Abstract
The JAX Diversity Outbred population is a new mouse resource derived from partially inbred Collaborative Cross strains and maintained by randomized outcrossing. As such, it segregates the same allelic variants as the Collaborative Cross but embeds these in a distinct population architecture in which each animal has a high degree of heterozygosity and carries a unique combination of alleles. Phenotypic diversity is striking and often divergent from phenotypes seen in the founder strains of the Collaborative Cross. Allele frequencies and recombination density in early generations of Diversity Outbred mice are consistent with expectations based on simulations of the mating design. We describe analytical methods for genetic mapping using this resource and demonstrate the power and high mapping resolution achieved with this population by mapping a serum cholesterol trait to a 2-Mb region on chromosome 3 containing only 11 genes. Analysis of the estimated allele effects in conjunction with complete genome sequence data of the founder strains reduced the pool of candidate polymorphisms to seven SNPs, five of which are located in an intergenic region upstream of the Foxo1 gene.
GENETICALLY defined mouse models provide a powerful experimental system for mapping and functional analysis of genetic variants in the context of a complex, living organism (Peters et al. 2007). It is widely recognized that most medically important traits are genetically complex. However, experimental strategies for complex trait analyses in the mouse have not kept pace with rapid developments in human genetic studies. To advance our understanding of complex genetic traits, new strategies for using model organisms are needed.
The Diversity Outbred (DO) is a newly developed mouse population derived from progenitor lines of the Collaborative Cross (Collaborative Cross Consortium 2012). Animals from incipient CC lines at early stages of inbreeding were used to establish the DO population, which is maintained by a randomized outbreeding strategy. Thus the DO and CC populations capture the same set of natural allelic variants derived from a common set of eight founder strains. There are approximately 45 million segregating SNPs—four times the number found in classical laboratory mouse strains (Yang et al. 2011). This genetic variation is maintained in two distinct population structures. Whereas each CC inbred strain represents a fixed and reproducible genotype, each DO animal is a unique individual with one of an effectively limitless combination of the segregating alleles. The DO is an ideal resource for high-resolution genetic mapping. The CC can provide predictive validation of mapping results obtained with the DO, as well as a source of reproducible genotypes for mechanistic studies. The potential applications of these resources have yet to be fully realized and their utility is likely to expand further when considered in combination with other newly developed tools for mouse genetics, such as the comprehensive knockout mouse resource (Skarnes et al. 2011).
The DO offers a number of advantages over classical approaches to mouse genetics. The DO can provide high mapping resolution that will continue to improve as outbreeding progresses. High-resolution mapping in the mouse has, until recently, proven to be impractical due to limited density of markers and the high cost of genotyping. However, the situation has changed with the development of high-density, low-cost genotyping platforms (Yang et al. 2009; Collaborative Cross Consortium 2012). DO mice are outbred. Complete inbreeding is an unusual genetic state for a mammal that does not accurately reflect the human genetic makeup. Inbred animals lack the natural buffering effects of heterozygosity, are prone to both early and late-life recessive allelic effects, and are noticeably less vigorous than their outbred counterparts. The genetic diversity of the DO is derived from naturally occurring allelic variants, unlike transgenic mouse models, which often represent extreme perturbations such as complete loss of function of a targeted gene. Recent studies of human diseases suggest that subtle variations, such as those affecting patterns of gene expression or conservative changes in polypeptides, are major contributors to phenotypic diversity (Stranger et al. 2011). Genetic variation in the DO is uniformly distributed with multiple allelic variants in the coding or regulatory regions of essentially all known genes. This is in contrast to classical inbred strains of mice, which have limited genetic diversity and large blind spots that are effectively devoid of variation (Yang et al. 2011). Thus, the DO provides an opportunity to study medically relevant traits in a powerful model system that more closely reflects the genetic mechanisms of human disease.
In this article we describe the breeding design of the DO and compare its properties to simulations. We describe phenotypic diversity in the DO and use genome-wide SNP data to examine allele frequencies and the distribution of recombination events. We describe analytical approaches for mapping complex trait loci in the DO and demonstrate that high-resolution mapping can be achieved with moderate numbers of DO mice.
Materials and Methods
Breeding strategy
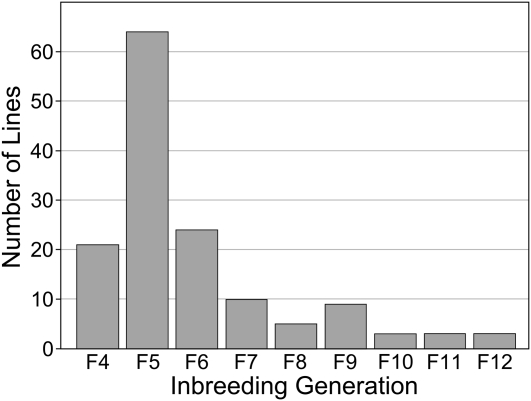
The founders of the DO were obtained in late 2008 through early 2009 from breeding lines of the Collaborative Cross at Oak Ridge National Laboratory (Chesler et al. 2008). The 144 partially inbred CC lines that contributed to the DO were at generations ranging from F4 to F12 of inbreeding (Figure 1). This strategy allowed us to capture recombination events that occurred in the early generations of CC breeding to effectively jump-start recombination density in the DO population. The first randomized outcrosses were carried out in a quarantine facility at The Jackson Laboratory (JAX). From these matings, four to six female and one to two male progeny, depending on availability, were transferred to the JAX importation facility for gamete harvest, in vitro fertilization (IVF), and embryo transfer. Progeny from IVF were brought across the specific pathogen-free (SPF) barrier into a production facility. The progeny of the IVF matings were designated as DO generation G1. Due to the bottleneck involved in the in vitro step, the production colony was initialized in three waves of 84, 55, and 28 matings each to achieve 167 breeding pairs. Each mating pair contributes one female pup and one male pup, selected at random from the first litter, to establish the next generation. The colony was expanded slightly and is now being maintained as a panel of 175 breeding pairs with three to four synchronized generations per year. This breeding strategy minimizes the effects of both drift and positive selection. Mating pairs that will produce the next generation are determined by randomization, with avoidance of sib mating. If a mating produces no offspring or produces offspring of only one sex, an alternative animal is selected at random from the available population.
Figure 1 .
Distribution of inbreeding generation number among the 144 partially inbred CC founder lines that contributed to the DO.
Animals and phenotyping
A pilot study was conducted using 150 early generation DO animals. Fourth (G4; n = 100; 50 females, 50 males)- and fifth (G5; n = 50; 25 females, 25 males)-generation DO animals were obtained from The Jackson Laboratory (Bar Harbor, ME). Animals were received at wean age (3 weeks old) and distributed into cages at a density of five same-sex animals per cage. Animals were housed in pressurized, individually ventilated cages (Thoren Caging Systems, Hazelton, PA) with pine bedding (Crobb Box, Ellsworth, ME) and had free access to standard rodent chow containing 6% fat by weight (LabDiet 5K52, LabDiet, Scott Distributing, Hudson, NH) and acidified water. Mice were subject to a 12 hr:12 hr light:dark cycle beginning at 06:00. Some DO mice (n = 50 G4; 25 females, 25 males) were fed a high-fat (HF) diet containing 22% fat by weight (TD.08811, Harlan Laboratories, Madison, WI) from wean age throughout the the study. All animal procedures were approved by the Animal Care and Use Committee at The Jackson Laboratory.
Blood was obtained from the retro-orbital sinus after administration of a topical anesthetic (tetracaine HCl) using a heparin-coated microcapillary tube and collected into a 1.5-ml eppendorf tube. To measure plasma components, approximately 150μl whole blood was collected into a tube containing 2μl of 10% sodium heparin, and plasma was separated by centrifugation at 10,000 rpm for 10 min at 4° and removed into a clean eppendorf tube. Plasma components (total cholesterol, HDL cholesterol, glucose, triglycerides) were measured using the Beckman Synchron DXC600Pro Clinical chemistry analyzer. For whole-blood analysis approximately 200 μl blood was collected into a tube containing 2 μl of 10% sodium EDTA. Hematological parameters were measured using the Siemens Advia 2120 haematology analyzer. Blood was collected in the morning; blood for plasma chemistry analysis was drawn after food was removed for a 4-hr period beginning at 07:00. Blood collections were separated by a period of 2 weeks to allow animals to fully recover blood volume and components. Plasma measurements were from animals at 8 weeks and 19 weeks of age and whole blood was analyzed from 10-week-old mice.
Body composition was assessed when mice were 12 weeks of age by dual energy X-ray absorptiometry (DEXA) using a Lunar PIXImus densitometer (GE Medical Systems) after mice were anesthetized intraperitoneally with tribromoethanol (0.2 ml 2% solution/10 g body weight). The skull is omitted from the DEXA analysis because it is so bone dense. Mice were weighed on an Ohaus Navigator scale with InCal calibration to accommodate animal movement.
Analysis of phenotype data
Phenotypes observed in the DO were compared to similar phenotypes measured among inbred strains. Data for inbred strains was obtained from the Mouse Phenome Database (MPD) using project data sets titled Naggert1 (Naggert et al. 2003), Peters1 (Peters and Barker 2001), and Paigen2 (Paigen et al. 2002). In these studies, baseline measurements were obtained from the inbred strains while they consumed standard chow, prior to being fed a HF diet. Because the HF diet fed to the inbred strains in the MPD projects differs in composition from that fed to the DO in this study, only measurements obtained from inbred mice on the chow diet were compared. Mice in the MPD projects were fed the identical standard chow as were the DO. Additionally, comparison groups are age matched.
Simulations of the DO breeding design
We used simulation to investigate the expected genetic composition of the DO population. For comparison we performed analogous simulations of CC and F2 breeding designs. Statistics were collected on 360 simulated trials per design. We simulated the autosomal mouse genome on the basis of the parameter of the standard mouse genetic map (Cox et al. 2009). Founders contributed distinct alleles at each (octa-allelic) marker such that descent in nonfounder generations was implied by genotypic state. Recombination was simulated as a continuous Poisson process following the Haldane model (Lynch and Walsh 1997). We simulated partially inbred CC mice following the funnel breeding design (Churchill et al. 2004) with randomized order of eight founders. Each simulated CC population was composed of 150 distinct CC funnels. We generated simulated DO mice in two steps starting from 150 sister–brother pairs of partially inbred CC lines at generation G2F6, i.e., with five generations of inbreeding. We then simulated rounds of mating in which mates were paired at random with the constraint that sib-matings are prohibited. The number of offspring per pair was constant at two. Intercross (F2) animals were simulated as in, for example, Valdar et al. (2006), also with a population size of 150.
Genotyping
DNA was prepared by standard methods from tail biopsies and genotyping was outsourced to GeneSeek (http://www.neogen.com/GeneSeek) for analysis using a Mouse Universal Genotyping Array (MUGA), a 7851 SNP array built on the Illumina Infinium platform. Markers on the MUGA are distributed genome-wide with an average spacing of 325 kb and standard deviation of 191 kb. The markers were selected such that they could uniquely identify any of the CC founders within a window of four to five SNPs. This gives the MUGA an average effective sampling sensitivity of just over 1 Mb. Thus, recombination segments smaller than 1 Mb may go undetected. After accounting for animals that did not complete the study and eliminating failed genotypes, a total of 141 genotypes were used for mapping and haplotype reconstruction.
Haplotype reconstruction
Haplotype reconstruction was based on Illumina's normalized MUGA intensity values rather than genotype calls. This is predicated from our observation that the allele clusters seen in a genotyping probe set can often be further subclustered according to the eight founders and the 28 possible F1s. This transforms the standard 3-state genotyping classification problem to one with 36 states for our population. The distributions of each genotype state at each marker were estimated using a combination of biological and technical replicates of each of the CC founders and F1s. These distributions are used as reference templates. We estimate the likelihood that a sample matches a particular template as a function of its 2D distance from each of the template's model. These probabilities were combined with a transition probability between genomically adjacent markers using a hidden Markov model (HMM). The transition probability parameters were selected so that evidence from approximately four sequential markers is necessary to change founder state. Moreover, the transition penalty varies depending on whether the current state and next state share a common founder or not. A dynamic programming algorithm was then utilized to calculate the maximum-likelihood founder assignment for each chromosome.
QTL mapping analysis
The genotyping HMM produced a matrix of 36 genotype state probabilities for each sample at each SNP. This matrix was condensed to produce an eight-founder state probability matrix by summing the probabilities contributed by each founder at each SNP. The eight-founder probabilities were used to fit a linear model, with sex and diet as additive covariates and variance components that account for genetic relatedness using QTLRel (http://www.palmerlab.org/software) (Cheng et al. 2011). Each phenotype was analyzed in this manner following a rank z-score transformation. The model was
where yi is the phenotype value for animal i, βs is the effect of sex, si is the sex of animal i, βd is the effect of diet, di is the diet for animal i, βj is the effect of founder allele j, gij is the founder probability for founder j in animal i, and γi is a random effect representing the polygenic influence of animal i.
The mapping statistic was the log of the odds ratio (LOD) from the linear model. Significance thresholds were determined by performing 100 permutations and fitting an extreme value distribution to the maximum LOD values (Dudbridge and Koeleman 2004). When this procedure indicated a significant result, we performed 1000 permutations using the above equation but without accounting for relatedness (Cheng et al. 2011).
Availability
The JAX:DO are available from The Jackson Laboratory (Bar Harbor, ME), as JAXMice stock number 009376. Sibling information at each generation is tracked and made available upon request. All data described in this report are available at http://cgd.jax.org/datasets/phenotype/SvensonDO.shtml. Please cite this article in any use of these data.
Results
Phenotypes
Plasma components (total cholesterol, HDL cholesterol, glucose, triglycerides) and body composition parameters (weight, percentage body fat) were measured for 100 DO mice from outcross generations G4 (n = 50; 25 females, 25 males) and G5 (n = 50; 25 females, 25 males) fed a standard chow diet (Table 1). Two measurements of body composition were made during the study: the first was made at 12 weeks of age and the second at 21 weeks of age. While females and males gained an average of 3.5 and 4.8 g, respectively, during the 9-week period between body composition measurements, data for both sexes reveals that some mice lost weight and some mice lost body fat, as reflected in minimum values for weight change and percentage body fat change, respectively. However, changes in weight were usually not associated with changes in body fat. Among females, only one animal lost weight (loss of 2.0 g) but gained 2.2% body fat (data not shown). All other females gained from 0.5 to 11.6 g of weight and showed wide variation (−0.6–9.8%) in the proportion of body fat gained (or lost) over the 9-week period between measurements (data not shown). Among males, only one animal lost weight, and this was accompanied by a loss in body fat. All other males gained weight, but 16/44 males lost body fat (loss of 0.1–7.9%) in the 9-week period (data not shown). Hematologic parameters were also measured for the same 100 mice (Table 2). Among females there is a 1.2- to 5.1-fold difference in minimum and maximum values for each trait, and among males there is 1.5- to 14.7-fold variation in minimum and maximum values for each trait. The wide range of phenotype values in this relatively small sample of animals illustrates the extent of phenotypic diversity in the DO population.
Table 1 .
Plasma lipids, glucose, and body composition in fourth- and fifth-generation DO mice fed standard chow
| Females |
Males |
|||||
| Phenotype | Mean ± SE | Min | Max | Mean ± SE | Min | Max |
| CHOL (mg/dl) | 77 ± 3 | 43 | 119 | 103 ± 3 | 62 | 163 |
| HDL (mg/dl) | 60 ± 2 | 32 | 97 | 78 ± 2 | 51 | 124 |
| TG (mg/dl) | 112 ± 6 | 35 | 219 | 167 ± 10 | 64 | 415 |
| GLU (mg/dl) | 160 ± 4 | 74 | 248 | 193 ± 4 | 125 | 279 |
| WT 1 (g) | 24.2 ± 0.5 | 18.5 | 36.8 | 31.8 ± 0.7 | 22.1 | 45.2 |
| WT 2 (g) | 27.8 ± 0.7 | 19.1 | 42.6 | 36.4 ± 0.8 | 25.7 | 53.4 |
| Change in WT (g) | 3.5 ± 0.4 | −2 | 11.6 | 4.8 ± 0.5 | −3.3 | 13 |
| % Fat 1 | 21.8 ± 0.6 | 15.1 | 31.1 | 19.5 ± 0.6 | 11.7 | 29.3 |
| % Fat 2 | 24.0 ± 0.8 | 14 | 37.7 | 20.9 ± 0.8 | 13.3 | 31.8 |
| Change in % Fat | 2.1 ± 0.5 | −4.6 | 9.8 | 1.2 ± 0.7 | −7.9 | 10.3 |
Values shown are means ± SE. Min, minimum value within a group for trait measured; Max, maximum value within a group for trait measured. CHOL, total plasma cholesterol; HDL, high-density lipoprotein cholesterol; TG, triglycerides; GLU, glucose. CHOL, HDL, TG, and GLU were obtained after food was removed from mice for 4 hr in the morning. WT 1, % Fat 1, body weight and % body fat at age 12 weeks; WT 2, % Fat 2, body weight and % fat at age 21 weeks. % Fat was obtained by dual-energy X-ray absorptiometry. Changes in weight and % fat were derived by subtracting value 1 from value 2 and are expressed as grams (g) and %, respectively. n = 50 females and 50 males for all measurements except WT 2, % Fat 2, change in WT and change in %Fat, for which n = 48 females and 44 males.
Table 2 .
Hematological parameters in fourth- and fifth-generation DO mice fed standard chow
| Females |
Males |
|||||
| Phenotype | Mean ± SE | Min | Max | Mean ± SE | Min | Max |
| WBC (×103/μl) | 9.1 ± 0.4 | 3.5 | 17.6 | 9.1 ± 0.4 | 5.2 | 18.1 |
| RBC (×103/μl) | 11.2 ± 0.09 | 9.4 | 12.4 | 10.7 ± 0.12 | 8 | 12.1 |
| HCT (%) | 54.5 ± 0.3 | 47.9 | 58.8 | 53 ± 0.6 | 39.8 | 60.2 |
| PLT (×103/μl) | 1264 ± 43 | 677 | 2042 | 1441 ± 32 | 925 | 1867 |
| LYM (%) | 82.5 ± 0.8 | 65.2 | 90.7 | 75.1 ± 1.3 | 48.2 | 87.9 |
| No. Retic (109 cells/liter) | 2.73 ± 0.11 | 0.98 | 5.01 | 3.25 ± 0.22 | 0.65 | 9.55 |
Values shown are averages ± SE. Min, minimum value within a group for trait measured; Max, maximum value within a group for trait measured. WBC, white blood cells; RBC, red blood cells; HCT, hematocrit; PLT, platelets; LYM, lymphocytes; Retic, reticulocytes. n = 50 females and 50 males.
Observation of early DO generations has revealed diversity in social behavior, use of bedding materials, and food handling. Home-cage behavior ranges from docile to highly aggressive. While both females and males show wildness behaviors and must be carefully handled to maintain containment in home cages, severe aggression with fighting was observed in only a few males. Of 75 males phenotyped, initially housed in pens of five animals each, one male from each of six pens was separated and housed individually due to aggressive behavior toward pen mates. Animals show diversity in use of bedding and compressed paper-enrichment materials, with some animals seemingly uninterested in the material and others shredding it and hiding under it. Some animals grind pelleted food from hoppers and pile it in a clean corner of the cage.
Coat colors in the DO are largely variations of agouti, with black and white appearing at expected proportions on the basis of the alleles of the CC founder strains. (A/J and NOD/ShiLtJ are albino. C57BL/6J and A/J both carry the recessive black allele at the agouti locus, but it is masked by albino in A/J.) The characteristic white forehead spot originating from CC founder strain WSB/EiJ is seen on the various agouti shades as well as on black fur. Skeletal dysmorphology is occasionally seen in the DO, notably a hunched back suggesting spinal aberration, and rarely DO mice show limb abnormalities, including complete absence of one limb.
Breeding in the DO is robust, suggesting little or no outcrossing depression in this population, a phenomenon that would be expected if allele incompatibilities were causing a decrease in fitness (Lynch and Walsh 1997). There is a small sex ratio bias toward the production of male progeny. At generation G4 of outcrossing, first litters averaged 7.5 pups/litter, producing 48% females and 52% males. Second G4 litters produced an average of 9 pups each, producing 47% females and 53% males. At generation G5 of outcrossing, first litters averaged 7.0 pups/litter, producing 47% females and 53% males. Second G5 litters averaged 8.0 pups/litter, producing 46% females and 54% males.
Comparison to phenome panels
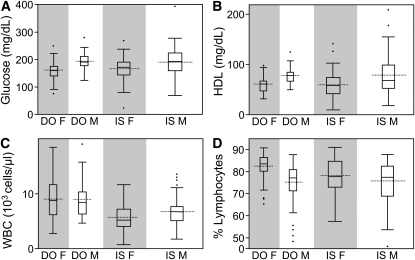
We examined how phenotypic variation among the relatively small number of DO mice presented in this report compared to that among the eight founder strains of the CC for similar phenotypic measurements (Figure 2). Inbred strain data were downloaded from the Mouse Phenome Database (http://www.jax.org/phenome) and includes measurements from 10 to 20 animals per sex per strain, comprising 215 individual animals. Only 100 of the 150 DO animals that we phenotyped were used in this analysis because they were fed the same standard chow diet as the inbred strains at the time of data collection. For most traits analyzed, average trait values are similar between DO and founders.
Figure 2 .
Comparisons of like phenotypes in CC founder inbred strains and 100 DO mice. Plasma parameters are (A) glucose and (B) HDL cholesterol, measured after a 4-hr period of food removal. Whole blood parameters are (C) white blood cells (WBC) and (D) lymphocytes. Lymphocytes are presented as percentage of total number of leukocytes. Data are separated by sex for each group (F, female; M, male). DO, Diversity Outbred (n = 100; 50 female, 50 male). IS, Inbred Strain founders of the Collaborative Cross (n = 215) collective inbred strain individuals: 109 females, 106 males. All mice consumed the same chow diet and are age matched.
Maintenance of allelic diversity
The mating strategy used to produce the DO is key to maintaining genetic diversity in a small population. Allowing only two offspring to be contributed by each mating pair to the next generation doubles the effective population size. The expected time to loss (or fixation) of a private allele (initial frequency 1/8) is 898 generations in the DO compared to 448 for random mating. The median time to fixation approximates the point at which we expect 50% of private alleles to be lost (or fixed). We estimated this to be 469 and 238 generations in the DO and random mating populations, respectively. These generational times far exceed the practical lifetime of a mouse colony. However, over the first 100 generations allelic loss is expected at 5% of loci in the DO compared to 22% in a random mating population of the same size. In both mating schemes allelic loss is minimal in the first 20 generations, but by 30 generations the random population will have lost 1% of private alleles. The DO reaches 1% loss at 60 generations, at which time the random population is expected to have lost 10% of private alleles. At smaller population sizes these differences are shifted proportionately toward earlier generation times. Considering that a number of outbred colonies have been maintained for more than 60 generations, with small and fluctuating population sizes, loss or fixation of alleles present in the founders is expected to occur at a substantial proportion of loci. The large and constant number of mating pairs used to maintain the DO population is critical to avoiding allelic loss and inbreeding.
Genotypes
We obtained genotypes at 7851 SNPs for 141 DO animals from outbreeding generations G4 (n = 94) and G5 (n = 47) using the MUGA platform. On average there are 30.3 SNPs between any pair of adjacent recombination events in the G4 and 28.9 SNPs in the G5 (Table 3). The observed distance between recombination events is 5.9 cM in G4 and 5.6 cM in G5 (Table 3), which is consistent with simulated results of 5.5 cM in G4 and 5.0 cM in G5 (Table 4). Even at early generations, this recombination spacing exceeds the average simulated spacing in F2 (22.6 cM) and CC (10.3 cM) breeding designs. The distance between recombinations is expected to decrease further to 3.5 cM in G10, providing even greater mapping resolution (Table 4).
Table 3 .
Statistical analysis of autosomal haplotype reconstructions for 141 DO samples
| G4 (n = 94) |
G5 (n = 47) |
|||||
| Mean | Median | SD | Mean | Median | SD | |
| % homozygosity (identity by state) | 60.5 | 60.2 | 1.6 | 61.1 | 60.9 | 1.6 |
| % heterozygosity (identity by state) | 39.5 | 39.8 | 1.6 | 38.9 | 39.1 | 1.6 |
| % homozygosity (identity by founder) | 9.3 | 9.2 | 1.8 | 10.2 | 10.2 | 2.1 |
| % heterozygosity (identity by founder) | 90.7 | 90.8 | 1.8 | 89.8 | 89.8 | 2.1 |
| Deviation of founder balance (%) | 2.3 | 1.9 | 1.7 | 2.4 | 2.0 | 1.8 |
| Proportion of each state (%) | 2.8 | 2.6 | 1.7 | 2.8 | 2.5 | 1.8 |
| Recombinations per sample | 223.8 | 223.0 | 14.5 | 235.0 | 234.5 | 19.5 |
| Distance between recomb. (Mb) | 9.9 | 6.6 | 10.2 | 9.5 | 6.4 | 9.7 |
| Distance between recomb. (cM) | 5.9 | 4.1 | 5.7 | 5.6 | 4.0 | 5.4 |
| SNPs between recomb. | 30.3 | 20.0 | 31.2 | 28.9 | 19.0 | 29.7 |
Samples have been divided into generations 4 and 5. Rows to be compared with the simulation results in Table 4 are set in bolded italics.
Table 4 .
Distance between recombination events and number of recombinations per sample from simulations in F2, CC and DO populations
| Distance between Recombinations (cM) | ||||||
| Population | F2 | CC | DO:G4 | DO:G5 | DO:G10 | DO:G20 |
| Mean | 22.6 | 10.3 | 5.5 | 5.0 | 3.5 | 2.2 |
| Median | 18.5 | 6.9 | 3.7 | 3.4 | 2.4 | 1.5 |
| SD | 17.8 | 10.6 | 5.5 | 5.0 | 3.5 | 2.2 |
| Recombinations per sample | ||||||
| Population | F2 | CC | DO:G4 | DO:G5 | DO:G10 | DO:G20 |
| Mean | 27.6 | 191.2 | 234.5 | 258.5 | 378.3 | 617.1 |
| Median | 27.0 | 191.0 | 234.0 | 258.0 | 378.0 | 617.0 |
| SD | 5.3 | 22.3 | 15.9 | 16.5 | 19.7 | 25.1 |
DO:G4 refers to the diversity outbred cross after four generations of outbreeding subsequent to initial mixing.
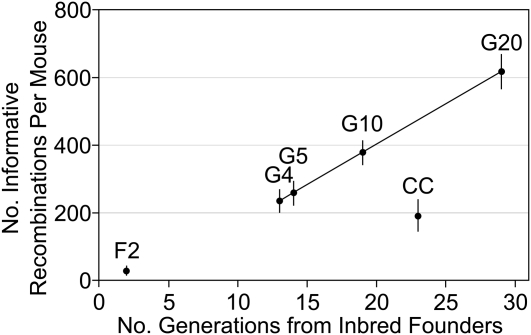
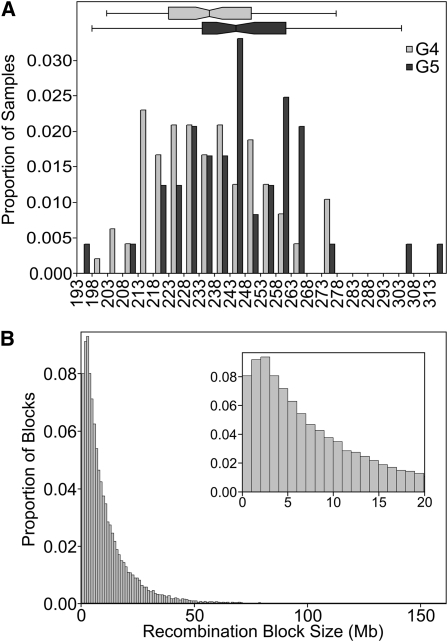
The number of informative recombination events per DO sample is expected to increase linearly by approximately 28 per generation. We observed an increase in average events per animal from 223.8 in G4 to 235 in G5 (Table 3). This is somewhat lower than the number of recombinations per sample found by simulation (234.5 in G4, 258.5 in G5, Table 4, Figure 3). The distribution of distances between recombination events shows an approximate exponential distribution with many small segments, a few large segments, and a coefficient of variation near 1 (Figure 4B and Table 3). However, segments of less than 1 Mb appear to be underrepresented. The average distance between SNPs on the MUGA is 325 Kb, and the HMM requires several informative SNPs to detect a change of genotype state. Therefore, segments of less than 1 Mb are near the detection limit of the array and are likely to be present in the population but remain undetected. This explains the decrease in number of recombinations per sample from the simulations, which count all recombinations, and the observed data, which undercounts closely located recombination events. In future studies of the DO, which will necessarily employ later generations, a higher density genotyping platform may be required.
Figure 3 .
Number of recombinations per mouse estimated by simulation for F2, CC, and DO mice at G4, G5, G10, and G20. G4 indicates four generations of outbreeding.
Figure 4 .
Recombination in the DO. (A) Distribution of the number of recombinations per DO sample. The generation plot is normalized by population size. Shaded bars are G4 (n = 94); solid bars are G5 (n = 47). (B) Distribution of recombination segment size among the DO. Inset details the distribution of segments ≤20 Mb.
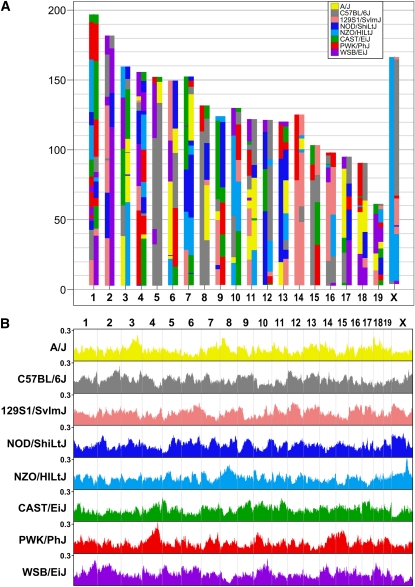
The typical DO genome at generation G4 is a mosaic of the eight founders of the CC and all eight founder alleles are represented across the entire genome (Figure 5A). The average heterozygosity, based on founder haplotypes, is about 90%, slightly higher in this sample than the expected 87.5%. Heterozygosity at the level of MUGA SNPs is about 40% (Table 3). Overall, the contribution from each of the founder strains ranged from 11.3 to 13.8%. The proportion of each founder allele varies somewhat across the genome (Figure 5B). Some of this variation is expected by chance, due to the small sample of genotyped animals, but some of the deviations are consistent with those seen in the CC population (Collaborative Cross Consortium 2012). These include an excess of WSB alleles on chromosome (chr) 2, an excess of PWK alleles on chr 4, a deficit of WSB alleles on chr 5, and a deficit of CAST alleles on chr X. In the future, with larger samples spanning multiple generations, it should be possible to confirm these deviations and observe trends in allele frequencies over time.
Figure 5 .
The DO is a mosaic of eight founder inbred strains. (A) Typical genome of a female DO at generation G4. Chromosome number is indicated on the x-axis. (B) Founder alleles represented across the genome. Chromosome number is at the top. For each SNP on the array (x-axis) the proportion of each founder allele across all samples is plotted on the y-axis. Inset in A provides color codes used to identify founder inbred strains.
Mapping
We performed QTL analysis by pooling the available animals across sexes and two dietary conditions. Sex, diet, and a random polygenic effect were included as covariates in the QTL analysis. We generated 113 phenotypes, some of which were calculated change between values measured at two time points in the phenotyping protocol. The number of animals with complete information varied across traits from 87 to 141. We identified significant QTL (genome-wide adjusted P < 0.05) for 11 traits, including 7 for hematological parameters, 2 for body composition, 1 for cardiac function, and 1 for plasma chemistries. These results represent an interim analysis of an ongoing study with a target sample size of 600 DO animals.
As a proof of principle, we mapped two coat color traits, albino and black. We coded each as binary traits and analyzed using linear regression as described above. We did not use any prior knowledge about their mode of inheritance. The albino trait is caused by a recessive allele in tyrosinase (Tyr) on mouse chr 7 (Russell and Russell 1948). We mapped albino to chr 7: 88–96 Mb (P < 0.001; Supporting Information, Figure S1A). Tyr lies at 94.6 Mb and falls within a two-LOD support interval. As expected, the effect plot (Figure S1B) shows that the two albino founders, A/J and NOD/ShiLtJ, have positive coefficients. The black trait is caused by a recessive allele in the nonagouti (a) gene on chr 2 (Markert and Silvers 1956). It mapped it to chr2: 152–160 Mb (P < 0.001; Figure S1D). The a gene lies at 154.7 Mb and falls within the two-LOD support interval. The effect plot (Figure S1E) shows that A/J and C57BL/6J both have positive coefficients. Although A/J is an albino mouse, if it did not carry the Tyrc mutation, it would be a black mouse. It is known that albino is epistatic to black and that both traits are recessive (Silvers 1979). The correct model to jointly map these loci should include dominance effects and allow for epistatic interactions. However, even though we fit a linear model without interactions between loci and only ∼10% of the mice exhibited each coat color trait, we were able to map both traits to the correct chromosomal location at high levels of statistical significance.
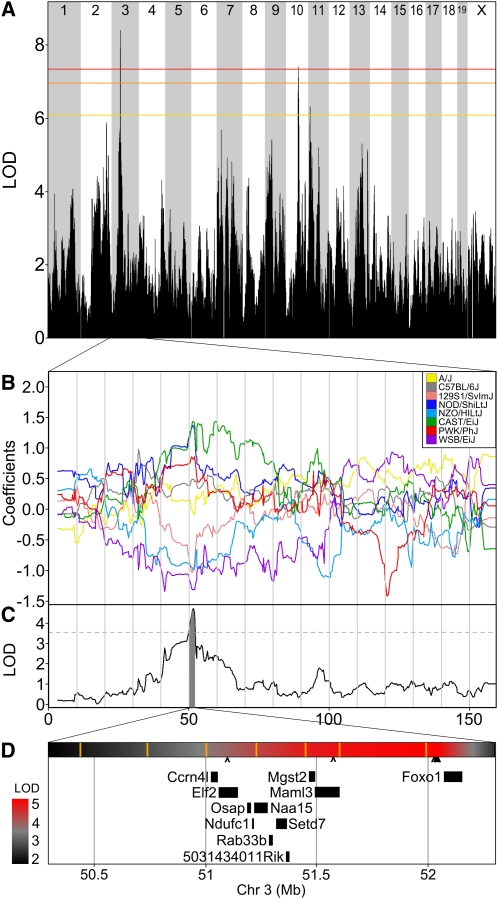
Among the clinical traits mapped in this study, we selected change in total plasma cholesterol from age 8 to 19 weeks to illustrate the potential precision of QTL mapping with the DO. A total of 91 DO animals had complete data for this trait. The genome-wide LOD profile identified a significant association on chromosome 3 (P = 0.014; Figure 6A). The allele effects plot (Figure 6B) indicates that founder haplotypes from strains 129Sv/ImJ, WSB/EiJ, and NZO/H1Lt/J are associated with a decrease in total plasma cholesterol over time. A two-LOD support interval identifies this QTL target region to be at 50.3–52.3 Mb (Figure 6C) spanning a region that contains 11 protein coding genes (Figure 6D). The 2-Mb regions encompasses 32,196 SNPs (Keane et al. 2011) of which only 7 are consistent with the allele effect pattern and, of these, 5 are located in the intergenic region upstream of Foxo1.
Figure 6 .
Change in plasma cholesterol has a significant QTL on chromosome 3. (A) Genome-wide scan for change in cholesterol from 7 to 18 weeks of age. Colored lines show permutation-derived significance thresholds at P = 0.05 (red), P = 0.10 (orange), and P = 0.63 (yellow). (B) The eight coefficients of the QTL model show the effects on the phenotype contributed by each founder haplotype on chr 3. (C) QTL plot for the chr 3 locus. Shading identifies a two-LOD support interval. Dashed line is maximum LOD −2. (D) Expansion of the two-LOD support interval containing 11 genes. A heatmap of the QTL P-value is shown above the gene locations with SNP locations indicated by orange vertical bars. The scale of significance (red most significant) is shown on the left. The seven Sanger SNPs that match the founder effect pattern are marked beneath the heat map with carats (^); five of these cluster upstream of Foxo1.
Discussion
The Diversity Outbred captures a vast background of allelic variants in a highly recombinant population. Each DO animal is unique and not reproducible, which precludes the use of certain study designs. However, the same allelic variants that are presented in the DO are also captured in the reproducible recombinant inbred strains of the Collaborative Cross. We anticipate that combined use of these two complementary populations will deliver the experimental power of inbred strain studies together with an unlimited diversity of allelic combinations and high precision mapping that are currently the exclusive domain of human genome-wide association studies.
Phenotypes of the DO animals measured in this study showed similar averages and ranges compared to a sample of inbred CC founder strains. However, we have observed many individual animals within the larger breeding colony that exhibit unexpected or extreme phenotypes. The effects of genetic variation on DO phenotypes may be buffered by heterozygosity when compared to a fully inbred strain panel. The range of phenotypic diversity is also likely to depend on the nature of the trait being studied. We expect that perturbations at the level of biochemical pathways related to medically relevant traits will contribute to our understanding of functional genetic variation and we plan to include in-depth biochemical phenotyping, such as gene expression profiling, in future studies of DO mice.
The DO breeding colony is maintained with a large number of mating pairs (currently 175) from which two offspring, one female and one male, per pair are selected at random to establish each subsequent generation. This mating design doubles the effective population size of the DO compared to a random mating population (Hartl and Clark 2007), minimizing the effects of drift and positive selection to ensure that existing genetic diversity will be maintained over the lifetime of the population.
The DO population is uniformly structured without major subdivisions. However, animals in any sample will be related to one another in varying degrees and it is important to account for these relationships in genetic mapping analysis (Cheng et al. 2010). We have implemented genome scans using a linear mixed model that includes a random polygenic term with a correlation structure reflecting kinship among animals in the study sample. We used the QTLRel software to compute an additive kinship matrix and to compute the genome scans (Cheng et al. 2011). For comparison, we also computed genome scans using a model without the polygenic term (data not shown). Although similar results were obtained in this study, we recommend the mixed model to minimize the risk of reporting spurious QTL. We further recommend that the kinship correlation matrix should be estimated from genotype data rather than from pedigree records. This provides a direct measure of relatedness, rather than an expectation based on ancestry. It also avoids the potential problem of errors in or missing breeding records but may be sensitive to SNP ascertainment bias.
Our genome scans employed a regression model with 8 degrees of freedom for the genetic effect of a locus. This model assumes additive intralocus effects such that the phenotypic mean of a heterozygote is intermediate between the expected means of the two corresponding homozygous genotypes. This model is convenient and effective, as we have demonstrated, but we risk overlooking loci with purely heterozygous effects. We used a Haley–Knott approximation (Haley and Knott 1992) by regressing directly on the estimated haplotype probabilities at each locus. In this scheme, a heterozygote has a score of 1/2 for each of the contributing alleles. In practice, the actual scores are spread across all eight alleles to reflect uncertainty in the genotype assignments. We used the same method to fit the full 36-state regression model, but found these scans to be unstable. Larger sample sizes are clearly needed to support a regression model with that many degrees of freedom. Alternative implementations that retain some degree of parsimony and, at the same time, allow for nonadditive intralocus effects are the subject of current research (Durrant and Mott 2010; Lenarcic et al. 2012). The additive genetic model provides a robust compromise for detecting genetic loci that have a marginal effect. For example, we successfully mapped two coat-color loci that are known to have recessive and epistatic effects, despite the simplicity of the analysis model (Figure S1).
Mapping analysis of the DO makes use of the recombination events that have arisen subsequent to the establishment of the founder strains. However, inbred founder strains of the CC, in particular the five classical strains, have an older history of shared ancestry that could potentially provide a finer grained mosaic for mapping. A companion publication (Collaborative Cross Consortium 2012) describes the local haplotype structure of the CC founders. This analysis reveals extensive shared ancestry, even extending to the wild-derived founders in some regions of the genome. However, we contend that little is to be gained in the few extra degrees of freedom from a reduced regression model based on shared ancestry of local haplotypes. The risk associated with overlooking a novel pattern of allele effects is substantial and since we must allow for the possibility of a private allele in any one of the founder strains, a model with 8 degrees of freedom is the minimum required. Nonetheless, as we illustrate in our analysis of the change-in-cholesterol QTL, the pattern of SNPs laid down by the ancestry of the founder strains can provide a remarkable level of resolution. In this example, the QTL is reduced to only seven candidate SNPs. This conclusion is contingent on the assumption that the QTL is due to a single SNP and that we have identified the correct pattern of allelic effects. These assumptions should and can be subject to validation. Obviously, our ability to resolve a QTL to the level of individual SNPs also relies on the availability of complete genomic sequence data of the founder strains (Keane et al. 2011).
We have identified a 2-Mb QTL interval on chromosome 3 containing 11 genes (Figure 6). Two of these genes have known functional relevance to cholesterol. Mgst2 encodes microsomal glutathione S-transferase, an enzyme involved in synthesis of leukotrienes (LT) via the 5-lipoxygenase (5-LO) pathway. Synthesis of LT through this pathway has been shown to be inhibited by cholesterol (Zagryagskaya et al. 2008). Foxo1 encodes forkhead box transcription factor O1 (FoxO1), a key regulator in bile acid biosynthesis and hence likely to affect subsequent catabolism of cholesterol (Li et al. 2009). Li and colleagues observed increased nuclear accumulation of FoxO1 and elevated plasma cholesterol in mice fed a HF diet similar to that used in the present study, suggesting an indirect role for FoxO1 in regulating cholesterol via its effect on bile acid synthesis. A QTL for coincident plasma cholesterol and HDL cholesterol (Chldq2) has been mapped near this chromosome 3 locus (Srivastava et al. 2006; Wergedal et al. 2007), supporting the likelihood that variants within this locus influence plasma cholesterol. Additional DO animals from our ongoing study will provide the opportunity to further refine the QTL location, to investigate potential interactions with diet, and to confirm the allele effects pattern that lead to the identification of candidate SNPs.
HDL cholesterol was among the traits examined in our study. A polymorphism in Apoa2 (chr 1, 173.155926 Mb) has been previously associated with elevated HDL levels in several mouse crosses. Specifically, a C to T transition results in the replacement of alanine with valine in the protein product (Wang et al. 2004). Among the eight CC founder strains, only 129S1/SvImJ (129S1) carries the T allele of this polymorphism. Therefore, we would expect that DO mice with the 129S1 genotype at this locus would have higher HDL levels. The homozygous 129S1 genotype occurred at this locus in only one DO animal from our study. However, we identified 37 animals that were heterozygous for the 129S1 allele. At the marker nearest the Apoa2 polymorphism (UNC010361641, chr 1, 173343908), we performed a Student's t-test and found a significant increase in HDL levels among strains containing the 129S1 allele (P = 0.022, one-sided test, unequal variance). However, due to the small number of homozygote 129S1 individuals in our study sample, this association was not significant in the genome-wide QTL scans. We expect that increased sample size would improve our prospects for rediscovery of this locus. The importance of the Apoa2 locus has been established in the literature. The DO provides an opportunity to discover new genes that affect HDL and numerous other traits that have been studied in classical mouse QTL crosses.
We envision many potential applications for DO mice that do not necessarily entail genetic mapping. Pharmaceutical development comes with a cost of approximately 800 million dollars and as long as 10 years to bring a product to market. Most of these drugs are well tolerated by the majority of patients. However, a small number of patients may experience unpredictable injury while taking a therapeutic dose and this can lead to removal of the drug from the market, thus denying the drug's benefit to the vast majority of patients. It has been hypothesized that genetic causes underlie many of these idiosyncratic adverse drug reactions (Wilke et al. 2007). If this is the case, then perhaps initial screening with a genetically diverse and heterogeneous model, such as DO mice, could allow for more realistic assessments of the risk of adverse effects and provide a more reliable route for generalization of findings from mouse to human populations.
The DO is a newly developed mouse resource that, even in early generations, is proving to be a robust tool for high-resolution mapping of complex traits. At outbreeding generations G4 and G5, using fewer than 150 mice, the mapping resolution was sufficient to identify QTL intervals of 2–8 Mb. We present the mapping of two Mendelian coat color traits to regions including genes known to regulate coat color and a complex metabolic trait to a region in which highly plausible candidate genes reside. These initial results demonstrate the utility of the DO and when used in concert with the Collaborative Cross these new coordinated resources will accelerate our discovery of the genetic basis for disease.
Acknowledgments
We thank Darla Miller for managing the production and transfer of partially inbred CC strains. We are especially grateful to Lisa Somes, Biomedical Technologist, and to the Importation Service at The Jackson Laboratory, for careful and competent management of specific breeding strategies to develop this population in its early phases. We thank Marge Strobel and Adam O'Neill at The Jackson Laboratory for continued dedication to current maintenance of the DO population. We thank Steven Ciciotte for preparing DNA. Finally, we thank our anonymous reviewers for comments that lead to improvements in the content and presentation of the manuscript. This work was supported by The Jackson Laboratory and National Institutes of Health (NIH) grants GM076468 and GM070683 to G.A.C., and by NIH grants R01DA021336, R21DA024845, and R01MH079103 and a Grant from the Schweppe Foundation to A.A.P.
Footnotes
Edited by Lauren M. McIntyre, Dirk-Jan de Koning, and 4 dedicated Associate Editors
Literature Cited
- Cheng R., Lim J. E., Samocha K. E., Sokoloff G., Abney M., et al. , 2010. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185: 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Abney M., Palmer A. A., Skol A., 2011. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19: 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium, 2012 The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401. [DOI] [PMC free article] [PubMed]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F., Koeleman B. P., 2004. Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Am. J. Hum. Genet. 75: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant C., Mott R., 2010. Bayesian quantitative trait locus mapping using inferred haplotypes. Genetics 184: 839–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Hartl D.L, Clark A. G., 2007. Principles of Population Genetics. Sinauer Associates, Sunderland, MA [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic A. B., Svenson K. L., Churchill G. A., Valdar W., 2012. A general Bayesian approach to analyzing diallel crosses of inbred strains. Genetics 190: 413–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Ma H., Park Y. J., Lee Y. K., Strom S., et al. , 2009. Forkhead box transcription factor O1 inhibits cholesterol 7alpha-hydroxylase in human hepatocytes and in high fat diet-fed mice. Biochim. Biophys. Acta 1791: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1997. Genetics and Analysis of Quantitative Traits, Sinauer Associates; Sunderland, MA [Google Scholar]
- Markert C. L., Silvers W. K., 1956. The effects of genotype and cell environment on melanoblast differentiation in the house mouse. Genetics 41: 429–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggert J. K., Svenson K. L., Smith R. V., Paigen B., Peters L. L., 2003 Diet effects on bone mineral density and content, body composition, and plasma glucose, leptin, and insulin levels. MPD:143, Mouse Phenome Database Website, The Jackson Laboratory, Bar Harbor, ME. (http://phenome.jax.org/)
- Paigen B., Svenson K. L., Peters L. L., 2002. Diet effects on plasma lipids and susceptibility to atherosclerosis (pathogen-free conditions). MPD:99, Mouse Phenome Database Website, The Jackson Laboratory, Bar Harbor, ME: (http://phenome.jax.org/) [Google Scholar]
- Peters L. L., Barker J. E., 2001 Hematology, clotting, and thrombosis MPD:62, Mouse Phenome Database Website, The Jackson Laboratory, Bar Harbor ME (http://phenome.jax.org/)
- Peters L. L., Robledo R. F., Bult C. J., Churchill G. A., Paigen B. J., et al. , 2007. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8: 58–69 [DOI] [PubMed] [Google Scholar]
- Russell L. B., Russell W. L., 1948. A study of the physiological genetics of coat color in the mouse by means of the dopa reaction in frozen sections of skin. Genetics 33: 237–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers W. K., 1979. The Coat Colors of Mice. Springer Verlag, New York [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., et al. , 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Mohan S., Masinde G. L., Yu H., Baylink D. J., 2006. Identification of quantitative trait loci that regulate obesity and serum lipid levels in MRL/MpJ × SJL/J inbred mice. J. Lipid Res. 47: 123–133 [DOI] [PubMed] [Google Scholar]
- Stranger B. E., Stahl E. A., Raj T., 2011. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 187: 367–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Wang X., Korstanje R., Higgins D., Paigen B., 2004. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 14: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wergedal J. E., Ackert-Bicknell C. L., Beamer W. G., Mohan S., Baylink D. J., et al. , 2007. Mapping genetic loci that regulate lipid levels in a NZB/B1NJxRF/J intercross and a combined intercross involving NZB/B1NJ, RF/J, MRL/MpJ, and SJL/J mouse strains. J. Lipid Res. 48: 1724–1734 [DOI] [PubMed] [Google Scholar]
- Wilke R. A., Lin D. W., Roden D. M., Watkins P. B., Flockhart D., et al. , 2007. Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat. Rev. Drug Discov. 6: 904–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagryagskaya A. N., Aleksandrov D. A., Pushkareva M. A., Galkina S. I., Grishina Z. V., et al. , 2008. Biosynthesis of leukotriene B4 in human polymorphonuclear leukocytes: regulation by cholesterol and other lipids. J. Immunotoxicol. 5: 347–352 [DOI] [PubMed] [Google Scholar]