Abstract
The classic diallel takes a set of parents and produces offspring from all possible mating pairs. Phenotype values among the offspring can then be related back to their respective parentage. When the parents are diploid, sexed, and inbred, the diallel can characterize aggregate effects of genetic background on a phenotype, revealing effects of strain dosage, heterosis, parent of origin, epistasis, and sex-specific versions thereof. However, its analysis is traditionally intricate, unforgiving of unplanned missing information, and highly sensitive to imbalance, making the diallel unapproachable to many geneticists. Nonetheless, imbalanced and incomplete diallels arise frequently, albeit unintentionally, as by-products of larger-scale experiments that collect F1 data, for example, pilot studies or multiparent breeding efforts such as the Collaborative Cross or the Arabidopsis MAGIC lines. We present a general Bayesian model for analyzing diallel data on dioecious diploid inbred strains that cleanly decomposes the observed patterns of variation into biologically intuitive components, simultaneously models and accommodates outliers, and provides shrinkage estimates of effects that automatically incorporate uncertainty due to imbalance, missing data, and small sample size. We further present a model selection procedure for weighing evidence for or against the inclusion of those components in a predictive model. We evaluate our method through simulation and apply it to incomplete diallel data on the founders and F1's of the Collaborative Cross, robustly characterizing the genetic architecture of 48 phenotypes.
THE diallel is one of the oldest designs in genetics and one whose analysis is notoriously complex. The premise is simple: given a set of J parents, generate and phenotype offspring from all J × J reciprocal crosses and from these data estimate genetic parameters that characterize how the parental genomes and sex influence phenotypic variation. Using this design one can estimate the average parental contribution to the phenotype and the effect of specific combinations with other parents. When the parents are inbred strains, one can also estimate parent-of-origin effects. Despite the potential wealth of information contained in a diallel, there has been much to discourage its use in practice. Controversies about the interpretation of estimated parameters have been inextricably confounded with controversies about the analysis methods themselves, and much of the discussant literature is steeped in terminology unfamiliar to potential users. Indeed, to the outsider, the diallel emerges as an arcane puzzle that is perhaps best avoided in favor of simpler designs.
The diallel originated in animal and plant breeding as an extension of the idea that, from a breeding perspective, you should judge the value of an individual by the phenotypes of its offspring (Christie and Shattuck 1992 and references therein). It was originally defined by Schmidt (1919) as the set of all possible J2 pairwise crosses and was later introduced into the mainstream genetics literature by Jinks and Hayman (1953). In the decade that followed, the diallel, whose definition quickly broadened to encompass any set of F1 crosses between J > 2 parents, caught the attention of an active group of quantitative geneticists who went on to develop a series of elaborations of the design and its analysis. Among the simplest and most popular analytic decompositions is that of Griffing (1956). If ηjk is the mean phenotype or predicted value for the cross of parent j with parent k, then the parental effects can be modeled as
| (1) |
where μ is the intercept, gj is the main effect of parent j, and sjk is the statistical interaction of j and k, that is, the deviation from the combined main effects induced by the specific pairing of parents j and k. Following the terminology introduced by Sprague and Tatum (1942) and used throughout diallel literature, gj is the generalized combining ability (GCA) of parent j whereas sjk is the specific combining ability (SCA) of the parents j and k. GCA captures aggregate effects of additive genetics whereas SCA reflects aggregate genetic effects that lead to departures from additivity, such as dominance and epistasis.
Numerous extensions to Griffing's model have been proposed to extract more subtle effects from the diallel. These include decomposing SCA into dominance, heterosis, and epistasis components (Hayman 1957; Gardner and Eberhart 1966), into reciprocal effects (Griffing 1956), their further decomposition into maternal and paternal effects (Cockerham and Weir 1977; Zhu and Weir 1996), and sex-linked variants thereof (Carbonell et al. 1983). Conversely, interest in obtaining GCAs with fewer than J2 crosses has motivated variants of the design such as the half-diallel (Griffing 1956) and the partial diallel (Kempthorne and Curnow 1961), among others (see Christie and Shattuck 1992; Lynch and Walsh 1998), which have themselves led to technical innovation (e.g., Greenberg et al. 2010).
Disagreement about the precise meaning of parameters estimated from a diallel cross has presented a theoretical stumbling block to their interpretation. The parents could be inbred lines, independent outbreeding populations (such as open-pollinating varieties of corn), or outbred individuals (Eberhart and Gardner 1966). They could have been chosen deliberately, sampled randomly, or some compromise of these. The intention of the experiment could be to draw inferences about the parents themselves, the populations each parent represents, or the broader population from which all parents were drawn. Joint consideration of such factors weaves through much of the foundational diallel literature from 1950 to 1970 and has continued to represent a source of controversy (Baker 1978; Wright 1985).
A more practical stumbling block arises from the difficulty of estimating parameters from diallel data that are incomplete, imbalanced, or contaminated with outliers. Although some diallel crosses are created deliberately, a significant number arise as a by-product of intermediate stages in a multiparent breeding program. Such accidental diallels can contain valuable information, but their often haphazard patterns of missingness make them an imperfect match for well-studied designs. For many incomplete diallels it has been unclear how to analyze the data without discarding observations, drastically reducing the scope of inference, or making other significant compromises.
Even when traditional analysis methods accommodate the design, choice about which parameters (e.g., explicit models of dominance, SCA, etc.) should be included in the model can alter the estimates and interpretation of the other parameters. The option of model selection by significance testing of individual terms, frequently proposed in older literature, provides some guidance but is unsatisfactory in that included parameters are estimated in a way that disregards uncertainty in model choice. The notion that any a priori plausible effect should be excluded from modeling seems to us artificial and out of step with modern approaches to applied statistical inference (e.g., Gelman and Hill 2007).
We propose a general and efficient method for the analysis of diallel crosses and applying it to a data set of 48 phenotypes collected from an incomplete eight-strain diallel that arose serendipitously from the establishment of the Collaborative Cross (Churchill et al. 2004; Chesler et al. 2008; Collaborative Cross Consortium 2012). Our methods of analysis provide an inferential framework that is robust to imbalance in the design, missing data, and outliers. We model a wide range of effects including additive, heterosis, epistatic, parent-of-origin, and sex-specific variants thereof. This structure accomplishes two important goals. The first is familiar and constant interpretation of parameters across models. The second is stable and coherent estimation and prediction that is achieved through hierarchical Bayesian shrinkage and model selection.
Statistical Models and Methods
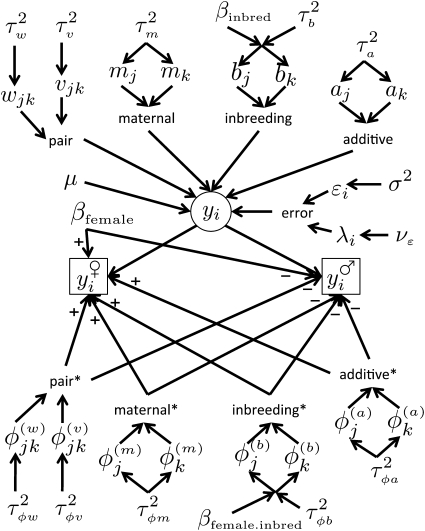
We describe our hierarchical decomposition of the diallel in stages. Starting with the simplest submodel, which describes only additive strain dosage effects, we present successive elaborations, building up to the model depicted in Figure 1 with parameters listed in Table 1. We then state the full model in compact form, detailing its efficient estimation and methods for choosing informative submodels. Models are described by a quoted string of characters in alphabetical order in the first column of Table 1 where, following marginality restrictions (Venables and Ripley 2002), some components imply the presence of others unless otherwise stated.
Figure 1 .
A directed acyclic graph depicting the hierarchy of the full model with outliers modeled at νε t-d.f., i.e., model “BsSasdsmsvswsOν”. Asterisks indicate components that represent sex-specific deviations from their nonasterisked counterparts, the latter representing an unsexed model.
Table 1 .
Summary of nomenclature
| Model component | Description | Model variables | Impliesa |
| B | Inbred penalty | βinbred | |
| a | Strain-specific additive | ||
| b | Strain-specific inbred | B | |
| m | Strain-specific maternal | ||
| v | Strain-specific symmetric | ||
| w | Strain-specific asymmetric | v | |
| S | Sex effect (female advantage) | βfemale | |
| Bs | Female inbred penalty | βfemale.inbred | B, S |
| as | Strain-specific female additive | a, S | |
| bs | Strain-specific female dominance | b, Bs | |
| ms | Strain-specific female maternal | M, S | |
| vs | Strain-specific female symmetric | v, S | |
| ws | Strain-specific female asymmetric | w, S | |
| Oν | Outlier model | ||
| (residual error) | Individual variation |
Components that would normally be included with this component (defined recursively).
The “a” model
Consider an incomplete diallel arising from crosses among J inbred strains. Index the mothers with j and the fathers with k, such that any mating pair is (j, k) ∈ {1, … , J}2. Let yi be the phenotype value of individual i and denote the mother, the father, and the mother–father pair relevant to individual i as j[i], k[i], and (j, k)[i]. For a continuous phenotype with normally distributed errors, we model
| (2) |
where μ is the intercept modeled as a fixed “overall” effect, motherj and fatherk are the contributions from mother and father, and εi ∼ N(0, σ2) is a deviation due to the specific individual (i.e., the residual). In this simplest model, each parental contribution amounts to a “dose” of the underlying strain; i.e.,
| (3) |
| (4) |
where aj is the dose of the parental strain j (equivalently for ak) and the effects are additive in the sense that two doses of strain j increase the expected phenotype value twice as much as one dose. The estimated effects are used for predicting phenotypes of unobserved crosses in the incomplete design. We consider an appropriate measure of discrepancy between estimated and true values of a to be the sum of squared errors (i.e., quadratic loss), . In decision problems involving the estimation of three or more effects under quadratic loss, shrinkage estimators typically dominate simple means (Parmigiani and Inoue 2009). Therefore, as in Zhu and Weir (1996), we model the aj's hierarchically, as if drawn from a common normal distribution,
| (5) |
where the variance parameter is given a weakly informative prior distribution. This model leads to an estimate of each aj that is dynamically shrunk toward zero to the extent that its supporting data are few and their dispersion is estimated to be small (Gelman and Hill 2007).
In the “a” model, aj coincides with the GCA for strain j described by Sprague and Tatum (1942). Nonetheless, we make further comparisons with GCA and SCA only parenthetically, as these constructs hinder interpretation of later models. When the “a” model is seen as an estimation problem with primary interest on a, the variance of additive effects does not require a biological interpretation; from a Bayesian perspective, it simply helps model how we would expect the data to appear. Nonetheless, anticipating the use of its estimate as a summary statistic, we explore its relation to the concept of heritability (Lynch and Walsh 1998) in Appendix A.
Accommodating outliers
When outliers are suspected, maybe as a result of erratic measurement error, it can be desirable to model the phenotype as being sampled from a distribution with heavier tails than the normal distribution. To simultaneously accommodate and detect outliers we model the individual deviations εi as if drawn from a scale mixture of normal densities,
| (6) |
| (7) |
where the scale factor λi is modeled as 1/νε times a draw from a chi-square distribution with νε d.f. As a result, εi is now t-distributed as εi | σ2, νε ∼ t(νε, 0, σ2). The scale λi for each data point has a prior mean E(λi | νε) = 1. A posteriori, λi acts as an indicator of the ith data point's outlier status, with E(λi | νε, Data) ≪ 1, suggesting a highly deviant observation. Setting νε → ∞ implies that residual errors closely reflect a normal distribution, while lower values of νε imply increasing probability of large outliers (West 1984; Carlin and Louis 2008). For νε we consider values of 15 (only slightly heavier tailed than the normal), 3 (large outliers likely; the lowest integer νε for which the t has finite variance), and 6 (an intermediate value, advocated in, e.g., Greenberg et al. 2010).
Inbreeding and dominance effects: the “Bab” model
Hybrid vigor, or heterosis, describes the change in phenotype value due to heterozygosity when crossing two inbred lines (Lynch and Walsh 1998). It is conventional to model this effect as a dominance term that describes the deviation of hybrid individuals from the expected average of homozygous phenotypes. However, we believe the diallel is more naturally modeled in a converse manner, with inbreds, not hybrids, as the deviant type. In a full diallel, inbreds are in the minority, corresponding to 1/Jth of the crosses. Even when the diallel is sparse, inbred crosses would seldom outnumber hybrids. We therefore define our predictive baseline as primarily modeling effects in hybrids, but accommodating inbred-specific effects through a deviation. First, Equation 2 is elaborated as
| (8) |
where pair(jk)[i] is a deviation specific to mother–father pair (jk). In the “Bab” model, this pair effect is then modeled as a strain-specific inbred penalty bj drawn from a common distribution centered at fixed effect βinbred; i.e.,
| (9) |
| (10) |
where implies that the inbred penalty is constant across strains. Modeling heterosis, either as an inbred or a dominance effect, inevitably changes the interpretation of aj. In our case, aj now estimates the dosage effect of strain j when combined with another sampled strain. Defining the additive effects this way results in more stable and precise estimates for all effects in the diallel.
Parent-of-origin effects: the “Babm” model
The full diallel includes reciprocal crosses of both motherj × fatherk and fatherj × motherk. This allows us to estimate strain-specific effects of maternity (which could include the uterine environment for mammals, mitochondrial effects, etc.) vs. paternity. We model these parent-of-origin effects as a symmetric deviation about the “Bab” model. If mj is the “maternal” contribution from mothers of strain j, then we revise Equations 3 and 4 to
with dispersion of maternal effects .
Symmetric and asymmetric effects: “Babmvw”
Departures from the “Babm” model corresponding to statistical interactions between pairs of strains can be represented through two additional layers of effects, revising Equation 9 to
| (11) |
| (12) |
| (13) |
Symmetric “v” effects model deviations that depend only on the strain labels and not how they are allocated between mother and father, whereas asymmetric “w” effects model deviations from the symmetric effect induced by differences between reciprocal crosses of the same strain pair.
Sex-specific effects
Sex-specific effects can add considerable complication to a statistical model because it can not only double the number of parameters but also change the meaning of the parameters if, for example, the effect is expressed as an offset for one sex. Here we expand our model while preserving the meaning of the existing parameters, modeling the effect of an individual being female vs. being male through a symmetric deviation about an unsexed mean. Let ψ encode femaleness,
such that for a parameter q, adding of the term ψ(sex) · q to the model pushes the expected phenotype up q/2 for females and down q/2 for males. We can view females and males as modeled by Equations 14 and 15, respectively,
| (14) |
| (15) |
with
specified for each sex. We define for each strain-specific effect variable qj ∈ {aj, bj, mj},
and for each strain pair-specific effect qjk ∈ {vjk, wjk},
Sexed effects are thus the regular unsexed effects plus a symmetric sex-specific deviation. For example, . We set for q ∈ {a, m} and , where for q ∈ {a, m, b}; and for q ∈ {v, w}, with constraints imposed as in Equations 12 and 13, where overall effects βfemale and βfemale.inbred are modeled as fixed effects, and each strain-specific sex-deviation component q is modeled as random with its own variance .
The full model
Including fixed covariates xi and R random-effect components , ∀r ∈ {1, … , R}, the “full model” (BsSasbsmsvsws) is
| (16) |
with νε ≠ ∞ described as the “full model with outliers” or “ ”.
Prior elicitation
We model prior belief about the fixed effects μ, βinbred, βfemale, and βfemale.inbred, using diffuse normal priors . For the variance parameters, which include σ2 and parameters controlling the dispersion of strain-specific effects in the components a, b, m, w, v, as, bs, ms, ws, vs, we model prior belief using inverted chi-square distributions of the form τ2 ∼ μτ × Inv – χ2(ντ) [equivalently described by the inverted gamma IG(ντ/2, μτ/2)], which is conjugate with our normal-likelihood and fixed-effect priors. To model vague prior information on reasonably scaled data, we apply weak priors with for the fixed effects and ντ = 0.02 and μτ = 2 for the variance parameters. The information in the variance priors is equivalent to adding 0.02 datapoints from an additional strain.
Setting shrinkage priors to beat the maximum-likelihood estimate
Within the class of unbiased regression estimators, the standard maximum-likelihood estimate is unbeatable (i.e., admissible) when judged by mean squared error (MSE) (). However, as famously observed by Stein (1955), once the number of estimated effects exceeds 2, it is uniformly beaten (i.e., dominated) by biased estimators that employ shrinkage (e.g., Parmigiani and Inoue 2009). In particular, ridge regression and generalized ridge regression will dominate the maximum-likelihood estimate (MLE) under MSE loss if
| (17) |
where X is a design matrix and Q is any generalized ridge shrinkage matrix (using Theorem 3.19 in Gross 2003). In our case, X specifies individuals’ parentage (see Appendix B), and Q is a diagonal matrix with values for strain effects based upon their membership groups. A good prior for is therefore one that makes Q−1 large enough for the above condition to likely hold. Thus, for large values of βj, one would want large parameters. In this case, “flat” priors (i.e., ∝ 1, as given by ντ = 2, μτ = 0, after, e.g., Gelman 2006, or our choice of a nonzero μτ) will often be preferable to the traditional Jeffreys prior [∝ 1/τ2; μτ = 0, ντ = 0 (Jeffreys 1946)], which includes strong prior weight around .
Posterior estimation
We estimate posterior distributions for all parameters, using an efficient Gibbs sampling scheme. Details are provided in Appendices B and C.
Posterior inference
We obtain raw posteriors by collecting sampled values for each parameter at each MCMC iteration. However, the Gibbs sampling scheme above gives marginal posteriors that appear overly vague for parameters that are grouped by a common variance [e.g., a = (a1, … , aj)]. This is due to the inherent lack of identifiability in a highly parameterized mixed model. For example, a range of alternative values for b, v, and w will produce identical predictions for yi. Hierarchical modeling means these alternatives are distinguished by their different posterior probabilities, but less so when the posteriors of , , and are vague. We note, however, that the purpose of the diallel experiment is most often to understand differences between strains and strain combinations, rather than estimate pure population means. Therefore, focusing on contrasts, at each iteration t = 1, … , T we calculate recentered versions of the grouped parameters
from the raw Gibbs samples, where q denotes the group of effects assigned to the same dispersion parameter , and |q| is the number of such effects. The resulting posterior intervals for are smaller and cover their true values at the desired 95% rate. To demonstrate an alternative method for fitting mixed models that is not (explicitly) Bayesian, we also fit Equation 16 using the “hglm” procedure of Ronnegard et al. (2010), which gives a point estimate by finding the maximum of the hierarchical likelihood [h-likelihood (Lee and Nelder 1996)] suggested by our model.
Posterior prediction
Phenotype predictions for unobserved individuals that both anticipate sampling variation and incorporate all posterior uncertainty are easily obtained from MCMC output. We predict the phenotype Y(j, k, s), for a future individual with mother j, father k, and sex s, by constructing the appropriate design matrix xjks and using the Gibbs samples to estimate the posterior predictive mean of all draws t ∈ 1, … , T. Prediction intervals for Y(j, k, s) are achieved from quantiles of a set of new draws , where σ2(t) is the tth draw of σ2 in the Gibbs sampler, but with Zt being a new independent draw from N(0, 1) or t(νε) random noise.
Model selection by information criteria
In comparing the suitability of different submodels estimated by Gibbs sampling, we consider the popular deviance information criterion (DIC) (Spiegelhalter et al. 2002), defined as , where θ collects all parameters, ℓ(·| y) is the log-likelihood, and is the mean over all T iterations. Lower DIC suggests a more parsimonious model containing fewer parameters that are poorly estimated.
Bayesian model selection by exclusionary Gibbs group sampling
With potentially >400 models of interest under consideration (resulting from different combinations of effects), it is impractical to use information criteria for model selection because each model requires its own Markov chain. Furthermore, although the DIC minimizer is often a successful predictor of future Y(j, k, s), it is seldom as parsimonious as the true model in simulation. In particular, our Bayesian adaptive shrinkage means that even the full model BsSasbsmsvsws can perform well against a model that is better informed. To deal with selection of parameter subsets q ∈ 1, … , Q in a way that better identifies valuable components, we consider a zero-inflated mixture prior (George and Foster 2000; Ishwaran and Rao 2005) on , namely
| (18) |
This corresponds to the full model but with the elaboration that each now has a prior probability 1 – πq of being inactive, that is, equal to zero and having all corresponding effects q equal to zero. We develop an algorithm to draw from the conditional distribution
This approach selects subsets of relevant parameters on the basis of our hierarchical model. Similarly, we model the selection of fixed effects l ∈ L as
We calculate the posterior model inclusion probability (MIP) for each parameter subset, using the Rao-Blackwellized estimate from the Gibbs samples (e.g., Guan and Stephens 2011). In this study, we set , and πr = πq = 0.5 for all r and q. More details of the algorithm are presented in Appendix D.
Experimental Materials and Methods
Phenotype data from a diallel of the Collaborative Cross founders
We collected data on multiple phenotypes (Supporting Information, File S1) in a diallel of eight inbred mouse strains (abbreviated names in parentheses), A/J (AJ), C57BL/6J (B6), 129S1/SvImJ (129), NOD/LtJ (NOD), NZO/H1LtJ (NZO), CAST/EiJ (CAST), PWK/PhJ (PWK), and WSB/EiJ (WSB), which are the founder strains of the Collaborative Cross (CC) (Churchill et al. 2004; Chesler et al. 2008; Collaborative Cross Consortium 2012). It would be expected that genetic effects present in the diallel will replicate in the CC itself, which motivates our interest in this population. Table 2 lists the phenotypes collected, transformations used to normalize each phenotype before diallel analysis, and the completeness of the data for each phenotype. All CC F1 animals had free access to standard laboratory chow containing 6% fat by weight (LabDiet 5K52) and acidified drinking water throughout the phenotyping protocol. Mice were on a 12-hr:12-hr light:dark cycle beginning at 6:00 am and were housed two to five animals per pen in pressurized, individually ventilated cages.
Table 2 .
Phenotypes collected on a diallel of the Collaborative Cross founders
| Summary |
Sample size |
Filled diallel cells (of 64) |
|||||||||
| Protocol | Code | Phenotype | Units | Transform | Mean | SD | Females | Males | Female | Male | All |
| ADVIA | BASO | % basophil | % | 0.23 | 0.16 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | CHCM | Red cell Hgb concentration mean | g/dl | 32.35 | 1.62 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | cHGB | Calculated Hgb | g/dl | 15.01 | 0.72 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | EOS | % Eosinophil | % | 2.80 | 2.20 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | HCT | Hematocrit | % | 46.47 | 2.60 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | HDW | Hgb concentration distribution width | g/dl | 1.93 | 0.39 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | LUC | % Large unstained cell | % | 0.54 | 0.26 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | LYM | % lymphocyte | % | 82.97 | 6.06 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | MCH | Mean cell hemoglobin content | pg | 15.60 | 0.71 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | MCHC | Mean cell hemoglobin concentration | g/dl | 33.67 | 1.22 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | MCV | Mean cell volume | fl | 46.37 | 2.19 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | mHGB | Measured Hgb | g/dl | 15.63 | 0.68 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | MONO | % Monocyte | % | 1.46 | 3.80 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | MPV | Mean platelet volume | fl | 5.81 | 1.83 | x−2 | 311 | 315 | 61 | 61 | 62 |
| ADVIA | NEUT | % neutrophil | % | 12.00 | 3.82 | log(x) | 311 | 315 | 61 | 61 | 62 |
| ADVIA | PLT | Platelet count | ×10E03 cells/liter | 1132.25 | 196.07 | log(x) | 311 | 315 | 61 | 61 | 62 |
| ADVIA | RBC | Red blood cell count | ×10E06 cells/liter | 10.03 | 0.54 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | RDW | Red cell distribution width | % | 14.74 | 1.30 | x | 311 | 315 | 61 | 61 | 62 |
| ADVIA | Retic | % reticulocyte | % | 2.79 | 1.15 | log(x) | 311 | 315 | 61 | 61 | 62 |
| ADVIA | WBC | White blood cell count | ×10E03 cells/liter | 7.01 | 1.91 | x | 311 | 315 | 61 | 61 | 62 |
| BP | PulseMean | Pulse rate | Beats/min | 577.35 | 99.47 | x | 89 | 99 | 20 | 24 | 24 |
| BP | SystolicMean | Systolic blood pressure | mmHg | 122.96 | 26.07 | x | 89 | 99 | 20 | 24 | 24 |
| CHEM | CHOL | Total cholesterol | mg/dl | 105.47 | 30.59 | log(x) | 320 | 311 | 62 | 61 | 62 |
| CHEM | GLU | Glucose | mg/dl | 185.86 | 33.42 | x | 320 | 311 | 62 | 61 | 15 |
| CHEM | HDL | HDL Cholesterol (as HDLD) | mg/dl | 8.21 | 7.39 | x1/4 | 320 | 311 | 62 | 61 | 62 |
| CHEM | TG | Triglycerides | mg/dl | 137.24 | 59.38 | log(x) | 320 | 311 | 62 | 61 | 62 |
| EKG | CV | Coefficient of variance | % | 2.68 | 3.04 | x−1/2 | 199 | 195 | 42 | 39 | 42 |
| EKG | HR | Heart rate | Beats/min | 768.10 | 65.33 | x/100 | 199 | 195 | 42 | 39 | 42 |
| EKG | HRV | Heart rate variability | Beats/min | 19.51 | 20.51 | x−1/2 | 199 | 195 | 42 | 39 | 42 |
| EKG | Rampl | Mean R amplitude | msec | 368.33 | 104.68 | x/100 | 199 | 195 | 42 | 39 | 42 |
| EKG | SRAmpl | Mean SR amplitude | msec | 416.10 | 165.92 | x/100 | 199 | 195 | 42 | 39 | 42 |
| EKG | PQ | PQ interval | msec | 21.58 | 3.04 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | PR | Interval between peak of P-wave and R-wave | msec | 28.56 | 3.49 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | QRS | Interval between start and end of QRS complex | msec | 10.52 | 1.00 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | QT | QT interval | msec | 41.61 | 4.55 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | QTD | Difference between smallest and largest QT interval | msec | 20.16 | 8.44 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | QTC | Rate corrected QT interval | msec | 46.85 | 3.47 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | QTCD | Rate corrected QT dispersion | msec | 22.31 | 8.72 | x | 199 | 195 | 42 | 39 | 42 |
| EKG | RR | R to R interval | msec | 78.93 | 8.08 | x | 199 | 195 | 42 | 39 | 42 |
| DEXA | B.Area | Bone area S | cm2 | 5.20 | 1.00 | x | 292 | 302 | 61 | 61 | 62 |
| DEXA | BMC | Bone mineral content | g/cm | 0.27 | 0.06 | 1/x | 292 | 302 | 61 | 61 | 62 |
| DEXA | BMD | Bone mineral density | g/cm2 | 0.06 | 0.01 | x | 292 | 302 | 61 | 61 | 62 |
| DEXA | LTM | Lean tissue mass | g | 9.57 | 3.08 | x | 292 | 302 | 61 | 61 | 62 |
| DEXA | RST | R-value of soft tissue | Unitless | 1.29 | 0.01 | x | 292 | 302 | 61 | 61 | 62 |
| DEXA | TTM | Total tissue mass | g | 12.82 | 5.22 | x | 292 | 302 | 61 | 61 | 62 |
| COMP | MLNA | Mouse length,nose to anus | cm | 26.71 | 8.89 | log(x) | 292 | 302 | 61 | 61 | 62 |
| COMP | Weight | Mouseweight | g | 26.71 | 8.89 | log(x) | 292 | 302 | 61 | 61 | 62 |
| COMP | PctFat | PctFat | % | 19.47 | 7.26 | log(x) | 292 | 302 | 61 | 61 | 62 |
ADVIA, blood composition; BP, blood pressure; CHEM, plasma chemistries; EKG, electrocardiography; DEXA, dual-energy X-ray absorptiometry; COMP, body composition.
Blood composition (ADVIA)
Whole blood was obtained in the morning from the retro-orbital sinus of nonfasted animals 7 weeks of age. Collection of 200 μl from each animal was performed using an EDTA-coated microhematocrit tube directed into a 1.5-ml microcentrifuge tube containing 2 μl of 10% EDTA. Samples were analyzed on the Bayer Advia 120 autoanalyzer within the 4 hr following collection.
Blood pressure
Animals were acclimated to a dedicated room for blood pressure (BP) measurement on the Friday before a 5-day testing period beginning the following Monday. Systolic blood pressure and pulse were measured using the BP-2000 tail-cuff system (Visitech Systems, Apex, NC). Four unanesthetized mice were placed on a warmed platform (37°) and each was held in place using a magnetic restraining cover. The tail is placed through a cuff and held with a magnetic sensor unit that detects when blood flow stops and starts. Each day 30 measurements are obtained per mouse. Mice were trained to the apparatus for the first 3 days of the testing period and data were collected for analysis in the last 2 days, for average values based upon 60 total measurements. Animals were 10 weeks of age when blood pressure was measured.
Plasma chemistries
Whole blood was obtained from 8-week-old animals after a 4-hr period of food removal in the morning (7:00–11:00 am). Collection of 150 μl from each animal was performed using a heparin-coated microcapillary tube inserted into the retro-orbital sinus and directed into a 1.5-ml microcentrifuge tube containing 2 μl of 1000 units/ml heparin. Samples were placed on ice prior to centrifugation at 10,000 rpm in a refrigerated microcentrifuge for 10 min. Plasma was collected into a clean tube for analysis on the Beckman (Fullerton, CA) CX-Delta5 Chemistry autoanalyzer.
Densitometry and body composition
Body composition was assessed when mice were 16 weeks of age by dual-energy X-ray absorptiometry (DEXA), using a Lunar PIXImus densitometer (GE Medical Systems) after mice were anesthetized intraperitoneally with tribromoethanol (0.2 ml 2% solution per 10 g body weight). Because the skull is so bone dense, it is omitted from the DEXA analysis. Mice were weighed using an Ohaus Navigator scale with InCal calibration to accommodate animal movement.
Electrocardiography
Unanesthetized mice aged 12 weeks were placed on the ECGenie (Mouse Specifics, Quincy, MA) for analysis of electrocardiogram parameters. Recording is initiated when the paws of the animal contact a 3-lead electrode plate. Data are analyzed using manufacturer's software.
Simulations
We assess the performance of our methods by simulation, evaluating their ability to infer genetic parameters and to predict future phenotypes on an 8 × 8 diallel of these inbred strains. Two genetic architectures are considered, one simple, with additive effects only, and one more complex, with additive, inbreeding, maternal, and sex-specific maternal effects. We refer to our general Bayesian model as “BayesDiallel” and to its associated model selection procedure as “BayesSpike”. See File S1.
Estimation and prediction of additive genetic effects in a simulated diallel
Gibbs sampler approaches can be difficult to compare with non-Bayesian methods or even with each other, given their indefinite approach to a point estimate. For the outlier model with low degrees of freedom, the posterior may possibly be multimodal. Furthermore, given the high dimensionality and structure of our decomposition of the diallel, not all parameters receive the same information content per complete diallel replicate, and whereas some parameters are better informed by incomplete diallels than others.
To start, we compare BayesDiallel and BayesSpike to estimates obtained from ordinary linear regression, using Griffing's model in Equation 1. We simulate an 8 × 8 complete diallel with five replicate individuals in each cell and assume all individuals are of the same sex. The first two columns of Table 3 (top section) list the values of the simulated parameters: eight additive strain effects (a1, … , a8), an intercept (μ), and noise variance (σ2). Sampling from a normal distribution, these are used to generate 5 × 64 = 320 simulated phenotypes for the diallel. Columns 3 and 4 give MLEs and 95% confidence intervals from linear regression using Griffing's model for GCA (Equation 1 without the sjk, which is directly competitive with the a model in Table 1) and for GCA + SCA (Equation 1 including sjk, which is directly competitive with the “av” model, as defined in Table 1). In this setting the GCA model is at a distinct advantage: it knows a priori the true architecture and can thus save degrees of freedom from fitting specious parameters. The misinformed GCA + SCA model, however, risks overfitting unless the point estimates for the SCA effects happen to be small. We then consider several options for analyzing the diallel within the (unsexed) BayesDiallel framework: the full model, which we fit with and without consideration of outliers, despite the fact that outliers are not simulated; the model that minimizes the DIC among all 26 = 64 unsexed models; the true model, which is a Bayesian version of the GCA; the full model fit by hglm, which maximizes the h-likelihood (see Statistical Models and Methods); and the component-switching BayesSpike, which attempts explicitly to learn components of value. For the BayesSpike models we report the marginal posterior median (after Meng 2008); for the hglm fit, we report the point estimate and 95% confidence interval; for all other BayesDiallel methods we report the posterior intervals, calculated as the central 95% quantiles of the posterior distribution (Carlin and Louis 2008). For the Bayesian methods, we consider their ability to select out parameters either through severe shrinking or through formal selection. In the “top false” row in Table 3, we report for the BayesSpike the largest MIP of untrue fixed and random components, assuming a (generous) prior MIP of 0.5 per component; for the remaining Bayesian methods, we report the estimated effect for the largest untrue fixed component and the τ2q estimate for the largest untrue random component q.
Table 3 .
Performance of traditional and Bayesian models in inference and prediction for a simulated complete eight-way diallel with five replicates and additive effects
| BayesDiallel |
||||||||||||
| Simulated parameters |
Griffing linear model |
Standard normal model |
Outlier model |
|||||||||
| Full model |
BayesSpike: | |||||||||||
| GCA: | GCA + SCA: | Full model: | Minimum DIC modela: | True model: | Full hglm fit: | BayesSpike: | d.f. = 3: | d.f. = 6: | d.f. = 15: | d.f. = 6: | ||
| Name | True value | “a” | “av” | “Bambvw” | “avw” | “a” | — | “Bambvw” | “Bambvw” | “Bambvw” | “Bambvw” | — |
| μ | 7 | 7.52b | 7.52b | 7.23c | 7.35c | 7.28c | 7.44 | 7.32d | 7.41c | 7.4c | 7.25c | 7.49d |
| (−4.94, 19.97) | (−90.92, 105.95) | (−2.72, 18.11) | (−3.65, 17.5) | (−3.46, 17.95) | (−2.51, 17.39) | (−2.6, 17.53) | (−3.91, 17.53) | (−3.71, 18) | (−2.84, 17.39) | (−2.14, 16.91) | ||
| a1 | −10 | −9.4 | −13.31 | −9.12 | −9.07 | −9.15 | −9.11 | −9.05 | −9.37 | −9.3 | −9.2 | −9.69 |
| (−11.61, −7.2) | (−17.8, −8.82) | (−11.43, −6.86) | (−11.18, −6.77) | (−11.5, −7.12) | (−14.6, −3.63) | (−11.28, −6.82) | (−11.75, −7.13) | (−11.68, −7.15) | (−11.52, −6.81) | (−11.5, −7.01) | ||
| a2 | −8 | −7.49 | −7.36 | −7.12 | −6.97 | −7.24 | −6.9 | −7.09 | −6.64 | −6.88 | −6.96 | −6.73 |
| (−9.69, −5.29) | (−11.85, −2.86) | (−9.5, −4.77) | (−9.37, −4.7) | (−9.3, −5.14) | (−12.38, −1.41) | (−9.38, −4.81) | (−8.88, −4.34) | (−9.2, −4.62) | (−9.1, −4.44) | (−9.11, −4.48) | ||
| a3 | −4 | −5.26 | −7.86 | −5.05 | −5 | −5.11 | −5 | −5.04 | −5.48 | −5.3 | −5.15 | −5.28 |
| (−7.46, −3.06) | (−12.35, −3.37) | (−7.21, −2.76) | (−7.1, −2.72) | (−7.26, −2.98) | (−10.48, 0.49) | (−7.4, −2.99) | (−7.96, −3.22) | (−7.56, −3.06) | (−7.68, −2.9) | (−7.57, −2.94) | ||
| a4 | −1 | −1.27 | −5.9 | −1.25 | −1.33 | −1.23 | −1.33 | −1.26 | −1.25 | −1.24 | −1.25 | −1.26 |
| (−3.47, 0.93) | (−10.39, −1.41) | (−3.52, 1) | (−3.63, 0.86) | (−3.3, 0.98) | (−6.81, 4.16) | (−3.43, 0.97) | (−3.55, 1.11) | (−3.53, 0.99) | (−3.61, 1.03) | (−3.42, 0.96) | ||
| a5 | 1 | 2.05 | −1.7 | 2 | 1.99 | 1.98 | 1.96 | 1.97 | 1.8 | 1.98 | 1.99 | 1.95 |
| (−0.15, 4.26) | (−6.2, 2.79) | (−0.27, 4.28) | (−0.26, 4.28) | (−0.23, 4.14) | (−3.53, 7.44) | (−0.23, 4.21) | (−0.6, 4.14) | (−0.21, 4.31) | (−0.25, 4.41) | (−0.31, 4.25) | ||
| a6 | 3 | 2.24 | −4.62 | 2.07 | 1.94 | 2.17 | 1.85 | 2.03 | 3.21 | 2.72 | 2.27 | 2.6 |
| (0.03, 4.44) | (−9.11, −0.13) | (−0.28, 4.3) | (−0.37, 4.22) | (0.14, 4.4) | (−3.63, 7.34) | (−0.21, 4.33) | (0.58, 5.74) | (0.18, 5.18) | (−0.21, 4.68) | (0.01, 4.83) | ||
| a7 | 7 | 7.86 | 5.46 | 7.61 | 7.65 | 7.63 | 7.69 | 7.59 | 6.95 | 7.21 | 7.41 | 7.16 |
| (5.66, 10.06) | (0.97, 9.95) | (5.22, 9.79) | (5.41, 9.9) | (5.61, 9.76) | (2.2, 13.17) | (5.22, 9.73) | (4.56, 9.29) | (4.8, 9.5) | (4.9, 9.6) | (4.86, 9.47) | ||
| a8 | 12 | 11.27 | 7.27 | 10.86 | 10.79 | 10.96 | 10.83 | 10.85 | 10.79 | 10.82 | 10.89 | 10.79 |
| (9.07, 13.47) | (2.78, 11.76) | (8.57, 13.21) | (8.39, 12.97) | (8.73, 13) | (5.35, 16.32) | (8.52, 13.07) | (8.38, 12.92) | (8.48, 13.1 | (8.56, 13.21) | (8.39, 12.98) | ||
| σ2 | 120 | 107.67 | 105.09 | 102.51 | 102.28 | 108.4 | 99.9h | 103.38 | 63.56 | 78.89 | 91.71 | 79.89 |
| (89.16, 126.17) | (86.05, 124.13) | (86.38, 118.92) | (85.76, 119.98) | (90.96, 125.84) | — | (86.16, 121.14) | (50.44, 77.27) | (64.87, 95.92) | (75.84, 107.49) | (66.35, 95.48) | ||
| Top false | Fixede | — | — | B (0.381) | — | — | B (0.376) | B (0.247) | B (0.233) | B (0.296) | B (0.34) | B (0.268) |
| Randomf | — | — | m (0.939) | m (1.02) | — | v (0.125) | m (0.832) | m (1.122) | m (1.047) | m (0.976) | m (0.87) | |
| Predict MSE | 119.66g | 122.5 | 324.5 | 124.7 | 124.6 | 122.5 | 126.3 | 124.3 | 125.9 | 125.2 | 124.8 | 124.9 |
| (111.28, 131.25) | (84.14, 166.45) | (243.59, 412.82) | (85.13, 169.98) | (85.35, 170.9) | (84.45, 166.14) | (86.96, 172.43) | (84.62, 169.96) | (85.89, 172.72) | (85.77, 171.32) | (85.42, 170.53) | (85.39, 170.81) | |
“avw” is the model that minimized the DIC.
Least-squares mean with 95% confidence interval in parentheses.
Posterior mean with 95% posterior interval in parentheses.
Posterior median with 95% posterior interval in parentheses.
For BayesDiallel component q ∈ {βinbreed, …, τ2φw}, “q(x)” denotes p(q ≠ 0 | Data) for BayesSpike and min[p(β > 0 | Data), p(β < 0 | Data)] otherwise.
Magnitude of the falsely included random-effects vector q averaged over T MCMC samples, as .
Mean and 95% confidence interval of MSE from 1000 simulations (see main text).
Confidence intervals were not available for the residual variance.
Diallel analysis should (at minimum) help the researcher predict phenotypes of new individuals from sampled crosses. We generate 1000 new simulated diallels on the basis of the same genetic architecture (i.e., using the parameters in Table 3, column 2). Separately we use the point estimates from each method to predict the expected phenotype of the new individuals in each cell and compare these predictions with each of the 1000 subsequently observed data sets. The bottom row in Table 3 compares predicted with observed values, reporting the MSE as the average over simulations (upper row) and as its 95% central quantile (lower row).
Athough μ tends to have a wide confidence/prediction interval relative to the additive effects, Table 3 shows that the BayesDiallel models can meet or improve on the MLE for GCA in terms of prediction error and point estimates, despite the fact that the full model has 82 parameters. Against the GCA + SCA MLE, however, any method using hierarchical shrinkage is twice as successful in forecasting new phenotypes. This advantage to hierarchical models would reduce with smaller σ2. The symmetric and asymmetric effects (v and w) tend to be the false components most likely to enter the model when using BayesSpike, although they are typically estimated with a much smaller magnitude than the additive effect.
Table 4 shows results from 100 simulation experiments, with 1000 test data sets per experiment. Values in this table measure the mean discrepancy in estimated effects and prediction MSE. Noise level, σ2 = 120, represents a lower limit on the performance of any estimator. We see that when the model is overspecified, as in the GCA + SCA model, lack of shrinkage severely affects the consistency of the MLE.
Table 4 .
Performance of traditional and Bayesian models in inference and prediction for 100 simulated complete eight-way diallels with five replicates and additive effects
| BayesDiallel |
||||||||||||
| Simulated component |
Griffing linear model |
Standard normal model |
Outlier model |
|||||||||
| Full model |
BayesSpike: | |||||||||||
| GCA: | GCA + SCA: | Full model: | Minimum DIC model: | True model: | Full hglm fit: | BayesSpike: | d.f. = 3: | d.f. = 6: | d.f. = 15: | d.f. = 6: | ||
| Name | Discrepancy | “a” | “av” | “Babmvw” | — | “a” | “Babmvw” | — | “Babmvw” | “Babmvw” | “Babmvw” | — |
| Additive (a) | 10.85 | 144.56 | 10.69 | 10.69 | 10.67 | 10.74 | 10.73 | 12.02 | 11.15 | 10.77 | 11.20 | |
| (2.5, 23.6) | (57, 266.2) | (2.7, 22.9) | (2.7, 23) | (2.8, 22.7) | (2.7, 24.7) | (2.8, 23.2) | (3.4, 28.3) | (2.8, 24.4) | (2.6, 23.2) | (2.8, 24.7) | ||
| Inbreeding (b) | 0.00 | 0.00 | 4.60 | 2.88 | 0.00 | 18.47 | 0.04 | 8.19 | 5.96 | 4.87 | 0.19 | |
| — | — | (0.3, 29.1) | (0, 24.3) | — | (0, 147.5) | (0, 0.1) | (0.4, 55) | (0.3, 31.3) | (0.3, 27.9) | (0, 0.1) | ||
| Maternal (m) | 0.00 | 0.00 | 1.37 | 0.91 | 0.00 | 0.78 | 0.22 | 1.73 | 1.53 | 1.41 | 0.27 | |
| — | — | (0.1, 5) | (0, 5.9) | — | (0, 7.1) | (0, 3) | (0.2, 6.8) | (0.2, 6) | (0.1, 5.8) | (0, 3.7) | ||
| Symmetric (v) | 0.00 | 2126.47 | 5.50 | 3.69 | 0.00 | 13.13 | 0.26 | 12.09 | 7.76 | 5.98 | 0.91 | |
| — | (860.2, 3899.1) | (0.9, 23.1) | (0, 24.5) | — | (0.1, 64.7) | (0, 0.8) | (1.3, 70.1) | (0.9, 37.1) | (0.9, 25.3) | (0, 10.3) | ||
| Prediction MSEa | 123.09 | 311.82 | 124.32 | 123.79 | 123.09 | 124.78 | 123.43 | 125.24 | 124.65 | 124.39 | 123.69 | |
| (120.5, 125.8) | (306.7, 316.7) | (121.7, 127) | (121.2, 126.5) | (120.6, 125.8) | (122.2, 127.4) | (120.9, 126.2) | (122.6, 128) | (122, 127.4) | (121.8, 127.1) | (121.2, 126.5) | ||
Mean and 95% confidence interval of MSEs from 50 independent simulation experiments.
Inferring a complex genetic architecture: a “BSabms” model
To investigate the performance of the Bayesian model when many effects are present, we simulated a more complex genetic architecture that included sex (“S”), maternal (“m”), sex-specific maternal (“ms”), and inbreeding (“Bb”). In this case the nonzero effects were a = (−7.21, −5.77, −2.88, −0.72, 0.72, 2.16, 5.05, 8.65), b = (4.12, −3.88, −3.88, 2.12, 3.12, −2.88, 0.12, 1.12), m = (1, 1.63, −2.54, 7.24, 9.76, −14.18, 0.19, −3.08), φ(m) = (3.75, 3.25, 4.25, −11.75, 0.75, −0.25, −4.75, 4.75), μ = 10, βfemale = 4, βinbreed = –4, and σ2 = 120. These were chosen so that ∼20% of variation was explained by the “a” component, 20% from “m”, 12% from “ms”, and 0.5% from “b”. We generated 295 simulated data sets of 8 × 8 diallels with five replicates of each sex (640 individuals per data set). To each diallel we applied all of the methods tested above, except model selection using DIC (which was intractable for multiple replications when the number of models was 400+ in sex-effect models). Table 5 provides a summary of prediction and estimation error analogous to Table 4. In this case, we see that all of the BayesDiallel models are able to adapt to the active components, producing acceptable prediction error. Predictions are weaker for strain-specific “b” and “ms” effects, reflecting the smaller amount of data that inform them. Griffing's models are shown for comparison, but as expected, perform poorly in this realm.
Table 5 .
Performance of traditional and Bayesian models in inference and prediction for 295 simulated “BSabms” models
| BayesDiallel |
|||||||||||
| Simulated component |
Griffing linear model |
Standard normal model |
Outlier model |
||||||||
| Full model |
BayesSpike: | ||||||||||
| GCA: | GCA + SCA: | Full model: | True model: | Full hglm fit: | BayesSpike: | d.f. = 3: | d.f. = 6: | d.f. = 15: | d.f. = 6: | ||
| Name | Discrepancy | “a” | “av” | “BsSasbs msvsws” | “BsSasbs msvsws” | “BsSasbs msvsws” | — | “BsSasbs msvswsO3” | “BsSasbs msvswsO6” | “BsSasbsms vswsO15” | — |
| Additive (a) | 9.42 | 288.41 | 7.25 | 6.85 | 6.73 | 7.31 | 8.20 | 7.60 | 7.36 | 7.67 | |
| (3.2, 18.3) | (177.5, 406.3) | (2, 15.3) | (1.9, 14.4) | (2, 14.7) | (2, 15.3) | (2, 17.4) | (1.8, 16.1) | (1.8, 15.5) | (1.8, 16.3) | ||
| Inbreeding (b) | 255.86 | —b | 113.20 | 110.82 | 91.21 | 144.65 | 118.66 | 116.03 | 113.85 | 147.12 | |
| —b | —b | (28.7, 216.6) | (27.8, 216.2) | (23.9, 202.1) | (31.8, 255.9) | (30.5, 221.7) | (30.4, 218.5) | (27.6, 218.3) | (33.9, 255.9) | ||
| Maternal (m) | 92.10 | —b | 5.45 | 5.33 | 5.33 | 5.43 | 6.05 | 5.63 | 5.47 | 5.60 | |
| —b | —b | (1.2, 11.9) | (1.1, 12) | (1.1, 11.8) | (1.2, 12.1) | (1.5, 12.3) | (1.4, 12) | (1.4, 12.3) | (1.3, 11.8) | ||
| Sex-specific maternal (ms) | 326.16 | —b | 20.35 | 20.13 | 20.14 | 20.46 | 22.69 | 21.09 | 20.46 | 21.29 | |
| —b | —b | (5.2, 47.2) | (5, 47.5) | (5, 46.6) | (5.4, 48.8) | (5, 51.8) | (6.2, 49.5) | (6.1, 48.9) | (6.1, 49.8) | ||
| Prediction MSEa | 173.68 | 326.71 | 126.60 | 125.95 | 126.79 | 131.72 | 127.71 | 126.99 | 126.68 | 132.03 | |
| (171.4, 175.9) | (323.2, 330) | (124.8, 128.3) | (124.3, 127.7) | (125, 128.6) | (130, 133.5) | (126, 129.5) | (125.3, 128.7) | (124.9, 128.5) | (130.3, 133.8) | ||
Mean and 95% confidence interval of MSEs from 300 independent simulation experiments.
These parameters are not explicitly modeled by the tested procedure.
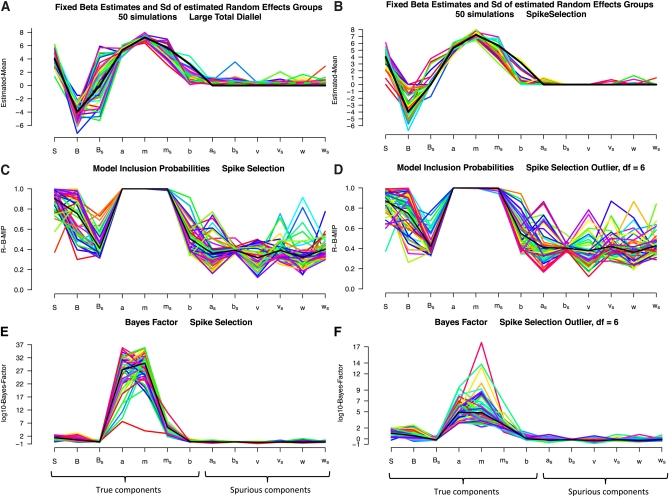
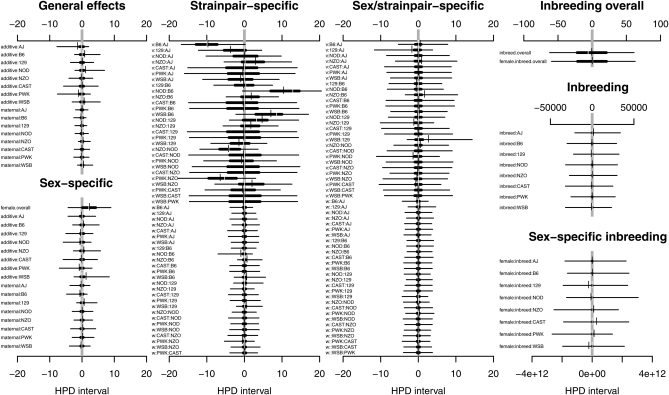
Figure 2 plots the results from fitting the Bayesian model to 50 of the simulated diallels described in Table 5. The black line in Figure 2, A and B, describes the set of parameters used for the simulation, plotting for each component q ∈ {Q, L} (i.e., among the Q random components or R fixed-effect components) the parameter value βq if it is a fixed effect or the SD of its vector q if it is a random effect. In Figure 2A, each colored line summarizes estimates from the full model applied to one simulated data set, plotting E(βq | Data) and SD(E(q|Data)), as appropriate. Figure 2B does the same for the BayesSpike, using median(βq | Data) and SD(median(q|Data)). Figure 2, A and B, shows that whereas the full model shrinks spurious components, BayesSpike forces them to zero. Figure 2C plots the posterior MIP, , estimated by BayesSpike starting with a componentwise prior of πq0 = 0.5, with the black line now indicating the median. Figure 2D shows MIPs for BayesSpike allowing for outliers at 6 d.f. and appears similar to Figure 2C. However, Figure 2 E and F, which plot the same data in a different way, reveal an important difference. They show the log10 of the Bayes factor, calculated as , which tracks the displacement of the posterior from the prior and thereby weighs the evidence provided by the data for or against a component's inclusion (Kass and Raftery 1995; Bernardo and Smith 2000). Accommodating outliers (Figure 2F) weakens the Bayes factors for inclusion of true components, illustrating the trade-off between increased robustness and reduced power.
Figure 2 .
(A–F) Summary statistics, posterior model inclusion probabilities, and Bayes factors for Bayesian models applied to 50 simulated diallels that share a complex genetic and parent-of-origin architecture. “True components” are group effects that were simulated; “spurious components” are group effects that were absent. Each colored line depicts results from a single simulation, with A and B showing the posterior mean for fixed-effect components (S, B, Bs) and the SD of sampled random intercepts for strain- and strain pair-specific components.
Application to Data
We apply our Bayesian models and BayesSpike procedure to data on 48 phenotypes, collected on mice from a diallel of founders of the Collaborative Cross. We start with an analysis of mouse weight, for which we were able to collect almost a full diallel. We then describe our semiautomated analysis of 48 phenotypes, commenting on select examples that demonstrate robustness and stability in the presence of sparsely sampled data, and richly characterize genetic and parent-of-origin architecture when data are abundant.
Analysis of weight data in a diallel of the Collaborative Cross founders
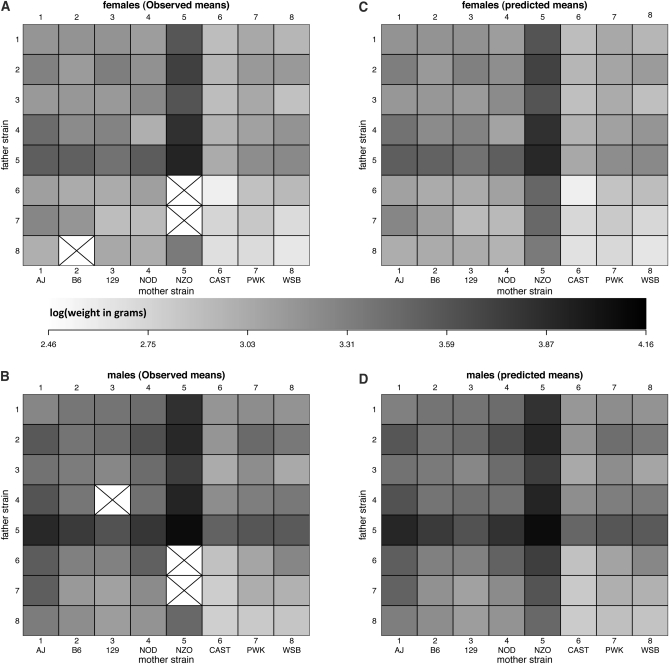
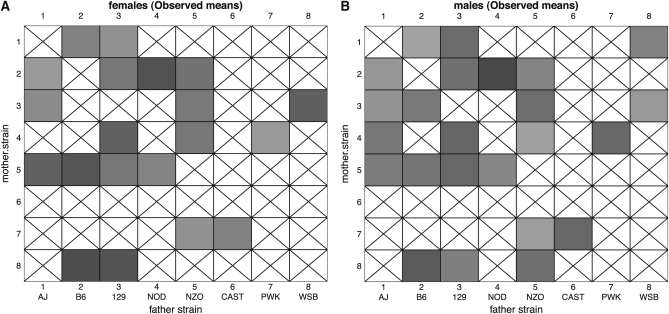
Body weight data were obtained for 292 female and 302 male mice (Figure 3). Shaded cells in Figure 3, A and B, represent observed phenotypes with the degree of shading indicating the average weight of mice in each group (log10 scale). Crossed boxes indicate the absence of phenotyped animals. Although mostly complete, this diallel is imbalanced: cells contained between 2 and 20 mice, with 1?13 male and 1?7 female. Visual inspection of this diallel reveals some striking trends. The dark banding in column and row 5 shows that the genome of the “New Zealand Obese” (NZO) mouse exerts a strong weight-inducing effect on its progeny (Taylor et al. 2001). The asymmetry of the banding, however, suggests this effect is transmitted more strongly from the mother than from the father. A similar asymmetry is apparent for strain AJ (row and column 1). Moreover, Figure 3, A and B, shows means only, unmoderated by shrinkage effects that would factor in the different numbers of mice that contribute to those estimates.
Figure 3 .
Weight data for 292 female and 302 male mice in an incomplete diallel of the Collaborative Cross founders. Shaded boxes indicate the average weight (on the log scale) of female (A) and male (B) mice, with crossed boxes showing missing data. C and D show the posterior predictive means after applying the full diallel model with outliers at 6 d.f.
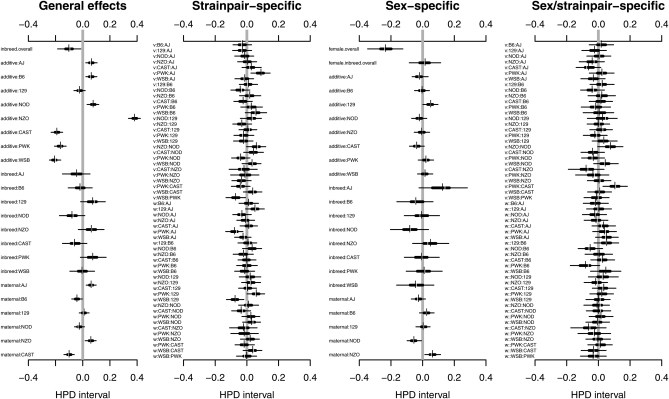
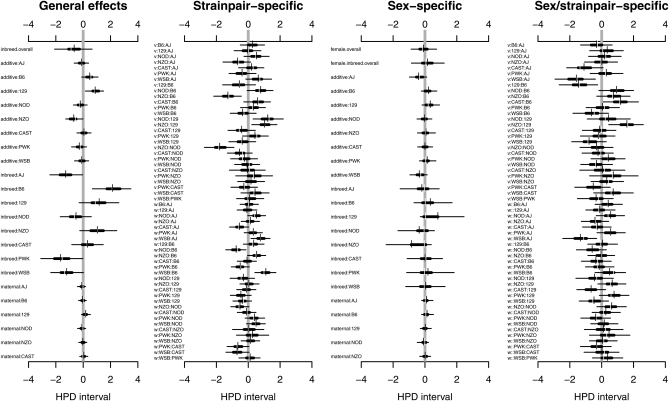
We applied the full model with outliers (BsSasbsmsvswsO6) to the body weight data. Highest posterior density (HPD) (Box and Tiao 1973) intervals of 163 effects parameters based on 8000 posterior samples from four independent Monte Carlo Markov chains are shown in Figure 4. Parameters are divided into four groups: general effects, which include the inbreeding penalty (“inbreed.overall”; B) and strain-specific effects of additive genetics (“additive”; a), inbreeding (“inbreed”; b), and parent-of-origin effects (“maternal”; m); strain pair-specific effects, which encompass effects peculiar to specific strain pairs (v and w); sex-specific effects, which include sex-specific deviations of the general effects (S, Bs, as, bs, ms); and sex/strain pair-specific effects, encompassing sex-specific deviations from the strain pair-specific effects (vs, ws). The general effects clearly show the high additive dosage effect of NZO (“additive:NZO”), plus some evidence for mothers transmitting this effect more strongly (“maternal:NZO”). A more striking parent-of-origin effect is evident in CAST (“maternal:CAST”), which has its 95% HPD most displaced from zero and indicates CAST mothers transmit low body weight more strongly than CAST fathers. The sex-specific effects include an expected drop in weight for females (“female.overall”) but few other strong deviations from zero. The strain pair-specific and sex/strain pair-specific effects are typically more vague. They represent fewer observations and so are more strongly subject to Bayesian adaptive shrinkage, which pulls extreme but sparsely supported means toward the middle. Figure 3, C and D, shows means of the (posterior) predictive distribution: that is, the average value of mice in a new diallel of the same strains that would be expected on the basis of the diallel model and the observed data. These Bayesian predictions incorporate all uncertainty due to finite sampling of the data and prior uncertainty about the parameters.
Figure 4 .
Highest posterior density (HPD) intervals for effects parameters fitted to weight data collected on 594 mice from a diallel of Collaborative Cross founders. Horizontal bars show for each parameter the region of highest posterior density that covers 50% (thick line) and 95% (thin line) of the posterior probability, with breaks indicating the posterior median and short vertical bars the posterior mean. The labels “additive”, “inbreed”, and “maternal” in the first graph refer to the a, b, and m effects in Table 1. In the third graph they refer to sex-specific effects as, bs, and ms. The “v” and “w” labels in the second and fourth graphs refer to symmetric and asymmetric effects (as in Table 1).
Semiautomated analysis of 48 phenotypes in a diallel of the CC founders
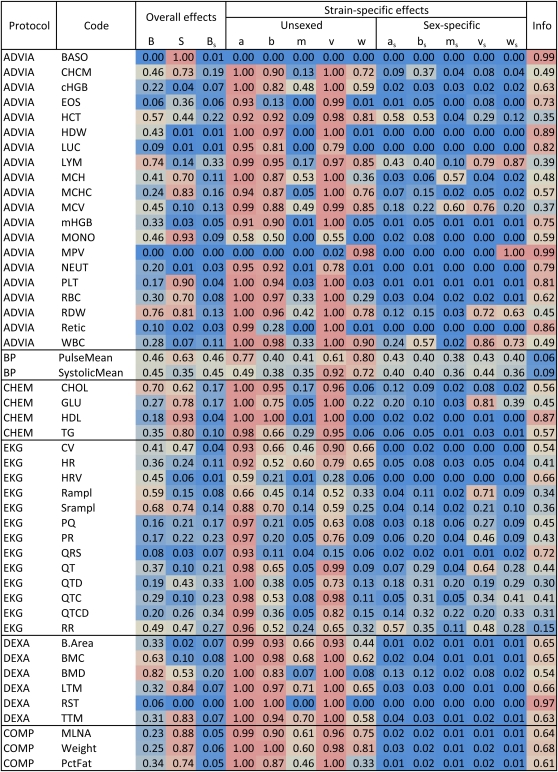
We applied the BayesSpike procedure for automated selection of diallel components to all 48 of the phenotypes in Table 2. Figure 5 lists the posterior MIPs for each diallel component, assuming a prior MIP in each case of . The “Info” column provides a measure between 0 and 1 that describes how much information the data have provided about model choice, defined by a scaled Kullback–Leibler divergence
where πq0 and πq are the prior and posterior MIPs for component q ∈ {Q, L} (where Q is the set of all effects grouped by a common variance, and L is the set of fixed effects), cQ = (2|{Q, L}|0)−1, cq = (–log[min(πq0, 1 – πq0)])−1, and ‖{Q, R}‖0 = 13 is the number of components considered in the selection. For each phenotype, the posterior MIPs (rounded to 2 decimal places) are based on 10,000 samples from five independent Markov chains.
Figure 5 .
Posterior model inclusion probabilities (MIPs) for genetic, sex, and parent-of-origin effects in 48 phenotypes measured in a diallel of the Collaborative Cross founders. All MIPs (colored columns except “Info”) are rounded to 2 d.p. and assume a prior of 0.5 for their components effect. Colors reflect the values and are scaled from blue (zero) to light red (one), with beige at 0.5 representing posterior belief about inclusion that is unmoved by the data. The “Info” column quantifies the gain in information about MIPs provided by the data, with values ranging from 1 (highly informative, red) to zero (uninformative, blue).
The numbers given for each effect in Figure 5 indicate how strongly, and in what direction, the data shift opinion about which components should be included. For example, the MIP values for systolic blood pressure (SystolicMean) are mostly near 0.5, indicating little posterior certainty and reflecting the fact that only 188 mice had measurements for this phenotype and these encompassed only 24 of 64 diallel combinations (Figure 6). Yet despite the sparsity of this data set, the BayesSpike returns stable, if vague, posterior opinion about inclusions, and the full model, fitted to the weight data above, provides stable, if vague, posterior distributions (Figure 7) for 175 effects and variance parameters, 188 outlier parameters, 2 × 64 predicted new crosses, and any further combination of parameters that is of interest. In particular, the data set contains no inbreds, which means that posterior information about inbreeding parameters (Figure 7, rightmost column) is almost as diffuse as the original priors, and BayesSpike probabilities indicate the absence of evidence for or against their inclusion. In contrast, HDL cholesterol (HDL), for which there are considerably more data (631 animals with 62/64 cells covered), provides strong evidence for an overall effect of sex (“S”), strain-specific effects of additive (“a”) and dominance (as inbreeding; “b”), and symmetric effects, i.e., effects that are specific to F1's between particular pairs of strains but for which the mother/father assignment does not matter. It also shows strong evidence against any sex-specific effects (e.g., sex-specific additive effects “as”) beyond that explained by an overall shift in mean.
Figure 6 .
(A and B) Systolic blood pressure collected on 188 mice in a sparsely sampled diallel of the Collaborative Cross founders. Shaded boxes indicate the average blood pressure in a group (darker equals higher), with crossed boxes showing missing data.
Figure 7 .
Highest posterior density (HPD) intervals for effects parameters fitted to systolic blood pressure data collected on 188 mice from an incomplete diallel of Collaborative Cross founders, with abbreviations and symbols as described in Figure 4. This diallel had no inbred animals. Posterior distributions for parameters relating to inbreeding (collected in the rightmost column) are therefore vague, reflecting mostly uninformative prior belief (note the x-axis scales). Nonetheless, they are stably estimated and do not disrupt estimation of the other effects.
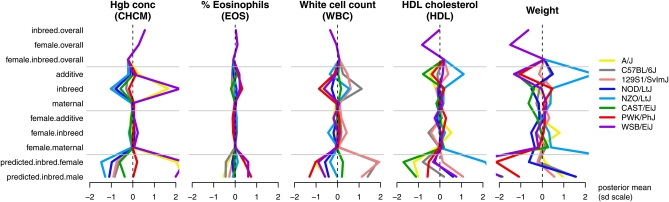
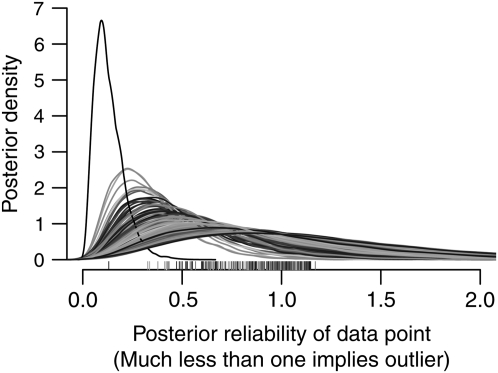
Strong evidence for a genetic effect need not imply that the effect exerts strong influence on the phenotype. Figure 8 plots strain-specific effects for some of the phenotypes listed in Figure 5. In this “straw plot”, each colored line tracks the posterior mean for the relevant CC founder strain, with predicted means of inbreds given in the bottom two rows (these contain a double dose of additive effects and so appear magnified). For ease of comparison, all values are shown on the scale of standard deviations of the transformed phenotype (transformations listed in Table 2). The percentage of eosinophils (EOS) (second straw plot) seems only moderately affected by additive genetics in this diallel. Nonetheless, as Figure 5 attests, the evidence for those effects is extremely strong, as is the evidence against substantial contributions from any other genetic, sex, or parent-of-origin effects, save a single symmetric increase attributed to the F1 combination of PWK and CAST (95% HPD is between 0.06 and 1.7, posterior mean = 0.88, posterior probability of being ≤0 = 0.99). The weight phenotype (rightmost straw plot) is the same as that described in Figures 3 and 4 and is marked by the effect of NZO escaping the ±2 SD axis boundaries. A similar, if more moderate effect of NZO is evident for HDL cholesterol (Figure 8, fourth plot). White blood cell count (WBC) is among the few phenotypes that show strong evidence for sex-specific effects, as well as unsexed effects (Figure 5, Figure 8). In particular, examination of its posterior HPD intervals (Figure 9) reveals that the sex specificity is confined to epistatic effects (vs, ws) involving strains AJ, WSB, and 129, with a wider range of epistatic effects existing irrespective of sex. These posteriors accommodate, through our outlier model the potential for erratic output from the measuring equipment. Figure 10 plots distributions of the posterior data weight (i.e., the datapoint reliability) attributed to white cell counts from each individual, distinguishing one observation so incongruous as to merit down-weighting to 1/10th of a data point on average. From this robust analysis, white blood cell count emerges as a phenotype that, although apparently free of parent-of-origin effects, has an otherwise complex genetic architecture.
Figure 8 .
A “straw plot” summarizing estimated effects and predicted values in five phenotypes collected on a diallel of the Collaborative Cross founders. Gray horizontal lines group posterior means for overall (B, S, Bs), strain-specific (a, b, m), and sex/strain-specific (as, bs, ms; as + dose for females, – dose for males) effects, as well as posterior predictive means for inbreds. For ease of comparison across phenotypes, x-axes are scaled to the SD of the transformed phenotype. Values for the NZO (light blue) strain in the HDL cholesterol and weight phenotypes are extreme enough to escape the 2 SD limits of the plot.
Figure 9 .
Highest posterior density (HPD) intervals for effects parameters fitted to white blood cell count (WBC) data collected on 626 mice from a diallel of Collaborative Cross founders, with abbreviations and symbols as described in Figure 4.
Figure 10 .
Simultaneous accommodation and detection of outliers in diallel data on white blood cell count (WBC), using the full model with outliers (at 6 d.f.). Each line represents the posterior distribution of the weight data (λi; see Models and Methods) for the phenotype measurement of individual i, with corresponding posterior means given as ticks on the x-axis and line shading for visualization only. On average, individuals will have weight at 1, but posterior distributions concentrated near 0, such as the high peak at 0.1, are outliers downweighted by the model.
Discussion
We describe an efficient and general framework for modeling effects of genetics, parent-of-origin, and sex on phenotypes collected for diallels of inbred strains. By deploying a fully Bayesian approach with conjugate priors, imbalance and missing data translate to vagueness in the posterior rather than instability of the estimates. By adopting an MCMC approach to estimation, we provide a flexible environment in which the posterior distribution of arbitrary combinations of parameters, including prediction of new data, can be easily obtained. Moreover, to satisfy all inferential tastes, we describe a rapid and powerful formulation of Bayesian model selection for weighing the evidence in support of each category of effect in the diallel.
Nonetheless, our approach is motivated by our bias: as geneticists focusing on model organisms, we are primarily concerned with characterizing the genetic architecture within the set of J2 genome combinations with a view to subsequent hypothesis-driven experiments. We less often seek to infer formally parameters relating to the superpopulation of individuals or species from which those inbred strains were drawn. Interestingly, traditional literature on diallel analysis has tended to oppose the use of random effects in our context. It espouses what we call the “random parents commonplace”: that modeling parental contributions as random effects is valid only if the parents have been drawn at random from a larger population and it is the variance parameters of this larger population that the investigator seeks to estimate. It further asserts that when the above conditions are not met, for example, if we are interested in parental effects present in the cross, or if the parents were chosen deliberately, then parental contributions should be modeled as fixed effects (e.g., Griffing 1956; Eberhart and Gardner 1966; Baker 1978). This commonplace persists in current literature, reiterated in, for example, Greenberg et al. (2010), who nonetheless develop an elegant Bayesian hierarchical model tailored to analyze a specific type of outbred diallel.
We consider this view in need of updating. Stein (1955) shocked the statistical world by showing that when simultaneously estimating three or more means as part of the same decision problem, fixed-effects modeling is dominated by biased methods that use shrinkage. Trading off errors in one dimension with those in another, the resulting estimates are drawn closer together when dimensions look similar, with useful shrinkage even when those dimensions are unrelated (Parmigiani and Inoue 2009). In the diallel, dimensions are related, making the argument for hierarchical modeling yet stronger. Although it is sometimes hard to concoct a rationale for parental effects coming from a common distribution, it should be easy to intuit that they lie on a common scale and that knowledge about a1, … , aj−1 provides information about how we would expect aj to appear. Bayesian updating provides a coherent rendering of this intuition, allowing information from the data and uncertainty from the priors to propagate through the hierarchy and inform posterior estimates of parameters and predictions of new effects (Bernardo and Smith 2000; Sorensen and Gianola 2004). To make our point, we demonstrate by simulation how a fixed-effects GCA model with 10 parameters is matched or beaten by Bayesian shrinkage models with n + 175 parameters in its own backyard (Tables 3 and 4).
When there is prior belief that phenotypic similarity will tend to follow overall genetic similarity (e.g., Kang et al. 2008), relatedness (e.g., Lynch and Walsh 1998), spatial distribution, or some other structure that can be incorporated into an expected J × J correlation matrix A, then this could aid estimation, potentially being incorporated by replacing Equation 4 with . Failing to do so, perhaps for convenience, makes inefficient use of available prior data, but discredits use of our hierarchy no more than it would for a fixed-effects model.
The use of a mixed model makes it tempting to interpret variance parameters as heritabilities. However, the very depth of genetic characterization afforded by the diallel, as well as legitimate aspects of the random parents commonplace described above, suggests a more reflective approach. Our decomposition of effects groups into a, b, m, v, w spans many possible reduced models for quantitative inheritance. When we further explore the heritability, or the amount of variance explained, by each group of effects, we find that, in a diallel breeding structure, the variance contributions of the groups interact. For an expedient method of calculating , the heritability attributable to effects group q, we promote a calculation using the estimated group dispersions (Appendix A).
In our model we do not treat explicit variance heterogeneity (Rodriguez et al. 1993). However, our outlier approach implicitly adapts to variance heterogeneity by assigning reduced weights λi to measurements it deems to be more variable. Postprocess exploration of may reveal strain-dependent variances. Algorithmically, an option for a parametric form of Var(εi | j, k, S) can be achieved by modeling 1/λi ∼ exp{xiT γ}, replacing the update for λi with Gibbs step
where drawing from a posterior using can be achieved by Metropolis–Hastings, to calculate the new λi values. Ideally, γ would have a reduced form including just additive, or sex-specific additive, components to retain model parsimony. See Rönnegård and Valdar (2011) for an example of such a double generalized linear model in practice.
Bayesian models are often slower to compute and more difficult to interpret, and can require a large and ill-defined set of prior choices. Although it is often the case that estimators using reasonable hyperpriors can outperform the MLE over a large parameter space, for most loss functions neither MLE, nor ridge, nor Bayes estimators completely dominate each other. Given a mixed-model decomposition of the diallel, including the possibility of outliers, unobserved diallel combinations, and unknown sparsity among the parameters, stable MLE estimates can be difficult to achieve. Penalized estimates [e.g., from group LASSO (Hastie et al. 2009)] represent an opportunity, but these can often be interpreted as a Bayesian prior hypothesis. We demonstrate the use of the h-likelihood (using hglm) for obtaining mostly accurate point estimates for our diallel model and in less computational time. Although useful for this purpose, the hglm approach does not easily provide the rich flexibility of our Gibbs sampler for, among other things, modeling outliers and uncertainty about component inclusions. We supply an algorithm with reasonable default priors, but users are still permitted to choose their own, including flat Gelman or Stein priors, Jeffreys priors, or heavily informed priors from the inverse gamma family. For the BayesSpike model, one might consider a priori that all components have an equal 50% chance of inclusion, that they have different anticipated prior weights, or that the prior proportion of active groups is itself unknown and must be learned in the model.
Approaches that model hierarchically, borrowing strength across data sources and explicitly defining higher-order components, are becoming essential to navigate the complex high-dimensional spaces created by high-throughput data of all types. However, such hierarchies should be designed to empower researchers, not intimidate or perplex them. We emphasize interpretability of all parameters in the model in the hope they can at some level be interpreted and critiqued by nonexperts. We provide a framework for high-, medium-, and low-level analysis of imperfectly sampled diallels. In doing so, we succinctly summarize results from a vast amount of original data on the genetic architecture of phenotypes in founders of the Collaborative Cross and their F1 hybrids.
Our diallel Gibbs sampler and BayesSpike software are provided free of charge as R packages R/BayesDiallel and R/BayesSpike as soon as practicably in File S1.
Acknowledgments
We thank Vasyl Zhabotynsky and Fred Wright for helpful discussions. The authors acknowledge partial support from the National Institutes of Health (NIH) Center of Excellence in Genome Sciences grants P50 MH090338 and P50 MH006582 (to A.B.L. and W.V.), from NIH grant GM076468 (to K.L.S. and G.A.C.), from The Jackson Laboratory (G.A.C.), and from the University of North Carolina Lineberger Comprehensive Cancer Center (A.B.L. and W.V.).
Appendix A: Heritability in the Diallel
Heritability is the proportion of phenotypic variance explained by genetic effects. Its exact definition depends on what effects are being considered and in which population phenotypic variance is measured. Consider any linear genetic model, yi = xiTβ + εi, where β ∈ ℝp is an explanatory vector of common effects, and εi ∼ N(0, σ2) are iid environmental effects, for independently sampled individuals i ∈ 1, …, N in the population. The vector xi ∈ ℝp×1 represents a draw from the genetic diversity of haplotypes/strains/alleles as distributed in the population, represented by stacked matrix . If both xi and β were randomly distributed and independent, and were the population mean, then the observed variability of Var(yi) = E[(yi – μY)2] has expectation that can be calculated
| (A1) |
Equation A1 provides a method to decompose observed variability of Y to the contributions of specific effects or groups of effects. If we assume that diallels of future strains will have effects distributed , then E(ββT) is a diagonal matrix with values for each group of factors. In a complete diallel experiment, E[xixiT] ∈ ℝp × p is a very structured design matrix. If we considered mother strain j and father strain k to be independent draws from a pool of J strains, and gender of the offspring to be an independent Bernoulli draw, then we can calculate explicit expected dosage amounts of each of the a, b, m, v, w, as, bs, ms, vs, and ws effects. For additive effect aj′, for strain j′, located at position {xi}a(j′) along the xi covariate vector, expected Since there are J separate aj′ terms, the variance contribution of additive effects can be approximated as . Further expectations are calculated in Table A1. With Table A1 we can then interpret the heritability contribution of additive effects to be
| (A2) |
in a diallel with no sex-specific effects. We see that heritability contributions of inbreeding effects b contribute at a reduced level of 1/J to the variability of the phenotype. This is because in a complete diallel, inbred subjects compose only 1/J of the population.
Table A1.
Expected heritability contribution of effects to Var(Yi) of the effects groups
| a | b | m | v | w | as | bs | ms | vs | ws |
Appendix B: Gibbs Sampling Scheme for the Full Model
Collecting all fixed-effect (e.g., βfemale) and strain-specific random-effect parameters (e.g., aj) in a single vector of pM regression coefficients β, we construct an n × pM design matrix X = [x1T … xnT]T and consider the regression problem
X is both sparse and highly structured in our diallel model. For instance, in the “av” model (additive and symmetric effects), pM = 1 + J + (J + 1) × J/2, and the last (J + 1) × J/2 positions in xi are mostly zeros with a single 1 at the position corresponding to mother/father pair (j, k)[i]. Let the parameter groups a, as, m … be enumerated as groups q ∈ 1, … , Q and (q) denote the group of coefficients qj assigned to the same shrinkage parameter . By introducing a Q matrix, as suggested from Equation 17, with diagonal terms for parameters grouped to , the multivariate posterior is
| (B1) |
which can be efficiently sampled by taking the Cholesky square root of XTX + σ2Q−1, where the diagonal of matrix XTX counts the number of subjects with relevant j, k pairs for each q group. We deploy Gibbs sampling (Geman and Geman 1984; Gelfand and Smith 1990; Casella and George 1992), using C-level BLAS (Dongarra 2002) code compiled in package format for R (R Development Core Team 2011), and find that the 2011 Macintosh vecLib LAPACK (Apple Developers) is sufficient to generate as many as a 8000 draws in 26.2 sec from the posterior, given that pM has an upper bound of 244 when the number of strains is 8. However, when pM gets larger, doing so as the number of strains increases, it is necessary to invert conditional subset groups of β. The advantage of a single Cholesky draw is to reduce autocorrelation of the Gibbs sampler. In this case, maximum autocorrelation at 1-lag for pM = 164 variables is 0.177, and max 20-lag is 0.086, with 95% of the 1-lag being between ±0.05. Given β, we draw as
where |q| is the count of members of group q, and draw σ2 as
Appendix C: Gibbs Sampling Scheme for the Full Model with Outliers
Computational complexity changes with the outlier model, where . In this case, if we draw λi as a weight for subject i from
then we can reweight by defining components and , in which case
becomes a new draw of β. Since the reweighting is an operation, it can slow down the algorithm when n > pM.
Appendix D: Exclusionary Gibbs Group Sampling in the Diallel
To sample efficiently from Equation 18, we consider the residual vector , which includes current information of all βj for j not in (q). Integrating out of this draw produces an unnormalized function . This function takes on a discrete probability at that we define as and also includes a continuous density with support on . If , then a Bernoulli draw of probability
provides a Gibbs decision to turn on or off parameters in group q. Moreover, to avoid potentially slow numerical integration, we construct a Metropolis–Hastings importance sample from , which approximates F+ in expectation.
Footnotes
Edited by Lauren M. McIntyre, Dirk-Jan de Koning, and 4 dedicated Associate Editors
Literature Cited
- Baker R. J., 1978. Issues in diallel analysis. Crop Sci. 18(834): 533–536 [Google Scholar]
- Bernardo J. M., Smith A. F. M., 2000. Bayesian Theory. John Wiley & Sons, Chichester, UK [Google Scholar]
- Box G. E. P., Tiao G. C., 1973. Bayesian Inference in Statistical Analysis, p. 608 Wiley-Interscience, New York [Google Scholar]
- Carbonell E. A., Nyquist W. E., Bell A. E., 1983. Sex-linked and maternal effects in the Eberhart-Gardner general genetics model. Biometrics 39(3): 607–619 [PubMed] [Google Scholar]
- Carlin B. P., Louis T. A., 2008. Bayesian Methods for Data Analysis, Ed. 3 Chapman & Hall/CRC, London/New York [Google Scholar]
- Casella G., George E. I., 1992. Explaining the Gibbs sampler. Am. Stat. 46(3): 167 [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19(6): 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie B. R., Shattuck V. I., 1992. The diallel cross: design, analysis, and use for plant breeders. Plant Breed. Rev. 9: 9–36 [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36(11): 1133–1137 [DOI] [PubMed] [Google Scholar]
- Cockerham C., Weir B., 1977. Quadratic analyses of reciprocal crosses. Biometrics 33(1): 187–203 [PubMed] [Google Scholar]
- Collaborative Cross Consortium, 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongarra J., 2002. Basic linear algebra subprograms technical forum standard. Int. J. High Performance Appl. Supercomput. 16: 115–199 [Google Scholar]
- Eberhart S., Gardner C., 1966. A general model for genetic effects. Biometrics 22(4): 864–881 [Google Scholar]
- Gardner C., Eberhart S., 1966. Analysis and interpretation of the variety cross diallel and related populations. Biometrics 22(3): 439–452 [PubMed] [Google Scholar]
- Gelfand A. E., Smith A. F. M., 1990. Sampling-based approaches to calculating marginal densities. J. Am. Stat. Assoc. 85(410): 398–409 [Google Scholar]
- Gelman A., 2006. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Int. Soc. Bayes. Anal. 1(3): 515–534 [Google Scholar]
- Gelman A., Hill J., 2007. Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge University Press, New York [Google Scholar]
- Geman S., Geman D., 1984. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Patt. Anal. Mach. Intell. 6(6): 721–741 [DOI] [PubMed] [Google Scholar]
- George E., Foster D., 2000. Calibration and empirical Bayes variable selection. Biometrika 87(4): 731–747 [Google Scholar]
- Greenberg A. J., Hackett S. R., Harshman L. G., Clark A. G., 2010. A hierarchical Bayesian model for a novel sparse partial diallel crossing design. Genetics 185(1): 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing B., 1956. Concept of general and specific combining ability in relation to diallel crossing systems. Aust. J. Biol. Sci. 9(4): 463–493 [Google Scholar]
- Gross J., 2003. Linear Regression. Springer-Verlag, Heidelberg, Germany/New York [Google Scholar]
- Guan Y., Stephens M., 2011. Bayesian variable selection regression for genome-wide association studies, and other large-scale problems. Ann. Appl. Stat. 5(3): 1780–1815 [Google Scholar]
- Hastie T., Tibshirani R., Friedman J., 2009. The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer Series in Statistics, Ed. 2). Springer-Verlag, New York [Google Scholar]
- Hayman B. I., 1957. Interaction, heterosis and diallel crosses. Genetics 42: 336–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H., Rao J. S., 2005. Spike and slab variable selection: Frequentist and Bayesian strategies. Ann. Stat. 33(2): 730–773 [Google Scholar]
- Jeffreys H., 1946. An invariant form for the prior probability in estimation problems. Proc. R. Soc. Lond. A Math. Phys. Sci. 186(1007): 453–461 [DOI] [PubMed] [Google Scholar]
- Jinks J. L., Hayman B. I., 1953. Analysis of diallel crosses. Maize Genet. Coop. News Lett. 27: 48–54 [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R., Raftery A., 1995. Bayes factors. J. Am. Stat. Assoc. 90(430): 773–795 [Google Scholar]
- Kempthorne O., Curnow R., 1961. The partial diallel cross. Biometrics 17(2): 229–250 [Google Scholar]
- Lee Y., Nelder J., 1996. Hierarchical generalized linear models. J. R. Stat. Soc. B 58(4): 619–678 [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Meng X.-L., 2008. Discussion: one-step sparse estimates in nonconcave penalized likelihood models: Who cares if it is a white cat or a black cat? Ann. Stat. 36(4): 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani G., Inoue L., 2009. Decision Theory: Principles and Approaches (Wiley Series in Probability and Statistics). John Wiley & Sons, New York [Google Scholar]
- R Development Core Team, 2011 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Rodriguez L. A., Fulker D. W., Cherny S. S., 1993. A maximum-likelihood model-fitting approach to conducting a Hayman analysis of diallel tables with complete or missing data. Behav. Genet. 23(1): 69–76 [DOI] [PubMed] [Google Scholar]
- Rönnegård L., Valdar W., 2011. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 188: 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L., Shen X., Alam M., 2010. hglm: a package for fitting hierarchical generalized linear models. R J. 2(2): 20–28
- Schmidt J., 1919. La valeur de l'individu à titre de génratéur appréciée suivant la méthode du croisement dialléle. C. R. Trav. Lab. Carlsberg 14(6): 1–33 [Google Scholar]
- Sorensen D., Gianola D., 2004. Likelihood, Bayesian and MCMC Methods in Quantitative Genetics. Statistics for Biology and Health. Springer-Verlag, New York [Google Scholar]
- Spiegelhalter D. J., Best N. G., Carlin B. P., van der Linde A., 2002. Bayesian measures of model complexity and fit. J. R. Stat. Soc. Ser. B Stat. Methodol. 64(4): 583–639 [Google Scholar]
- Sprague G., Tatum L., 1942. General vs. specific combining ability in single crosses of corn. J. Am. Soc. Agron. 34: 923–932 [Google Scholar]
- Stein C., 1955. Inadmissibility of the usual estimator for the mean of a multivariate normal distribution, pp. 197–206 in Proceedings of the Third Berkeley Symposium on Mathematical Statistics and Probability, Vol. 1, Issue 4. University of California Press, Berkeley [Google Scholar]
- Taylor B. A., Wnek C., Schroeder D., Phillips S. J., 2001. Multiple obesity QTLs identified in an intercross between the NZO (New Zealand obese) and the SM (small) mouse strains. Mamm. Genome 12(2): 95–103 [DOI] [PubMed] [Google Scholar]
- Venables W., Ripley B., 2002. Modern Applied Statistics with S. Springer-Verlag, New York [Google Scholar]
- Wright A. J., 1985. Diallel designs, analyses, and reference populations. Heredity 54: 307–311 [DOI] [PubMed] [Google Scholar]
- Zhu J., Weir B., 1996. Mixed model approaches for diallel analysis based on a bio-model. Genet. Res. 68(3): 233–240 [DOI] [PubMed] [Google Scholar]