Abstract
The Collaborative Cross Consortium reports here on the development of a unique genetic resource population. The Collaborative Cross (CC) is a multiparental recombinant inbred panel derived from eight laboratory mouse inbred strains. Breeding of the CC lines was initiated at multiple international sites using mice from The Jackson Laboratory. Currently, this innovative project is breeding independent CC lines at the University of North Carolina (UNC), at Tel Aviv University (TAU), and at Geniad in Western Australia (GND). These institutions aim to make publicly available the completed CC lines and their genotypes and sequence information. We genotyped, and report here, results from 458 extant lines from UNC, TAU, and GND using a custom genotyping array with 7500 SNPs designed to be maximally informative in the CC and used a novel algorithm to infer inherited haplotypes directly from hybridization intensity patterns. We identified lines with breeding errors and cousin lines generated by splitting incipient lines into two or more cousin lines at early generations of inbreeding. We then characterized the genome architecture of 350 genetically independent CC lines. Results showed that founder haplotypes are inherited at the expected frequency, although we also consistently observed highly significant transmission ratio distortion at specific loci across all three populations. On chromosome 2, there is significant overrepresentation of WSB/EiJ alleles, and on chromosome X, there is a large deficit of CC lines with CAST/EiJ alleles. Linkage disequilibrium decays as expected and we saw no evidence of gametic disequilibrium in the CC population as a whole or in random subsets of the population. Gametic equilibrium in the CC population is in marked contrast to the gametic disequilibrium present in a large panel of classical inbred strains. Finally, we discuss access to the CC population and to the associated raw data describing the genetic structure of individual lines. Integration of rich phenotypic and genomic data over time and across a wide variety of fields will be vital to delivering on one of the key attributes of the CC, a common genetic reference platform for identifying causative variants and genetic networks determining traits in mammals.
GENETIC reference populations (GRPs) are defined as sets of individuals with fixed and known genomes that can be replicated indefinitely. Typically they consist of dozens to hundreds of inbred lines related by descent from a set of common ancestors (i.e., the founders). GRPs have been developed for many organisms, including yeast, plants, flies, and mammals (Bailey 1971; Crow 2007; Buckler et al. 2009; Ayroles et al. 2009; Kover et al. 2009; Cubillos et al. 2011). GRPs are popular for the study of complex traits and biological systems in both medical and life science applications because genotyping is required only once (described as the “genotype once, phenotype many times” paradigm); replicate individuals can be produced with the same genotype allowing for optimal case/control and gene-by-environment designs, and custom analysis tools can be developed to pave the way for the use of these resources by nonexperts (Wang et al. 2003; Chesler et al. 2004; Kang et al. 2008). GRPs are also attractive because over time the phenotypic, genetic, and genomic data associated with each line becomes richer, making possible the integration of data from distinct biological fields that support a more holistic view of biological processes.
Most mouse GRPs are collections of inbred lines derived from pairs of inbred strains. In mice, these include panels of chromosome substitutions strains (i.e., consomics), recombinant inbred lines (RIL), and subcongenics (Bailey 1971; Taylor et al. 1971; Hudgins et al. 1985; Demant and Hart 1986; Nadeau et al. 2000). Alternative GRPs include panels of extant inbred lines with complex population structures and nonuniform genetic relationships among the lines, such as the Laboratory Strain Diversity Panel derived from the Mouse Phenome Project (Paigen and Eppig 2000) and combinations of diversity panels and pairwise panels (Bennett et al. 2010). Key parameters that determine the usefulness of GRPs for the analysis of complex traits are the number of lines; the density, distribution, and functional significance of the genetic variation present in the GRP; the number and distribution of unique recombination sites; the presence of population structure; and the level of inbreeding and genetic drift.
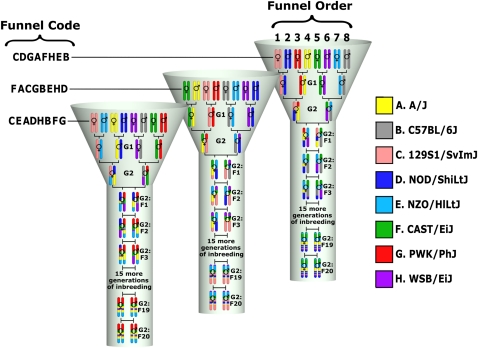
The Collaborative Cross (CC) concept of a multiparental RIL panel was proposed in 2002, as a project aimed at generating a common platform for mammalian complex traits genetics that overcomes the limitations of existing resources (Threadgill et al. 2002) and that can advance the field beyond complex trait analyses toward systems genetics (Threadgill 2006). The final eight-way RIL design of the CC was community driven (Churchill et al. 2004) and included founders from five classical inbred strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, and NZO/HlLtJ) and three wild-derived strains that were selected to represent three Mus musculus subspecies (CAST/EiJ, PWK/PhJ, and WSB/EiJ). The CC lines were generated via a funnel breeding scheme that combined the eight founder genomes in three outbreeding generations prior to repeated generations of inbreeding through sibling mating (Figure 1). The eight founder strains capture a much greater level of genetic diversity than existing RIL panels or other extant mouse GRPs, and the genetic variants are more uniformly distributed across the genome than in other GRPs (Roberts et al. 2007; Keane et al. 2011; Yalcin et al. 2011; Yang et al. 2011). In the absence of selection and errors, the breeding design predicts that the captured genetic variation will be randomly distributed among the CC lines with each line being independent (i.e., CC lines do not share recombination events and local founder contributions). Therefore, the use of the CC should not result in spurious associations in mapping studies that frequently occur in other GRPs (Manenti et al. 2009).
Figure 1 .
Breeding scheme of CC lines. The figure shows the breeding scheme for three independent CC lines. Each line has a funnel section followed by an inbreeding section. The eight founder strains are arranged in different positions (1–8) in each line, and this order determines the funnel code on the basis of a single letter code for each line. Founder order is randomized and not repeated across lines. The colors used for founder strains are seen throughout this article. Each mouse is represented by a pair of homologous autosomes and a symbol denoting its sex.
Because of practical and budgetary constraints, breeding of CC lines started simultaneously in 2004 at different locations from common founder lines. The US lines were started at Oak Ridge National Laboratory (ORNL) in Tennessee and were subsequently relocated to the University of North Carolina in 2009 (hereafter referred to as the CC-UNC). A second set of CC lines was started at the International Livestock Research Institute (ILRI) in Kenya and relocated to Tel Aviv University (Israel) in 2006 (hereafter referred to as CC-TAU). A third set of CC lines was started in Western Australia by Geniad Ltd. (hereafter referred to as CC-GND). The combined CC-UNC, CC-TAU, and CC-GND populations are the focus of this study. Initial status reports for each of these populations were published in 2008 (Chesler et al. 2008; Iraqi et al. 2008; Morahan et al. 2008) with subsequent publications detailing breeding, simulation, and statistical modeling (Broman 2005, 2007, 2012a, 2012b; Valdar et al. 2006; Teuscher and Threadgill et al. 2011; Gong and Zou 2012; Lenarcic et al. 2012; Zhang et al. 2012). Phenotypic and mapping results for a variety of traits using incompletely inbred CC lines are available (pre-CC) (Mathes et al. 2010; Aylor et al. 2011; Durrant et al. 2011; Philip et al. 2011; Kelada et al. 2012). These pivotal proof-of-concept studies used various subsets of pre-CC lines from either the CC-UNC or CC-TAU populations, some of which have since become extinct. The previous analyses of subsets of lines from each of the three populations provided only a limited view into the combined genome architecture of the “final” CC population as a whole due to use of different genotyping platforms, haplotype reconstruction methods, and analytical pipelines. Furthermore, most of these studies did not incorporate recent results on the subspecific origin and haplotype diversity present in the founder strains (Yang et al. 2011), nor the whole genome sequence of the eight founder inbred strains recently completed by the Mouse Genome Project from the Wellcome Trust/Sanger Institute (Keane et al. 2011; Yalcin et al. 2011). This project reported the presence of at least 36,155,524 SNPs in the founder strains of the CC. Initial analyses in CC founders and incompletely inbred lines indicate that the high level of genetic diversity is responsible for the vast number and strength of differences in gene expression in the CC (Aylor et al. 2011; Sun et al. 2012). Finally, The Jackson Laboratory is leading an ongoing effort to create a complementary resource, the Diversity Outcross (DO), derived from partially inbred CC lines originating from the CC-UNC population (Svenson et al. 2012).
Here, we report the joint genetic analysis of all three populations. This study was conducted by the Collaborative Cross Consortium and in what we expect will be an ongoing community effort to popularize this resource. We focused only on extant lines that will be part of the final CC population and conducted the analysis to provide the research community with a more complete picture of the genome architecture expected to appear in the set of CC lines that are publicly available. All genotypes are available (Supporting information, Table S1) and use of these data should cite this publication as a reference. Genotypes will also be available at a dedicated website (http://csbio.unc.edu/CCstatus/). We have created a novel genome browser inspired by the Mouse Phylogeny Viewer (MPV; Yang et al. 2011; Wang et al. 2011) to facilitate visualization and interaction with the genomes of any given CC line (http://csbio.unc.edu/CCstatus/?run=CCV). Finally, we provide details of a Material Transfer Agreement that ensures availability of the CC population for use by the research community.
Materials and Methods
Mice and DNA
CC-TAU lines are bred and maintained in the small animal facility at The Sackler Faculty of Medicine, TAU. Mice are housed on hardwood chip bedding in open-top cages and are given tap water and rodent chow ad libitum. CC-UNC lines are bred and maintained under specific pathogen-free conditions in the Genetics Medicine Vivarium at UNC, where rodent chow and tap water are provided ad libitum and mice are maintained on bed-o'cobs with a nestlet placed in each breeding cage. CC-GND lines are bred and maintained at the Animal Resources Centre (ARC) in Western Australia and are housed under specific pathogen-free conditions with tap water and chow ad libitum. The Institutional Animal Care and Use Committees of TAU, UNC, and ARC have approved all experimental protocols at their respective institutions. During the generation of the CC population, CC-UNC lines are named with the prefix OR (that stands for the two first letters of the Oak Ridge National Laboratory) followed by a number with two to four digits. CC-TAU lines are named IL (which represents the first two letters of the International Livestock Research Institute) followed by a number with two to four digits. CC-GND lines have unique names followed by a two-letter code reflecting the strain located in positions 1 and 8 of the funnel (Figure 1 and also see Chesler et al. 2008; Aylor et al. 2011; Threadgill et al. 2011). Once the CC lines are deemed complete (>97% inbred), they will be renamed in accordance with the rules of the International Nomenclature Committee (see Discussion). Specifically, each line will be named CC#/@, where # are four digits from a consecutive sequence across all three CC populations and @ is the location from whence the line originated (Unc, US lines; Tau, Israeli lines; and Geni, Geniad lines). For example, the first completed line, OR867, is now CC0001/Unc and the second line, IL6211, is CC0002/Tau.
DNA isolation and genotyping
Tail clips were used to isolate DNA using Qiagen Gentra Puregene blood kits from 458 lines (199 from CC-UNC, 214 from CC-TAU, and 45 from the CC-GND lines at the most advanced generations of inbreeding that were available at the time of analysis from approximately 230 extant lines). DNA was resuspended in water and 15-μl aliquots at concentrations ranging from 50 to 200 ng/μl were sent in 96-well plates to Neogen's GeneSeek division for genotyping. Genotyping was conducted using our custom designed Mouse Universal Genotyping Array (MUGA). MUGA is a 7851-SNP marker genotyping array built on the Illumina Infinium platform. SNP markers are distributed throughout the mouse genome with an average spacing of 325 kb (SD 191 kb). The markers were chosen to be maximally informative and maximally independent for the eight founder strains of the CC. This combination was achieved by selecting SNPs with high minor-allele frequencies (maximizing entropy) and low local pairwise linkage disequilibrium (minimizing mutual information). The design criteria make the platform optimal for detecting heterozygous regions, while in homozygous regions they allow for optimal discrimination between haplotypes. These optimization criteria are population dependent. All genotypes are available in Table S1. (If you use these genotypes or the updated genotypes available on the Collaborative Cross Consortium website, http://www.csbio.unc.edu/CCstatus/index.py/, we request that you also cite this article.)
CC founder haplotype inference
Existing techniques for minimizing recombination breakpoints (Zhang et al. 2009), and for haplotype inference such as in GAIN (Liu et al. 2010) and HAPPY (Mott et al. 2000), use four discrete genotype calls as input (homozygous allele 1, homozygous allele 2, heterozygous, or no-call). Rather than using discrete genotype calls, our haplotype reconstructions directly use Illumina's normalized intensity values. This is based on our observation that the allele clusters seen in a genotyping probe set can often be further subclustered according to the intensity values of the eight founders and the 28 possible F1's (Figure S1). This subclustering within genotype clusters can be attributed to subtle differences in the genomic sequence, such as unreported genetic variants within or nearby probes. Our use of subclusters from intensity values transforms the standard 4-state genotyping classification problem to one with 36 possible states for the CC population. The most likely founder at each position is assigned using a hidden Markov model (HMM) similar to the one used in GAIN, a genotype call-based method designed for pedigrees with inbreeding such as the CC (Liu et al. 2010).
The founder states are based on 2D distributions of intensity clusters of biological and technical replicates of CC founders and F1's at each marker (163 replicates in total: 8 replicates for each founder except C57BL/6J, which has 9, and 3.5 replicates on average for each of the 28 F1's). These distributions are then used as reference models for each founder and F1 combination. We estimate the likelihood that a test sample fits a particular model as a function of the test sample's probe intensities Euclidean 2D distance from the model's mean. These distance-derived probabilities are combined with a transition probability between adjacent markers using an HMM. The transition probability parameters were selected so that evidence of sufficient distance from approximately three sequential markers is necessary to change founder state. Moreover, the transition penalty varies depending on the number of shared founders between adjacent states, with the highest penalty assigned to adjacent states with no shared founders. A dynamic programming algorithm was then used to calculate the maximum-likelihood founder assignment for each genomic position.
Identification of related lines and lines with breeding errors
Related IDs (for example, IL1912 and IL3912 or IL51 and IL551) were purposely used to identify cousin lines in the CC-TAU population, as well as mice from CC-TAU lines that were shipped from TAU to UNC for accelerated completion through marker-assisted inbreeding (MAI; Welsh and McMillan 2012). Note that samples from CC-TAU lines used for MAI at UNC are renamed with the OR prefix for colony-management purposes (Table S2). The cousin lines were segregated from the original lines between 6 and 11 generations of the inbreeding process (Figure 1). We used shared recombination events to confirm the identity of related lines. Shared recombination events are defined as those involving the same two strains in the same proximal-to-distal orientation at the same chromosome position. We determined the number of shared recombination events in the autosomes between all pairwise combinations of the 458 genotyped CC samples. Events that are fixed in a strain were counted only once. As expected, most pairs of lines do not share any recombination events (mean 0.0653 ± 0.7552) but a subset of pairs had a significantly higher rate of shared events (Figure S2). All known related lines have at least three shared events, while not a single pair of independent lines with three shared recombination events exists, and only 5% of 47,278 pairwise combinations between independent lines have one or two shared events (Figure S2). We identified 99 related CC samples that define 46 sets of related lines (Table S3). For each set we retained the sample with the lowest heterozygosity for further analyses.
Among the 405 independent lines, only 330 have alleles from each of the eight founder strains present in the autosomes (Table S2 and >Table S3). Based on the simulation of 7 million CC lines, we estimate that 0.05% will have <1% of any given founder. The rate of CC lines missing one or more founders was significantly higher than the results of the simulation, and we eliminated any line with more than one founder missing. Finally, eight CC-UNC lines were eliminated because they represent four pairs of lines, with each pair missing one founder strain caused by the incorrect use of one of four G1 males (Figure 1) that were likely not hybrids between the expected two CC founder lines. Twenty CC lines with one missing founder were retained in the analyses (Table S2). The 350 independent lines passing these quality metrics were used to analyze genome architecture in the CC populations.
Transmission ratio distortion (TRD)
In the autosomes, the frequency of the haplotypes inherited from each CC founder strain should be ∼12.5% (one out of eight equally likely founders) in the final CC population (as well as in the individual populations). To determine the significance of local distortion in founder frequency we simulated the inbreeding of mouse genomes (19 autosomes + 2 sex chromosomes) using the same breeding scheme as the CC with a Haldane recombination model including interference (Welsh and McMillan 2012). We simulated 20,000 independent sets of 350 lines and tabulated the founder contribution over all haplotype segments within each set (a haplotype segment is the region from one recombination breakpoint to the next in any of the 350 lines). Each simulation used the same funnel code (Figure 1) as the actual CC-UNC, CC-TAU, and CC-GND populations when available. A random funnel code was used for lines where that line's funnel code was unknown or inconsistent with the genotypes. The funnel code reflects the position of the founder strains in the funnel (Figure 1). This position has consequences for the inheritance of mitochondrial genome (inherited from the strain in position 1), chromosome (chr) Y (inherited from the strain in position 8), and chr X. For chr X, the expected contribution of each CC founder depends on the funnel order. Founders in positions 4, 7, and 8 cannot contribute a chr X to the line while the founder in position 3 has double the opportunity to contribute compared with the rest of the positions. Finally, after the G1 generation (Figure 1) no CC mouse can be heterozygous for alleles from founder strains located in positions 1 and 2, 3 and 4, 5 and 6, and 7 and 8 in that line. We found that reported funnel codes did not match the expectations in many CC-TAU lines.
Expectations for the founder contribution for chr X (and estimation of the TRD significance) would be best achieved by simulations based on the actual funnel codes of the 350 independent CC lines. However, given the issues with the funnel codes of the CC-TAU population, the significance of local distortion in founder allele frequency was modeled on the basis of equal contribution from each founder in the CC-TAU population. Actual contributions for the CC-UNC and CC-GND populations are provided in Table S4.
Finally, we assigned the subspecific origin of each CC line using the subspecific assignments of each CC founder (Yang et al. 2011) overlaid on the inferred CC haplotype mosaics.
Linkage disequilibrium
We partitioned the genome into 5295 nonoverlapping 500-kb windows and binned all of the previously reported mouse diversity array (MDA; Yang et al. 2011) SNPs into these windows. We then computed the maximum linkage disequilibrium (LD) value, on the basis of the r2 metric (Pearson correlation squared), among all SNP pairs within each pair of windows.
The genotypes of CC lines were imputed at MDA resolution by assembling MDA founder genotypes according to the haplotype mosaics inferred from the founder assignment algorithm described previously. For each recombination we defined a recombination interval flanked by the most distal SNP assigned to the proximal haplotype and the proximal SNP assigned to the distal haplotype. We used the midpoint of these recombination intervals as the dividing point between the founder haplotypes. Each chromosome was imputed separately, giving two haplotype sequences per sample. We modeled a final predicted genome of each inbred CC line by randomly choosing one of the two haplotypes associated with a given line in each chromosome.
The comparative analyses with a panel of 88 inbred strains required matching population sizes. Therefore, we randomly chose an equal number (n = 88) of CC lines to compute the LD for the panel using the same metric. We repeated the random selection of 88 haplotypes 100 times and then found the average maximum LD value for each window pair. We considered all SNPs with fewer than 5% H or N calls across all samples, and of the SNPs considered, we calculated LD for only those SNPs with a minor allele frequency of 5% or higher.
The panel of classical inbred strains includes the following 88 inbred strains: 129P1/ReJ, 129P3/J, 129S1SvlmJ, 129S6, 129T2/SvEmsJ, 129X1/SvJ, A/J, AEJ/GnLeJ, AEJ/GnRkae/ae, AKR/J, ALR/LtJ, ALS/LtJ, BALB/cByJ, BDP/J, BPH/2J, BPL/1J, BPN/3J, BTBR T+tf/J, BUB/BnJ, BXSB/MpJ, C3H/HeJ, C3HeB/FeJ, C57BL/10J, C57BL/6J, C57BLKS/J, C57BR/cdJ, C57L/J, C58/J, CBA/CaJ, CBA/J, CE/J, CHMU/LeJ, DBA/1J, DBA/1LacJ, DBA/2HaSmnJ, DBA/2J, DDK/Pas, DDY/JclSidSeyFrkJ, DLS/LeJ, EL/SuzSeyFrkJ, FVB/NJ, HPG/BmJ, I/LnJ, IBWSR2, ICOLD2, IHOT1, IHOT2, ILS, ISS, JE/LeJ, KK/HlJ, LG/J, LP/J, LT/SvEiJ, MRL/MpJ, NOD/ShiLtJ, NON/ShiLtJ, NONcNZO10/LtJ, NONcNZO5/LtJ, NOR/LtJ, NU/J, NZB/BlNJ, NZM2410/J, NZO/HlLtJ, NZW/LacJ, P/J, PL/J, PN/nBSwUmabJ, RF/J, RHJ/LeJ, RIIIS/J, RSV/LeJ, SB/LeJ, SEA/GnJ, SEC/1GnLeJ, SEC/1ReJ, SH1/LeJ, SI/Col Tyrp1, Dnahc11/J, SJL/J, SM/J, ST/bJ, STX/Le, SWR/J, TALLYHO/JngJ, TKDU/DnJ, TSJ/LeJ, YBR/EiJ, ZRDCT Rax+ChUmd. This set of strains represents the largest panel of classical inbred strains genotyped with the MDA after excluding substrains that are identical by descent (IBD) genome wide (Yang et al. 2011; Wang et al. 2012). The panel overlaps significantly with the strains of the Mouse Phenome Project (Paigen and Eppig 2000) and the Hybrid Mouse Diversity Panel (Bennett et al. 2010). All genotypes have been reported previously (Yang et al. 2011).
Ancestral haplotype diversity in the CC founders
We generated compatible intervals on the basis of the four-gamete rule (Hudson and Kaplan 1985) for the five classical founder inbred strains of the CC using MDA genotypes (Wang et al. 2010; Yang et al. 2011). We then generated the intersection between these intervals and the transitions between subspecific origin in one or more of the eight CC founder strains (Yang et al. 2011). Among strains with the same subspecific origin we estimated the number of haplotypes on the basis of MDA genotype similarity, using a threshold of 97% to identify regions that are IBD among CC founders. The rationale for this threshold has been described in a recent study of haplotype diversity in a large panel of laboratory strains (Yang et al. 2011), and it is supported by validation of large-scale SNP genotype imputation in mouse inbred strains (Wang et al. 2012) and the mouse genome sequencing project (Keane et al. 2011).
CC viewer
We have developed a web-based genome browser for visualizing genomic data over multiple CC lines to aid in comparative analysis. This tool is freely available online at http://csbio.unc.edu/CCstatus/?run=CCV. Available data includes 458 incipient CC lines. We visualize subspecific origin, founder haplotype, and haplotype identity mosaics as stacked horizontal tracks to align coincident features. Our tool includes dynamic panning and zooming, which allows for intuitive navigation about the genome. It also has dynamic interaction features that are applied to the various data sets, including sample sorting based upon similar features as a selected locus. The tool also automatically generates stacked histograms that show the distribution of subspecific origin and founder contribution for a user-selected subset of lines.
Results
Breeding, extinction, and reproductive performance in the CC
Although this report focuses on extant lines, data on all initiated lines in the CC-UNC population are provided to frame our results within the larger context of the CC project. Importantly, the value of our characterization of the genome landscape of the CC resource depends on whether a given CC line that is extant today eventually survives the inbreeding process and becomes available to the research community.
In the CC-UNC population, we included only CC lines that bore a litter within 6 months of this study's starting date (December 2010). The extinction rate in the UNC arm of the CC project was 73.04% (199 extant lines out of 738 lines started at ORNL). The high rate of extinction is consistent with previous reports (Chesler et al. 2008; Philip et al. 2011). Since the last status report, we have attempted to reduce loss of lines due to colony management, and we started MAI of the most advanced lines (Welsh and McMillan 2012). We also relocated the project to the University of North Carolina upon closure of the Mouse Genetics Program at ORNL (Threadgill et al. 2011). We determined the reproductive performance of the extant lines on the basis of average litter size per generation and time between generations (Figure S3). As expected, reproductive performance decreases significantly during inbreeding but stabilizes after generation G2:F7. On the basis of data available for the most advanced generations of inbreeding (>12), the “final” CC lines will have reproductive performances within the range observed in the founder CC strains. The CC lines and corresponding reproductive performance data will be available at the Collaborative Cross Consortium website (http://www.csbio.unc.edu/CCstatus/index.py). (Please cite this article when using this information.)
Genotyping and haplotype reconstruction
We selected a single male from 458 CC lines for genotyping, 199 from the CC-UNC population, 214 from the CC-TAU population, and 45 from the CC-GND population (Table S3). The genotyped male either belonged to the most advanced generation of each line at the time of sample collection or was the most inbred male in the case of lines with multiple males genotyped (i.e., lines actively undergoing MAI). All samples passed the initial QC step on the basis of the fraction of SNP genotypes called (Table S1).We then performed founder assignment (see Materials and Methods) and determined the contribution of each founder strain to each CC line (Table S2). Unexpectedly, we found that numerous CC lines had fewer than eight CC founders’ alleles in their genome. This result could be explained by breeding errors (the missing founder was never present in the line), selection against a given CC founder genome, or chance. Given that one of our main goals seeks to compare the genome composition of the final CC population to what may be expected based on the genome of the CC founders, we established a set of criteria to identify CC lines with breeding errors and to identify related lines in the CC-TAU population (“cousin” lines and sister lines of CC-TAU lines sent to UNC for MAI). These criteria include the frequency of shared recombination events between pairs of samples and the number of missing founders (Materials and Methods). We identified 55 samples with more than one CC founder missing. Eight of these lines belong to the CC-UNC population, 44 to the CC-TAU population, and 3 to the CC-GND population (Table S2 and Table S3). Among the remaining 403 samples, 99 are related and represent 46 independent lines. Related lines are denoted as rCC while incomplete lines are denoted as iCC in Table S2. After these quality-control steps, our final sample set for analysis consists of 350 independent CC lines, 191 CC-UNC lines, 117 CC-TAU lines, and 42 CC-GND lines.
For each line we estimated the residual heterozygosity as the fraction of the genome for which a line has contributions from two different CC founders (Table S2). Average heterozygosity was 25.38% in the CC population genotyped for this study, but the range varied between 0.21% and 66.96% (Figure S4). Note that most of the CC lines have progressed between one and three generations since the mice were genotyped. The distribution of residual heterozygosity was as expected for the number of generations of inbreeding (Broman, 2012a; Welsh and McMillan 2012) and both the CC-UNC and CC-TAU populations having two waves of production that started 3–4 years apart.
Founder contribution
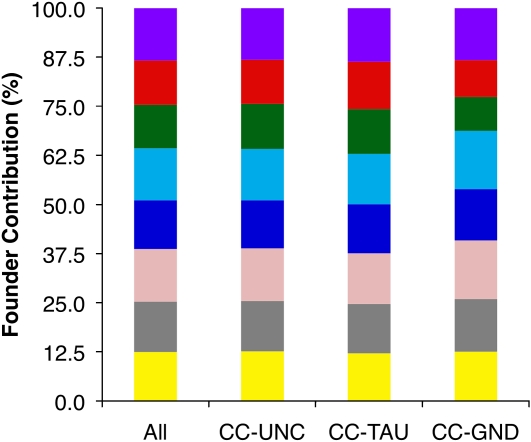
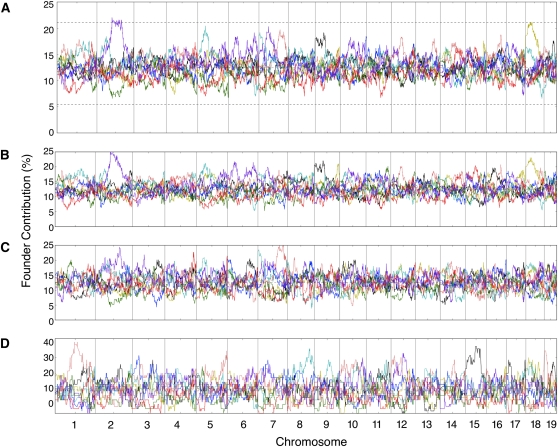
Overall the eight founder strains’ alleles were similarly represented when averaged across the autosomes of the CC lines (Figure 2), and their contribution varied between 11.06% for CAST/EiJ and 13.40% for 129S1/SvImJ (Table S2). The lower contribution of CAST/EiJ holds true for all three populations, CC-TAU, CC-GND, and CC-UNC (Figure 2), and becomes more pronounced when chr X is included (see below). On the other hand, founder contribution varied significantly along the autosomes (Figure 3A). In general, deviation from the expected 12.5% contribution resulted from an overrepresentation of a single founder strain, while a similar level of underrepresentation of a founder was less frequent.
Figure 2 .
Overall contribution of the eight CC founder strains to the autosomes of the CC lines. The stacked columns show the founder contribution to the overall CC, CC-UNC, CC-TAU, and CC-GND populations.
Figure 3 .
Local founder strain contribution along the autosomes. (A) The CC population, (B) CC-UNC population, (C) CC-TAU population, and (D) CC-GND population. The percentage contribution from each founder is represented as a continuous line using the color schema shown in Figure 1. The dotted lines represent the threshold for TRD at P = 0.05 adjusted for genome-wide significance.
Notably, there is a significant (P < 0.05, corrected for genome-wide significance) excess of WSB/EiJ alleles spanning a 51.6-Mb genomic region (73.25–124.85 Mb) on chr 2 in the overall set. Similar levels of distortion were observed in the independent CC-UNC, CC-TAU, and CC-GND populations (Figure 3, B–D). This region overlaps with a putative region of TRD in favor of WSB/EiJ reported previously in the pre-CC experiments (Aylor et al. 2011; Durrant et al. 2011). There are 66 CC-UNC lines in common between one of the pre-CC experiments (Aylor et al. 2011) and the 191 CC-UNC lines in this study; the level of distortion among the animals used in the pre-CC experiments is not significantly different than the final CC set (23.5%, 31 WSB/EiJ chromosomes out 132 total: two chromosomes × 66 samples). Therefore, we conclude that the distortion in favor of WSB/EiJ at this locus is a general feature of the CC rather than simply a chance event. The large size of the region and the shape of the TRD peak on the overall population (Figure 3A) strongly suggest the involvement of multiple loci.
Three additional regions of distortion are consistent between CC-UNC and CC-TAU populations (the CC-GND population was not considered in this analysis because its smaller size leads to highly variable allele frequencies; Figure 2D): overrepresentation of NZO/HlLtJ on chr 5 and overrepresentation of WSB/EiJ and 129S1/SvImJ on chr 7 (Figure 3). Multiple examples of strong deviation from expectations are population specific. For example, an excess of WSB/EiJ, C57BL/6J, and A/J is found on chrs 6, 9, and 18, respectively, in the CC-UNC population. There is an excess of WSB/EiJ, CAST/EiJ, and NOD/ShiLtJ on chrs 3, 4, and 6, respectively, in the CC-TAU population. Whether these findings are due to differential selection based on differences in husbandry between the two sites or due to chance is not known.
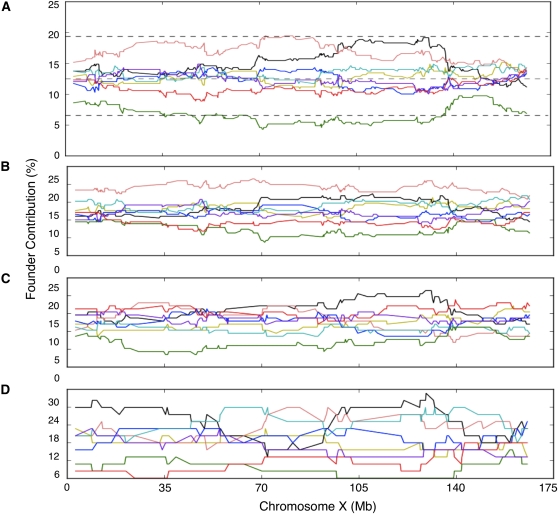
In contrast with the situation in the autosomes, we observed consistent underrepresentation of founder strains’ alleles on chr X (Figure 4). The most striking observation is a significant (P < 0.05, corrected for genome-wide significance) underrepresentation of the CAST/EiJ contribution for much of chr X in all populations. TRD spans at least a 100-Mb region (35–135 Mb) that includes the center of chr X (Figure 4). Estimation of TRD significance was based on assuming equal contribution of each founder rather than the actual contribution dictated by the frequency at which each founder was at each position in the funnel (funnel order, see Materials and Methods and Figure 1). However, the actual contribution for 233 known CC lines (Table S4) indicates that underrepresentation of chr X from CAST/EiJ at the initial generations of the CC can not be responsible for the observed TRD.
Figure 4 .
Local founder strain contribution on chromosome X. (A) Final CC population, (B) CC-UNC population, (C) CC-TAU, and (D) CC-GND population. The percentage contribution from each founder is represented as a continuous line using the color schema shown in Figure 1. The dotted lines represent the threshold for TRD at P = 0.05 adjusted for genome-wide significance.
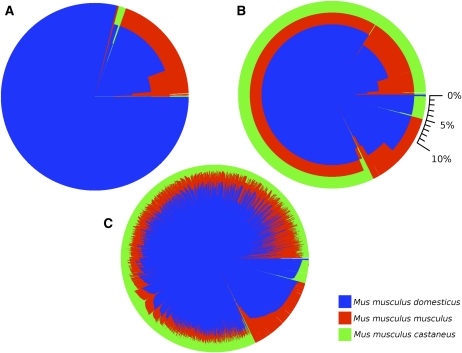
We have recently assigned each region of the genome of the eight CC founders to one of three M. musculus subspecies (Yang et al. 2011). On the basis of this assignment we determined the subspecific origin of each CC line (Figure S5). When the CC founder strains were selected, an important consideration was the inclusion of three wild-derived strains thought to be pure representatives of three major M. musculus subspecies (Chesler et al. 2008). We now know, however, that in two of the wild-derived strains, CAST/EiJ (assumed to be M. m. castaneus) and PWK/PhJ (assumed to be M. m. musculus), a significant amount of their genome originates from M. m. domesticus due to intersubspecific introgression (Yang et al. 2007, Yang et al. 2011). Furthermore, classical inbred strains have little contribution of subspecies other than M. m. domesticus and that contribution is not randomly distributed across the genome. The impact of inclusion of wild-derived strains and the overall representation of the three subspecies is shown in Figure 5; the representation of each subspecies in the individual CC lines varies dramatically (Figure S5). Although the overall subspecies representation is not dramatically distorted, a small excess of M. m. domesticus exists compared to simulations. This conclusion is based on comparing the subspecies distribution observed in the extant CC lines with the anticipated subspecies distribution of founder strains in simulations of the generation of similar number of independent CC lines.
Figure 5 .
Subspecific contribution to the genome of the CC lines. Each pie chart depicts the fraction of the genome that has a given pattern of subspecific contribution in each set of lines. (A) Subspecific contribution in the five CC founder strains that are classified as classical (A/J, 129S1SvImJ, C57BL/6J, NOD/ShiLtJ, and NZO/HILtJ). (B) Subspecific contribution in the eight CC founders. (C) Subspecific contribution in the 308 lines that represent the combined CC-UNC and CC-TAU populations. Blue represents M. m. domesticus, red represents M. m. musculus, and green represents M. m. castaneus. A scale in percentage is provided in B.
Linkage and gametic disequilibrium
We determined the extent and strength of LD and gametic disequilibrium (GD), which is also known as long-range LD, in the CC. LD decays rapidly in the final population (Figure S6 and Figure S7).
More interestingly for users of mouse GRPs, we compared LD and GD between the CC population and a large panel of 88 classical inbred strains (see Materials and Methods). To facilitate comparisons between these two GRPs, we subsampled the CC to ensure the same population size (n = 88). We further selected only one representative among recently derived substrains (Yang et al. 2011). Figure 6 shows the striking differences in genome-wide LD/GD between these two populations. The genome-wide LD/GD in the entire set of 350 CC lines is shown in Figure S8.
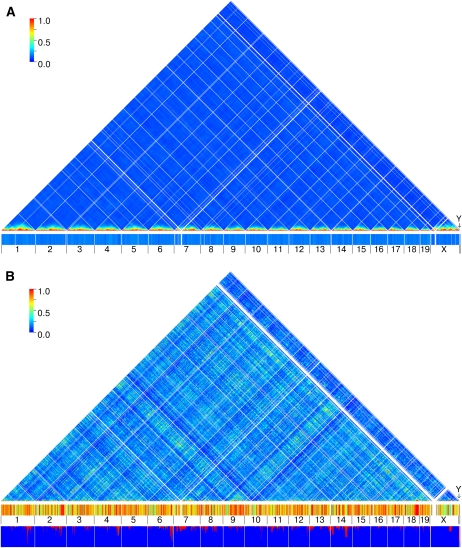
Figure 6 .
Linkage and gametic disequilibrium in mouse GRPs. Chromosomes are arranged in sequential order in the horizontal axis and the color of each pixel represents the maximum level of LD at that pair. The tick boxes denote the maximum level of gametic disequilibrium found genome-wide for each 500-kb window. (A) Mean level of maximum LD in 100 random sets of 88 CC lines. (B) A panel of 88 mouse inbred strains. The additional box at the bottom of the panel represents the cumulative contribution of the subspecies to the panel 88 of inbred strains. Blue represents M. m. domesticus, red represents M. m. musculus, and green represents M. m. castaneus.
In the CC, high LD is observed only between SNP loci that are in close physical proximity, and we see no evidence of significant GD among any unlinked markers. In contrast, the panel of classical inbred strains shows limited local LD but high GD is pervasive throughout the genome. The LD decay is considerably different in these two populations (Figure S6). In the panel of classical inbred strains LD decays very rapidly, but at distances over 20 Mb it stabilizes at 0.17. In the CC, LD decay is initially slower but it continues to decrease over longer distances. At distances over 55 Mb (∼27 cM) LD is substantially lower in the CC than in the classical inbred panel (Figure S6). For example, at 80 Mb the mean LD in 88 CC lines is approximately two-thirds that of the LD observed in the classical inbred panel (and less than one-third in the complete set of 350 CC lines compared to the panel of classical inbred strains).
We estimated the mean and the maximum GD between unlinked markers (>100 Mb that represents 50 cM on average in the mouse) (Figure S6). The mean GD in both populations has a unimodal distribution but with very different means and variance. In the panel of classical inbred strains, the mean is 0.1733 but we observe wide variance. In the CC the mean is 0.0968, and the variance is low. The distribution of maximum GD shows similar but more extreme features (Figure S6). The most striking result is the number of 500-kb windows that have at least one SNP locus very high LD (>0.75) with unlinked SNP loci in the panel of classical inbred strains (Figure 6 and Figure S7).
Discussion
We provide the first comprehensive view of the genetic architecture of the extant CC breeding populations, a framework for future use of this resource, and the ways it complements ongoing research and related resources such as the DO (Svenson et al. 2012). This study has the advantage of combining the three populations (CC-UNC, CC-TAU, and CC-GND) that will be publicly available. We also focus on lines that are most likely to survive inbreeding and, therefore, will be used in future research.
Our analysis also benefits from consistency in genotyping and analyses; the MUGA genotyping platform was primarily designed as a tool to help accelerate inbreeding and detect breeding errors during the generation of the CC population. However, MUGA was not designed to provide a definitive resolution description of the genome of CC lines. During MUGA development, containing costs, a reasonable turnaround time, and operational simplicity were the main considerations. The number of SNP loci was dictated by the price and real estate of the Illumina Infinium platform. The average number of SNPs required to infer founder–strain origin dictates that we will not have resolution under 1 Mb. This is confirmed by the fact that the number of recombination events and segments per CC line (Figure S9) is lower than predicted by simulations (Broman 2005; Teuscher and Broman 2007; Welsh and McMillan 2012) and observed in the pre-CC (Aylor et al. 2011; Durrant et al. 2011), which used the much denser MDA (Yang et al. 2009). The average number of segments in our analysis (92.1 ± 12.8) is 30–50% lower that these estimates. This is explained in part by a marked reduction in the number of small segments under 2 Mb in CC founder haplotype reconstructions (Figure S9). However, LD and TRD distortion analyses should be largely unaffected by resolution of founder haplotype assignments.
Founder–strain contribution varies widely among the 350 CC lines included in this study (Figure S10). TRD is common in mouse crosses (Eversley et al. 2010) and can be due to multiple causes (Pardo-Manuel de Villena and Sapienza 2001). Our results suggest the operation of both positive and negative selection during the generation of the CC. Positive selection for the WSB/EiJ haplotype on chr 2 occurred at the expense of all other founder strains (Figure 3) and is observed uniformly over a wide range of generations of inbreeding, suggesting that it operated in the outcross generations and/or the earliest generations of inbreeding. TRD in favor of WSB/EiJ alleles is also observed in the early generations of the DO (Svenson et al. 2012).
Conversely, our results suggest that negative selection against the CAST/EiJ haplotype is responsible for the distortion on chr X. The involvement of the sex chromosomes in TRD in populations derived from multiple mouse subspecies is not unexpected (Payseur et al. 2005; Mihola et al. 2009; White et al. 2011) and may provide an elegant model for speciation. However, we believe that the selection against the M. m. castaneus X chromosome in a population that is mostly M. m. domesticus is novel. Furthermore, we expect that most TRD in the CC will involve epistatic interactions between multiple loci. Because of the wide range of heterozygosity in the current CC population (Figure S4), we did not attempt to perform analyses involving more than one locus. When the CC population is fully inbred, such analyses should be conducted.
Among the most important characteristics of the CC as a GRP is the presence of multiple haplotypes and the high minor allele frequencies for every SNP. We have shown previously that the use of eight allele models (representing the eight founder strains) can improve mapping (Valdar et al. 2006; Aylor et al. 2011) compared to standard biallelic SNP models (Zhang et al. 2012). However, the founder strains of the CC have their own population history and structure (Yang et al. 2011). Therefore, it is important to determine the number of founder haplotypes on a local scale. For more discussion of the eight alleles model, see Svenson et al. (2012). Almost half of the genome has six distinct haplotypes represented in the eight founder strains (Table S5). Most of the remaining genome has four to eight haplotypes while almost none have fewer than four haplotypes. The regions of consistent haplotypes are dictated by the historical recombinations in the founder strains (Yang et al. 2011) and on average are 371 kb long but vary widely across the genome. Comparison with the distribution of haplotypes in the five classical founder strains clearly demonstrates the value of including the three wild-derived strains. The spatial variation in ancestral haplotype diversity is reflected in the CC genome browser. Ultimately, we plan to determine haplotype diversity on the basis of whole-genome sequence of the founders and the new recombination intervals created during the generation of the CC.
One major finding in our analysis of the CC compared to extant classical inbred strain panels is the difference in long range LD (GD), particularly across chromosomes. Existing classical inbred strains have high levels of long-range LD, likely due to their complicated breeding histories and limited founder populations. High GD in essence creates a situation in which association mapping has high type I error rates (false positives). This has been previously noted (Burgess-Herbert et al. 2009), although the mechanism responsible for the high false-positive rate was unknown. Here we show that this is due to extensive long-range LD in extant inbred strain panels that, while partially overcome by taking population structures into account (Kang et al. 2008), will still lead to extraordinarily high rates of false positives. In contrast, because of the independent inheritance of all genomic intervals, the independent breeding lines of the CC are devoid of long-range LD and present an ideal population for association studies.
The pattern of long-range LD observed in our panel of classical inbred strains is very similar to the one reported previously in laboratory strains (Petkov et al. 2005) despite the differences in strain composition, marker density, and ascertainment bias. These results combined with our more complete understanding of the origin of the genome of the laboratory mouse strongly suggest that history rather than selection was the major driving force in setting these patterns. In fact, there is no evidence that long-range LD in the panel of classical inbred strains is driven by combination of alleles from different subspecies (Figure 6b). However, the high extinction rates observed during the derivation of the CC (Chesler et al. 2008; Iraqi et al. 2008; Morahan et al. 2008; Threadgill et al. 2011), the presence of replicable and significant TRD, and the reduction in breeding performance (Figure S3) indicates that the role of biological selection in shaping the CC resource needs to be explored in the future.
The long-range LD structure in extant classical inbred lines negatively affects other QTL mapping studies. Biological systems analyses based on correlation structures are predicted to contain multiple erroneous correlations when using extant classical inbred lines because of the preexisting genomic correlations. Because the CC lacks long-range LD and thus preexisting correlation structures, the CC is also optimally suited for systems-level analyses.
To ensure unfettered community access to the CC, a Material Transfer Agreement (MTA) was executed between all parties who developed this new resource. This MTA will promote efficient distribution and use of the CC. The five institutions involved in developing the CC include The Jackson Laboratory, the University of North Carolina, Tel Aviv University, Oxford University, and Geniad Ltd. and are parties to an MTA that establishes policies for distribution of CC mice. CC mice, regardless of where they were originally developed, as well as services for their use, are available from any of the MTA parties. Conditions of use (COU) for the mice are based on community standards and are identical to the COU currently covering mice from The Jackson Laboratory. To promote use of the CC population, genotypes of the sampled CC lines will be made publicly available (http://www.csbio.unc.edu/CCstatus). Because the MTA also aims to preserve the genetic integrity of the CC lines, distribution centers will repopulate from a common source of mice or embryos. UNC and TAU will act as distribution centers for CC mice in the United States and Europe. Furthermore, the U.S. center at UNC has established an external advisory board to provide guidance and advice on completion, archiving, and distribution of CC mice (Table S6). As CC lines are deemed inbred, they will be cryopreserved and rederived by the UNC Mutant Mouse Regional Repository Center and the Wellcome Trust into a vendor-quality health status. Finally, The Genome Institute at Washington University is carrying out an ongoing effort to sequence the genomes of each CC line as the line is completed. Full genome sequence information for each line will also be publicly available.
Supplementary Material
Acknowledgments
This work is published under a consortium authorship (listed on opening page) that includes mouse breeders, tool developers, and users of the resources. We acknowledge the following members of the consortium who have made special significant contributions to generation of mice, genotyping, and analysis reported in this manuscript: Fuad A. Iraqi, Mustafa Mahajne, Yasser Salaymah, Hanna Tayem, Karin Vered, Hani Sandovski, Richard Mott, Caroline Durrant, David L. Aylor, Ryan J. Buus, John P. Didion, Chen-Ping Fu, Terry J. Gooch, Stephanie D. Hansen, Leonard McMillan, Kenneth F. Manly, Darla R. Miller, Fernando Pardo-Manuel de Villena, Ginger D. Shaw, Jason S. Spence, David W. Threadgill, Jeremy Wang, Catherine E. Welsh, Grant Morahan, Lois Balmer, Ken Pettit, and Michael Hall. This work was supported by grants from the National Institutes of Health U01CA134240, P50MH090338, P50HG006582, and U54AI081680; Ellison Medical Foundation grant AG-IA-0202-05, National Science Foundation grants IIS0448392, IIS0812464, the Australian Research Council grant DP-110102067, and the Wellcome Trust grants 085906/Z/08/Z, 083573/Z/07/Z, and 090532/Z/09/Z. Essential support was provided by the Dean of the University of North Carolina (UNC) School of Medicine, the Lineberger Comprehensive Cancer Center at UNC, and the University Cancer Research Fund from the state of North Carolina. We also thank Tel-Aviv University for their core funding and technical support.
Footnotes
Edited by Lauren M. McIntyre, Dirk-Jan de Koning, and 4 dedicated Associate Editors
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D. W., 1971. Recombinant-inbred strains: an aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11: 325–327 [DOI] [PubMed] [Google Scholar]
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman, K. W., 2012a Genotype probabilities at intermediate generations in the construction of recombinant inbred lines. Genetics 190: 403–412. [DOI] [PMC free article] [PubMed]
- Broman, K. W., 2012b Haplotype probabilities in advanced intercross populations. G3: Genes, Genomes, Genetics 2: 199–202. [DOI] [PMC free article] [PubMed]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Burgess-Herbert S. L., Tsaih S. W., Stylianou I. M., Walsh K., Co A. J., et al. , 2009. An experimental assessment of in silico haplotype association mapping in laboratory mice. BMC Genet. 10: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., Lu L., Wang J., Williams R. W., Manly K. F., 2004. WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 7: 485–486 [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19: 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Crow J. F., 2007. Haldane, Bailey, Taylor and recombinant-inbred lines. Genetics 176: 729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F. A., Billi E., Zörgö E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Demant P., Hart A. A., 1986. Recombinant congenic strains: a new tool for analyzing genetic traits determined by more than one gene. Immunogenetics 24: 416–422 [DOI] [PubMed] [Google Scholar]
- Durrant C., Tayem H., Yalcin B., Cleak J., Goodstadt L., et al. , 2011. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 21: 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eversley C. D., Clark T., Xie Y., Steigerwalt J., Bell T. A., et al. , 2010. Genetic mapping and developmental timing of transmission ratio distortion in a mouse interspecific backcross. BMC Genet. 11: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y., and F. Zou, 2012 Varying coefficient models for mapping quantitative trait loci using recombinant inbred intercrosses. Genetics 190: 475–486. [DOI] [PMC free article] [PubMed]
- Hudgins C. C., Steinberg R. T., Klinman D. M., Reeves M. J., Steinberg A. D., 1985. Studies of consomic mice bearing the Y chromosome of the BXSB mouse. J. Immunol. 134: 3849–3854 [PubMed] [Google Scholar]
- Hudson R., Kaplan N., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi F. A., Churchill G., Mott R., 2008. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm. Genome 19: 379–381 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada, S. N. P., D. L. Aylor, B. C. E. Peck, J. F. Ryan, U. Tavarez et al., 2012 Genetic analysis of hematological parameters in incipient lines of the Collaborative Cross. G3: Genes, Genomes, Genetics 2: 157–165. [DOI] [PMC free article] [PubMed]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic, A. B., K. L. Svenson, G. A. Churchill, and W. Valdar, 2012 A general Bayesian approach to analyzing diallel crosses of inbred strains. Genetics 190: 413–435. [DOI] [PMC free article] [PubMed]
- Liu E. Y., Zhan Q., McMillan L., de Villena F. P., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti G., Galvan A., Pettinicchio A., Trincucci G., Spada E., et al. , 2009. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive Loci. PLoS Genet. 5: e1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes W., Aylor D., Miller D., Churchill G., Chesler E., et al. , 2010. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am. J. Physiol. 300: E1124–E1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375 [DOI] [PubMed] [Google Scholar]
- Morahan G., Balmer L., Monley D., 2008. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm. Genome 19: 390–393 [DOI] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine-mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J. H., Singer J. B., Matin A., Lander E. S., 2000. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 24: 221–225 [DOI] [PubMed] [Google Scholar]
- Paigen K., Eppig J. T., 2000. A mouse phenome project. Mamm. Genome 11: 715–717 [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F., Sapienza C., 2001. Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome 12: 331–339 Review [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Krenz J. G., Nachman M. W., 2005. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078 [DOI] [PubMed] [Google Scholar]
- Petkov P. M., Graber J. H., Churchill G. A., DiPetrillo K., King B. L., et al. , 2005. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 1: e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip V. M., Sokoloff G., Ackert-Bicknell C. L., Striz M., Branstetter L., et al. , 2011. Genetic analysis in the Collaborative Cross breeding population. Genome Res. 21: 1223–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D., 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data. Mamm. Genome 18: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W., S. Lee, V. Zhabotynsky, F. Zou, F. A. Wright , 2012 Transcriptome atlases of mouse brain reveals differential expression across brain regions and genetic backgrounds. G3: Genes, Genomes, Genetics 2: 203–211. [DOI] [PMC free article] [PubMed]
- Svenson, K. L., D. M. Gatti, W. Valdar, C. E. Welsh, R. Cheng et al., 2012 High resolution genetic mapping using the mouse Diversity Outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed]
- Taylor B. A., Meier H., Myers D. D., 1971. Host-gene control of C-type RNA tumor virus: inheritance of the group-specific antigen of murine leukemia virus. Proc. Natl. Acad. Sci. USA 68: 3190–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuscher F., Broman K. W., 2007. Haplotype probabilities for multiple-strain recombinant inbred lines. Genetics 175: 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., 2006. Meeting report for the 4th annual Complex Trait Consortium meeting: from QTLs to systems genetics. Mamm. Genome 17: 2–4 [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., Hunter K. W., Williams R. W., 2002. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome 13: 175–178 [DOI] [PubMed] [Google Scholar]
- Threadgill D. W., Miller D. R., Churchill G. A., Pardo-Manuel de Villena F., 2011. The Collaborative Cross: recombinant inbred panels in the systems genetics era. ILAR J. 52: 24–31 [DOI] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R., 2006. Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Williams R. W., Manly K. F., 2003. WebQTL: web-based complex trait analysis. Neuroinformatics 1: 299–308 [DOI] [PubMed] [Google Scholar]
- Wang J., Pardo-Manuel de Villena F., Moore K. J., Wang W., Zhang Q., et al. , 2010. Genome-wide compatible SNP intervals and their properties. Proceedings of ACM International Conference on Bioinformatics and Computational Biology, Niagara Falls, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pardo-Manuel Villena F., McMillan L., 2011. Dynamic visualization and comparative analysis of multiple collinear genomic data. Proceedings of ACM International Conference on Bioinformatics and Computational Biology, Chicago, IL: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. R., F. Pardo-Manuel de Villena, H. A. Lawson, J. M. Cheverud, G. A. Churchill et al., 2012 Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics 190: 449–458. [DOI] [PMC free article] [PubMed]
- Welsh, C. E., and L. McMillan, 2012 Accelerating the inbreeding of multi-parental recombinant inbred lines generated by sibling matings. G3: Genes, Genomes, Genetics 2: 191–198. [DOI] [PMC free article] [PubMed]
- White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Wong K., Agam A., Goodson M., Keane T. M., et al. , 2011. Sequence-based characterization of structural variation in the mouse genome. Nature 477: 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Bell T. A., Churchill G. A., Pardo-Manuel de Villena F., 2007. On the origin of the laboratory mouse. Nat. Genet. 39: 1100–1107 [DOI] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods 6: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., et al. , 2011. Genome-wide maps of subspecific origin and identity by descent in the laboratory mouse. Nat. Genet. 43: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Wang W., McMillan L., Pardo-Manuel de Villena F., Threadgill D., 2009. Inferring genome-wide mosaic structure. Proc. PSB 14: 150–161 [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., X. Zhang, and W. Wang, 2012 HTreeQA: using semi-perfect phylogeny trees in quantitative trait loci study on genotype data. G3: Genes, Genomes, Genetics 2: 175–189. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.