Abstract
Dosage-sensitive modifier screening is a powerful tool for linking genes to biological processes. Use of chromosomal deletions permits sampling the effects of removing groups of genes related by position on the chromosome. Here, we explore the use of inducible microRNA transgenes as a complement to deficiency-based modifier screens. miRNAs are predicted to have hundreds of targets. miRNA overexpression provides an efficient means to reduces expression of large gene sets. A collection of transgenes was prepared to allow overexpression of 89 miRNAs or miRNA clusters. These transgenes and a set of genomic deficiencies were screened for their ability to modify the bristle phenotype of the cell-cycle regulator minus. Sixteen miRNAs were identified as dominant suppressors, while the deficiency screen uncovered four genomic regions that contain a dominant suppressor. Comparing the genes uncovered by the deletions with predicted miRNA targets uncovered a small set of candidate suppressors. Two candidates were identified as suppressors of the minus phenotype, Cullin-4 and CG5199/Cut8. Additionally, we show that Cullin-4 acts through its substrate receptor Cdt2 to suppress the minus phenotype. We suggest that inducible microRNA transgenes are a useful complement to deficiency-based modifier screens.
GENETIC modifier screens have proven to be a powerful means with which to identify genes in a common biological process. Modifier screens can be carried out with high efficiency by chemical mutagenesis, but recombination mapping to identify the affected loci can be tedious. In Drosophila, a popular alternative has been to use a collection of deletion mutant strains known as the “deficiency kit” (St Johnston 2002), which consists of mapped chromosomal deletions that remove large blocks of genes. This approach allows sampling of the effects of removing one copy each of hundreds of genes in a single genetic cross. The deficiency kit currently available from the Bloomington Drosophila Stock Center covers 97.8% of annotated euchromatic genes (Cook et al. 2010). The advantage of the approach is that screens for autosomal modifiers can be performed with fewer than 400 crosses. An offsetting disadvantage is that genetic heterogeneity in these strains can affect the results.
In this context we decided to explore the possibility of using microRNAs (miRNAs) as an alternative means of downregulating hundreds of genes concurrently. miRNAs are small noncoding RNAs that post-transcriptionally silence gene expression. Target prediction algorithms suggest the existence of hundreds of targets per miRNA (reviewed in Bartel 2009; Thomas et al. 2010), recently including sites in protein-coding regions of the genes (Schnall-Levin et al. 2010). miRNA overexpression can cause simultaneous reduction of the expression levels of hundreds of genes (Lim et al. 2005; Easow et al. 2007; Baek et al. 2008; Selbach et al. 2008). Potential advantages of the use of miRNAs include the following:
miRNA targets may allow access to genes not covered by the deficiency kit, as well as facilitating access to genes on the X chromosome. There are 176 annotated Drosophila miRNAs (Kozomara and Griffiths-Jones 2011).
Overexpression studies can be conducted in a spatiotemporally controlled manner through the use of the GAL4–UAS system (Brand and Perrimon 1993). Potential drawbacks of the miRNA approach include: (a) miRNAs do not target all protein coding genes and (b) miRNAs do not regulate those they target with equal efficiency.
Identification of biologically significant targets is imperfect due to limitations in target prediction algorithms.
To explore the potential of the miRNA-based screening approach we made use of the minus mutant, which exhibits a “small bristle” phenotype that is sensitive to genetic background (Szuplewski et al. 2009). Drosophila adult mechanosensory bristles have been used as a model system to study cell determination and asymmetric cell division (Abdelilah-Seyfried et al. 2000; Mummery-Widmer et al. 2009). Each bristle is composed of four cells: the neuron, its sheath cell, and two external cells forming the shaft and socket. Growth of the outer cells requires endoreplication, a variant cell cycle in which cells become polyploid through DNA replication without subsequent cell division (Weng et al. 2003). Therefore, bristles provide a model to study endoreplication. We reported previously that Minus influences cyclin E degradation and that reduction of Cyclin E gene dosage could rescue the minus mutant bristle endoreplication defect (Szuplewski et al. 2009). This phenotype does not affect viability or fertility of the fly, so minus mutants provide a suitable genetic background for modifier screens to identify regulators of the endocycle.
Material and Methods
Molecular biology
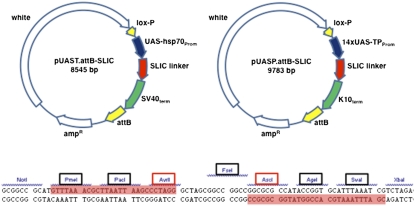
pUAST.attB-SLIC (sequence and ligation-independent cloning) and pUASP.attB-SLIC were produced as follows: Two synthetic oligonucleotides (SLIC-fw, 5′-GCGGCCGCATGTTTAAACGCTTAATTAAGCCCTAGGGCTAGCGGCCGGCCGGCGCGCCATACCGGTGCATTTAAATCGTCTAGA-3′, and SLIC-rv, 5′- TCTAGACGATTTAAATGCACCGGTATGGCGCGCCGGCCGGCCGCTAGCCCTAGGGCTTAATTAAGCGTTTAAACATGCGGCCGC-3′) were annealed, treated with Taq polymerase for 15 min at 72°, and cloned into PCR2.1-TOPO. The insert (SLIC-linker) was excised using NotI/XbaI and ligated into NotI/XbaI-digested pUASTattB (Bischof et al. 2007) to generate pUAST.attB-SLIC and into NotI/XbaI-digested pUASP2 (Rorth 1998) to generate pUASP-SLIC. The pUASP-cassette was PCR amplified from pUASP-SLIC (forward primer, 5′−CTAGCGGATCCGGGAATTGGGAATTCTAGAATTGGCCGCTCTAGCC-3′; reverse primer, 5′-AGTGGATCTCTAGAGGTACCCTCGAGTCCGTGGGGTTTGAATTAAC-3′) and inserted into EcoRI/XhoI-digested pattB using the SLIC to generate pUASP.attB-SLIC (Bischof et al. 2007; Li and Elledge 2007). To generate UAS–microRNA overexpression vectors, genomic fragments containing the miRNA stem loop plus ∼100 nucleotides upstream and downstream were PCR amplified using primers with extensions homologous to the SLIC-linker sequence (extension of the forward primer, 5′GTTTAAACGCTTAATTAAGCCCTAGG-3′; extension of the reverse primer, 5′- CGATTTAAATGCACCGGTATGGCGC GCC-3′) and subsequently SLIC recombined (Li and Elledge 2007) into AvrII/AscI-digested pUAST.attB-SLIC and pUASP.attB-SLIC to generate RT–miR and RP–miR transgenes, respectively. Primer sequences are available upon request. The subset of miRNAs chosen for the miRNA overexpression library is in Supporting Information, Table S1. This collection was completed by cloning some additional miRNAs in pUAS-T-DSred2 for random insertion into fly genome (Table S1; Stark et al. 2003).
The Cul-4 3′-UTR luciferase reporter was generated by cloning the 551-bp 3′-UTR from Cul-4 into a vector directing firefly luciferase under control of the tubulin promoter. The Cul-4 3′-UTR reporter with miR-5 sites mutated was generated by PCR amplification using primers designed to incorporate the mutations. miRNA overexpression vectors for S2 cell transfections were generated by cloning genomic DNA fragments containing the miR-5 hairpin into pJB25 (tub > MCS_SV40 polyA) (Stark et al. 2005) to generate pJB25-miR-5. The UAS–Cul-4 transgene lacking the 3′-UTR binding site was generated by PCR amplification using the following primers: aaaaagcggccgcGGCGTCGAAGGAACCCCA and tttggtaccTCGGTGTTCGGATTTCAC.
Drosophila stocks and genetics
The following stocks and all the deficiencies were obtained from Bloomington Stock Center: CycEAr95, UAS–GFP, sca–GAL4, pnr–GAL4, Cul-4KG02900 (Table 1 and Table S2). UAS–RNAi-Rac2 (construct 17536), UAS–RNAi-CG5199 and UAS–RNAi-minus lines were from the Vienna Drosophila RNAi center. Cul411L, Cul46AP were provided by R. J. Duronio (Hu et al. 2008) and Cul4G1-3, Cul4L2-1, Cul4G3-5 by C.-T. Chien (Lin et al. 2009). l(2)dtlc02261 was from the Exelixis Collection at Harvard Medical School. 11295R-1 and 11295R-4 were from NIG–FLY. Transgenic strains were prepared carrying RT–miR and RP–miR constructs integrated into defined landing-sites (see Table S2 for insertion sites), all obtained from the Bloomington Stock Center. To perform the screen, at least 5 males from each UAS–miRNA line were crossed to at least 10 sca–GAL4, UAS–RNAi-mi/CyO virgin females. All crosses were performed at 25°. Phenotypes were screened in female flies, which are larger and have longer bristles. A deficiency or miRNA overexpression transgene was scored as a suppressor when at least 50% of the progeny had obviously wild-type or near wild-type bristles.
Table 1. Deletions used to locate dominant suppressor of minus loci.
| Region | Deficiency | Breakpoints | Suppression |
|---|---|---|---|
| 35D1; 35E2 | Df(2L)TE35BC-24 | 35B4; 35E2 | Su(mi) |
| Df(2L)r10 | 35D1; 36A6–7 | Su(mi) | |
| 44A4; 44C1 | Df(2R)cn9 | 42E; 44C1 | No suppression |
| Df(2R)H3C1 | 43F; 44D8 | Su(mi) | |
| Df(2R)H3E1 | 44D1; 44F12 | No suppression | |
| Df(2R)BSC267 | 44A4; 44C4 | Su(mi) | |
| 59D11; 59F8 | Df(2R)vir130 | 59B; 59D8–E1 | No suppression |
| Df(2R)or-BR6 | 59D5; 60B8 | Su(mi) | |
| Df(2R)Chig230 | 60A3–60A7; 60B4 | No suppression | |
| Df(2R)bw-HB132 | 59D11; 59F8 | Su(mi) | |
| 77C3; 77D1 | Df(3L)ED4858 | 76D3; 77C1 | No suppression |
| Df(3L)rdgC-co2 | 77A1; 77D1 | Su(mi) | |
| Df(3L)ri-79c | 77B–C; 77F–78A | No suppression | |
| Df(3L)BSC796 | 77C3; 77E4 | Su(mi) |
Quantitative PCR (qPCR)
Ten pairs of salivary glands were dissected from prewandering third instar larvae expressing either an UAS–GFP or an UAS–miR-4-5 transgene driven by patched-Gal4 (ptc-Gal4). RNA was isolated using TRIZOL reagent (Invitrogen) and DNaseI treated (Promega) using manufacturer’s recommendations. After quantification, 800 ng total RNA per sample was reverse transcribed using Superscript III reverse transcriptase (Invitrogen). qPCR was performed on two biological replicates with three independent primer pairs covering different exon junctions in cullin-4 mRNA and RP49-specific primers for normalization.
Cell transfection and luciferase assays
S2 cells were transfected in 24-well plates with 200 ng empty pJB25 or pJB25-miR-5 vector, 80 ng firefly luciferase with or without intact or mutant Cul-4 3′−UTR, and 80 ng Renilla luciferase DNA as transfection control. Dual luciferase assays were performed 60 hr post-transfection with the Dual-Glo luciferase kit (Promega).
Results
Generation of UAS–miRNA strains
To generate a library of fly strains for conditional overexpression of microRNAs under GAL4 control, we assembled two vectors suitable for recombination-based SLIC cloning (Li and Elledge 2007): pUAST.attB-SLIC and pUASP.attB-SLIC (Figure 1). Both vectors allow site-specific integration into the genome (Bischof et al. 2007) and contain a common SLIC polylinker for recombination-based cloning. In pUAST.attB-SLIC, the polylinker is flanked by the hsp70 promoter and the SV40 3′-UTR. Because pUAST-based constructs express poorly in the germ line, pUASP.attB-SLIC was based on the germ line-competent pUASP vector (Rorth 1998). Genomic fragments spanning miRNA hairpins by ∼100 bp upstream and downstream were amplified and cloned into the UAS vectors to generate RT–miR and RP–miR plasmids (Table S1). The UAS–miRNA collection includes several lines described previously and others cloned into pUAST-DsRed2 (Stark et al. 2003) for random insertion into the genome. In total 109 miRNAs were overexpressed (Table S1).
Figure 1 .
The miRNA overexpression vectors. Schematic representation of pUAST.attB-SLIC for somatic miRNA overexpression and pUASP.attB-SLIC optimized for germ line miRNA overexpression. The sequence of the SLIC linker is indicated below. All restriction sites of the SLIC linker are unique in pUAST.attB-SLIC. Unique sites for both vectors are boxed. When AvrII and AscI are used to linearize the vectors for SLIC cloning (Li and Elledge 2007), the sequences marked in red should be added to the forward (AvrII) and reverse (AscI) primers.
Characterization of a sensitive minus background
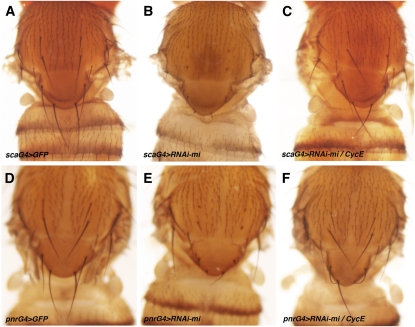
minus mutants show a reduction in the size of the large thoracic mechanosensory bristles (Szuplewski et al. 2009). Cyclin E (CycE) mutants behave as dominant suppressors of the minus bristle phenotype in that they restore bristle size to normal in a minus mutant background. We designed a genetic modifier screen to identify suppressors with the aim of uncovering regulators of endoreplication. To simplify the screen, we sought to reduce minus mRNA levels by expression of a UAS–RNAi transgene. This would allow a F1 screens for the effects of coexpressing UAS–miRNA transgenes or introduction of a genomic deficiency. Expression of UAS–RNAi-minus under control of scabrous-GAL4 (scaG4) or pannier-GAL4 (pnrG4) (Klaes et al. 1994; Calleja et al. 1996) recapitulated the minus mutant phenotype (Figure 2, A, B, D, and E). In both combinations, removing one copy of CycE suppressed the RNAi phenotype, as observed previously with the minus mutant (Compare Figure 2, B, C, E, and F). On this basis, we chose the scaG4 > UAS–RNAi-mi background for an F1 miRNA modifier screen.
Figure 2 .
minus RNAi phenotype sensitive to CycE dosage. Images of the dorsal thorax of adult female flies of the indicated genotypes. (A) sca–GAL4/UAS–GFP, scaG4 control. (B) sca–GAL4, UAS–RNAi-mi/+ caused a size reduction of thoracic macrochaete compared to control. (C) sca–GAL4, UAS–RNAi-mi/CycEAr95: reduction of cyclin E gene dosage rescued the RNAi phenotype. (D) pnrG4 control: UAS–GFP/+; pnr–GAL4/+ (E) pnr–GAL4: UAS–RNAi-mi/+ caused a size reduction of thoracic macrochaete compared to control. (F) CycEAr95/+; pnr–GAL4, UAS–RNAi-mi/+: reduction of cyclin E rescued the phenotype.
Results of the screens
We first screened 111 deletions on the second chromosome and 139 deletions on the third chromosome (Table S2). Four chromosomal regions that suppressed the minus bristle phenotype were uncovered. Table 1 shows the deletions that were used to narrow down the locations of the suppressor loci. On the second chromosome two newly identified loci were located in the intervals 44A4–44C1 and 59D11–59F8, in addition to the region where the previously identified suppressor, CycE, is located (35D1–35E2). One new suppressor was identified in the interval 77C3–77D1 on the third chromosome.
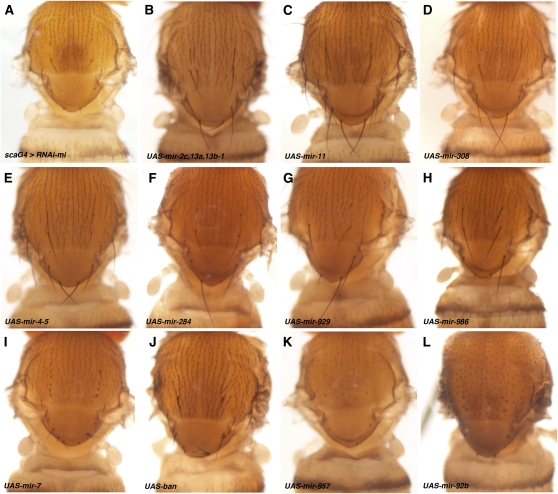
The UAS–miRNA screen identified 13 transgenes that suppressed the minus phenotype (Table 2 and Table S1). Members of the miR-2 seed family figure prominently in this group. The miR-2 family consists of 8 genes at six loci (Table 2). All six loci proved to be effective suppressors of minus when the miRNAs were overexpressed (Table 2 and Figure 3, A–D). Overexpression of the miR-4-5 cluster, miR-284, miR-929, or miR-986 also suppressed the minus phenotype (Figure 3, E–H).
Table 2. List of miRNAs whose overexpression suppressed minus bristle phenotype.
| Su(mi) | Cytogenetic location |
|---|---|
| miR-2a-1, miR-2a-2, miR-2b-2 | 37F2 |
| miR-2b-2 | 37F2 |
| miR-2c, miR-13a, miR-13b-1 | 88F4 |
| miR-6-1, miR-6-2, miR-6-3 | 56E1 |
| miR-11 | 93E9 |
| miR-308 | 50E4 |
| miR-4, miR-5 | 56E1 |
| miR-284 | 87C1 |
| miR-929 | 82A1 |
| miR-986 | 44D1 |
Figure 3 .
Examples of phenotypes observed in the screen. Images of the dorsal thorax of adult female flies of the indicated genotypes. (A) sca–GAL4, UAS–RNAi-mi/+ phenotype shown for comparison. (B) UAS–mir-2c,13a,13b/+; sca–GAL4, UAS–RNAi-mi/+. (C) sca–GAL4, UAS–RNAi-mi/+; UAS–mir-11/+ (note the extra macrochaete on the notum for B and C). (D) sca–GAL4, UAS–RNAi–mi/UAS–mir-308 (note the extra scutellar bristles in B–E). (E) sca–GAL4, UAS–RNAi-mi/UAS–miR-4, 5. (F) sca–GAL4, UAS–RNAi-mi/+; UAS–mir-284/+. (G) sca–GAL4, UAS–RNAi-mi/+; UAS–mir-929/+. (H) sca–GAL4, UAS–RNAi-mi/+; UAS–mir-986/+. (I and J) Absence of suppression was oberved following miR-7 or bantam overexpressison: (I) UAS–mir-7/+; sca–GAL4, UAS–RNAi-mi/+. (J) sca–GAL4, UAS–RNAi-mi/+; UAS–ban/+. (K) Absence of suppression and loss of bristles, sometimes associated with an absence of sockets, was observed following miR-957 overexpression: sca–GAL4, UAS–RNAi-mi/UAS–mir-957. (L) Enlarged shaft and bristle cells of macro- and microchaete were caused by miR-92b overexpressison. Genotype: sca–GAL4, UAS–RNAi-mi/+; UAS–mir-92b/+.
In addition to modifiers of the UAS–RNAi-mi phenotype this screen revealed bristle phenotypes associated with miRNA overexpression that were unrelated to the sensitized background (Table S1). Overexpression of miR-2 family miRNAs resulted in occasional additional bristles, as reported previously (Lai et al. 2005). This phenotype was suggested to reflect regulation of Notch activity and asymmetric division of the progenitors, a process distinct from endoreplication. Similarly, overexpression of miR-7 and bantam have been previously reported to result in additional bristles (Abdelilah-Seyfried et al. 2000; Lai et al. 2005). We observed additional bristles when miR-7 and bantam were coexpressed with UAS–RNAi-mi, but all the bristles had the small size caused by depletion of minus, suggesting that there was no suppression of the minus phenotype by these two miRNAs (Figure 3, I and J). We also observed cases in which miRNA overexpression caused bristle loss, which precluded analysis of the minus phenotype. miR-957, miR-92a, or miR-92b appeared to enhance the UAS–RNAi-mi phenotype (Figure 3, K and L); however, bristle loss was observed when these miRNAs were expressed alone under scaG4 control. miR-9a overexpression also results in a loss of scutellar bristles when expressed under apterous-GAL4 control (Li et al. 2006). Similarly, a number of UAS–miRNA transgenes were lethal when coexpressed with the UAS–RNAi-mi transgene under scaG4 control, precluding analysis of their potential as modifiers in this screen (Table S1).
Comparison of the two screens
The screens each identified many interaction candidates: genes located in a deletion and those predicted to be miRNA targets (Table S3). The Microcosm (Griffiths-Jones et al. 2008) and TargetScan (Ruby et al. 2007) databases were searched for predicted targets of the miRNAs of interest (Table 2). Genes uncovered by the relevant deletions and predicted as miRNA targets are listed in Table 3. This approach identified seven candidate genes for the interval 35D1–35E2, eight candidates for 44A4–44C1, nine candidates for 59D11–59F8, and only four candidates for 77C3–77D1 (Table S4). Overlapping the putative miRNA targets therefore reduced the number of candidates to 10–15% of the number identified on the basis of the overlapping deficiency intervals alone (Table 3).
Table 3. Genes in the four genomic regions that are targeted by a suppressor miRNA.
| Interval | Deficiency | No. genes deleted | miR targets in overlap | % target/overlap |
|---|---|---|---|---|
| 35D1–35E2 | Df(2L)TE35BC-24 | 127 | ||
| Df(2L)r10 | 122 | |||
| Overlap | 68 | 7 | 10.29 | |
| 44A4–44C1 | Df(2R)H3C1 | 136 | ||
| Df(2R)BSC267 | 53 | |||
| Overlap | 53 | 8 | 15.09 | |
| 59D11–59F8 | Df(2R)or-BR6 | 187 | ||
| Df(2R)bw-HB132 | 65 | |||
| Overlap | 65 | 9 | 13.85 | |
| 77C3–77D1 | Df(3L)rdgC-co2 | 87 | ||
| Df(3L)BSC796 | 35 | |||
| Overlap | 31 | 4 | 12.90 | |
| Total candidates | (Overlap) | 217 | 28 | 12.90 |
The interval 35D1–35E2 contains Cyclin E, an already known suppressor of minus; therefore we focused on the other three regions. We noted, however, that none of the miRNAs identified as suppressors are predicted to target Cyclin E.
44A4–44C1: Identification of miR-5 target Cul-4 as a suppressor of minus
Most of the deficiencies identified as suppressors in our initial screen are large and have ill-defined endpoints. Molecularly characterized deficiencies that allow more precise definition of the intervals containing candidate suppressors are now available (Cook et al. 2010). In the 44A–C region, the small deficiency Df(2R)Exel7094 suppressed the UAS–RNAi-mi phenotype. This deletion uncovers 20 genes, of which 4 are predicted suppressor miRNA targets (Table S4): CG8713 (miR-5), CG11210 (miR-2 family), SOCS44A (miR-4), and Cullin-4 (miR-5).
In light of its role in protein turnover, Cullin-4 (Cul-4) stood out as an obvious candidate for further analysis. Cul-4 encodes a scaffold protein, which is a member of the CRL-4 family of Cullin-4/RING E3 ubiquitin ligases (Jackson and Xiong 2009). The CRL-4 complex consists of a Cullin-4 scaffold, a RING protein catalyzing target ubiquitination, the linker protein DDB1, and a substrate receptor that recruits the target of the ubiquitin ligase complex. Cul-4 is an attractive candidate to be a suppressor of minus because E3 ubiquitin ligases based on this scaffold protein are required for degradation of cell-cycle regulators (Abbas and Dutta 2011; Havens and Walter 2011).
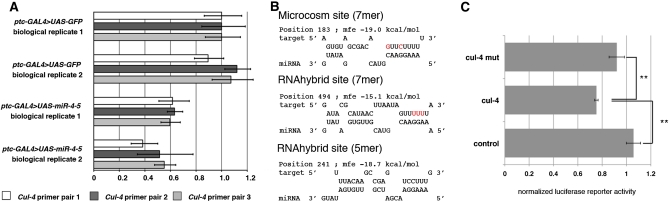
To assess whether Cul-4 might be the miR-5 target that interacts with minus, we asked whether miR-5 overexpression could regulate Cul-4 in vivo. miRNAs often reduce the transcript level of their targets. For this experiment we made use of larval salivary glands, which like the bristles use endoreplication to increase cell growth. Overexpression of miR-4-5 in salivary glands under ptc-Gal4 control led to a reduction of Cul-4 mRNA level, measured by quantitative RT–PCR (Figure 4A), suggesting that miR-5 can regulate Cul-4.
Figure 4 .
Cul-4 is a miR-5 target. (A) Cul-4 RNA levels measured using three sets of Cul-4 primer pairs. RNA was extracted from salivary glands expressing GFP (ptc-GAL4 > UAS–GFP) or miR-4-5 cluster (ptc-GAL4 > UAS–miR-4-5). Data represent mean ±SD for three technical replicates. Results of two independent experiments are shown (1 and 2). (B) Predicted miR-5 target sites in the Cul-4 3′−UTR. Minimal free energy (mfe) is calculated by RNAhybrid. Nucleotides changed to generate the target site mutant UTR are in red. The predicted RNAhybrid 5mer shown is the example with the highest mfe in this category. (C) Luciferase assays showing regulation of a Cul-4 3′-UTR reporter. Luciferase reporter activity is normalized to the Renilla transfection control and to empty miRNA overexpression vector control. Data represent mean ±SD. (**) P < 0.01 (Student's t-tests).
To explore this relationship further, we examined the regulation of Cul-4 mRNA by miR-5 in cell-based assays. The Microcosm algorithm predicts one miR-5 site in the Cul-4 3′−UTR (Figure 4B). Further analysis using RNAhybrid (Kruger and Rehmsmeier 2006) revealed another putative miR-5 site featuring a 7mer seed match and four sites with 5mer seed matches but extensive 3′ pairing (Figure 4B). To test whether Cul-4 is a direct miR-5 target, we transfected S2 cells with luciferase reporter constructs that carried the intact Cul-4 3′-UTR or a version with mutations in the two 7mer seed sites. Coexpression with miR-5 reduced Cul-4 3′-UTR reporter activity to ∼70% of control levels, indicating that miR-5 can downregulate Cul-4 (Figure 4C). The mutant version of the Cul-4 3′-UTR reporter was less susceptible to downregulation by miR-5. The difference between the degree of downregulation of intact and site mutant UTRs was statistically significant (P < 0.01), indicating that the predicted target sites contribute to regulation of Cul-4 by miR-5. The remaining capacity of miR-5 to act on the double site mutant UTR reporter likely reflects the presence of four additional weaker sites. Together these data provide evidence that Cul-4 mRNA can be directly targeted by miR-5.
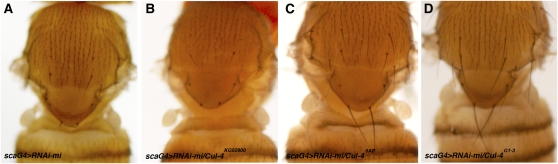
Identification of Cul-4 as a miR-5 target prompted us to ask whether reducing Cul-4 levels by an independent genetic means could suppress the minus phenotype. Cul-4 mutant alleles were introduced into the scaG4 > UAS–RNAi-mi background. Introducing the hypomorphic allele Cul-4KG02900 had little or no effect on the severity of the scaG4 > UAS–RNAi-mi phenotype (Figure 5, A and B). However, Cul-4 null alleles proved to be effective (Figure 5, C and D). The Cul-46AP and Cul-4G1-3 alleles were produced by imprecise excision of independent P element insertions so their genetic background is different (Hu et al. 2008; Lin et al. 2009). Both Cul-4 null alleles also behaved as suppressors of the pnrG4 > RNAi-mi sensitized background (Figure S1). No suppression was observed for an RNAi bristle loss phenotype on the basis of a different transgene (Figure S1), suggesting that the effect is specific for the minus RNAi phenotype.
Figure 5 .
Cul-4 dosage reduction suppresses minus bristle phenotype. Images of the dorsal thorax of adult female flies of the indicated genotypes. (A) sca–GAL4, UAS–RNAi-mi/+. (B) sca–GAL4, UAS–RNAi-mi/Cul-4KG02900. (C) sca–GAL4, UAS–RNAi-mi/Cul-46AP. (D) sca–GAL4, UAS–RNAi-mi/Cul-4G1-3. The minus mutant bristle phenotype was not suppressed by the hypomorphic allele Cul-4KG02900 but was suppressed by two independent Cul-4 null alleles.
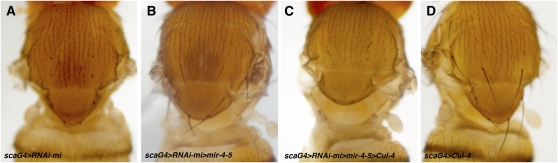
To further evaluate the specificity of Cul-4 as the mediator of the interaction between minus and miR-5 we asked whether restoring Cul-4 expression could offset the suppressive effects of miR-5. To enable this test to be done, we generated a UAS–Cul-4 transgene in which the endogenous 3′−UTR was replaced by the SV40 3′-UTR. Use of this transgene ensured that Cul-4 could be coexpressed with miR-5, without being subject to downregulation by miR-5. It also alleviates the concern that overexpressing a target would serve as a miRNA sponge sequestering the miRNA and thereby reducing its effectiveness for reasons independent of Cul-4 protein coding sequence. The modified UAS–Cul-4 transgene was introduced into the scaG4 > UAS–RNAi-mi; UAS–miR-4-5 background and was found to abrogate the suppressive effect of UAS–miR-4-5 on scaG4 > UAS–RNAi-mi (Figure 6, A–C). Four independent UAS–Cul-4 insertions produced equivalent results, arguing against an effect of integration site. Expression of the UAS–Cul-4 transgene on its own had little or no effect on bristle size (Figure 6D). These results provide evidence that downregulation of Cul-4 is a major contributor to the ability of miR-5 to suppress the minus phenotype and identify Cul-4 as a bona-fide modifier of the minus bristle phenotype.
Figure 6 .
Cul-4 downregulation is responsible for the Suppressor of minus behavior caused by miR-5 overexpression. Images of the dorsal thorax of adult female flies of the indicated genotypes. (A) sca–GAL4, UAS–RNAi-mi/+, and (B) sca–GAL4, UAS–RNAi-mi/UAS–miR-4-5, show the minus RNAi phenotype and the suppressed phenotype as controls for C. (C) sca–GAL4, UAS–RNAi-mi/UAS–miR-4-5; UAS–Cul-4/+. Coexpression of Cul-4 and the miR-4-5 cluster in the sca–UAS–RNAi-mi sensitized background restored the mutant phenotype. (D) sca–GAL4/+; UAS–Cul-4/+. Expression of Cul-4 alone with sca–GAL4 caused only a slight reduction of some bristles.
59D11–59F8: Reduced l(2)dtl dosage suppresses the minus bristle phenotype
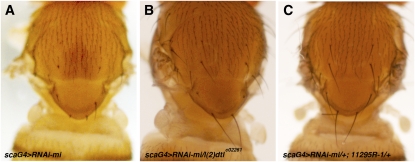
A subset of CRL-4 family E3 ligases recognize their targets through the substrate receptor Cdt2 and these CRL-4–Cdt2 complexes seem to function exclusively in S phase of the cell cycle (Abbas and Dutta 2011; Havens and Walter 2011). Interestingly, the Drosophila Cdt2 ortholog l(2)dtl is located in the genetic interval 59D11–59F8, which includes a suppressor of minus (Table 1). We therefore asked whether l(2)dtl behaves as suppressor of minus. The l(2)dtlc02261 allele contains a piggyBac transposon inserted in l(2)dtl coding sequence, and may be a functional null allele (Thibault et al. 2004). Introducing one copy of l(2)dtlc02261 suppressed the bristle phenotypes of scaG4 > UAS–RNAi-mi (Figure 7, A and B) and pnrG4 > UAS–RNAi-mi (Figure S1, A and C). In addition, coexpression with a UAS–RNAi transgene targeting l(2)dtl suppressed the scaG4 > UAS–RNAi-mi (Figure 7C) and pnrG4 > UAS–RNAi-mi (Figure S1, A and C) phenotypes. The control for effects on the RNAi machinery again proved negative (Figure S1, G and H). Thus, reduction of CRL4-Cdt2 activity specifically suppressed the minus phenotype.
Figure 7 .
l(2)dtl dosage reduction. Images of the dorsal thorax of adult female flies of the indicated genotypes. (A) sca–GAL4, UAS–RNAi-mi/+; shows the minus RNAi phenotype as a control for the suppression shown by reduction of l(2)dtl in B. (B) sca–GAL4, UAS–RNAi-mi/ l(2)dtlc02261. (C) sca–GAL4, UAS–RNAi-mi/11295R-1 shows the effect of depleting l(2)dtl by RNAi.
77C3-77D1: miR-284 target CG5199/Cut8 is a suppressor of minus
The genomic region defined by the overlap of the deficiencies Df(3L)rdgC-co2 and Df(3L)BSC796 contains 31 genes, of which four are predicted targets of the suppressor miRNAs: CG5199 and CG5969 by miR-284, CG13250 by the miR-2 family, CG5104 by miR-4 (Table 3 and Table S4). CG5199 is related to fission yeast Cut8 (Takeda and Yanagida 2005). In yeast, Cut8 is conditionally required for destruction of a mitotic cyclin (Cdc130) and in both organisms mediates localization of the proteasome to the nuclear periphery (Tatebe and Yanagida 2000). As minus was demonstrated to influence cyclin E degradation (Szuplewski et al. 2009) and as the identification of the CRL4–Cdt2 implicates proteasomal degradation as relevant to minus function, we asked whether CG5199 was the relevant suppressor in 77C3–77D1. Coexpression with a UAS–RNAi transgene targeting CG5199, P{GD11574}v27372 suppressed the scaG4 > UAS–RNAi-mi phenotype, suggesting that CG5199 is the relevant suppressor within the 77C3–77D1 genomic region. No additional CG5199 alleles or RNAi lines are available, so we could not evaluate this interaction further.
Discussion
microRNA overexpression screens for genetic modifiers
This study illustrates the potential of a genetic modifier screen based on inducible miRNA-expressing transgenes. The utility of this approach lies in the fact that there are many predicted miRNA targets, so the probability of intersection between the target list of any miRNA and the list of genes uncovered by any deletion is quite high, approaching 50%. Most of the genes that are uncovered by an interacting deletion are unlikely to be responsible for the interaction phenotype. Similarly, most of the predicted targets of a miRNA are unlikely to be causally linked to the genetic interaction with the miRNA. This study identified four deletion intervals and six miRNA families that interacted with minus. Use of the predicted targets of these six miRNAs narrowed down the number of candidate genes to 10–15% of the total number of genes in the four deletion intervals to produce a short list for further analysis (Table 3). Together with CycE, which was known previously to interact with minus, the predicted miRNA targets Cul-4, and very likely CG5199/Cut8, each account for a dominant suppressor locus in two of the four chromosomal regions identified by the deficiency interaction screen. Similarity in function to these, two allowed for identification of l(2)dtl as an interacting locus in the fourth deficiency, although l(2)dtl was not a predicted miRNA target.
A limitation inherent in the miRNA overexpression approach is its dependence on miRNA target prediction. Current algorithms differ in their specificity and sensitivity and often show limited overlap in the targets they predict (See Table S3; Thomas et al. 2010). Improvements in these methods should enhance the utility of the miRNA-based screening approach in future. A second inherent limitation to this approach is that miRNAs do not target all genes in the genome, nor do they act with comparable efficiency on those they do target. A third limitation is that some miRNAs produce phenotypes that may preclude their use in a particular screen. We have highlighted a few examples for screens involving bristle size and morphology. In the most extreme form, this limitation is exemplified by miRNAs that were lethal when overexpressed. This problem might be solved by using the temperature-sensitive inhibitor of GAL4 (McGuire et al. 2003), Gal80ts, in combination with a suitable Gal4 driver to control when and where the transgenes are expressed.
Reduction of CRL4–Cdt2 activity suppresses the minus mutant phenotype
The finding that reduction of CRL4–Cdt2 activity suppresses the minus mutant phenotype suggests a contribution of elevated of E3 ubiquitin ligase activity. Three substrates for CRL4–Cdt2 have ben identified in Drosophila: the cyclin-dependent kinase inhibitor Dacapo (Dap, the ortholog of mammalian p21), the Drosophila orthologs of Cdt1 replication factor, Double parked (Dup), and the Drosophila E2F oncogene ortholog (Higa et al. 2006a,b; Shibutani et al. 2008; Lee et al. 2010). All three are cell-cycle regulators involved in G1 or S phase that possess a PCNA-interacting polypeptide box degron required for their CRL4–Cdt2-dependent proteasomal degradation. Therefore, it seems reasonable to assume that reduced CRL4–Cdt2 activity leads to elevated levels of Dap, Dup, and E2F. However, introduction of mutant alleles of dap, dup, or E2F did not modify the bristle phenotype in the l(2)dtl / sca–GAL4 > UAS–RNAi-mi background (data not shown). This suggests that excessive downregulation of these genes is unlikely to explain the effects of elevated CRL4–Cdt2 activity in the minus mutant. However, the possibility exists that concurrent downregulation of all three might produce a different experimental outcome than from removing one at a time. An alternative hypothesis is that CRL4–Cdt2 may act via another, yet to be identified, substrate in this context.
Acknowledgments
We thank Gregory Somers, Daniel Kirilly, Fengwei Yu, and William Chia for the deficiency collection covering the second and the third chromosomes. We thank Cheng-Tin Chien, Robert Duronio, Eric Lai, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, the NIG-FLY, and the Exelixis Collection at Harvard Medical School for fly strains, and Kah Junn Tan for technical support.
Footnotes
Communicating editor: N. Perrimon
Literature Cited
- Abbas T., Dutta A., 2011. Crl4cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 10: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelilah-Seyfried S., Chan Y. M., Zeng C., Justice N. J., Younger-Shepherd S., et al. , 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F. D., Gygi S. P., et al. , 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P., 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific Phic31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Calleja M., Moreno E., Pelaz S., Morata G., 1996. Visualization of gene expression in living adult Drosophila. Science 274: 252–255 [DOI] [PubMed] [Google Scholar]
- Cook K. R., Parks A. L., Jacobus L. M., Kaufman T. C., Matthews K. A., 2010. New research resources at the Bloomington Drosophila Stock Center. Fly (Austin) 4: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G., Teleman A. A., Cohen S. M., 2007. Isolation Of microRNA targets by miRNP immunopurification. RNA 13: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H. K., Van Dongen S., Enright A. J., 2008. Mirbase: tools for microRNA genomics. Nucleic Acids Res. 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens C. G., Walter J. C., 2011. Mechanism Of Crl4cdt2, a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 25: 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa L. A., Banks D., Wu M., Kobayashi R., Sun H., et al. , 2006a. L2dtl/Cdt2 interacts with the Cul4/Ddb1 complex and PCNA and regulates Cdt1 proteolysis in response to DNA damage. Cell Cycle 5: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Higa L. A., Yang X., Zheng J., Banks D., Wu M., et al. , 2006b. Involvement of Cul4 ubiquitin E3 ligases in regulating Cdk inhibitors dacapo/P27kip1 and cyclin E degradation. Cell Cycle 5: 71–77 [DOI] [PubMed] [Google Scholar]
- Hu J., Zacharek S., He Y. J., Lee H., Shumway S., et al. , 2008. Wd40 protein Fbw5 promotes ubiquitination of tumor suppressor Tsc2 by Ddb1-Cul4-Roc1 ligase. Genes Dev. 22: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S., Xiong Y., 2009. Crl4s: the Cul4-ring E3 ubiquitin ligases. Trends Biochem. Sci. 34: 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaes A., Menne T., Stollewerk A., Scholz H., Klambt C., 1994. The ETS transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell 78: 149–160 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S., 2011. Mirbase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J., Rehmsmeier M., 2006. Rnahybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34: W451–W454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., Tam B., Rubin G. M., 2005. Pervasive regulation of Drosophila Notch target genes by Gy-Box-, Brd-Box-, and K-Box-Class microRNAs. Genes Dev. 19: 1067–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. O., Zacharek S. J., Xiong Y., Duronio R. J., 2010. Cell type-dependent requirement for Pip Box-regulated Cdt1 destruction during S phase. Mol. Biol. Cell 21: 3639–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Z., Elledge S. J., 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4: 251–256 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J. A., Gao F. B., 2006. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20: 2793–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., et al. , 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773 [DOI] [PubMed] [Google Scholar]
- Lin H. C., Wu J. T., Tan B. C., Chien C. T., 2009. Cul4 and Ddb1 regulate Orc2 localization, Brdu incorporation and Dup stability during gene amplification in Drosophila follicle cells. J. Cell Sci. 122: 2393–2401 [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer J. L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., et al. , 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78: 113–118 [DOI] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., et al. , 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall-Levin M., Zhao Y., Perrimon N., Berger B., 2010. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc. Natl. Acad. Sci. USA 107: 15751–15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., et al. , 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shibutani S. T., De La Cruz A. F., Tran V., Turbyfill W. J., 3rd, Reis T., et al. , 2008. Intrinsic negative cell cycle regulation provided by Pip Box- and Cul4cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188 [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R. B., Cohen S. M., 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R. B., Cohen S. M., 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Szuplewski S., Sandmann T., Hietakangas V., Cohen S. M., 2009. Drosophila Minus is required for cell proliferation and influences cyclin E turnover. Genes Dev. 23: 1998–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Yanagida M., 2005. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122: 393–405 [DOI] [PubMed] [Google Scholar]
- Tatebe H., Yanagida M., 2000. Cut8, essential for anaphase, controls localization of 26s proteasome, facilitating destruction of cyclin and Cut2. Curr. Biol. 10: 1329–1338 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and Piggybac. Nat. Genet. 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Thomas M., Lieberman J., Lal A., 2010. Desperately seeking microRNAa targets. Nat. Struct. Mol. Biol. 17: 1169–1174 [DOI] [PubMed] [Google Scholar]
- Weng L., Zhu C., Xu J., Du W., 2003. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 22: 3865–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]