. 2006, 2011). In addition, the levels of the complex sphingolipids, which are derived from PI, are reduced when cells are deprived of inositol and increase when inositol is supplied (Becker and Lester 1977; Hanson and Lester 1980; Jesch et al. 2010). The changes that occur in phospholipid synthesis and composition in response to exogenous serine (Kelley et al. 1988) and choline (Gaspar et al. 2006) are much less dramatic than the effects of exogenous inositol. However, when inositol is present in the growth medium, serine (Homann et al.1987), ethanolamine, and choline (Poole et al. 1986) result in the reduction of the activities of PS synthase and CDP-DAG synthase at the transcriptional level (see below). Exogenous choline also results in a dramatic change in the rate and mechanism of PC turnover, leading to a switch from a phospholipase D to a phospholipase B-mediated route (Dowd et al. 2001). The phospholipase responsible for turnover of PC under these conditions is Nte1 (Zaccheo et al. 2004).
CTP is derived from UTP by the cytosolic URA7- and URA8-encoded CTP synthetase enzymes (Ozier-Kalogeropoulos et al. 1991, 1994).
The nucleotide CTP plays a critical role in phospholipid synthesis as the direct precursor of the high-energy intermediates CDP-DAG, CDP-choline, and CDP-ethanolamine that are used in the CDP-DAG and Kennedy pathways (Figure 2) (Chang and Carman 2008). It is also used as the phosphate donor for the synthesis of PA by DAG kinase (Han et al. 2008b). CTP synthetase (Ozier-Kalogeropoulos et al. 1991, 1994) that produces CTP is allosterically inhibited by the product (Yang et al. 1994; Nadkarni et al. 1995), and this regulation ultimately determines the intracellular concentration of CTP (Yang et al. 1994; McDonough et al. 1995). CTP inhibits the CTP synthetase activity by increasing the positive cooperativity of the enzyme for UTP, and at the same time by decreasing the affinity for UTP (Yang et al. 1994; Nadkarni et al. 1995). However, CTP synthetases containing an E161K mutation are less sensitive to CTP product inhibition (Ostrander et al. 1998), and cells expressing the mutant enzymes exhibit a 6- to 15-fold increase in the CTP level (Ostrander et al. 1998). They also show alterations in the synthesis of membrane phospholipids, which include a decrease in the amounts of PS and increases in the amounts of PC, PE, and PA (Ostrander et al. 1998). The decrease in the amount of PS results from direct inhibition of PS synthase activity by CTP (McDonough et al. 1995), and this inhibition favors the synthesis of phospholipids by the Kennedy pathway (Figure 4). The increase in PC synthesis is ascribed to a higher utilization of the CDP-choline branch of the Kennedy pathway due to the stimulation of choline P cytidylyltransferase activity (McDonough et al. 1995; Ostrander et al. 1998) by increased substrate availability of CTP (McDonough et al. 1995; Kent and Carman 1999). Likewise, the increase in PE synthesis could be attributed to stimulation of ethanolamine-P cytidylyltransferase activity. The increase in PA content may result from the stimulation of DAG kinase activity by increased availability of its substrate CTP (Han et al. 2008b). CTP also inhibits the activity of Pah1 (Wu and Carman 1994), another factor that contributes to an elevation of PA content. The cells expressing the E161K mutant CTP synthase excrete inositol (Ostrander et al. 1998), a characteristic phenotype that typifies the misregulation of UASINO-containing phospholipid synthesis genes (see below) in cells that accumulate an excess of PA (Carman and Henry 2007). It is unclear whether UASINO-containing genes in the CDP-DAG and Kennedy pathways are derepressed in CTP overproducing cells, but the overriding regulation that governs the utilization of the two pathways appears to be biochemical in nature.
AdoHcy is a product of the AdoMet-dependent methylation reactions that are catalyzed by Cho2 and Opi3 in the CDP-DAG pathway (Figure 2). SAH1-encoded AdoHcy hydrolase (Malanovic et al. 2008) requires NADH for the hydrolytic breakdown of AdoHcy to adenosine and homocysteine (Takata et al. 2002). AdoHcy (Figure 4) is a competitive inhibitor of the phospholipid methyltransferase enzymes (Gaynor and Carman 1990). Thus, down-regulation of the AdoHcy hydrolase causes the accumulation of AdoHcy and the inhibition of PC synthesis, which leads to an increase in PA content and consequently, as described below, to the derepression of UASINO-containing genes (Malanovic et al. 2008). Although the effects of AdoHcy on phospholipid composition have not been addressed, its accumulation causes an increase in TAG synthesis and LD content (Malanovic et al. 2008), further underscoring the metabolic interconnection between phospholipid and TAG homeostasis discussed above.
Cell Biology of Lipids
In yeast, most subcellular membranes are composed of similar glycerophospholipid classes but the quantitative phospholipid composition of subcellular organelles differs substantially (Schneiter et al. 1999). Moreover, as described above, multiple cellular organelles and compartments contribute to glycerolipid metabolism (Figure 2, Tables 1–3). While multiple compartments contribute to lipid synthesis in yeast (Zinser et al. 1991; Natter et al. 2005), most reactions are confined to single membrane compartments. Thus, extensive regulated flux of lipids, across and among individual membranes and organelles, is required to enable the cell to adjust lipid composition in specific compartments under changing growth conditions. A number of mechanisms involved in these complex interactions have been identified.
For example, a set of membrane-bound flippases are known to govern transmembrane movement of phospholipids (Pomorski et al. 2004; Holthuis and Levine 2005). Intermembrane transfer of lipids is facilitated by a family of proteins, termed oxysterol-binding protein (OSBP) related proteins (ORP), of which seven members, Osh1–7, with overlapping functions, exist in yeast (Beh and Rine 2004). Some of the yeast Osh proteins have been localized to membrane contacts sites (Levine and Loewen 2006; Raychaudhuri and Prinz 2010). While none of these genes individually are essential, simultaneous deletion of all seven OSH genes is lethal and conditional alleles, following a shift to restrictive conditions, result in pleiotropic effects on vesicular trafficking and phospholipid and sterol composition of membranes, as do combinations of individual deletions (Beh and Rine 2004; Fei et al. 2008). The six members of the Sec14 superfamily constitute another major class of proteins involved in sensing and regulating membrane lipid composition (Griac 2007; Bankaitis et al. 2010). The five Sec14 homologs (Sfh1–5) share 28–76% similarity to Sec14 (Griac 2007) and localize to multiple subcellular organelles (Schnabl et al. 2003). Sec14, which performs an essential function at the Golgi, was originally identified as a PI/PC transfer protein, but its role in establishing phospholipid homeostasis is complex (Bankaitis et al. 2010).
In some instances, movement of lipids between membrane compartments is required to carry out a sequence of reactions. For example, PS is synthesized in the ER, but the major PS decarboxylase, Psd1, is located in the mitochondria (Figure 2, Table 1). Subsequently, the PE produced by Psd1 must be transported back to the ER to undergo methylation leading to PC. PS is transported into the mitochondria by an ubiquitin-regulated process that is insensitive to disruption of vesicular trafficking and involves specialized regions of apposition of mitochondria and ER known as the mitochondria-associated ER or MAM (Clancey et al. 1993; Trotter et al. 1993; Trotter and Voelker 1995; Achleitner et al. 1999; Voelker 2003; Osman et al. 2011). An ER–mitochondria tethering complex potentially involved in movement of phospholipids between the ER and mitochondria has been described (Kornmann et al. 2009).
The complex relationship between phospholipid metabolism in the ER and the synthesis and breakdown of cytosolic LD represents another example of the intricate interaction among cellular compartments that occurs in the course of lipid metabolism. LD are not merely storage depots for TAG and steryl esters. They are metabolically active; harboring multiple enzymes involved in lipid metabolism (Athenstaedt et al. 1999a; Rajakumari et al. 2008; Goodmann 2009; Kohlwein 2010b) (See also chapter: Lipid Droplets and Peroxisomes). Interaction of LD with the ER, mitochondria, and peroxisomes has been reported (Goodmann 2009). Pah1 clearly plays a critical role in TAG synthesis, LD formation, and the balance between nuclear/ER membrane proliferation and LD formation (Carman and Han 2009a; Adeyo et al. 2011). Several enzymes involved in TAG synthesis show a dual localization to the ER and LD (Figure 2, Table 1), including acyl-DHAP reductase (Ayr1), glycerol-3-P acyltransferase (Gpt2), and acyl-CoA–dependent DAG acyltransferase (Dga1). Formation of LD is believed to occur between the leaflets of the ER membrane, but alternative models such as vesicular trafficking have also been suggested (Thiele and Spandl 2008; Farese and Walther 2009; Guo et al. 2009). The close interaction of LD with mitochondria and peroxisomes also underscores the important role of LD in metabolism (Goodmann 2009). TAG levels fluctuate drastically during cellular growth and division, and multiple conditions contribute to the level of TAG storage in LD in stationary phase cells (Kurat et al. 2006; Czabany et al. 2007; Daum et al. 2007; Rajakumari et al. 2008; Zanghellini et al. 2008). As cells exit stationary phase, TAG hydrolysis supplies precursors for membrane lipid synthesis. As active cellular growth progresses, de novo FA synthesis increases, satisfying cellular requirements for lipid synthesis but even while TAG turnover is still in progress, de novo formation of TAG and LD is initiated (Kurat et al. 2006; Zanghellini et al. 2008). Methods for imaging-based screening of mutant libraries have been designed to identify factors involved in LD content, morphology, and mobilization (Wolinski and Kohlwein 2008; Wolinski et al. 2009) and screens using such methods have identified >200 genes that potentially influence these processes (Szymanski et al. 2007; Fei et al. 2008). However, there is surprisingly little overlap between the mutants identified in the published screens, indicating that current screens are far from being saturated.
Roles of lipids in organelle function and morphology
Due to the ubiquitous presence of most phospholipids in all subcellular membranes it can be difficult to assess specific phospholipid functions, in particular organelles. However, quite specific functions have been attributed to several lipids in the mitochondrion. For example, PE synthesized in the mitochondria by Psd1 plays an important role in stabilizing mitochondrial protein complexes (Birner et al. 2003; Osman et al. 2009a). Consequently, the psd1Δ mutant is nonviable in conjunction with defects in components of the prohibitin complex, which functions as a protein and lipid scaffold and ensures the integrity and functionality of the mitochondrial inner membrane (Osman et al. 2009b; Potting et al. 2010; van Gestel et al. 2010; Osman et al. 2011). In another example, CL, which is present only in the mitochondrion (Li, G. et al. 2007), plays an important role in mitochondrial genome stabilization and higher order organization of components of the mitochondrial respiratory chain (Koshkin and Greenberg 2002; Zhang et al. 2003; Zhong et al. 2004; Mileykovskaya et al. 2005; Joshi et al. 2009). However, CL function reaches beyond the mitochondria, indirectly regulating morphology and acidification of the vacuole through the retrograde (RTG2) signaling pathway and the NHX1-encoded vacuolar Na+/H+ exchanger (Chen et al. 2008). Lack of CL in crd1Δ mutants is in part compensated by an upregulation of the PE content, which is dependent on the mitochondrial synthesis through Psd1 (Gohil et al. 2005). Defects in Crd1 are also, in part, compensated by an increase in the precursor lipid PGP. Accordingly, mutation of the PGS1 gene encoding PGP synthase leads to more pronounced mitochondrial defects and a petite negative phenotype (Janitor et al. 1996; Chang et al. 1998a).
Alterations in organelle morphology and/or function observed in response to changes in lipid composition due to mutation or pharmacological inhibition of enzymes (Schneiter and Kohlwein 1997) provide tantalizing clues as to the roles of specific lipids. Notably, the pah1Δ mutant exhibits the expansion of the nuclear/ER membrane and up-regulation of phospholipid biosynthesis genes (Santos-Rosa et al. 2005). As discussed previously, dephosphorylation of PA by Pah1 provides DAG for the synthesis of TAG, as well as PE and PC in the Kennedy pathway. Consequently, the pah1Δ mutant has elevated PA levels, which result in up-regulation of phospholipid biosynthesis (see below) and membrane proliferation (Han et al. 2005; Carman and Henry 2007; Carman and Han 2009b). A number of mutants defective in FA metabolism also exhibit morphological abnormalities. For example, conditional mutants defective in FA desaturation (ole1/mdm2) show defective mitochondrial morphology and distribution upon cell division (Stewart and Yaffe 1991; Tatzer et al. 2002). The finding that several dozen proteins species are required to sustain cell viability in the presence of unsaturated FA suggests an enormously complex network of processes and interactions to maintain membrane homeostasis and function (Lockshon et al. 2007). In another example, a conditional acc1 mutant defective in FA de novo synthesis has impaired morphology of the vacuole due to reduced acylation of Vac8 that is involved in stabilizing the vacuolar membrane and vacuole inheritance (Schneiter et al. 2000). Mutants defective in FA elongation show similar fragmentation of the vacuole due to impaired synthesis of sphingolipids, which are important for the stability of the vacuolar ATPase complex (Chung et al. 2003).
Due to their important roles in protein modification, myristic and palmitic acid affect the membrane association of numerous proteins and consequently influence signaling, membrane function and organelle morphology (Dietrich and Ungermann 2004). NMT1 encodes the essential N-myristoyl transferase that attaches coenzyme A-activated myristic acid (C14-CoA) to a glycine residue close to the N-terminus of target proteins, resulting in cleavage of the peptide bond and N-terminal glycine acylation (Towler et al. 1987). Although the myristoyl residue is shorter than the typical C16 or C18 chain length found in membrane phospholipids and, therefore may not fully interdigitate into a membrane leaflet, it serves an essential function in promoting membrane association of proteins (Duronio et al. 1989). Palmitoylation of internal cysteine residues within the peptide chain plays an important role in post-translational modification of some 50 proteins in yeast (Roth et al. 2006), modulating membrane association, folding and activity. Protein palmitoylation is catalyzed by seven members of the “Asp-His-His-Cys-cystein-rich“ (DHHC-CRD) domain family of proteins, and includes the Erf2/Shr5 complex, Akr1, Akr2, Pfa3, Pfa4, Pfa5, and Swf1. Among the substrates for palmitoylation are Vac8, SNAREs, and Ras2, indicating important regulatory functions for this FA modification of proteins in multiple pathways involved in membrane trafficking and signaling.
Mechanisms of compartmentalization and localization of enzymes of lipid metabolism
Many enzymes involved in glycerolipid metabolism have transmembrane domains (Tables 1–3), which anchor the proteins in their specific membrane environment and promote access to hydrophobic lipid substrates. However, several membrane-associated enzymes lack defined transmembrane spanning domains and membrane interaction is mediated, perhaps in a regulated manner, with specific membrane-resident anchor proteins. For example, PIK1-encoded PI 4-kinase binds to the membrane through Frq1 and the VPS34-encoded PI 3-kinase binds through Vps15 (Strahl and Thorner 2007). In addition, enzymes may bind to the membrane through specific interaction with lipids in the membrane, such as PI 3-P, PI 4-P, PI 3,5-P2 and PI 4,5-P2 through PH, PX, FYVE or ENTH domains (Strahl and Thorner 2007). A number of enzymes involved in lipid biosynthesis utilize water soluble lipid precursors, including the choline and ethanolamine kinases and the enzymes of the FAS complex, involved in long chain FA synthesis, and are localized to the cytosol. Acyl-CoA, generated by the FAS complex, has a highly amphiphilic structure and easily associates with membrane surfaces, facilitating the subsequent incorporation of the acyl chains into lipids by membrane-bound enzymes. However, Pah1 is an example of an enzyme that is associated with both cytosolic and membrane compartments. PA, the substrate for Pah1, is present in the membrane, but the largest pool of Pah1 is cytosolic, highly phosphorylated and inactive. Pah1 must be dephosphorylated by the Nem1-Spo7 complex in order to be functional in vivo (Santos-Rosa et al. 2005; Han et al. 2006; Carman and Han 2009a; Choi et al. 2011). Dephosphorylation of Pah1 by the Nem1-Spo7 complex leads to its membrane association and its activation (Choi et al. 2011), leading to the production of DAG and TAG and the lowering of PA levels, which in turn affects regulation of phospholipid biosynthetic genes (see below) (Carman and Henry 2007; Carman and Han 2009b).
Regulation of lipid metabolism in response to membrane function
Several regulatory circuits have been uncovered that govern, by different mechanisms, the cross-talk between membrane function and lipid synthesis. Proteins involved in this regulation may sense specific changes in lipid composition, membrane charge, fluidity, or curvature. One such mechanism controls the expression of the fatty acid desaturase, Ole1, by the ER membrane-bound transcription factors Mga2 and Spt23, which are cleaved and released from the membrane in response to changes in membrane fluidity or permeability (Hoppe et al. 2000). Both proteins are synthesized as larger, ER membrane-bound proteins that are ubiquitinated and cleaved by the activity of the 26S proteasome under low oxygen conditions, in conjunction with the ubiquitin-selective chaperone CDC48UFD1/NPL4. This cleavage detaches the soluble fragments from their membrane anchors, and allows them to enter the nucleus to control transcription of OLE1 and several other genes through the low oxygen response (LORE) promoter element. Proteasomal cleavage of Mga2 and Spt23 requires the essential E3 ubiquitin ligase, Rsp5, and lack of this activity results in a cellular requirement for unsaturated FA. Cleavage of Spt23 and Mga2 in the ER is believed to be regulated by the membrane environment, adjusting membrane lipid composition through the control of FA desaturation. Under normoxic conditions, OLE1 expression is activated by the oxygen-responsive transcription factor Hap1. Addition of unsaturated FA, including linoleic acid (C18:2) or arachidonic acid (C20:4), which are normally not present in yeast, results in a drastic and rapid reduction of OLE1 mRNA expression and stability. However, low oxygen tension leads to a massive induction of OLE1 expression through the LORE element present in its promoter. This induction is believed to increase efficiency of FA desaturation to support cellular growth under oxygen limiting (hypoxic) conditions (Martin et al. 2007; see also the chapter on Lipid Droplets and Peroxisomes for more discussion of fatty acid metabolism and regulation). Mga2 (but not Spt23) was also shown to be responsible for the rapid, but transient, up-regulation of OLE1 and other LORE-containing genes observed when inositol is added to cultures of wild-type yeast growing logarithmically in the absence of inositol (Jesch et al. 2006). Regulation of the phospholipid biosynthetic genes containing the UASINO promoter element also occurs in response to changes in the lipid composition in the ER membrane, through the binding of the Opi1 repressor to PA (see below). The ER is also the locus of complex regulatory cross-talk involving membrane expansion, the secretory pathway, the unfolded protein response (UPR) pathway, and phospholipid metabolism (Cox et al. 1997; Travers et al. 2000; Block-Alper et al. 2002; Chang et al. 2002, 2004; Hyde et al. 2002; Brickner and Walter 2004; Jesch et al. 2006; Gaspar et al. 2008; Schuck et al. 2009).
Another example of regulation of lipid metabolism in yeast in response to changing membrane conditions was discovered in the course of analysis of a cho2Δopi3Δ mutant strain completely defective in PC formation via the PE methylation pathway (Boumann et al. 2006). When this mutant is deprived of choline, PC levels decline and as this process progresses, concomitant changes in acyl-chain distribution occur in PC and other phospholipids, especially PE. However, similar changes were not observed in the neutral lipid fraction. The shortening and increased saturation of PE acyl chains decreases the bilayer forming potential of PE and Boumann et al. (2006) suggest that phospholipid remodeling under these conditions may provide a mechanism for maintaining intrinsic membrane curvature. The nature of such a mechanism has not yet been defined, but yeast has a number of proteins harboring a BAR domain, including Rvs161 and Rvs167, which bind to liposomes in a curvature-dependent manner and promote tubule formation in vitro. In vivo studies in yeast indicate inappropriate regulation of phosphoinositide and sphingolipid metabolism impinges on Rsv161 and Rsv167 function (Ren et al. 2006; Youn et al. 2010). The Rim101 pathway, which is involved in regulating cellular pH in response to alkaline conditions and cell wall biogenesis, also appears to be involved in sensing membrane curvature (Mattiazzi et al. 2010).
Genetic Regulation of Glycerolipid Synthesis
In wild-type cells, both glycerolipid composition and the expression of lipid biosynthetic genes are influenced by a wide variety of growth conditions, including growth phase, temperature, pH, and the availability of nutrients such as carbon, nitrogen, phosphate, zinc, and lipid precursors. Transcript levels of certain lipid biosynthetic genes in yeast are also regulated by message stability. A number of viable mutants defective in genes encoding lipid metabolic enzymes, transporters, and regulatory factors exhibit changes in lipid composition. These complex and interrelated topics have been the subjects of numerous reports and reviews (Henry 1982; Carman and Henry 1989; Paltauf et al. 1992; Henry and Patton-Vogt 1998; Carman and Henry 1999; Gardocki et al. 2005; Jesch and Henry 2005b; Santos-Rosa et al. 2005; Carman and Henry 2007; Chen et al. 2007a; Daum et al. 2007; Gaspar et al. 2007; Li, G. et al. 2007; Patton-Vogt 2007; Gaspar et al. 2008; Schuiki et al. 2010; Young et al. 2010; Carman and Han 2011), as discussed below with a focus on the transcriptional regulation of lipid biosynthetic genes in response to PA.
Regulation of transcript abundance by mRNA degradation
The transcript abundance of some glycerolipid metabolic genes is regulated at the level of mRNA degradation. Genes that exhibit this level of regulation include CHO1 (Choi and Carman 2007) and OLE1 (Gonzalez and Martin 1996; Vemula et al. 2003; Martin et al. 2007). In wild-type cells, CHO1 mRNA is moderately stable with a half-life of 12 min when compared with other S. cerevisiae mRNAs that have half-lives ranging from 1 to 60 min (Herrick et al. 1990). However, CHO1 mRNA is greatly stabilized with a half-life >45 min in respiratory mutants (Choi and Carman 2007). This results in increased levels of the PS synthase protein and its associated activity (Choi and Carman 2007). Given that CHO1 mRNA decays by the primary 5′–3′ decay pathway when cells are respiratory sufficient (Parker and Song 2004), it is reasonable to predict that the rate of deadenylation and/or decapping may be reduced when respiration is blocked. The OLE1 transcript is destabilized when cells are supplemented with unsaturated FA (Gonzalez and Martin 1996; Vemula et al. 2003; Martin et al. 2007). This FA-regulated decay of OLE1 mRNA occurs through both the 5′–3′ general pathway and via exosomal 3′–5′ degradation activities (Martin et al. 2007).
Transcriptional regulation by inositol and choline
Expression of the INO1 gene (Figure 3, Table 2) is repressed by ≥100-fold in the presence of inositol. Exogenous choline results in an additional severalfold reduction in INO1 transcript level but only if inositol is also present. However, in the absence of exogenous inositol, choline has little or no effect on INO1 expression (Hirsch and Henry 1986; Griac et al. 1996; Jesch et al. 2005a). Other genes encoding enzymes of lipid metabolism, including many involved in PC biosynthesis, show similar patterns of regulation in response to inositol and choline (Carman and Henry 1989, 1999, 2007; Paltauf et al. 1992; Henry and Patton-Vogt 1998; Jesch and Henry 2005; Jesch et al. 2005; Chen et al. 2007a; Carman and Han 2011). However, no other gene in the yeast genome shows as great a repression ratio in the presence of inositol and choline as INO1 (Santiago and Mamoun 2003; Jesch et al. 2005, 2006).
The regulation (Figure 3) controlling the expression of INO1 and coregulated genes of lipid metabolism has been the subject of a number of recent comprehensive reviews (Carman and Henry 2007; Chen et al. 2007a; Carman and Han 2011). Expression of these genes is controlled by the cis-acting element, consensus 5′CATGTGAAT3′ (Bachhawat et al. 1995; Greenberg and Lopes 1996), known as the inositol sensitive upstream activating element (UASINO), or alternatively as the inositol/choline responsive element (ICRE) (Schuller et al. 1992). The Ino2-Ino4 heterodimer, which is required for activation of transcription of UASINO-containing genes, binds to this site (Ambroziak and Henry 1994). Genome-wide location analysis confirmed the sequence of the UASINO element and the binding of the Ino2-Ino4 heterodimer to the element across the genome (Lee et al. 2002; Harbison et al. 2004). Since both Ino2 and Ino4 are required for activation of expression of INO1, ino2, and ino4 mutants exhibit inositol auxotrophy (Ino− phenotype). However, since many genes involved in lipid metabolism in addition to INO1 contain the UASINO promoter element (Greenberg and Lopes 1996), the ino2 and ino4 mutants exhibit altered phospholipid compositions including reduced PC content, even when growing in the presence of inositol (Loewy and Henry 1984). The fact that the INO2 regulatory gene also contains a UASINO element in its promoter, and is auto-regulated in response to inositol, adds an additional layer of complexity to this regulatory mechanism (Ashburner and Lopes 1995, Chen et al. 2007a). The INO1 locus has also recently been reported to localize at the nuclear periphery when it is being actively transcribed (Brickner and Walter 2004), through a chromatin-mediated mechanism that persists for several generations following addition of inositol (Brickner et al. 2007; Brickner 2009).
Repression of UASINO-containing genes in response to the presence of exogenous inositol involves the Opi1 repressor (Hirsch and Henry 1986; White et al. 1991) (Figure 3), which interacts with transcriptional activation domains in Ino2 (Wagner et al. 2001; Dietz et al. 2003). Opi1 has also been reported to interact with a number of other regulatory proteins (Chen et al. 2007a), including the pleiotropic corepressors, Sin3 and Ssn6 (Jaschke et al. 2011). Deletion of the OPI1 gene renders the cell incapable of repressing UASINO genes in response to inositol, and leads to expression of UASINO-containing genes at levels that exceed their normal derepressed levels by severalfold, whether inositol or choline are present or not (White et al. 1991; Santiago and Mamoun 2003; Jesch et al. 2005). Such high levels of INO1 expression lead to over-production of inositol (the Opi− phenotype for which the opi1 mutants are named) and the excess inositol excreted from Opi− mutants can be detected in a plate assay (Greenberg et al. 1982a,b). In addition, even when growing in the absence of inositol, opi1Δ cells have elevated levels of PI and levels of other lipids that resemble those of wild-type cells growing in the presence of inositol (Klig et al. 1985).
Role of PA in regulation of UASINO-containing genes
Opi− phenotypes are also associated with mutants (cho1, cho2, and opi3) defective in the synthesis of PC via the CDP-DAG pathway (Greenberg et al. 1983; Letts et al. 1983; Summers et al. 1988; McGraw and Henry 1989) (Figure 2). Similar to the opi1 mutant, cho2 and opi3 mutants overexpress INO1 and fail to repress INO1 and other UASINO-containing genes in response to inositol. However, in contrast to opi1 mutants, the constitutive overexpression of INO1 and Opi− phenotypes of opi3 and cho2 mutants are suppressed when they are grown in the presence of exogenous soluble precursors that restore PC biosynthesis by entering the Kennedy pathway downstream of their respective genetic lesions in PE methylation (Figure 2). Thus, the Opi− phenotype is eliminated and PC biosynthesis is restored in cho2 mutants grown in the presence of monomethylethanolamine (MME), dimethylethanolamine (DME), or choline (Summers et al. 1988), while these phenotypes are corrected only by DME and choline in opi3 mutants (McGraw and Henry 1989). Such observations led to the hypothesis that the mechanism by which the yeast cell senses the presence of exogenous inositol, choline, and other phospholipid precursors, such as serine, ethanolamine, MME, and DME, involves their effect on phospholipid synthesis (Carman and Henry 1989).
Further support for this hypothesis came from studies involving strains carrying temperature-sensitive mutations of the essential PI/PC transport protein, Sec14. While a full discussion of the biology of Sec14 is beyond the scope of this review article, the role it has played in discovery that PA is the signaling molecule controlling INO1 expression will be covered in brief here. The growth and secretory phenotypes of sec14ts mutants are suppressed by mutations in any one of the three genes encoding enzymes in the Kennedy pathway for PC biosynthesis (i.e., cki1, pct1, and cpt1) (Cleves et al. 1991). Such strains (for example: sec14tscki1Δ) have Opi− phenotypes and excrete choline (Opc− phenotype) (Patton-Vogt et al. 1997). These phenotypes are due to increased PA levels resulting from elevated turnover of PC, catalyzed by the SPO14-encoded phospholipase D (Sreenivas et al. 1998; Xie et al. 1998). These observations led to the identification of PA as the probable signaling lipid responsible for derepression of INO1 and coregulated genes in the absence of exogenous inositol (Griac et al. 1986; Summers et al. 1988; McGraw and Henry 1989; Henry and Patton-Vogt 1998). This model is consistent with the Opi− phenotypes of mutants defective in reactions in the CDP-DAG pathway for PC synthesis and the fact that these phenotypes are suppressed by supplying these mutants with precursor that enters the Kennedy pathway downstream of the defect in the CDP-DAG pathway (Henry and Patton-Vogt 1998). Interruption of any of the reactions downstream of PA via the CDP-DAG pathway leads to a buildup of PA and overexpression of INO1, which raises endogenous inositol production. This leads to excretion of inositol and renders the cell insensitive to exogenous inositol (Henry and Patton-Vogt 1998). Metabolites that enter the Kennedy pathway downstream of the specific metabolic lesion counteract PA build up by increasing the use of DAG in the Kennedy pathway (Figure 2). Also consistent with this model is the Opi− phenotype observed in cells engineered to have reduced expression of PIS1, thereby slowing PI synthesis and raising PA levels (Jani and Lopes 2009).
The mechanism by which PA regulates INO1 expression was elucidated when Loewen et al. (2004) showed that Opi1 contains PA binding domains that facilitate its interaction with PA in the perinuclear ER in cells growing in the absence of inositol (Figure 3). Opi1 also contains a motif known as FFAT (two phenylalanines in an acidic tract) that binds to Scs2, an integral ER membrane protein (Loewen et al. 2003). To remain localized to the ER membrane in the absence of inositol, Opi1 must interact with both Scs2 and PA. In wild-type yeast cells, addition of inositol to the growth medium leads to a dramatic increase in the rate of synthesis of PI, resulting in consumption of both PA and CDP-DAG. As PA levels drop, Opi1 is released from the ER and enters the nucleus where it represses expression of INO1 and other UASINO-containing genes (Loewen et al. 2004). Consistant with this model, mutations in Opi1 that interfere with its interaction with either PA or Scs2 result in Ino− phenotypes (Loewen et al. 2004) and scs2 mutants also exhibit an Ino− phenotype (Nikawa et al. 1995). Increased PC synthesis via the Kennedy pathway is also characteristic of scs2 mutants, a phenotype that has not been fully explored, and mutations blocking PC synthesis by this route suppress both the Ino− phenotype and increased synthesis of PC in the scs2 mutant (Kagiwada and Zen 2003). Since, as described above, blocking the reuse of choline via the Kennedy pathway should reduce the draw on PA and thus enhance INO1 expression, these phenotypes are consistent with the model of Loewen et al. (2004) concerning Opi1 function (Figure 3).
Regulation of gene expression by zinc
The expression of several phospholipid synthesis genes is also regulated in coordination with mechanisms that control zinc homeostasis (Figure 3) (Carman and Han 2007; Eide 2009). Zinc is an essential nutrient required for the growth and metabolism of S. cerevisiae (Eide 2009). It is a cofactor for key metabolic enzymes and a structural component of a diverse set of proteins that include chaperons, lipid binding proteins, and transcription factors (Vallee and Falchuk 1993; Schwabe and Klug 1994; Ellis et al. 2004; Eide 2009). Cells grown in medium lacking zinc exhibit the induced expression of plasma membrane and vacuolar membrane zinc transporters (encoded by ZRT1, ZRT2, ZRT3, and FET4) to maintain the cytosolic levels of zinc (Guerinot and Eide 1999; Eide 2003, 2009). At the same time, a reduction in zinc causes changes in membrane phospholipid composition that are brought about by changes in the expression of phospholipid synthesis enzyme activities (Han et al. 2004, 2005; Iwanyshyn et al. 2004; Kersting and Carman 2006; Carman and Han 2007). It is postulated that changes in phospholipid composition may govern the insertion, topology, and/or function of the membrane-associated zinc transporters (Carman and Han 2007).
The regulation of phospholipid synthesis genes by zinc availability (Figure 3) involves the control of PA content through the activation of PI synthase function. This regulation occurs in the absence of inositol supplementation and is mediated by the zinc-sensing and zinc-inducible transcriptional activator Zap1 and the zinc-responsive cis-acting element (UASZRE) (Carman and Han 2007). Zinc depletion causes an increase in the synthesis of PI through increased expression of PI synthase (Iwanyshyn et al. 2004; Han et al. 2005). This regulation is controlled by the interaction of Zap1 with a UASZRE in the PIS1 promoter (Iwanyshyn et al. 2004; Han et al. 2005). As discussed above, increased PI synthesis causes a decrease in PA content, which results in the Opi1-mediated repression of UASINO-containing genes and a decrease in the activities of the CDP-DAG pathway enzymes (Iwanyshyn et al. 2004). The major effects of zinc depletion on phospholipid composition include an increase in PI and a decrease in PE (Iwanyshyn et al. 2004). Although levels of enzymes in the CDP-DAG pathway are reduced by zinc depletion, the amount of PC is not significantly affected (Iwanyshyn et al. 2004). This is attributed to the Zap1-mediated inductions of choline kinase and ethanolamine kinase for PC synthesis via the Kennedy pathway (Kersting and Carman 2006; Soto and Carman 2008). Like PIS1, the CKI1 and EKI1 genes contain a UASZRE in their promoters that interact with Zap1 for gene activation (Kersting and Carman 2006; Soto and Carman 2008). Opi1-mediated regulation of CKI1 and EKI1 are overcome by their derepression via Zap1 (Kersting and Carman 2006; Soto and Carman 2008).
Among the genes in lipid metabolism that contain the UASZRE, DPP1 is the most highly regulated by zinc availability (Lyons et al. 2000; Carman and Han 2007; Eide 2009). The DPP1 gene encodes DGPP phosphatase, an enzyme associated with the vacuole membrane (Wu et al. 1996; Toke et al. 1998; Han et al. 2001). This enzyme catalyzes the removal of the β-phosphate from DGPP, a minor phospholipid in yeast, to form PA, followed by the dephosphorylation of PA to form DAG (Wu et al. 1996). The zinc-mediated regulation of DPP1 expression correlates with the metabolism of DGPP and PA in the vacuole membrane (Han et al. 2004). In zinc-replete medium, DGPP and PA account for 0.6 mol% and 1.4 mol% of the total phospholipids in vacuole membranes, but in zinc-depleted medium, the amounts of DGPP and PA are decreased to an undetectable level and 0.3 mol%, respectively (Han et al. 2004). The function of DPP1-encoded DGPP phosphatase is still unclear. However, it is speculated that the enzyme controls the levels of DGPP and PA in vacuolar membranes, which in turn mediates the cellular functions occurring in response to zinc depletion (Carman and Han 2007).
Regulation by phosphorylation
Phosphorylation is a major covalent post-translational modification by which the activity of an enzyme or a transcription factor is regulated (Karin and Hunter 1995; Calkhoven and Ab 1996; Hung et al. 1997; Kaffman et al. 1998; Komeili and O'Shea 1999; Liu et al. 2000). Global analyses of protein phosphorylation indicate that several enzymes and transporters of glycerolipid metabolism are subject to phosphorylation (Tables 1–3). Some proteins have many sites of phosphorylation, whereas others have only a few. It is also telling that many enzymes and transporters have no sites of phosphorylation, and thus the function of these proteins might be regulated by other mechanisms (e.g., substrate availability). The identity of the protein kinases involved and the physiological consequences of their phosphorylations have only been determined for a few proteins in glycerolipid metabolism (Carman and Han 2011). The protein kinases known to regulate the function of catalytic and regulatory proteins in glycerolipid metabolism include AMP-activated protein kinase, protein kinases A and C, casein kinase II, and cyclin-dependent kinase. Glycerolipid enzymes known to be bona fide substrates of protein kinases and regulated by phosphorylation include CTP synthetase, choline kinase, PS synthase, PA phosphatase, and TAG lipase. The repressor Opi1 is also regulated by phosphorylation. A discussion of these phosphorylations may be found in a recent review by Carman and Han (2011).
Regulation of PA phosphatase
Pah1 is one of the most highly regulated enzymes in lipid metabolism. Its activity is governed by several of the biochemical mechanisms discussed above including phosphorylation, enzyme location, and modulation by components of lipid metabolism. As discussed above, the DAG generated in the Pah1 reaction is used for the synthesis of TAG (Han et al. 2006) and for the synthesis of PE and PC via the Kennedy pathway (Carman and Han 2006, 2008). The enzyme also plays a major role in controlling the cellular concentration of its substrate PA (Figure 3) (Han et al. 2006), the precursor of phospholipids that are synthesized via the CDP-DAG pathway (Carman and Zeimetz 1996; Carman and Henry 1999; Carman and Han 2008). In addition, the substrate PA plays a signaling role (see above) in the transcriptional regulation of phospholipid synthesis genes (Carman and Henry 2007). pah1Δ mutants exhibit a >90% reduction in TAG content, as well as derepression of phospholipid synthesis genes and massive expansion of the nuclear/ER membrane (Santos-Rosa et al. 2005; Han et al. 2007). Thus, the regulation of Pah1 activity governs the synthesis of TAG, the pathways by which phospholipids are synthesized, PA signaling, and the growth of the nuclear/ER membrane (Carman and Han 2008). Pah1 is associated with the cytosolic and membrane fractions of the cell, and its association with the membrane is peripheral in nature (Han et al. 2006). Chromatin immunoprecipitation analysis indicates that Pah1 may also be localized in the nucleus (Santos-Rosa et al. 2005).
The association of Pah1 with the membrane where its substrate PA resides is essential to its function in vivo, and membrane association is largely governed by the phosphorylation state of the enzyme (Karanasios et al. 2010; Choi et al. 2011). Phosphorylation favors a cytosolic association, whereas dephosphorylation favors a membrane association (Choi et al. 2011). The important sites of phosphorylation that govern this regulation include the seven Ser/Thr-Pro targets for CDC28-encoded and PHO85-encoded cyclin-dependent kinases. Pah1 is dephosphorylated by the Nem1-Spo7 phosphatase complex that is associated with the nuclear/ER membrane (Siniossoglou et al. 1998; Santos-Rosa et al. 2005). In the absence of Nem1-Spo7, wild-type Pah1 is enriched in the cytosol where it is physiologically inactive, whereas a nonphosphorylatable mutant of Pah1 with alanine substitutions of Cdc28 and Pho85 target sites is enriched in the membrane and is physiologically active (Choi et al. 2011). The requirement of Nem1-Spo7 indicates that Pah1 is recruited to the membrane for its physiological function. Indeed, the Nem1-Spo7–dependent membrane localization of Pah1 is enhanced by elevated levels of PA (Karanasios et al. 2010). Once the enzyme is dephosphorylated, it anchors onto the nuclear/ER membrane via a short N-terminal amphipathic helix allowing for the production of DAG for TAG synthesis (Karanasios et al. 2010).
Under normal physiological conditions (i.e., presence of the Nem1-Spo7 complex), the level of wild-type Pah1 detected on the membrane is very low. In fact, microscopic analysis of live S. cerevisiae cells expressing Pah1-GFP show a cytosolic localization without a detectable fluorescence signal associated with the nuclear/ER membrane unless PA levels are elevated (Karanasios et al. 2010). Yet we know that Pah1 is physiologically active with respect to lipid metabolism throughout cell growth (Han et al. 2006). Purified Pah1 has a relatively high catalytic efficiency when compared with other enzymes of lipid metabolism (Lin and Carman 1989), and the lethal phenotype of cells that overexpress Nem1-Spo7 (Santos-Rosa et al. 2005) indicates that an excess of Pah1 function is detrimental to cell physiology. Indeed, suppression of this phenotype by overexpression of Pah1 (Santos-Rosa et al. 2005) led to the discovery of Pah1 function (Han et al. 2007). Thus, under normal physiological conditions, the amount of Pah1 associated with membranes must be small for its physiological function to be controlled, and this regulation must be mediated by the amount of the Nem1-Spo7 on the membrane. In support of this hypothesis, the expression level of Nem1 is 10-fold lower when compared with Pah1 (Ghaemmaghami et al. 2003).
Pah1 activity is also modulated by cytosolic- and membrane-associated factors. The nucleotides ATP and CTP, which are precursors for the synthesis of phospholipids (Carman and Henry 1999), are inhibitors of Pah1 activity (Wu and Carman 1994). Indeed high levels of ATP favor elevated PA content and phospholipid synthesis, whereas low levels of ATP favor reduced PA content and an increase in the synthesis of TAG (Wu and Carman 1994). As discussed elsewhere, elevated CTP content favors an increase in PA content and derepression of UASINO-containing phospholipid synthesis genes (Ostrander et al. 1998). The patterns of regulation of PA phosphatase activity by ATP and by CTP are consistent with the regulation of lipid synthesis observed in cells that have fluctuations in ATP and CTP (Wu and Carman 1994; Ostrander et al. 1998).
Membrane lipids modulate Pah1 activity. For example, CDP-DAG, PI, and CL enhance activity (Wu and Carman 1996), whereas the sphingoid bases phytosphingosine and sphinganine inhibit activity (Wu et al. 1993). The major effect of the lipid activators is to decrease the Km of Pah1 for PA. Sphinganine antagonizes the activation of PA phosphatase activity by CL and PI, whereas it causes an increase in the cooperativity of CL activation (Wu and Carman 1996). Conversely, sphinganine has little effect on the cooperativity of PI activation, but causes an increase in the activation constant for PI (Wu and Carman 1996). On the basis of the activation/inhibitor constants and cellular concentrations for these lipid effector molecules, their regulatory roles on Pah1 should be physiologically relevant (Wu et al. 1993; Wu and Carman 1996).
DAG kinase counteracts PA phosphatase in regulating PA levels
Dgk1 is a unique CTP-dependent nuclear/ER membrane-associated enzyme that catalyzes the formation of PA from DAG (Han et al. 2008a,b). Dgk1 counteracts Pah1 in controlling PA content and consequently, transcriptional regulation of UASINO-containing genes (Han et al. 2008a,b). DGK1 overexpression causes an increase in PA content, derepression of UASINO-containing genes, and abnormal nuclear/ER membrane expansion (Han et al. 2008a) like those that occur in the pah1Δ mutant (Santos-Rosa et al. 2005; Han et al. 2007). Also consistent with the regulatory role of PA, DGK1 overexpression suppresses the inositol auxotrophy caused by PAH1 overexpression (Han et al. 2008a), whereas the dgk1Δ mutation suppresses the phenotypes caused by the pah1Δ mutation (Han et al. 2008a,b).
Ino− and Opi− phenotypes are associated with mutations affecting many functions
To date, the Ino− and Opi− phenotypes of mutants affecting lipid metabolism, which have been examined in detail for correlation of changes in lipid metabolism with changes in INO1 expression, have been consistent with predictions based on the role of PA as discussed above (Carman and Henry 2007). However, mutants defective in a wide variety of other cellular functions have also been reported to have these phenotypes (Henry and Patton-Vogt 1998) and several genome-wide screens for mutants with Opi− or Ino− phenotypes have been conducted (Hancock et al. 2006; Young et al. 2010; Villa-Garciá et al. 2011).
The screen of the Matα viable yeast deletion collection conducted by Hancock et al. (2006), using the Opi− plate test, as well as an INO1-LacZ reporter construct, identified 89 Opi− mutants. Among these mutants were opi1, cho2, opi3, ume6, rpd3, sin3, and reg1, which had previously been shown to affect INO1 expression (Henry and Patton-Vogt 1998; Shirra and Arndt 1999; Ouyang et al. 1999; Elkhaimi et al. 2000). Hancock et al. (2006) also reported Opi− phenotypes for the first time in mutants affecting functions such as the NuA4 histone acetyl transferase, vacuolar protein sorting, and other aspects of membrane trafficking and the UPR.
Young et al. (2010) identified 231 mutants from the yeast haploid deletion collection that showed measurable growth reduction at 37°, when grown in medium lacking inositol. The major focus of this study was on the discovery of pH sensitive Ino− phenotypes associated with mutants defective in all of the subunits of the vacuolar adenosine triphosphatase (V-ATPase) complex, as well as factors in the ER responsible for the assembly of this complex. The pma1-007 mutant, defective in the essential P-type ATPase of the plasma membrane, which regulates cellular pH, was also found to have an Ino− phenotype, as was trk1Δ, defective in a K+ transporter that activates Pma1. The Ino− phenotypes of these mutants were observed at pH 3 but were suppressed at pH 4 and 5 and this pH sensitivity was attributed to pH dependence of binding of Opi1 to PA. The binding of PA to Spo20, a yeast SNARE (Nakanishi et al. 2004), was also shown to be pH sensitive, suggesting that binding of proteins to PA is affected by cellular pH. Changes in the ionization state of PA affect binding of Opi1 to PA in vitro, supporting the electrostatic/hydrogen bond switch model of Kooijman et al. (2007) for interaction of proteins with PA. Repression of INO1 in response to glucose starvation also correlates to cellular pH, but not to PA levels. Young et al. (2010) have concluded that the Reg1-mediated mechanism that regulates INO1 expression via the glucose response pathway (Ouyang et al. 1999; Shirra and Arndt 1999) is most likely not dependent on the absolute level of PA, but rather on the effect of pH on the ionization state of PA.
The screen for Ino− mutants of the diploid viable yeast deletion collection conducted by Villa-Garciá et al. (2011) has resulted in the identification of 419 genes, which when deleted confer the Ino− phenotype under one or more growth conditions. This screen involves comparing growth in the presence and absence of inositol at two growth temperatures, 30° and 37°, in the presence and absence of choline. Choline sensitive Ino− phenotypes and the strengthening of weak Ino− phenotypes by choline are potentially attributable to reduction in PA levels due to the consumption of DAG produced from PA when PC is synthesized via the Kennedy pathway (see above) using exogenous choline (Carman and Henry 2007). Growth temperature has also been shown to influence lipid metabolism in wild-type and mutant strains. Growth at 37° results in an increased rate of synthesis of PI in wild-type cells growing in the presence of inositol (Gaspar et al. 2008), as well as an increase in PC synthesis and turnover (Dowd et al. 2001). Among the gene ontology (GO) categories enriched among the Ino− mutants identified by Villa-Garciá et al. (2011) are: response to stress, protein modification, chromosome organization, response to chemical stimulus, cellular carbohydrate metabolism, psuedohyphal growth, and transcription. Mutants in the category response to stress include a number with defects in stress response pathways that had previously been reported to be associated with Ino− phenotypes, including the protein kinase C-cell wall integrity (PKC-CWI) (Nunez et al. 2008; Jesch et al. 2010), the UPR (Cox et al. 1997; Chang et al. 2002) and the glucose response pathway (Shirra et al. 2001, 2005). However, Ino− phenotypes have also been observed in mutants affecting signaling pathways not previously reported to be associated with such phenotypes including: target of rapamycin (TOR), high osmolarity glycerol (HOG), cAMP-dependent protein kinase, calcineurin and filamentous growth and cell cycle regulation (Villa-Garciá et al. 2011).
Causes of Ino− phenotypes
It is often assumed that an Ino− phenotype provides evidence that a mutant has impaired INO1 expression or, alternatively, that a signaling pathway and/or a transcription factor defective in the mutant in question, is directly involved in regulating INO1 transcription. For example, the Ino− phenotypes of the ire1Δ and hac1Δ mutants, defective in UPR activation, have been attributed to a role for the Hac1 transcription factor in activating expression of INO1 (Cox et al. 1997). The evidence for this interpretation includes the observations that ire1Δ and hac1Δ mutants fail to sustain expression of INO1 when shifted to medium lacking inositol and that the opi1Δ mutation supresseses their Ino− phenotypes (Cox et al. 1997). Cox et al. (1997) also reported that growth in the absence of inositol activates the UPR in wild-type cells and that INO1 expression is activated in wild-type cells treated with tunicamycin in the presence of normally repressing levels of inositol. However, activation of INO1 expression in response to tunicamycin in the presence of inositol was not observed by Chang et al. (2002) and INO1 is repressed with normal kinetics in response to addition of inositol in cells in which the UPR was constitutively induced by constitutive expression of activated Hac1 (Jesch et al. 2006). The reasons for these divergent results with respect to the role of Hac1 in INO1 transcription are not clear.
A number of mutants in the PKC-CWI pathway have Ino− phenotypes that are intensified at 37° and in the presence of choline (Nunez et al. 2008; Fernandez-Murray et al. 2009; Jesch et al. 2010; Villa-Garciá et al. 2011). Futhermore, the Ino− phenotypes of these mutants are suppressed by opi1Δ. It is clear, however, that the PKC pathway has no direct role in regulating INO1 expression. The PKC pathway mutant, slt2Δ/mpk1Δ, exhibits normal INO1 derepression in response to a shift to inositol medium even when choline is present at 37°, a condition under which growth ceases and the mutant begins to lose viability within 4–5 hr (Nunez et al. 2008). The stt4ts and mss4ts mutants, defective in the PI kinases that are responsible for producing the PI 4-P and PI 4,5-P2 pools on the plasma membrane and are essential for PKC-CWI signaling during heat stress (Audhya and Emr 2002; Audhya and Emr 2003), also exhibit Ino− phenotypes at semipermissive growth temperatures (Jesch et al. 2010). However, INO1 undergoes normal derepression immediately following a shift to medium lacking inositol in the stt4ts and mss4ts mutants, even at the temperature at which they exhibit an Ino− phenotype (Jesch et al. 2010). Significantly, the PKC-CWI pathway is activated in wild-type cells grown in the absence of inositol (Nunez et al. 2008) in response to reduction in synthesis of inositol-containing sphingolipids (Jesch et al. 2010). Thus, it is clear that the Ino− phenotypes of PKC-CWI mutants are not attributable to a defect in INO1 expression, but rather to failure to mount a stress response that is essential for survival of stress caused by growth in the absence of inositol (Nunez et al. 2008; Jesch et al. 2010; Villa et al. 2011).
Supression of an Ino− phenotype by opi1Δ is also often cited as evidence that the mutant in question is defective in a functon that regulates INO1. However, the opi1Δ mutation also suppresses the Ino− phenotypes of slt2Δ and other PKC pathway mutants (Nunez et al. 2008), which, as discussed above, are not attributable to failure to express INO1. The explanation for this observation lies in the fact that the opi1Δ mutation does not simply restore INO1 expression to the level seen in wild-type cells growing in the absence of inositol. Instead, the deletion of the OPI1 gene completely eliminates the ability of the cell sense inositol, whether endogenously produced or exogenously supplied, leading expression of INO1 at levels as high as 5- to 6-fold higher than normal derepressed levels (Bachhwat et al. 1995). As a consequence of the resulting overproduction of inositol, opi1 cells have levels of PI and other lipids resembling the lipid composition of wild-type cells grown in the presence of high levels of exogenous inositol (Klig et al. 1985). Indeed, opi1Δ mutants excrete so much inositol that they support growth of any Ino− mutant in their vicinity, the characteristic used in the original bioassay used in the isolation of Opi− mutants (Greenberg et al. 1982a). Microarray experiments showed that, in contrast to wild-type cells, opi1Δ cells do not exhibit activation of UPR target genes when grown in the absence of inositol (Jesch et al. 2005). This is presumably due to the fact that overexpression of the INO1 gene in opi1Δ cells produces sufficient inositol to repress expression of all genes that are normally activated in absence of inositol, except of course the UASINO-containing genes, which are directly dependent on Opi1p for repression in the presence of inositol (Jesch and Henry 2005). Thus, suppression by opi1Δ of an Ino− phenotype conferred by mutation of a given gene does not consitute sufficient evidence on its own that the gene product in question is involved in regulation of INO1.
Microarray studies have revealed that the transcript levels of hundreds of genes in yeast are affected by the availability of inositol and/or inositol plus choline in the medium (Santiago and Mamoun 2003; Jesch et al. 2005, 2006; Nunez et al. 2008). Most of these “inositol-regulated” genes are not under the control of Opi1 and/or involved in lipid metabolism and a number of these genes are known targets of the stress response pathways that are associated with Ino− phenotypes, including the UPR and PKC pathways (Jesch et al. 2005, 2006; Nunez et al. 2008; Jesch et al. 2010; Villa et al. 2011). As discussed above, expression of INO1 at the level observed in wild-type cells growing in the absence of inositol does not suffice to support the rate of PI synthesis observed in the same cells growing in the presence of exogenous inositol (Kelley et al. 1988; Gaspar et al. 2006). Moreover, in wild-type cells, the changes in lipid metabolism that occur in response to removal of inositol and/or its resupply have been shown to affect signaling associated with PA (Loewen et al. 2004; Young et al. 2010), and with inositol containing sphingolipids and the pools of PI 4-P and PI 4,5-P2 in the plasma membrane (Jesch et al. 2010).
As the exploration of the regulation and cell biology of lipid metabolism in yeast accelerates in response to genomic approaches, additional examples of lipid-mediated signaling influenced by the availability of lipid precursors are likely to be discovered. A major advantage of yeast in such studies is the fact, as described in this YeastBook chapter, that its lipid metabolism can readily be manipulated by changing the supply of exogenous precursors and/or by introducing mutations affecting specific steps in lipid metabolism.
Perspectives
Monumental advancements have been made in determining gene enzyme relationships and elucidating the fundamental biochemistry, regulatory mechanisms, and cellular roles of lipids in the 20 years since the publication of the last edition of the Molecular Biology of the Yeast Saccharomyces: Gene Expression in 1992. Yet while the identities of many of the genes encoding enzymes, transporters, and regulatory factors controlling lipid metabolism in yeast have been discovered, described, and analyzed, there remain many gaps in our knowledge. The functions of many genes, genome-wide, have yet to be clarified and among them are certainly a considerable number of “missing links” in yeast lipid metabolism and its regulation. For example, the molecular organization of lipid biosynthetic pathways that ensures metabolic channeling of critical compounds, enzyme topology, and the integration of soluble and membrane-bound intermediates in these pathways is only beginning to be addressed. Furthermore, we do not have complete knowledge of the mechanisms by which lipids synthesized in one compartment are trafficked to other locations within the cell or the mechanisms by which the precise lipid compositions of specific membrane compartments are established and maintained. High-resolution lipidomic analyses that have uncovered some 100 different molecular species of glycero- and sphingolipids have added another level of complexity to this question. Moreover, our understanding of the regulatory networks that link lipid metabolism to other aspects of cell biology and metabolism, including energy metabolism, membrane biogenesis, and trafficking, signaling, and cell division, remains limited. However, the increasing availability of archived genetic and biochemical data on a wide variety of eukaryotic organisms and the extensive homologies that exist among eukaryotes are opening new avenues for pursuing these important challenges. Due to its genetic tractability and comparably “simple” lipid complement, yeast continues to be among the most attractive and important model systems for such research.
Acknowledgments
We are indebted to Maria Laura Gaspar for her collaboration in preparing Figure 2 and to Yu-Fang Chang for providing fluorescence microscopy images of Opi1 localization for Figure 3. We also acknowledge our colleagues Yu-Fang Chang, Maria Laura Gaspar, Harald Hofbauer, Gil-Soo Han, Stephen Jesch, and Manuel Villa for critical reading of the manuscript and many helpful discussions. Work in the Henry laboratory is supported by National Institutes of Health (NIH) grant GM-19629. Work in the Kohlwein laboratory is supported by grants from the Austrian Science Funds (project F30 LIPOTOX), the Ph.D. program Molecular Enzymology, and the Austrian Federal Government for Science and Research (Project GOLD in the framework of the Austrian Genome Program GEN-AU). Work in the Carman laboratory is supported by NIH grants GM-28140 and GM-50679.
Footnotes
Communicating editor: A. G. Hinnebusch
Literature Cited
- Achleitner G., Zweytick D., Trotter P. J., Voelker D. R., Daum G., 1995. Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 270: 29836–29842 [DOI] [PubMed] [Google Scholar]
- Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S. D., et al. , 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264: 545–553 [DOI] [PubMed] [Google Scholar]
- Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., et al. , 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Feel W., Chirala S. S., Wakil S. J., 1992. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA 89: 4534–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroziak J., Henry S. A., 1994. The INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269: 15344–15349 [PubMed] [Google Scholar]
- Ashburner B. P., Lopes J. M., 1995. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol. Cell. Biol. 15: 1709–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G., 1997. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J. Bacteriol. 179: 7611–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G., 2000. 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. 2000. J. Biol. Chem. 275: 235–240 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G., 2003. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278: 23317–23323 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Daum G., 2005. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280: 37301–37309 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G., 1999a Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181: 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., Weys S., Paltauf F., Daum G., 1999b Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J. Bacteriol. 181: 1458–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson K. D., Jensen B., Kolat A. I., Storm E. M., Henry S. A., et al. , 1980a Yeast mutants auxotropic for choline or ethanolamine. J. Bacteriol. 141: 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson K., Fogel S., Henry S. A., 1980b Yeast mutant defective in phosphatidylserine synthesis. J. Biol. Chem. 255: 6653–6661 [PubMed] [Google Scholar]
- Audhya A., Emr S. D., 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2: 593–605 [DOI] [PubMed] [Google Scholar]
- Audhya A., Emr S. D., 2003. Regulation of PI4,5P2 synthesis by nuclear-cytosolic shuttling of the Mss4 lipid kinase. EMBO J. 22: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat N., Ouyang Q., Henry S. A., 1995. Functional Characterization of an Inositol-sensitive Upstream Activation Sequence in Yeast. A cis-Regulatory Element Responsible for Inositol-Choline Mediated Regulation of Phospholipid Biosynthesis. J. Biol. Chem. 270: 25087–25095 [DOI] [PubMed] [Google Scholar]
- Bae-Lee M. S., Carman G. M., 1984. Phosphatidylserine synthesis in Saccharomyces cerevisiae. Purification and characterization of membrane-associated phosphatidylserine synthase. J. Biol. Chem. 259: 10857–10862 [PubMed] [Google Scholar]
- Bankaitis V. A., Mousley C. J., Schaaf G., 2010. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 35: 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh C. T., Rine J., 2004. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117: 2983–2996 [DOI] [PubMed] [Google Scholar]
- Benghezal M., Roubaty C., Veepuri V., Knudsen J., Conzelmann A., 2007. SLC1 and SLC4 encode partially redundant Acyl-Coenzyme A 1-Acylglycerol-3-phosphate O-Acyltransferases of budding yeast. J. Biol. Chem. 282: 30845–30855 [DOI] [PubMed] [Google Scholar]
- Beranek A., Rechberger G., Knauer H., Wolinski H., Kohlwein S. D., et al. , 2009. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J. Biol. Chem. 284: 11572–11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Nebauer R., Schneiter R., Daum G., 2003. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae. Mol. Biol. Cell 14: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black P. N., Dirusso C. C., 2007. Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim. Biophys. Acta 1771: 286–298 [DOI] [PubMed] [Google Scholar]
- Block-Alper L., Webster P., Zhou X., Supekova L., Wong W. H., et al. , 2002. INO2, a positive regulator of lipid biosynthesis, is essential for formation of inducible membranes in yeast. Mol. Biol. Cell 13: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., et al. , 2008. PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26: 1339–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie M. A., Martin C. E., 1989. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J. Bacteriol. 171: 6409–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumann H. A., Gubbens J., Koorengevel M. C., Oh C. S., Martin C. E., et al. , 2006. Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol. Biol. Cell 17: 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K., Mick D. U., Frazier A. E., Taylor R. D., Meisinger C., et al. , 2005. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome. Mol. Biol. Cell 16: 5202–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner, D.G., I. Cajigas, Y. Fondufe-Mittendorf, S. Ahmed, P. Lee, 2007 H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PloS Biol. 5: 0704–0716. [DOI] [PMC free article] [PubMed]
- Brickner J. H., Walter P., 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner J. H., 2009. Transcriptional memory at the nuclear periphery. Curr. Opin. Cell Biol. 21: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkhoven C. F., Ab G., 1996. Multiple steps in the regulation of transcription-factor level and activity. Biochem. J. 317: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G. S., 2006. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G. S., 2007. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim. Biophys. Acta 1771: 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G. S., 2009a Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284: 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G.-S., 2009b Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50: S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Han G.-S., 2011. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80: 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A., 1989. Phospholipid biosynthesis in yeast. Annu. Rev. Biochem. 58: 635–669 [DOI] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A., 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38: 361–399 [DOI] [PubMed] [Google Scholar]
- Carman G. M., Henry S. A., 2007. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G. M., Zeimetz G. M., 1996. Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271: 13293–13296 [DOI] [PubMed] [Google Scholar]
- Carter J. R., Kennedy E. P., 1966. Enzymatic synthesis of cytidine diphosphate diglyceride. J. Lipid Res. 7: 678–683 [PubMed] [Google Scholar]
- Chang H. J., Jones E. W., Henry S. A., 2002. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H. J., S. A. Jesch, M. L. Gaspar and S. A. Henry, 2004 Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168: 1899–1913. [DOI] [PMC free article] [PubMed]
- Chang S. C., Heacock P. N., Clancey C. J., Dowhan W., 1998a The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273: 9829–9836 [DOI] [PubMed] [Google Scholar]
- Chang S. C., Heacock P. N., Mileykovskaya E., Voelker D. R., Dowhan W., 1998b Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273: 14933–14941 [DOI] [PubMed] [Google Scholar]
- Chang Y.-F., Carman G. M., 2008. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 47: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Hancock L. C., Lopes J. M., 2007a Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim. Biophys. Acta 1771: 310–321 [DOI] [PubMed] [Google Scholar]
- Chen Q., Kazachkov M., Zheng Z., Zou J., 2007b The yeast acylglycerol acyltransferase LCA1 is a key component of Lands cycle for phosphatidylcholine turnover. FEBS Lett. 581: 5511–5516 [DOI] [PubMed] [Google Scholar]
- Chen S., Tarsio M., Kane P. M., Greenberg M. L., 2008. Cardiolipin mediates cross-talk between mitochondria and the vacuole. Mol. Biol. Cell 19: 5047–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Wu J. L., Padmanabha R., Glover C. V., 1988. Isolation, sequencing, and disruption of the CKA1 gene encoding the alpha subunit of yeast casein kinase II. Mol. Cell. Biol. 8: 4981–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala S. S., 1992. Coordinated regulation and inositol-mediated and fatty acid-mediated repression of fatty acid synthase genes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 10232–10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala S. S., Kuziora M. A., Spector D. M., Wakil S. J., 1987. Complementation of mutations and nucleotide sequence of FAS1 gene encoding beta subunit of yeast fatty acid synthase. J. Biol. Chem. 262: 4231–4240 [PubMed] [Google Scholar]
- Cho R. J., Campbell M. J., Winzeler E. A., Steinmetz L., Conway A., et al. , 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65–73 [DOI] [PubMed] [Google Scholar]
- Choi H.-S., Carman G. M., 2007. Respiratory deficiency mediates the regulation of CHO1-encoded phosphatidylserine synthase by mRNA stability in Saccharomyces cerevisiae. J. Biol. Chem. 282: 31217–31227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., et al. , 2011. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae. Identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286: 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Martin C. E., 1999. The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J. Biol. Chem. 274: 4671–4683 [DOI] [PubMed] [Google Scholar]
- Chung J. H., Lester R. L., Dickson R. C., 2003. Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J. Biol. Chem. 278: 28872–28881 [DOI] [PubMed] [Google Scholar]
- Clancey C. J., Chang S. C., Dowhan W., 1993. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268: 24580–24590 [PubMed] [Google Scholar]
- Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., et al. , 1991. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart L. A., Obeid L. M., 2007. Yeast sphingolipids: recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta 1771: 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Chapman R. E., Walter P., 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T., Athenstaedt K., Daum G., 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta 1771: 299–309 [DOI] [PubMed] [Google Scholar]
- Daum G., Wagner A., Czabany T., Athenstaedt K., 2007. Dynamics of neutral lipid storage and mobilization in yeast. Biochimie 89: 243–248 [DOI] [PubMed] [Google Scholar]
- Dean-Johnson M., Henry S. A., 1989. Biosynthesis of inositol in yeast. Primary structure of myo- inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J. Biol. Chem. 264: 1274–1283 [PubMed] [Google Scholar]
- Deng L., Fukuda R., Kakihara T., Narita K., Ohta A., 2010. Incorporation and remodeling of phosphatidylethanolamine containing short acyl residues in yeast. Biochim. Biophys. Acta 1801: 635–645 [DOI] [PubMed] [Google Scholar]
- Denic V., Weissman J. S., 2007. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677 [DOI] [PubMed] [Google Scholar]
- Dickson R. C., 2008. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 49: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., 2010. Roles for sphingolipids in Saccharomyces cerevisiae. Adv. Exp. Med. Biol. 688: 217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Lester R. L., 2002. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583: 13–25 [DOI] [PubMed] [Google Scholar]
- Dietrich L. E., Ungermann C., 2004. On the mechanism of protein palmitoylation. EMBO Rep. 5: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz M., Heyken W. T., Hoppen J., Geburtig S., Schuller H. J., 2003. TFIIB and subunits of the SAGA complex are involved in transcriptional activation of phospholipid biosynthetic genes by the regulatory protein Ino2 in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 48: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Henry S. A., 1981. Myoinositol-1-phosphate synthetase: Characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256: 7077–7085 [PubMed] [Google Scholar]
- Dowd S. R., Bier M. E., Patton-Vogt J. L., 2001. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J. Biol. Chem. 276: 3756–3763 [DOI] [PubMed] [Google Scholar]
- Duronio R. J., Towler D. A., Heuckeroth R. O., Gordon J. I., 1989. Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science 243: 796–800 [DOI] [PubMed] [Google Scholar]
- Duronio R. J., Knoll L. J., Gordon J. I., 1992. Isolation of a Saccharomyces cerevisiae long chain fatty acyl:CoA synthetase gene (FAA1) and assessment of its role in protein N-myristoylation. J. Cell Biol. 117: 515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J., 2003. Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J. Nutr. 133: 1532S–1535S [DOI] [PubMed] [Google Scholar]
- Eide D. J., 2009. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284: 18565–18569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., et al. , 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhaimi M., Kaadige M. R., Kamath D., Jackson J. C., Biliran H., Jr, et al. , 2000. Combinatorial regulation of phospholipid gene expression by the UME6, SIN3 and RPD6 genes. Nucleic Acids Res. 28: 3160–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C. D., Wang F., MacDiarmid C. W., Clark S., Lyons T., et al. , 2004. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman N. J., Black P. N., Zhao X. D., Knudsen J., DiRusso C. C., 2001. The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization. J. Biol. Chem. 276: 37051–37059 [DOI] [PubMed] [Google Scholar]
- Faergeman N. J., Feddersen S., Christiansen J. K., Larsen M. K., Schneiter R., et al. , 2004. Acyl-CoA-binding protein, Acb1p, is required for normal vacuole function and ceramide synthesis in Saccharomyces cerevisiae. Biochem. J. 380: 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakas S., Konstantinou C., Carman G. M., 2011. DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286: 1464–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese R. V., Jr, Walther T. C., 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner A. J., Chen X., Rush J., Horazdovsky B., Waechter C. J., et al. , 1999. The LPP1 and DPP1 gene products account for most of the isoprenoid phosphatase activities in Saccharomyces cerevisiae. J. Biol. Chem. 274: 14831–14837 [DOI] [PubMed] [Google Scholar]
- Fei W., Alfaro G., Muthusamy B. P., Klaassen Z., Graham T. R., et al. , 2008. Genome-wide analysis of sterol-lipid storage and trafficking in Saccharomyces cerevisiae. Eukaryot. Cell 7: 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Wang H., Fu X., Bielby C., Yang H., 2009. Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae. Biochem. J. 424: 61–67 [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray J. P., McMaster C. R., 2005a. Glycerophophocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J. Biol. Chem. 280: 38290–38296 [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray J. P., McMaster C. R., 2005b Nte1p-mediated deacylation of phosphatidylcholine functionally interacts with Sec14p. J. Biol. Chem. 280: 8544–8552 [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray J. P., McMaster C. R., 2007. Phosphatidylcholine synthesis and its catabolism by yeast neuropathy target esterase 1. Biochim. Biophys. Acta 1771: 331–336 [DOI] [PubMed] [Google Scholar]
- Fernandez-Murray J. P., Gaspard G. J., Jesch S. A., McMaster C. R., 2009. NTE1-encoded phosphatidylcholine phospholipase b regulates transcription of phospholipid biosynthetic genes. J. Biol. Chem. 284: 36034–36046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl A. S., Carman G. M., 1983. Phosphatidylinositol biosynthesis in Saccharomyces cerevisiae: purification and properties of microsome-associated phosphatidylinositol synthase. J. Bacteriol. 154: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E., Almaguer C., Holic R., Griac P., Patton-Vogt J., 2005. Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280: 36110–36117 [DOI] [PubMed] [Google Scholar]
- Flanagan C. A., Schnieders E. S., Emerick A. W., Kunisawa R., Admon A., et al. , 1993. Phosphatidylinositol 4-kinase: Gene structure and requirement for yeast cell viability. Science 262: 1444–1448 [DOI] [PubMed] [Google Scholar]
- Flick J. S., Thorner J., 1993. Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 5861–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Jigami Y., 2008. Lipid remodeling of GPI-anchored proteins and its function. Biochim. Biophys. Acta. 1780: 410–420 [DOI] [PubMed] [Google Scholar]
- Fyrst H., Oskouian B., Kuypers F. A., Saba J. D., 1999. The PLB2 gene of Saccharomyces cerevisiae confers resistance to lysophosphatidylcholine and encodes a phospholipase B/lysophospholipase. Biochemistry 38: 5864–5871 [DOI] [PubMed] [Google Scholar]
- Gaigg B., Neergaard T. B., Schneiter R., Hansen J. K., Faergeman N. J., et al. , 2001. Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Padamsee M., Wilcox L., Oelkers P. M., D’Ambrosio D., et al. , 2009. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid mediated cell death. J. Biol. Chem. 184: 30994–31005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bustos J. F., Marini F., Stevenson I., Frei C., Hall M. N., 1994. PIK1, an essential phosphatidylinositol 4-kinase associated with the yeast nucleus. EMBO J. 13: 2352–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardocki M. E., Jani N., Lopes J. M., 2005. Phosphatidylinositol biosynthesis: biochemistry and regulation. Biochim. Biophys. Acta 1735(2): 89–100 [DOI] [PubMed] [Google Scholar]
- Gary J. D., Sato T. K., Stefan C. J., Bonangelino C. J., Weisman L. S., et al. , 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell 13: 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M. L., Aregullin M. A., Jesch S. A., Henry S. A., 2006. Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281: 22773–22785 [DOI] [PubMed] [Google Scholar]
- Gaspar M. L., Aregullin M. A., Jesch S. A., Nunez L. R., Villa-Garcia M., et al. , 2007. The emergence of yeast lipidomics. Biochim. Biophys. Acta 1771: 241–254 [DOI] [PubMed] [Google Scholar]
- Gaspar M. L., Jesch S. A., Viswanatha R., Antosh A. L., Brown W. J., et al. , 2008. A block in endoplasmic reticulum-to-Golgi trafficking inhibits phospholipid synthesis and induces neutral lipid accumulation. J. Biol. Chem. 283: 25735–25751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar M. L., Hofbauer H. F., Kohlwein S. D., Henry S. A., 2011. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 286: 1696–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor P. M., Carman G. M., 1990. Phosphatidylethanolamine methyltransferase and phospholipid methyltransferase activities from Saccharomyces cerevisiae. Enzymological and kinetic properties. Biochim. Biophys. Acta 1045: 156–163 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gohil V. M., Thompson M. N., Greenberg M. L., 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280: 35410–35416 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. I., Martin C. E., 1996. Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. J. Biol. Chem. 271: 25801–25809 [DOI] [PubMed] [Google Scholar]
- Goodmann J. M., 2009. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50: 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. L., Lopes J. M., 1996. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. L., Reiner B., Henry S. A., 1982a Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol excreting mutants. Genetics 100: 19–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M., Goldwasser P., Henry S., 1982b Characterization of a regulatory mutant constitutive for inositol-1-phosphate synthetase. Mol. Gen. Genet. 186: 157–163 [DOI] [PubMed] [Google Scholar]
- Greenberg M. L., Klig L. S., Letts V. A., Shicker-Loewy B., Henry S. A., 1983. Yeast mutant defective in phosphatidylcholine synthesis. J. Bacteriol. 153: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griac P., 2007. Sec14 related proteins in yeast. Biochim. Biophys. Acta 1771: 737–745 [DOI] [PubMed] [Google Scholar]
- Griac P., Swede M. J., Henry S. A., 1996. The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J. Biol. Chem. 271: 25692–25698 [DOI] [PubMed] [Google Scholar]
- Gu Z., Valianpour F., Chen S., Vaz F. M., Hakkaart G. A., et al. , 2004. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol. Microbiol. 51: 149–158 [DOI] [PubMed] [Google Scholar]
- Guan X. L., Wenk M. R., 2006. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast 23: 465–477 [DOI] [PubMed] [Google Scholar]
- Guerinot M. L., Eide D., 1999. Zeroing in on zinc uptake in yeast and plants. Curr. Opin. Plant Biol. 2: 244–249 [DOI] [PubMed] [Google Scholar]
- Guo S., Stolz L. E., Lemrow S. M., York J. D., 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274: 12990–12995 [DOI] [PubMed] [Google Scholar]
- Guo Y., Cordes K. R., Farese R. V., Jr, Walther T. C., 2009. Lipid droplets at a glance. J. Cell Sci. 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeler G., Natter K., Thallinger G. G., Crawford M. E., Kohlwein S. D., et al. , 2002. YPL.db: the Yeast Protein Localization database. Nucleic Acids Res. 30: 80–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H. J., Rho H. J., Shin S. K., Yoon H. J., 2010. The TGL2 gene of Saccharomyces cerevisiae encodes an active acylglycerol lipase located in the mitochondria. J. Biol. Chem. 285: 3005–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Gable K., Kohlwein S. D., Beaudoin F., Napier J. A., et al. , 2002. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J. Biol. Chem. 277: 35440–35449 [DOI] [PubMed] [Google Scholar]
- Han G.-S., Johnston C. N., Chen X., Athenstaedt K., Daum G., et al. , 2001. Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J. Biol. Chem. 276: 10126–10133 [DOI] [PubMed] [Google Scholar]
- Han G.-S., Audhya A., Markley D. J., Emr S. D., Carman G. M., 2002. The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J. Biol. Chem. 277: 47709–47718 [DOI] [PubMed] [Google Scholar]
- Han G.-S., Johnston C. N., Carman G. M., 2004. Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J. Biol. Chem. 279: 5338–5345 [DOI] [PubMed] [Google Scholar]
- Han G.-S., Wu W.-I., Carman G. M., 2006. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281: 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-S., Siniossoglou S., Carman G. M., 2007. The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282: 37026–37035 [DOI] [PubMed] [Google Scholar]
- Han G.-S., O'Hara L., Carman G. M., Siniossoglou S., 2008a An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283: 20433–20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-S., O'Hara L., Siniossoglou S., Carman G. M., 2008b Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J. Biol. Chem. 283: 20443–20453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.-H., Han G.-S., Iwanyshyn W. M., Carman G. M., 2005. Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 280: 29017–29024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock L. C., Behta R. P., Lopes J. M., 2006. Genomic analysis of the Opi- phenotype. Genetics 173: 621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. A., Lester R. L., 1980. Effects of inositol starvation on phospholipid and glycan syntheses in Saccharomyces cerevisiae. J. Bacteriol. 142: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., et al. , 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harington A., Schwarz E., Slonimski P. P., Herbert C. J., 1994. Subcellular relocalization of a long-chain fatty acid CoA ligase by a suppressor mutation alleviates a respiration deficiency in Saccharomyces cerevisiae. EMBO J. 13: 5531–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasslacher M., Ivessa A. S., Paltauf F., Kohlwein S. D., 1993. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 268: 10946–10952 [PubMed] [Google Scholar]
- Heier C., Taschler U., Rengachari S., Oberer M., Wolinski H., et al. , 2010. Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801: 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., 1982. Membrane lipids of yeast: biochemical and genetic studies, pp. 101–158 The Molecular Biology of the Yeast Saccharomyces. Metabolism and Gene Expression, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Henry S. A., Patton-Vogt J. L., 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog.Nucleic Acid Res. 61: 133–179 [DOI] [PubMed] [Google Scholar]
- Herman P. K., Emr S. D., 1990. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A., 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen J. K., Autio K. J., Schonauer M. S., Kursu V. A., Dieckmann C. L., et al. , 2010. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 1797: 1195–1202 [DOI] [PubMed] [Google Scholar]
- Hirsch J. P., Henry S. A., 1986. Expression of the Saccharomyces cerevisiae inositol-1-P synthetase gene (INO1) gene is regulated bt factors that affect phospholipid synthesis. Mol. Cell. Biol. 6: 3320–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M., 1987. Mutants of Saccharomyces cerevisiae defective in sn-1,2- diacylglycerol cholinephosphotransferase: Isolation, characterization, and cloning of the CPT1 gene. J. Biol. Chem. 262: 3909–3917 [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M., 1988. The sn-1,2-diacylglycerol ethanolaminephosphotransferase of Saccharomyces cerevisiae. Isolation of mutants and cloning of the EPT1 gene. J. Biol. Chem. 263: 19748–19757 [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M., 1990. The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J. Biol. Chem. 265: 1755–1764 [PubMed] [Google Scholar]
- Hjelmstad R. H., Bell R. M., 1991. sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae. Nucleotide sequence of the EPT1 gene and comparison of the CPT1 and EPT1 gene products. J. Biol. Chem. 266: 5094–5103 [PubMed] [Google Scholar]
- Holthuis J. C., Levine T. P., 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6: 209–220 [DOI] [PubMed] [Google Scholar]
- Homann M. J., Bailis A. M., Henry S. A., Carman G. M., 1987. Coordinate regulation of phospholipid biosynthesis by serine in Saccharomyces cerevisiae. J. Bacteriol. 169: 3276–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., et al. , 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586 [DOI] [PubMed] [Google Scholar]
- Hosaka K., Kodaki T., Yamashita S., 1989. Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli. J. Biol. Chem. 264: 2053–2059 [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., et al. , 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585 [DOI] [PubMed] [Google Scholar]
- Hung W., Olson K. A., Breitkreutz A., Sadowski I., 1997. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur. J. Biochem. 245: 241–251 [DOI] [PubMed] [Google Scholar]
- Hyde M., Block-Alper L., Felix J., Webster P., Meyer D. J., 2002. Induction of secretory pathway components in yeast is associated with increased stability of their mRNA. J. Cell Biol. 156: 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanyshyn W. M., Han G. S., Carman G. M., 2004. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 279: 21976–21983 [DOI] [PubMed] [Google Scholar]
- Jacquier N., Schneiter R., 2010. Ypk1, the yeast orthologue of the human serum- and glucocorticoid-induced kinase, is required for efficient uptake of fatty acids. J. Cell Sci. 123: 2218–2227 [DOI] [PubMed] [Google Scholar]
- Jain S., Stanford N., Bhagwat N., Seiler B., Costanzo M., et al. , 2007. Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282: 30562–30569 [DOI] [PubMed] [Google Scholar]
- Jandrositz A., Petschnigg J., Zimmermann R., Natter K., Scholze H., et al. , 2005. The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1735: 50–58 [DOI] [PubMed] [Google Scholar]
- Jani N. M., Lopes J. M., 2009. Regulated transcription of the Saccharomyces cerevisiae phosphatidylinositol biosynthetic gene, PIS1, yields pleiotropic effects on phospholipid synthesis. FEM. Yeast Res. 9: 552–564 [DOI] [PubMed] [Google Scholar]
- Janitor M., Subik J., 1993. Molecular cloning of the PEL1 gene of Saccharomyces cerevisiae that is essential for the viability of petite mutants. Curr. Genet. 24: 307–312 [DOI] [PubMed] [Google Scholar]
- Janitor M., Obernauerova M., Kohlwein S. D., Subik J., 1996. The pel1 mutant of Saccharomyces cerevisiae is deficient in cardiolipin and does not survive the disruption of the CHO1 gene encoding phosphatidylserine synthase. FEMS Microbiol. Lett. 140: 43–47 [DOI] [PubMed] [Google Scholar]
- Jaschke Y., Schwarz J., Clausnitzer D., Muller C., Schuller H. J., 2011. Pleiotropic corepressors Sin3 and Ssn6 interact with repressor Opi1 and negatively regulate transcription of genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 285: 91–100 [DOI] [PubMed] [Google Scholar]
- Jesch, S. A., and S. A. Henry, 2005 Yeast inositol lipids: synthesis, regulation, and involvement in membrane trafficking and lipid signaling, pp. 105–242 in Cell Biology and Dynamics of Yeast Lipids, Vol. 36/661, edited by G. Daum. Research Signpost, Kerala, India.
- Jesch S. A., Zhao X., Wells M. T., Henry S. A., 2005. Genome wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280: 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch S. A., Liu P., Zhao X., Wells M. T., Henry S. A., 2006. Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J. Biol. Chem. 281: 24070–24083 [DOI] [PubMed] [Google Scholar]
- Jesch S. A., Gaspar M. L., Stefan C. J., Aregullin M. A., Henry S. A., 2010. Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J. Biol. Chem. 285: 41947–41960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Rizavi H. S., Greenberg M. L., 1997. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol. Microbiol. 26: 481–491 [DOI] [PubMed] [Google Scholar]
- Johnson D. R., Knoll L. J., Levin D. E., Gordon J. I., 1994a Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J. Cell Biol. 127: 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. R., Knoll L. J., Rowley N., Gordon J. I., 1994b Genetic analysis of the role of Saccharomyces cerevisiae acyl-CoA synthetase genes in regulating protein N-myristoylation. J. Biol. Chem. 269: 18037–18046 [PubMed] [Google Scholar]
- Joshi A. S., Zhou J., Gohil V. M., Chen S., Greenberg M. L., 2009. Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank N. M., O'Neill E. M., Huang L. S., O'Shea E. K., 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486 [DOI] [PubMed] [Google Scholar]
- Kagiwada S., Zen R., 2003. Role of the yeast VAP homolog, Scs2p, in INO1 expression and phospholipid metabolism. J. Biochem. 133: 515–522 [DOI] [PubMed] [Google Scholar]
- Karanasios E., Han G.-S., Xu Z., Carman G. M., Siniossoglou S., 2010. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA 107: 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Hunter T., 1995. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5: 747–757 [DOI] [PubMed] [Google Scholar]
- Kelley M. J., Carman G. M., 1987. Purification and characterization of CDP-diacylglycerol synthase from Saccharomyces cerevisiae. J. Biol. Chem. 262: 14563–14570 [PubMed] [Google Scholar]
- Kelley M. J., Bailis A. M., Henry S. A., Carman G. M., 1988. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J. Biol. Chem. 263: 18078–18085 [PubMed] [Google Scholar]
- Kent C., Carman G. M., 1999. Interactions of pathways for phosphatidylcholine metabolism, CTP synthesis, and secretion through the Golgi apparatus. Trends Biochem. Sci. 24: 146–150 [DOI] [PubMed] [Google Scholar]
- Kersting M. C., Carman G. M., 2006. Regulation of the Saccharomyces cerevisiae EKI1-encoded ethanolamine kinase by zinc depletion. J. Biol. Chem. 281: 13110–13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Sakuraba H., Ikeda M., Denpoh A., Igarashi Y., 2008. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J. Biol. Chem. 283: 11199–11209 [DOI] [PubMed] [Google Scholar]
- Kim K., Kim K.-H., Storey M. K., Voelker D. R., Carman G. M., 1999. Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J. Biol. Chem. 274: 14857–14866 [DOI] [PubMed] [Google Scholar]
- Kiyono K., Miura K., Kushima Y., Hikiji T., Fukushima M., et al. , 1987. Primary structure and product characterization of the Saccharomyces cerevisiae CHO1 gene that encodes phosphatidylserine synthase. J. Biochem. 102: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Klig L. S., Henry S. A., 1984. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc. Natl. Acad. Sci. USA 81: 3816–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig L. S., Homann M. J., Carman G. M., Henry S. A., 1985. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: Pleiotropically constitutive opi1 mutant. J. Bacteriol. 162: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T., Yamashita S., 1987. Yeast phosphatidylethanolamine methylation pathway: Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262: 15428–15435 [PubMed] [Google Scholar]
- Kodaki T., Yamashita S., 1989. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur. J. Biochem. 185: 243–251 [DOI] [PubMed] [Google Scholar]
- Koffel R., Tiwari R., Falquet L., Schneiter R., 2005. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell. Biol. 25: 1655–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S. D., 2010a Obese and anorexic yeasts: experimental models to understand the metabolic syndrome and lipotoxicity. Biochim. Biophys. Acta 1801: 222–229 [DOI] [PubMed] [Google Scholar]
- Kohlwein S. D., 2010b Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285: 15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S. D., Eder S., Oh C. S., Martin C. E., Gable K., et al. , 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., O'Shea E. K., 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284: 977–980 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., et al. , 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman E.E., Tieleman D.P., Testerink C., Munnik T., Rijkers D. T., et al. , 2007. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282: 11356–11364 [DOI] [PubMed] [Google Scholar]
- Koshkin V., Greenberg M. L., 2002. Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem. J. 364: 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchler K., Daum G., Paltauf F., 1986. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., et al. , 2002. Subcellular localization of the yeast proteome. Genes Dev. 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., et al. , 2009. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33: 53–63 [DOI] [PubMed] [Google Scholar]
- Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., et al. , 2006. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281: 491–500 [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., Chalmers J. A. J., Douglas M. G., Hitzeman R. A., Mattick J. S., et al. , 1983. Molecular cloning of fatty acid synthase genes from Saccharomyces cerevisiae. J. Biol. Chem. 258: 11648–11653 [PubMed] [Google Scholar]
- Kvam E., Gable K., Dunn T. M., Goldfarb D. S., 2005. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol. Biol. Cell 16: 3987–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guedard M., Bessoule J. J., Boyer V., Ayciriex S., Velours G., et al. , 2009. PSI1 is responsible for the stearic acid enrichment that is characteristic of phosphatidylinositol in yeast. FEBS J. 276: 6412–6424 [DOI] [PubMed] [Google Scholar]
- Lee K. S., Patton J. L., Fido M., Hines L. K., Kohlwein S. D., et al. , 1994. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269: 19725–19730 [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., et al. , 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Letts V. A., Klig L. S., Bae-Lee M., Carman G. M., Henry S. A., 1983. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc. Natl. Acad. Sci. USA 80: 7279–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T., Loewen C., 2006. Inter-organelle membrane contact sites: through a glass, darkly. Curr. Opin. Cell Biol. 18: 371–378 [DOI] [PubMed] [Google Scholar]
- Li G., Chen S., Thompson M. N., Greenberg M. L., 2007. New insights into the regulation of cardiolipin biosynthesis in yeast: implications for Barth syndrome. Biochim. Biophys. Acta 1771: 432–441 [DOI] [PubMed] [Google Scholar]
- Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., et al. , 2007. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 6: 1190–1197 [DOI] [PubMed] [Google Scholar]
- Lin Y.-P., Carman G. M., 1989. Purification and characterization of phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 264: 8641–8645 [PubMed] [Google Scholar]
- Liu C., Yang Z., Yang J., Xia Z., Ao S., 2000. Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J. Biol. Chem. 275: 31972–31978 [DOI] [PubMed] [Google Scholar]
- Lockshon D., Surface L. E., Kerr E. O., Kaeberlein M., Kennedy B. K., 2007. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 175: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J. R., Roy A., Levine T. P., 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22: 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C. J. R., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., et al. , 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Loewy B. S., Henry S. A., 1984. The INO2 and INO4 loci of yeast are pleiotropic regulatory genes. Mol. Cell. Biol. 4: 2479–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., et al. , 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97: 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanovic N., Streith I., Wolinski H., Rechberger G., Kohlwein S. D., et al. , 2008. S-adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J. Biol. Chem. 283: 23989–23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. E., Oh C. S., Jiang Y., 2007. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771: 271–285 [DOI] [PubMed] [Google Scholar]
- Mattiazzi M., Jambhekar A., Kaferle P., Derisi J. L., Krizaj I., et al. , 2010. Genetic interactions between a phospholipase A2 and the Rim101 pathway components in S. cerevisiae reveal a role for this pathway in response to changes in membrane composition and shape. Mol. Genet. Genomics 283: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr J. A., Kohlwein S. D., Paltauf F., 1996. Identification of a novel, Ca(2+)-dependent phospholipase D with preference for phosphatidylserine and phosphatidylethanolamine in Saccharomyces cerevisiae. FEBS Lett. 393: 236–240 [DOI] [PubMed] [Google Scholar]
- McDonough V. M., Stukey J. E., Martin C. E., 1992. Specificity of unsaturated fatty acid-regulated expression of the Saccharomyces cerevisiae OLE1 gene. J. Biol. Chem. 267: 5931–5936 [PubMed] [Google Scholar]
- McDonough V. M., Buxeda R. J., Bruno M. E. C., Ozier-Kalogeropoulos O., Adeline M.-T., et al. , 1995. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by CTP. J. Biol. Chem. 270: 18774–18780 [DOI] [PubMed] [Google Scholar]
- McGraw P., Henry S. A., 1989. Mutations in the Saccharomyces cerevisiae OPI3 gene: Effects on phospholipid methylation, growth, and cross pathway regulation of phospholipid synthesis. Genetics 122: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster C. R., Bell R. M., 1994. Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J. Biol. Chem. 269: 28010–28016 [PubMed] [Google Scholar]
- Merkel O., Fido M., Mayr J. A., Pruger H., Raab F., et al. , 1999. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 274: 28121–28127 [DOI] [PubMed] [Google Scholar]
- Merkel O., Oskolkova O. V., Raab F., El-Toukhy R., Paltauf F., 2005. Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae. Biochem. J. 387: 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E., Zhang M., Dowhan W., 2005. Cardiolipin in energy transducing membranes. Biochemistry (Mosc.) 70: 154–158 [DOI] [PubMed] [Google Scholar]
- Min-Seok R., Kawamata Y., Nakamura H., Ohta A., Takagi M., 1996. Isolation and characterization of ECT1 gene encoding CTP:phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae. J. Biochem. 120: 1040–1047 [DOI] [PubMed] [Google Scholar]
- Morash S. C., McMaster C. R., Hjelmstad R. H., Bell R. M., 1994. Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J. Biol. Chem. 269: 28769–28776 [PubMed] [Google Scholar]
- Muccioli G. G., Sia A., Muchowski P. J., Stella N., 2009. Genetic manipulation of palmitoylethanolamide production and inactivation in Saccharomyces cerevisiae. PLoS ONE 4: e5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. J., Vance J., 1999. Mechanisms for lipid-body formation. Trends Biochem. Sci. 24: 109–115 [DOI] [PubMed] [Google Scholar]
- Murray M., Greenberg M. L., 2000. Expression of yeast INM1 encoding inositol monophosphatase is regulated by inositol, carbon source and growth stage and is decreased by lithium and valproate. Mol. Microbiol. 36: 651–661 [DOI] [PubMed] [Google Scholar]
- Nadkarni A. K., McDonough V. M., Yang W.-L., Stukey J. E., Ozier-Kalogeropoulos O., et al. , 1995. Differential Biochemical Regulation of the URA7- and URA8-encoded CTP synthetases from Saccharomyces cerevisiae. J. Biol. Chem. 270: 24982–24988 [DOI] [PubMed] [Google Scholar]
- Nakanishi H., de los Santos P., Neiman A. M., 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15: 1802–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natter K., Leitner P., Faschinger A., Wolinski H., McCraith S., et al. , 2005. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol. Cell. Proteomics 4: 662–672 [DOI] [PubMed] [Google Scholar]
- Nikawa J., Yamashita S., 1984. Molecular cloning of the gene encoding CDP-diacylglycerol- inositol 3-phosphatidyl transferase in Saccharomyces cerevisiae. Eur. J. Biochem. 143: 251–256 [DOI] [PubMed] [Google Scholar]
- Nikawa J., Tsukagoshi Y., Yamashita S., 1986. Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J. Bacteriol. 166: 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Kodaki T., Yamashita S., 1987a Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J. Biochem. 262: 4876–4881 [PubMed] [Google Scholar]
- Nikawa J., Tsukagoshi Y., Kodaki T., Yamashita S., 1987b Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase. Eur. J. Biochem. 167: 7–12 [DOI] [PubMed] [Google Scholar]
- Nikawa J., Tsukagoshi Y., Yamashita S., 1991. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266: 11184–11191 [PubMed] [Google Scholar]
- Nikawa J., Murakami A., Esumi E., Hosaka K., 1995. Cloning and sequence of the SCS2 gene, which can suppress the defect of INO1 expression in an inositol auxotrophic mutant of Saccharomyces cerevisiae. J. Biochem. (Tokyo) 118: 39–45 [DOI] [PubMed] [Google Scholar]
- Nunez L. R., Jesch S. A., Gaspar M. L., Almaguer C., Villa-Garcia M., et al. , 2008. Cell wall integrity MAPK pathway is essential for lipid homeostasis. J. Biol. Chem. 283: 34204–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeyer T., Fraisl P., DiRusso C. C., Black P. N., 2007. Topology of the yeast fatty acid transport protein Fat1p: mechanistic implications for functional domains on the cytosolic surface of the plasma membrane. J. Lipid Res. 48: 2354–2364 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., et al. , 2000. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275: 15609–15612 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J. T., Sturley S. L., 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277: 8877–8881 [DOI] [PubMed] [Google Scholar]
- Oh C. S., Toke D. A., Mandala S., Martin C. E., 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272: 17376–17384 [DOI] [PubMed] [Google Scholar]
- Oshiro J., Han G.-S., Carman G. M., 2003. Diacylglycerol pyrophosphate phosphatase in Saccarhomyces cerevisiae. Biochim. Biophys. Acta 1635: 1–9 [DOI] [PubMed] [Google Scholar]
- Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., et al. , 2009a The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Merkwirth C., Langer T., 2009b Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122: 3823–3830 [DOI] [PubMed] [Google Scholar]
- Osman C., Voelker D. R., Langer T., 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Haag M., Wieland F. T., Brugger B., Langer T., 2010. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander D. B., O'Brien D. J., Gorman J. A., Carman G. M., 1998. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 273: 18992–19001 [DOI] [PubMed] [Google Scholar]
- Ouyang Q., Ruiz-Noriega M., Henry S. A., 1999. The REG1 gene product is required for repression of INO1 and other inositol-sensitive upstream activating sequence-containing genes of yeast. Genetics 152: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozier-Kalogeropoulos O., Fasiolo F., Adeline M.-T., Collin J., Lacroute F., 1991. Cloning, sequencing and characterization of the Saccharomyces cerevisiae URA7 gene encoding CTP synthetase. Mol. Gen. Genet. 231: 7–16 [DOI] [PubMed] [Google Scholar]
- Ozier-Kalogeropoulos O., Adeline M.-T., Yang W.-L., Carman G. M., Lacroute F., 1994. Use of synthetic lethal mutants to clone and characterize a novel CTP synthetase gene in Saccharomyces cerevisiae. Mol. Gen. Genet. 242: 431–439 [DOI] [PubMed] [Google Scholar]
- Paltauf F., Kohlwein S. D., Henry S. A., 1992. Regulation and compartmentalization of lipid synthesis in yeast, pp. 415–500 The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, edited by Jones E. W., Pringle J. R., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., et al. , 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell 11: 2445–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Song H., 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Patton-Vogt J., 2007. Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim. Biophys. Acta 1771: 337–342 [DOI] [PubMed] [Google Scholar]
- Patton-Vogt J. L., Henry S. A., 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149: 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton-Vogt J. L., Griac P., Sreenivas A., Bruno V., Dowd S., et al. , 1997. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J. Biol. Chem. 272: 20873–20883 [DOI] [PubMed] [Google Scholar]
- Paulus H., Kennedy E. P., 1960. The enzymatic synthesis of inositol monophosphatide. J. Biol. Chem. 235: 1303–1311 [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., et al. , 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., et al. , 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284: 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A., 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771: 405–420 [DOI] [PubMed] [Google Scholar]
- Pomorski T., Holthuis J. C., Herrmann A., van Meer G., 2004. Tracking down lipid flippases and their biological functions. J. Cell Sci. 117: 805–813 [DOI] [PubMed] [Google Scholar]
- Poole M. A., Homann M. J., Bae-Lee M., Carman G. M., 1986. Regulation of phosphatidylserine synthase from Saccharomyces cerevisiae by phospholipid precursors. J. Bacteriol. 168: 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potting C., Wilmes C., Engmann T., Osman C., Langer T., 2010. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 29: 2888–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G., 2010a Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Daum G., 2010b Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285: 15769–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Grillitsch K., Daum G., 2008. Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47: 157–171 [DOI] [PubMed] [Google Scholar]
- Rajakumari S., Rajasekharan R., Daum G., 2010. Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801: 1314–1322 [DOI] [PubMed] [Google Scholar]
- Rattray J. B., Schibeci A., Kidby D. K., 1975. Lipids of yeast. Bacteriol. Rev. 39: 197–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Prinz W. A., 2010. The diverse functions of oxysterol-binding proteins. Annu. Rev. Cell Dev. Biol. 26: 157–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. S., Singh A. K., Rajasekharan R., 2008. The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J. Biol. Chem. 283: 8846–8854 [DOI] [PubMed] [Google Scholar]
- Ren G., Vajjhala P., Lee J. S., Winsor B., Munn A. L., 2006. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol. Mol. Biol. Rev. 70: 37–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof W. R., Wu J., Gijon M. A., Zarini S., Murphy R. C., et al. , 2007a Lysophosphatidylcholine Metabolism in Saccharomyces cerevisiae: The role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 282: 36853–36861 [DOI] [PubMed] [Google Scholar]
- Riekhof W. R., Wu J., Jones J. L., Voelker D. R., 2007b Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282: 28344–28352 [DOI] [PubMed] [Google Scholar]
- Rijken P. J., Houtkooper R. H., Akbari H., Brouwers J. F., Koorengevel M. C., et al. , 2009. Cardiolipin molecular species with shorter acyl chains accumulate in Saccharomyces cerevisiae mutants lacking the acyl coenzyme A-binding protein Acb1p: new insights into acyl chain remodeling of cardiolipin. J. Biol. Chem. 284: 27609–27619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenfeller P., Ring J., Muschett V., Beranek A., Buettner S., et al. , 2010. Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle 9: 2836–2842 [DOI] [PubMed] [Google Scholar]
- Rose K., Rudge S. A., Frohman M. A., Morris A. J., Engebrecht J., 1995. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 92: 12151–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., et al. , 2006. Global analysis of protein palmitoylation in yeast. Cell 125: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba J. D., Nara F., Bielawska A., Garrett S., Hannun Y. A., 1997. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1- phosphate lyase. J. Biol. Chem. 272: 26087–26090 [DOI] [PubMed] [Google Scholar]
- Santiago T. C., Mamoun C. B., 2003. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278: 38723–38730 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S., 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24: 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnewski M., Pongdontri P., Mora G., Hoppert M., Fulda M., 2008. Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 275: 2765–2778 [DOI] [PubMed] [Google Scholar]
- Schjerling C. K., Hummel R., Hansen J. K., Borsting C., Mikkelsen J. M., et al. , 1996. Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 271: 22514–22521 [DOI] [PubMed] [Google Scholar]
- Schnabl M., Oskolkova O. V., Holic R., Brezna B., Pichler H., et al. , 2003. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270: 3133–3145 [DOI] [PubMed] [Google Scholar]
- Schneiter R., Kohlwein S. D., 1997. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell 88: 431–434 [DOI] [PubMed] [Google Scholar]
- Schneiter R., Brugger B., Sandhoff R., Zellnig G., Leber A., et al. , 1999. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 146: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., Guerra C. E., Lampl M., Tatzer V., Zellnig G., et al. , 2000. A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol. Cell. Biol. 20: 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D., et al. , 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260: 88–91 [DOI] [PubMed] [Google Scholar]
- Schuck S., Prinz W. A., Thorn K. S., Voss C., Walter P., 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuiki I., Schnabl M., Czabany T., Hrastnik C., Daum G., 2010. Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801: 480–486 [DOI] [PubMed] [Google Scholar]
- Schuller H. J., Hahn A., Troster F., Schutz A., Schweizer E., 1992. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 11: 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller H. J., Schutz A., Knab S., Hoffmann B., Schweizer E., 1994. Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur. J. Biochem. 225: 213–222 [DOI] [PubMed] [Google Scholar]
- Schwabe J. W., Klug A., 1994. Zinc mining for protein domains. [news; comment] Nat. Struct. Biol. 1: 345–349 [DOI] [PubMed] [Google Scholar]
- Schweizer M., Roberts L. M., Holtke H. J., Takabayashi K., Hollerer E., et al. , 1986. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol. Gen. Genet. 203: 479–486 [DOI] [PubMed] [Google Scholar]
- Shelton S. N., Barylko B., Binns D. D., Horazdovsky B. F., Albanesi J. P., et al. , 2003. Saccharomyces cerevisiae contains a Type II phosphoinositide 4-kinase. Biochem. J. 371: 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Heacock P. N., Clancey C. J., Dowhan W., 1996. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271: 789–795 [DOI] [PubMed] [Google Scholar]
- Shirra M. K., Arndt K. M., 1999. 1999 Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics 152: 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra M. K., Patton-Vogt J., Ulrich A., Liuta-Tehlivets O., Kohlwein S. D., et al. , 2001. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 5710–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra M. K., Rogers S. E., Alexander D. E., Arndt K. M., 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169: 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simockova M., Holic R., Tahotna D., Patton-Vogt J., Griac P., 2008. Yeast Pgc1p (YPL206c) controls the amount of phosphatidylglycerol via a phospholipase C-type degradation mechanism. J. Biol. Chem. 283: 17107–17115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E., 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17: 6449–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D., Daum G., 2002. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D., Athenstaedt K., Hrastnik C., Daum G., 2004. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 279: 31190–31196 [DOI] [PubMed] [Google Scholar]
- Soto A., Carman G. M., 2008. Regulation of the Saccharomyces cerevisiae CKI1-encoded choline kinase by zinc depletion. J. Biol. Chem. 283: 10079–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivas A., J. L. Patton-Vogt, V. Bruno, P. Griac, and S. A. Henry, 1998 A role for phospholipase D (Pld1p) in growth, secretion and regulation of membrane lipid synthesis in yeast. J. Biol. Chem. 273: 16635–16638. [DOI] [PubMed]
- Stålberg K., Neal A. C., Ronne H., Stahl U., 2008. Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae. J. Lipid Res. 49: 1794–1806 [DOI] [PubMed] [Google Scholar]
- Stewart L. C., Yaffe M. P., 1991. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J. Cell Biol. 115: 1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz L. E., Huynh C. V., Thorner J., York J. D., 1998a Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148: 1715–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T., Thorner J., 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771: 353–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey J. E., McDonough V. M., Martin C. E., 1989. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264: 16537–16544 [PubMed] [Google Scholar]
- Summers E. F., Letts V. A., McGraw P., Henry S. A., 1988. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics 120: 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W. P., et al. , 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA 104: 20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata Y., Yamada T., Huang Y., Komoto J., Gomi T., et al. , 2002. Catalytic mechanism of S-adenosylhomocysteine hydrolase. Site-directed mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190. J. Biol. Chem. 277: 22670–22676 [DOI] [PubMed] [Google Scholar]
- Tamaki H., Shimada A., Ito Y., Ohya M., Takase J., et al. , 2007. LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282: 34288–34298 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Fukuda R., Ono Y., Eguchi H., Nagasawa S., et al. , 2008. Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1781: 391–399 [DOI] [PubMed] [Google Scholar]
- Tatzer V., Zellnig G., Kohlwein S. D., Schneiter R., 2002. Lipid-dependent subcellular relocalization of the acyl chain desaturase in yeast. Mol. Biol. Cell 13: 4429–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. S., Maehama T., Dixon J. E., 2000. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 97: 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O., Scheuringer K., Kohlwein S. D., 2007. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 1771: 255–270 [DOI] [PubMed] [Google Scholar]
- Testet E., Laroche-Traineau J., Noubhani A., Coulon D., Bunoust O., et al. , 2005. Ypr140wp, 'the yeast tafazzin', displays a mitochondrial lysophosphatidylcholine (lyso-PC) acyltransferase activity related to triacylglycerol and mitochondrial lipid synthesis. Biochem. J. 387: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele C., Spandl J., 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20: 378–385 [DOI] [PubMed] [Google Scholar]
- Toke D. A., Martin C. E., 1996. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271: 18413–18422 [DOI] [PubMed] [Google Scholar]
- Toke D. A., Bennett W. L., Dillon D. A., Chen X., Oshiro J., et al. , 1998. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273: 3278–3284 [DOI] [PubMed] [Google Scholar]
- Toke D. A., Bennett W. L., Oshiro J., Wu W. I., Voelker D. R., et al. , 1999. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273: 14331–14338 [DOI] [PubMed] [Google Scholar]
- Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., et al. , 1987. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. USA 84: 2708–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., et al. , 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Voelker D. R., 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270: 6062–6070 [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Pedretti J., Voelker D. R., 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268: 21416–21424 [PubMed] [Google Scholar]
- Trotter P. J., Pedretti J., Yates R., Voelker D. R., 1995. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J. Biol. Chem. 270: 6071–6080 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi Y., Nikawa J., Yamashita S., 1987. Molecular cloning and characterization of the gene encoding cholinephosphate cytidylyltransferase in Saccharomyces cerevisiae. Eur. J. Biochem. 169: 477–486 [DOI] [PubMed] [Google Scholar]
- Tuller G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., et al. , 1998. YDL142c encodes cardiolipin synthase (Clslp) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421: 15–18 [DOI] [PubMed] [Google Scholar]
- Zaccheo O., Dinsdale D., Meacock P. A., Glynn P., 2004. Neuropathy target esterase and its yeast homolog degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 279: 24024–24033 [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Falchuk K. H., 1993. The biochemical basis of zinc physiology. Physiol. Rev. 73: 79–118 [DOI] [PubMed] [Google Scholar]
- van Gestel R. A., Rijken P. J., Surinova S., O'Flaherty M., Heck A. J., et al. , 2010. The influence of the acyl chain composition of cardiolipin on the stability of mitochondrial complexes; an unexpected effect of cardiolipin in alpha-ketoglutarate dehydrogenase and prohibitin complexes. J. Proteomics 73: 806–814 [DOI] [PubMed] [Google Scholar]
- Vicinanza, M., G. D’Angelo, A. Di Campli, and M. A. De Matteis 2008 Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol. Life Sci. 65: 2833–2841. [DOI] [PMC free article] [PubMed]
- Villa-García M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., et al. , 2011. Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285: 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemula M., Kandasamy P., Oh C. S., Chellappa R., Gonzalez C. I., et al. , 2003. Maintenance and regulation of mRNA stability of the Saccharomyces cerevisiae OLE1 gene requires multiple elements within the transcript that act through translation-independent mechanisms. J. Biol. Chem. 278: 45269–45279 [DOI] [PubMed] [Google Scholar]
- Voelker D. R., 2003. New perspectives on the regulation of intermembrane glycerophospholipid traffic. J. Lipid Res. 44: 441–449 [DOI] [PubMed] [Google Scholar]
- Wagner C., Dietz M., Wittmann J., Albrecht A., Schuller H. J., 2001. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41: 155–166 [DOI] [PubMed] [Google Scholar]
- Waksman M., Eli Y., Liscovitch M., Gerst J. E., 1996. Identification and characterization of a gene encoding phospholipase D activity in yeast. J. Biol. Chem. 271: 2361–2364 [DOI] [PubMed] [Google Scholar]
- Watkins P. A., Lu J. F., Steinberg S. J., Gould S. J., Smith K. D., et al. , 1998. Disruption of the Saccharomyces cerevisiae FAT1 gene decreases very long-chain fatty acyl-CoA synthetase activity and elevates intracellular very long-chain fatty acid concentrations. J. Biol. Chem. 273: 18210–18219 [DOI] [PubMed] [Google Scholar]
- Wenz P., Schwank S., Hoja U., Schuller H. J., 2001. A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product. Nucleic Acids Res. 29: 4625–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. J., Hirsch J. P., Henry S. A., 1991. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem. 266: 863–872 [PubMed] [Google Scholar]
- Wiradjaja F., Ooms L. M., Whisstock J. C., McColl B., Helfenbaum L., et al. , 2001. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail: regulation of secretion from the endoplasmic reticulum. J. Biol. Chem. 276: 7643–7653 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Kohlwein S. D., 2008. Microscopic analysis of lipid droplet metabolism and dynamics in yeast. Methods Mol. Biol. 457: 151–163 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Natter K., Kohlwein S. D., 2009. The fidgety yeast: focus on high-resolution live yeast cell microscopy. Methods Mol. Biol. 548: 75–99 [DOI] [PubMed] [Google Scholar]
- Woods A., Munday M. R., Scott J., Yang X., Carlson M., et al. , 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269: 19509–19515 [PubMed] [Google Scholar]
- Wu W.-I., Carman G. M., 1994. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by nucleotides. J. Biol. Chem. 269: 29495–29501 [PubMed] [Google Scholar]
- Wu W.-I., Carman G. M., 1996. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by phospholipids. Biochemistry 35: 3790–3796 [DOI] [PubMed] [Google Scholar]
- Wu W.-I., Lin Y.-P., Wang E., Merrill A. H., Jr, Carman G. M., 1993. Regulation of phosphatidate phosphatase activity from the yeast Saccharomyces cerevisiae by sphingoid bases. J. Biol. Chem. 268: 13830–13837 [PubMed] [Google Scholar]
- Wu W.-I., Liu Y., Riedel B., Wissing J. B., Fischl A. S., et al. , 1996. Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 271: 1868–1876 [DOI] [PubMed] [Google Scholar]
- Xie Z., Fang M., Rivas M. P., Faulkner A. J., Sternweis P. C., et al. , 1998. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. USA 95: 12346–12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., DeWald D. B., Boronenkov I. V., Anderson R. A., Emr S. D., et al. , 1995. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell 6: 525–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bard M., Bruner D. A., Gleeson A., Deckelbaum R. J., et al. , 1996. Sterol esterification in yeast: a two-gene process. Science 272: 1353–1356 [DOI] [PubMed] [Google Scholar]
- Yang W.-L., McDonough V. M., Ozier-Kalogeropoulos O., Adeline M.-T., Flocco M. T., et al. , 1994. Purification and characterization of CTP synthetase, product of the URA7 gene in Saccharomyces cerevisiae. Biochemistry 33: 10785–10793 [DOI] [PubMed] [Google Scholar]
- Yoko-o T., Matsui Y., Yagisawa H., Nojima H., Uno I., et al. , 1993. The putative phosphoinositide-specific phospholipase C gene, PLC1, of the yeast Saccharomyces cerevisiae is important for cell growth. Proc. Natl. Acad. Sci. USA 90: 1804–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ohya Y., Goebl M., Nakano A., Anraku Y., 1994a A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem. 269: 1166–1172 [PubMed] [Google Scholar]
- Yoshida S., Ohya Y., Nakano A., Anraku Y., 1994b Genetic interactions among genes involved in the STT4–PKC1 pathway of Saccharomyces cerevisiae. Mol. Gen. Genet. 242: 631–640 [DOI] [PubMed] [Google Scholar]
- Youn J. Y., Friesen H., Kishimoto T., Henne W. M., Kurat C. F., et al. , 2010. Dissecting BAR Domain Function in the Yeast Amphiphysins Rvs161 and Rvs167 during Endocytosis. Mol. Biol. Cell 21: 3054–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B. P., Shin J. J., Orij R., Chao J. T., Li S. C., et al. , 2010. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 329: 1085–1088 [DOI] [PubMed] [Google Scholar]
- Zaccheo O., Dinsdale D., Meacock P. A., Glynn P., 2004. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J. Biol. Chem. 279: 24024–24033 [DOI] [PubMed] [Google Scholar]
- Zanghellini J., Natter K., Jungreuthmayer C., Thalhammer A., Kurat C. F., et al. , 2008. Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J. 275: 5552–5563 [DOI] [PubMed] [Google Scholar]
- Zhang M., Su X., Mileykovskaya E., Amoscato A. A., Dowhan W., 2003. Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278: 35204–35210 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Zou J., 2001. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J. Biol. Chem. 276: 41710–41716 [DOI] [PubMed] [Google Scholar]
- Zhong Q., Gohil V. M., Ma L., Greenberg M. L., 2004. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279: 32294–32300 [DOI] [PubMed] [Google Scholar]
- Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., et al. , 1991. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173: 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]