Abstract
In relating genotypes to fitness, models of adaptation need to both be computationally tractable and qualitatively match observed data. One reason that tractability is not a trivial problem comes from a combinatoric problem whereby no matter in what order a set of mutations occurs, it must yield the same fitness. We refer to this as the bookkeeping problem. Because of their commutative property, the simple additive and multiplicative models naturally solve the bookkeeping problem. However, the fitness trajectories and epistatic patterns they predict are inconsistent with the patterns commonly observed in experimental evolution. This motivates us to propose a new and equally simple model that we call stickbreaking. Under the stickbreaking model, the intrinsic fitness effects of mutations scale by the distance of the current background to a hypothesized boundary. We use simulations and theoretical analyses to explore the basic properties of the stickbreaking model such as fitness trajectories, the distribution of fitness achieved, and epistasis. Stickbreaking is compared to the additive and multiplicative models. We conclude that the stickbreaking model is qualitatively consistent with several commonly observed patterns of adaptive evolution.
ADAPTIVE evolution is challenging to understand because it depends on a rich array of biological properties. Among those receiving recent theoretical and experimental attention are the magnitude and distribution of mutational fitness effects, the length of adaptive walks, the rate of fitness increase, and the population dynamics that drive it (e.g., Gerrish and Lenski 1998; de Visser et al. 1999; Orr 2002, 2003; Rozen et al. 2002; Cowperthwaite et al. 2005; Barrett et al. 2006; Desai et al. 2007; Eyre-Walker and Keightley 2007; Joyce et al. 2008; Rokyta et al. 2008; Barrick et al. 2009; Betancourt 2009; Burch and Chao 1999; Kryazhimskiy et al. 2009; Schoustra et al. 2009). Equally important are epistasis, pleiotropy, parallelism, mutation order, and the number of beneficial mutations available (e.g., Wichman et al. 1999, 2005; Holder and Bull 2001; Kim and Orr 2005; Weinreich et al. 2006; Silander et al. 2007; Rokyta et al. 2009, 2011; Chou et al. 2011; Khan et al. 2011; Kvitek and Sherlock 2011; Miller et al. 2011). Note that the latter features of adaptation are more meaningful when the identities of the mutations are known and when we consider adaptation as a process subject to replication. For example, epistasis occurs when specific mutations have different effects on different genetic backgrounds (Bonhoeffer et al. 2004; Sanjuán et al. 2004). The rise of genomic sequencing technologies is having a dramatic effect on the ability of researchers to know the identity of mutations occurring during adaptation.
Knowing the identities of adaptive mutations expands the types of questions that can be addressed, but also creates new challenges. All models of adaptation must assign fitness values to genotypes that have arisen through mutation. In connecting genotype and fitness, a model must have the following property: if the wild-type background acquires mutations A1, A2, and A3 to yield a genotype with fitness w1,2,3, every possible order of these mutations must also result in fitness w1,2,3. As the number of fixed mutations grows, the number of possible pathways grows in a factorial manner. We call this consistency requirement the bookkeeping problem.
At least two groups of population genetic models address the bookkeeping problem: one maps genotype change (i.e., mutation) directly onto fitness (GF models), and the second maps genotype onto phenotype and then phenotype onto fitness (GPF models). Here we focus on the simpler GF models. Among these, the additive model assumes mutations have an additive effect on fitness. To be more precise, the fitness after mutations A1 and A2 occur on the wild-type background is w1,2 = wwt + Δw1 + Δw2, where Δw1 and Δw2 are the intrinsic effects expressed as fitness differences of mutations A1 and A2. The bookkeeping problem is solved by the commutative property of addition (i.e., Δw1 + Δw2 = Δw2 + Δw1). Under the multiplicative model, the intrinsic effects are selection coefficients affecting fitness in a multiplicative fashion: w1,2 = wwt(1 + s1)(1 + s2), where s1 and s2 are the intrinsic effects of mutations A1 and A2. Multiplication also has the same commutative property [i.e., (1 + s1)(1 + s2) = (1 + s2)(1 + s1)] and thus solves the bookkeeping problem. Both of these solutions to the bookkeeping problem are simple to simulate and test on real data.
Another solution, implicit in the uncorrelated landscape model (Gillespie 1991; Orr 2002; Joyce et al. 2008), is to assume that the set of mutations arising in an adaptive walk can arise in only one order because each mutation is beneficial on exactly one background. This occurs because the probability of a mutation being beneficial on more than one highly fit background is small enough to be ignored. Thus under the uncorrelated model, once replicate adaptive walks depart from each other, they are 100% divergent. Since the bookkeeping problem involves convergence, the bookkeeping problem is avoided. However, the uncorrelated model makes the extreme prediction for real data that no mutation will be beneficial on two different backgrounds.
Another set of models that avoids the bookkeeping problem is those that assume the number of beneficial mutations on any background is effectively infinite. Under this assumption, the probability of convergent evolution is zero and the bookkeeping problem does not arise. Examples of models that make this assumption include Gerrish and Lenski (1998), Rozen et al. (2002), Desai et al. (2007), and Kryazhimskiy et al. (2009).
The NK model (Kauffman 1993) is unusual among GF models in that it can produce landscapes with intermediate levels of epistasis. In the NK model, N is the number of sites and K is the number of other sites each site interacts with. When K = 0, it is the additive model and when K = N − 1 it is equivalent to the uncorrelated model. When 0 < K < N − 1, the interaction terms mean that the mutational effects are no longer background independent. The interactions bring more biological realism and allow richer patterns of epistasis, but at the expense of model simplicity. Simulating data when 0 < K < N − 1, while ensuring the bookkeeping criteria are met, is computationally challenging because it requires assigning fitnesses to the entire fitness landscape. The interactions also pose a problem for analyzing real data because they introduce a large number of parameters that must be estimated.
Kryazhimskiy et al. (2009) have also developed a flexible GF modeling framework where the uncorrelated and additive models arise as special cases. These models allow different types of epistasis and decelerating fitness trajectories to be produced. However, because the fitness of beneficial mutations in such models depends only on the current fitness, they do not solve the bookkeeping problem.
Thus there is an array of GF models. Among those that offer simple solutions to the bookkeeping problem (additive, multiplicative, and uncorrelated), they generally fail to predict several commonly observed properties of real adaptation. Specifically, in laboratory adaptations parallel evolution is not uncommon, most fitness gain occurs early in a walk, and epistasis is common (Lenski and Travisano 1994; Bull et al. 1997; Elena and Lenski 1997; Wichman et al. 1999, 2005; Cooper and Lenski 2000; Burch et al. 2003; Sanjuán et al. 2004; Cowperthwaite et al. 2005; Woods et al. 2006; Barrick et al. 2009; Betancourt 2009; Rokyta et al. 2009, 2011; Chou et al. 2011; Khan et al. 2011).
This leads us to propose a novel GF model for combining mutational effects that we call stickbreaking. The stickbreaking model is premised on the familiar idea that mutations have intrinsic effects. But rather than assuming fitness differences are background independent (like the additive model) or that differences scale by background fitness (like the multiplicative model), differences in the stickbreaking model scale by how near the current background is to a hypothesized upper fitness boundary. For example, if mutation A1 has stickbreaking coefficient u1 and the fitness distance from the wild type to the boundary is d, then the mutation will increase fitness by the amount du1 (Figure 1). We use theory and simulations to show that stickbreaking both solves the bookkeeping problem and produces some qualitative features commonly observed in adaptive evolution.
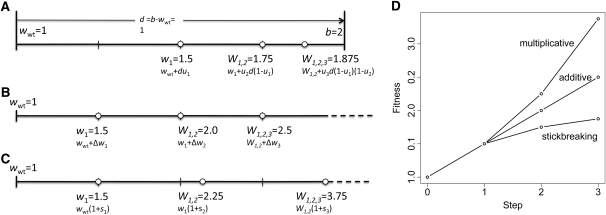
Figure 1 .
Simple comparison of (A) stickbreaking, (B) additive, and (C) multiplicative models when all three begin with the same initial fitness (wwt = 1) and fix three mutations of the same intrinsic effect (0.5) (i.e., u1 = u2 = u3 = Δw1 = Δw2 = Δw3 = s1 = s2 = s3 = 0.5). In A–C, the thick horizontal lines are fitness and the circles are genotypes with the first, second, and third mutations to fix. In the stickbreaking model (A), each mutation closes the distance from the current fitness to the boundary by the mutation’s intrinsic effect. D displays the same information as A–C by plotting step vs. fitness and better illustrates how stickbreaking produces diminishing effects and a decelerating fitness trajectory.
Models
Stickbreaking
We begin by introducing the stickbreaking model and compare it to the additive and multiplicative GF models. Suppose the maximum fitness achievable in the current environment is wmax while the current fitness is wwt. Let d = wmax − wwt be the maximum possible fitness gain through adaptation. Let ui be the stickbreaking coefficient of Ai such that its fitness on the wild-type background, wi, is given by wi = wwt + dui, where ui ≤ 1. In the stickbreaking model, stickbreaking coefficients are assumed to be background independent. If a second mutation, Aj, with stickbreaking coefficient uj occurs on the Ai background, its fitness is given by, wi,j = wwt + d(u1 + u2(1 − u1)). To see why, note that after the first mutation fixes, the remaining distance to the boundary is d(1 − ui) and the second mutation therefore increases the fitness by ujd(1 − ui). Adding this increase to the fitness of the first mutation, wwt + dui + ujd(1 − ui), and simplifying gives wwt + d(u1 + u2(1 − u1)). But since u1 + u2(1 − u1) = 1 − (1 − u1)(1 − u1), we can rewrite the fitness of the double mutant as wi,j = wwt + d(1 − (1 − ui)(1 − uj)). In general, if m mutations with identities A1, A2, … , Am and stickbreaking coefficients u1, u2, … , um accumulate on the wild-type background, the fitness is given by
| (1) |
The intrinsic effect of each mutation Ai thus closes the distance between the current background and the fitness limit by a proportion ui. This process is analogous to a stickbreaking exercise. With a stick of length d laid along a number line, the first mutation dictates where, in a fractional sense, it is broken. Setting the left portion of the stick aside, the next mutation determines where the remaining right portion is broken. The process continues with subsequent mutations breaking the remaining right portion into ever smaller pieces. Unless a stickbreaking coefficient of 1 is available, fitness will never actually reach the fitness maximum.
The stickbreaking model solves the bookkeeping problem because, as Equation 1 shows, the final fitness depends on the product of intrinsic effects and is therefore order independent. Note that mutations with intrinsic effects between 0 and 1 are beneficial. It is less obvious that intrinsic effects may be zero or negative, representing neutral and deleterious mutations, respectively. We also note that the stickbreaking metaphor appears in other modeling contexts, for example, to describe niche partitioning and species abundance in ecology (MacArthur 1957; Patil and Taillie 1977) and in population genetics to derive the distribution of age-ordered alleles under the infinite-alleles model (Donnelly and Joyce 1989). To our knowledge, stickbreaking has not previously been applied to the subject of adaptive evolution.
Stickbreaking compared to additive and multiplicative models
Because of the mathematical similarities between the stickbreaking, additive, and multiplicative models, it is possible to assess when they yield similar results and when they do not. Fitness effects are expressed as fitness differences (Δw) in the additive model, selection coefficients (s) in the multiplicative model, and stickbreaking coefficients (u) in the stickbreaking model. In each case, the model’s respective fitness effects are assumed to be background independent. More precisely, if b is the genetic background and i is the arising mutation, then Δwi|b = wi,b − wb, si|b = (wi,b − wb)/wb, and ui|b = (wi,b − wb)/(wmax − wb).
Under the additive model, the fitness after A1, A2, … , Am mutations with fitness differences Δw1, Δw2, … , Δwm have accumulated on the wild-type background is
| (2) |
Under the multiplicative model, the fitness after m mutations with selection coefficients s1, s2, … , sm have accumulated is given by
| (3) |
The stickbreaking, additive, and multiplicative models converge to the same model when effect sizes are small and walks are not too long. This occurs when the product of effect size and walk length is small. Note that if the product in Equation 3 is expanded and all higher-order terms are assumed to be zero, then fitness under the multiplicative model is approximated by a sum,
| (4) |
Similarly, if Equation 1 is expanded and higher-order terms are ignored, then fitness under stickbreaking is also approximated by a sum,
| (5) |
Combining Equations 4, 5, and 2 gives
| (6) |
If fitness effects are small and walks not long, it implies that wwtsi ≈ dui ≈ Δwi.
Definitions of fitness
Before continuing, it is important to clarify our approach to defining fitness. We have denoted and continue to denote fitness in a generic sense as w. Fitness is more precisely defined in two ways that we call Darwinian and Malthusian fitness. Darwinian fitness is λ in a discrete population growth model, Nt = N0λt, where N0 and Nt are the population sizes at time 0 and time t. Malthusian fitness is r in the continuous growth model, Nt = N0ert. One can be easily transformed to the other by λ = er or ln(λ) = r. They can also be defined in relative terms where the change in frequency of a mutant to a reference type gives the ratio of growth rates (Hartl and Clark 1997); their meaning and log relationship are the same.
In this article, the definition of fitness is important when we consider (i) how fitnesses arise during an adaptive walk and (ii) what type of fitness is measured when a walk is “observed”. In modeling walks (i), we maintain generality by considering mutations acting in an additive, multiplicative, or stickbreaking manner on either Darwinian or Malthusian fitness. This yields six combinations. Note, because multiplicative effects on λ and additive effects on r are equivalent, there are actually five different models. For clarity, however, we describe them as a set of six models. After an adaptive walk occurs, we imagine measuring fitness (ii). Throughout this article we measure Malthusian, but not Darwinian, fitness to simplify our results and because Malthusian fitness is the predominant definition used in the experimental evolution literature.
Fitness trajectories
The predicted fitness under the additive, multiplicative, and stickbreaking models after m steps can be approximated if we assume the pool of beneficial mutations (M) is large enough that sampling is effectively done with replacement (i.e., M ≫ m). Then, under strong selection, weak mutation (SSWM) conditions, the expected effect of a mutation that arises, escapes drift, and sweeps to fixation is given by (Gillespie 1991), where xi represents the intrinsic effect under either of the three models (i.e., Δwi, si, or ui). We therefore replace Δwi, si, and ui in Equations 1, 2, and 3 with ν. Note that when mutations affect λ, but we measure r, a log transformation is necessary. These approximations as well as model abbreviations are given in Table 1.
Table 1 . Expected fitness after m steps given the model generating fitness effects.
| Model | Model abbreviation | Expected fitness (r) |
|---|---|---|
| Additive on λ | Add on λ | ln(λwt+ mν) |
| Multiplicative on λ | Mult on λ | ln(λwt) + m ln(1 + ν) |
| Stickbreaking on λ | Stick on λ | ln[λwt + d(1 − (1 − ν)m)] |
| Additive on r | Add on r | rwt + mν |
| Multiplicative on r | Mult on r | rwt(1 + ν)m |
| Stickbreaking on r | Stick on r | λwt + d(1 − (1 − ν)m) |
Fitness is measured on the Malthusian, r, scale. Expected fitness is obtained by replacing mutational effects in Equations 1–3 with the mean intrinsic effect of a fixing mutation, ν. The second column gives model abbreviations used throughout this article.
Distributions of fitness during replicate walks
We want to know the distribution of fitness achieved at step m when the total number of beneficial mutations available is M under each of the three models. Note that this differs from the familiar distribution of fitness effects and the distribution of fitnesses across the landscape; rather, it is the distribution of fitness achieved among replicate walks after m steps when all walks begin at the same genotype. The details of this derivation are provided in the Appendix. Denote the intrinsic effect of mutation i as xi, where xi = Δwi, xi = si, and xi = ui under the three models. Assume the xi values are drawn from a distribution and replicate walks occur using this fixed set of mutations (i.e., on a fixed landscape). Let Yj be the intrinsic effect of the mutation that fixes at step j. Note that si and ui differ from Δwi by a scaling factor that cancels when calculating the scale-free quantity Yj. If M is large and m is an order of magnitude smaller, such that as both M → ∞ and m → ∞, m ln(M)/M → 0, then Y1, Y2, … , Ym will be approximately independent and identically distributed with for j = 1, 2, … , m. On the basis of the central limit theorem, this implies that the distribution of will be approximately normal, will be approximately log normal, and will be approximately negative log normal. Thus, when M is large and m is small, but not extremely small (i.e., when the pool of beneficial mutations is large and the number to have fixed is moderately small), fitness of replicate walks under the additive, multiplicative, and stickbreaking models follows the normal, log-normal, and negative log-normal distributions with density functions and parameter values provided in the Appendix. These limiting distributions can be obtained as a function of time, not mutational step, using a scale transformation.
Epistasis
Epistasis occurs when a mutation has different fitness effects in different genetic backgrounds. One way to measure epistasis is therefore to assess the fitness effect of a mutation across different backgrounds. A second way to examine epistasis is as a deviation of observation from prediction: (i) measure the fitness effects of two or more mutations on the same genetic background, (ii) predict their combined fitness effect under an assumed model on the basis of their individual effects, (iii) measure their combined fitness effect, and (iv) define epistasis as the disparity between predicted (ii) and observed (iii). The first approach is more intuitive, and the latter is more commonly used in the literature as a measure of epistasis. We pursue both here.
Epistasis as different effects of the same mutation across backgrounds:
For any mutation, we specifically wish to know how its fitness effects change across the steps of a walk beginning with the wild type and continuing until the mutation actually fixed. Following convention, we define fitness effects as differences in r. As above, we consider data arising under each of six models. We assume the pool of beneficial mutations is large and SSWM conditions operate such that the expected fitness effect of a mutation at each step is given by ν. An adaptive walk of length m − 1 occurs. If we imagine a mutation of average (fixed) effect, ν, is then inserted (i.e., genetically engineered) as the mth mutation on the m − 1 background, the expected value of Δr that results is contained in Table 2.
Table 2 . Expected fitness effects for a mutation fixing after m − 1 steps.
| Model | Expected fitness effect (Δr) |
|---|---|
| Additive on λ | ln[(λwt + mν)/(λwt + mν − ν)] |
| Multiplicative on λ | ln(1 + ν) |
| Stickbreaking on λ | ln[(λwt + d − d(1 − ν)m)/(λwt + d − d(1 − ν)m−1)] |
| Additive on r | ν |
| Multiplicative on r | rwtν(1 + ν)m−1 |
| Stickbreaking on r | νd(1 − ν)m−1 |
The left column gives the model under which data arise. Fitness effects (right column) are defined as the expected fitness differences in r as a consequence of the mth mutation.
Epistasis as departure of observed from predicted effects of combined mutations:
An alternative way to measure epistasis is as a departure of observation from prediction: ε = robs − rpred. Predicted values are based on additivity on r while observed data arise according to one of the six models. We are interested in how the disparity between observed and predicted fitness depends on the model under which fitness effects arise and the number of mutations considered, m. Again, we assume SSWM conditions and a large pool of beneficial mutations such that the expected effect of a randomly fixing mutation is ν. Table 3 gives the expected values for ε for each of the six models.
Table 3 . Expected deviations from additivity on r (ε).
| Model | Expected ε |
|---|---|
| Additive on λ | ln(λwt + mν) − n ln(1 + ν/λwt) |
| Multiplicative on λ | 0 |
| Stickbreaking on λ | ln[(λwt + d − d(1 − ν)m] − rwt − m ln(1 + dν/λwt) |
| Additive on r | 0 |
| Multiplicative on r | rwt (1 + ν)m − rwt (1 + mν) |
| Stickbreaking on r | d − d(1 − ν)m − mdν |
ε is defined as robs − rpred. The left column gives the model under which data arise while the right column gives the expected value of ε as a function of the number of fixed mutations (m) and the mean intrinsic effect of mutations that fix (ν).
Simulations
Overview
Simulations written in R (R Development Core Team 2009) were used to study the patterns of fitness trajectory, distribution of fitness effects, and epistasis and to compare these to the theoretical results derived above. All simulations were done in the following basic framework. First, we assumed SSWM dynamics (Gillespie 1991) such that the population is described by a procession of fixed beneficial mutations. Second, a fitness landscape was defined by a relatively small number of beneficial mutations (M = 50) with fitness effects, x, randomly drawn from a distribution. Neither the pool of mutations nor their inherent effects change as adaptive walks proceed. Third, the time until the next mutation fixed was simulated by drawing random exponential waiting times for all M − m available mutations with rate Nμbπ(si), where N was set at 105, the per site per generation beneficial mutation rate, μb, was set to 2 × 10−7, and the fixation probability for mutation Ai, π(si), is given by (Kimura 1962), where si is the selection coefficient of Ai as traditionally defined [i.e., fractional changes in λ or differences in r (Chevin 2010)]. The mutation that fixed was that with the shortest waiting time.
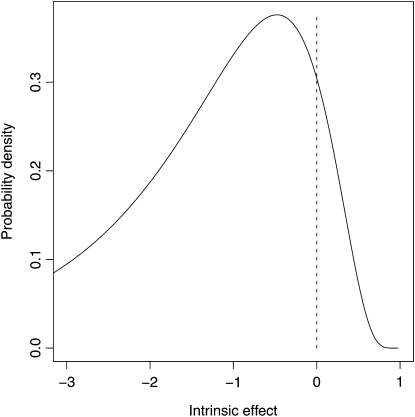
In conducting simulations, we had to decide whether to conduct replicate walks on one landscape or single walks on replicate landscapes. In other words, should we average over replicate walks or replicate landscapes? We argue that conducting replicate walks on the same landscape is more analogous to experimental evolution where these models may ultimately be tested empirically. Consequently, we simulated a single landscape and ran 1000 replicate walks on this landscape, collecting and summarizing relevant information. We then repeated this entire process over several landscapes and confirmed that the observed qualitative patterns that are our focus here do not depend on the particular landscape (results not shown). To generate a landscape, 50 beneficial mutations were drawn from the positive region of a negative log normal (Appendix). If X ∼ Normal (μ, σ), then 1 − eX is a sample from the negative log normal. Parameters for the negative log normal (μ = 0.75, σ = 0.6) were chosen so that 10% of the probability is positive (Figure 2). This distribution was used because it produces values ≤1 as required by the stickbreaking model while also being consistent with the additive and multiplicative models. Once it was generated, we used this single set of 50 values to simulate replicate walks under the six models: additive, multiplicative, and stickbreaking affecting Darwinian or Malthusian fitness. For all models the initial fitness was set at 1, and for both stickbreaking models, the fitness boundary was set at 2 such that d = 1. Walks were simulated until all 50 beneficial mutations fixed.
Figure 2 .
Negative log normal distribution from which effects are drawn. All mutations are drawn from 10% of the distribution >0 (indicated by dashed line).
Analysis of simulations
Three analyses of simulated data were conducted. First, we compared the mean fitness trajectory for each of the six models. Because final fitness differs dramatically between models, trajectories were rescaled for every simulated walk to range from zero to one. Second, to assess the distribution of fitness, we sampled fitness for each of the 1000 walks at steps 5, 10, 20, and 30 and generated histograms from the results. Third, epistasis was measured in the same two ways we quantify it in the Epistasis section above: (i) as fitness effects and (ii) as a departure from additivity on r. In approach i, we took a mutation that arose later in a walk, simulated engineering it into each of the preceding backgrounds, and measured its resulting fitness effect. We arbitrarily used the mutation fixing 10th and we defined fitness effect as the difference in r. In the latter approach (ii), we compared observed fitness with predicted fitness on the r scale. For each simulated walk, we considered the first m mutations that fixed for m = 2, 3, … , 10. We then imagined measuring the effect of each of these m mutations on the wild-type background (i.e., as first-step mutations) yielding Δr1|wt, Δr2|wt, … , Δrm|wt. Under the additive model, the predicted fitness when all m mutations are combined is just r1,2,…,m(pred) = rwt + Δr1|wt + Δr2|wt + … Δrm|wt. Epistasis, as a function of the number of mutations, is then εm = r1,2,…,m(obs) − r1,2,…,m(pred).
Results and Discussion
Our objective in this work is to propose and explore a new model of combining mutational effects, which we call stickbreaking. Stickbreaking is premised on the idea that, in the current environment and on short evolutionary timescales, there is a fitness boundary imposed by the laws of biochemistry and by restrictions on how radically the architecture of the genome can be altered by mutation. This limits how dramatically phenotype can be changed over a short evolutionary time span. Within the scope of available phenotypes, the optimal one corresponds to the fitness boundary. For example, if a set of mutations affects the rate a virus attaches to its host, the accumulation of many such mutations will not indefinitely push the attachment rate higher; rather, a boundary on attachment and therefore fitness will be imposed by the kinetics of collisions of objects in random motion. Such boundaries help provide a basic rationale behind the stickbreaking model.
Stickbreaking may also arise when organisms are moderately redundant such that they may solve a given problem multiple ways. Once substantial progress is made toward one solution (through mutation), pursuing alternative solutions to the same problem may be beneficial, but not nearly as much as the first. In the attachment example above, we might imagine multiple residues where binding can occur to the host; a virus that attaches poorly requires a mutation at only one of these residues to dramatically increase attachment. Subsequent mutations offering alternative ways to bind will provide diminishing beneficial effects. Conversely, when an organism is very near the optimal fitness because it has found several, semiredundant solutions to a problem, a deleterious mutation that disrupts one solution will have a relatively small negative effect on fitness. It is also noteworthy that patterns qualitatively similar to stickbreaking can emerge from metabolic control theory (Kacser and Burns 1981). When a mutation changes the activity of an enzyme in a pathway, its effect on the pathway’s flux is smaller than on the enzyme itself and it diminishes the nearer the pathway is to the maximum flux.
In stickbreaking, these biological assumptions of a boundary and diminishing effects are translated mathematically by allowing mutations to further and further subdivide the distance to the boundary in a multiplicative manner (Equation 1, Figure 1). Because it involves a product, stickbreaking has the commutative property and, like the additive and multiplicative models, thereby solves the bookkeeping problem. However, this process of subdivision leads to different walk properties from those models.
Fitness trajectory
Different models lead to dramatically different trajectories of fitness as a function of mutational step over an adaptive walk (Figure 3A). When mutations affect r, the trajectories for the additive, multiplicative, and stickbreaking models are approximately linear, exponential, and rapidly decelerating, respectively. When mutations instead affect λ, the trajectories are shifted: additive becomes modestly decelerating, multiplicative becomes approximately linear, and deceleration under stickbreaking becomes very slightly exaggerated. Note that the theoretical expectations from Table 1 (Figure 3A, shaded lines) are qualitatively correct; the disparities between them and the simulations (Figure 3A, solid lines) reflect the limited pool of beneficial mutations and the biased nature in which selection fixes mutations.
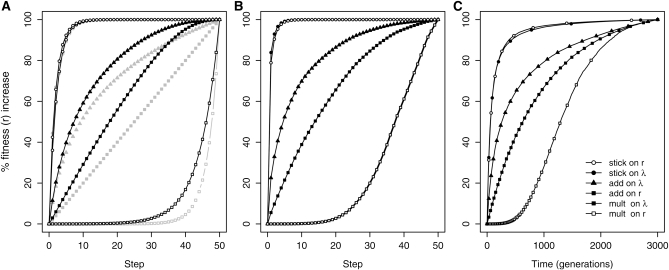
Figure 3 .
Mean fitness trajectory scaled as percentage of final fitness achieved under the six models indicated in the inset in C (see Table 1 for model abbreviations). Symbols on trajectories identify model and denote mutations. Note “add on r” and “mult on λ” are the same model with the same trajectory. (A) Mean fitness trajectories by mutational step based on simulations (solid lines) and theoretical predictions from Table 1 that assume a constant mean effect for all mutations (shaded lines). SSWM conditions are assumed. (B) Simulated trajectories under extreme clonal interference where mutations fix in order of descending effect size. (C) Same fitness trajectories as in A except plotted by time, not step, for the first 3000 generations.
A survey of the experimental evolution literature indicates that, in most cases, the observed fitness trajectory decelerates as adaptation proceeds. This result has been observed in Escherichia coli (Lenski and Travisano 1994; de Visser et al. 1999; Barrick et al. 2009), the DNA bacteriophages ϕX174 and G4 (Bull et al. 1997; Wichman et al. 1999; Holder and Bull 2001), RNA bacteriophage (Burch and Chao 1999; Betancourt 2009), and the animal RNA virus, vesicular stomatitis virus (VSV) (Elena et al. 1998). The exceptions we are aware of are approximately linear trajectories in Saccharomyces cerevisiae (Desai et al. 2007) and in one study on VSV (Novella et al. 1995). Of the models considered here, both stickbreaking models show rapidly decelerating trajectories and the additive model on λ shows a moderately decelerating trajectory.
This suggests that one of these three models is likely nearer the truth than the model most commonly assumed in the literature, additivity on r (multiplicative on λ) with its approximately linear trajectory. There are at least two reasons to be somewhat cautious regarding this conclusion. First, our results are based on SSWM dynamics while many experimental and real world systems involve interference dynamics with more than one mutation contending simultaneously. Under interference dynamics, selection is more efficient at fixing bigger-effect mutations early in a walk compared to SSWM conditions (Rozen et al. 2002; Barrett et al. 2006). We can obtain a bound on this effect by assuming the pool of contending mutations is the entire pool of beneficial mutations and selection therefore fixes them in descending order from the largest to the smallest. Figure 3B shows this trajectory. As expected, interference shifts all the trajectories toward a decelerating pattern although the effect is modest.
Second, trajectories are affected by whether fitness is considered a function of mutational step (as we have done thus far) or time. Plotting fitness against time instead of step bends most of the trajectories toward a more concave, decelerating shape (Figure 3C). Under all models, there is a tendency to fix mutations from larger to smaller intrinsic effect. When all else is equal, this leads to selection coefficients (as traditionally defined, see Simulations) tending from large to small and, therefore, to waiting times between fixation events tending from short to long. In the “add on r” model, this is the only effect, and the trajectory decelerates moderately. In the “add on λ” model there is also the effect that as fitness grows in an additive way, the proportional effect of each added mutation (the selection coefficient) becomes smaller. The stickbreaking models are most dramatically affected by the timescale because as they approach their boundary, selection coefficients become very small and waiting times very long. At the other extreme lies the “mult on r” model where selection coefficients actually get larger as the walk proceeds, causing the walk to accelerate in time for most of its duration. We leave a statistical treatment of trajectory data for later work and here emphasize three things: (i) most of the models show decelerating trajectories, (ii) the slowdown is exaggerated both by clonal interference and by using time rather than step as the explanatory variable, and (iii) with or without these influences, the stickbreaking models show much more dramatic decelerating effects than the other models.
Distribution of fitness over replicate walks
When mutations affect Malthusian fitness, r, and fitness is measured as r, the theoretical distributions from replicate walks (Appendix) are log normal, normal, and negative log normal for the additive, multiplicative, and stickbreaking models (solid lines in Figure 4, A–C). When mutations affect λ instead, these qualitative patterns are only slightly changed with heavier left tails (Figure 4, D and E). These predictions are based on asymptotic assumptions that (i) the total number of beneficial mutations, M, is large, (ii) the step where fitness is measured is far smaller than the number of beneficial mutations, m ≪ M, and (iii) m is large enough for the law of large numbers to apply. In reality, M will often be modest (e.g., 10 < M < 100) and m may be relatively small (e.g., ≤30). The simulations shed light on what effect violating these assumptions has.
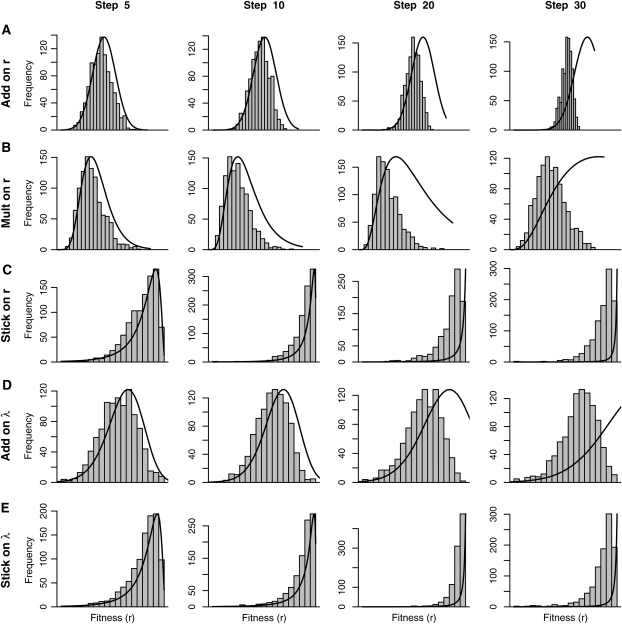
Figure 4 .
(A–E) Distributions of fitness achieved at the indicated step (top) under the six models (left) where the equivalent “add on r” and “mult on λ” are consolidated. Shaded bars show frequency of fitness over 1000 walks on the same landscape while solid lines are predicted distributions based on asymptotic results in the Appendix. The x-axis (Malthusian fitness) is the same for all panels. When mutations affect r, the additive, multiplicative, and stickbreaking models (A–C) yield normal, log-normal, and negative log-normal distributions of fitness.
Early in a walk (m ≤ 10) there is good agreement between the observed and predicted distributions (Figure 4) in terms of both mean and variance. As a walk approaches its midpoint, observed means are notably smaller than the predicted means because the theory assumes constant effect sizes while, in simulated walks, fitness increase slows as large-effect mutations are removed from the available pool. Still, the shapes of the distributions remain the same even when m is large. The different models make qualitatively different predictions about the distribution of fitness during replicate adaptive walks: both stickbreaking models predict heavy left tails, the multiplicative on r model a heavy right tail, and both additive models an approximately normal distribution. Whether mutations affect r and λ is relatively minor. Note also that the distributions are in terms of number of mutations fixed (steps), not time elapsed. As shown in the Fitness trajectory subsection above, different models fix mutations in different lengths of time (Figure 3C) and will therefore achieve the distributions shown in Figure 4 at different rates (see the Appendix for details).
Epistasis
Epistasis occurs when the fitness effect of a mutation depends on the genetic background. We investigate epistasis in two ways: first as the effect of a single mutation across a procession of backgrounds and second as departures from additivity when a set of single mutations is combined. For the first approach, we simulate replicate walks of 10 mutational steps under each model on a single landscape. We then imagine taking the mutation that fixed 10th and engineering it into each of the preceding backgrounds in the walk. (Our choice of the 10th mutation is arbitrary, but using other stop points does not change the qualitative patterns observed; data not shown).
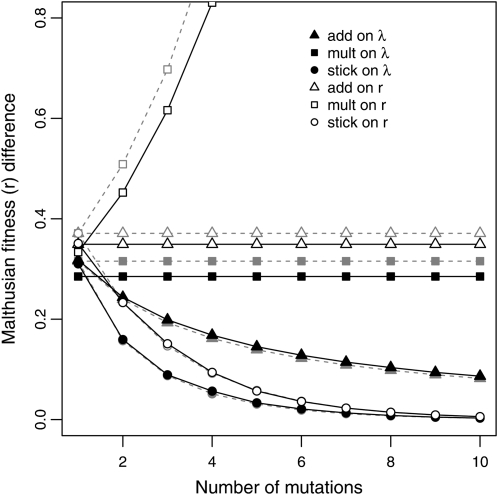
The solid lines in Figure 5 show the means of simulation results when fitness effects are defined as differences in r while the shaded lines give the theoretical relationships (Table 2). The results show how the observed fitness effects change along the walk under the different models for the same mutation (or as the intrinsic effect is held constant). Effect sizes grow exponentially for the mult on r model, are constant for the add on r (mult on λ) model, decay moderately for add on λ, and show rapidly diminishing effects for both stickbreaking models. Of course, these patterns closely reflect the previously discussed fitness trajectories. Here we are considering how the vertical distance (fitness) between steps qualitatively changes along a walk when the intrinsic effect is held constant. It is also noteworthy that because differences in r are, in fact, selection coefficients, Figure 5 illustrates how selection coefficients change across a walk under each model. As discussed above, this, in turn, explains how waiting times between mutations change across a walk (Figure 3C).
Figure 5 .
Mean fitness effect of the mutation fixing at step 10 inserted into the procession of preceding backgrounds beginning with the wild type. Shaded dashed lines are theoretical predictions from Table 2. Stickbreaking on either r or λ and, to a lesser extent, additivity on λ produce diminishing returns epistasis.
In the literature, epistasis is more commonly quantified as the departure from additivity when single mutations are combined. We again simulated replicate walks under each model on a single landscape. For the first m mutations that fixed, we imagined engineering each into the wild type and measuring their fitness effects (as difference in r). In keeping with the literature, we predicted fitness on the basis of the additivity of the r model (i.e., summing fitness effects). Epistasis is then defined as ε = robs − rpred. For beneficial mutations ε < 0 and ε > 0 are termed antagonistic and synergistic epistasis, respectively.
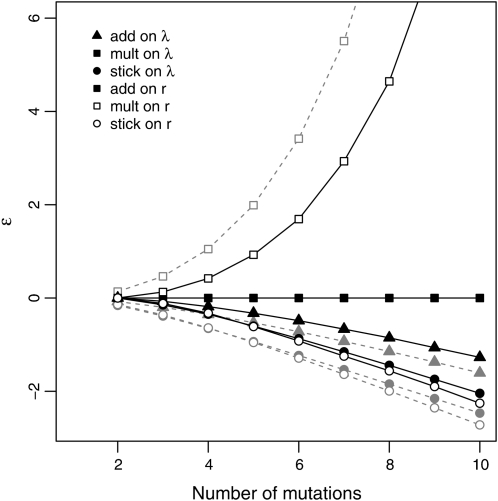
The patterns of ε (Figure 6) are similar to those observed in Figure 5. The stickbreaking models show strong antagonistic epistasis, add on λ shows moderate antagonistic epistasis, add on λ (mult on r) shows no epistasis (by definition), and mult on r shows strong synergistic epistasis. In fact, it is easy to understand why ε (Figure 6) and fitness effect (Figure 5) must follow the same basic pattern. Consider two mutations, A1 and A2. If ε < 0 (antagonistic epistasis), then robs < rpred. Letting Δr denote fitness effect on r and rwt denote the wild-type fitness, this implies that rwt + Δr1|wt + Δr2|1 < rwt + Δr1|wt + Δr2|wt, which implies Δr2|1 < Δr2|wt, or a diminishing effect. Similar arguments can be made for ε = 0 and ε > 0.
Figure 6 .
Mean epistasis (ε = robs − rpred) as a function of the number of mutations (m) composing the measured genotype. Predicted values assume additivity on r. Genotypes are always formed from the first m mutations in a walk. Shaded dashed lines are theoretical predictions from Table 3.
In the experimental evolution literature, the commonly observed patterns of epistasis are (1) diminishing effects, where the same mutation has smaller effects on more fit backgrounds and conversely larger effects on less fit ones, and (2) antagonistic epistasis is more frequent than synergistic epistasis (Burch et al. 2003; Sanjuán and Elena 2006). For example, Bull et al. (2000) found that the fitness effect of one mutation (1727T) in the bacteriophage ϕX174 decreased across four backgrounds of increasing fitness. Recently, Chou et al. (2011), Khan et al. (2011), and Kvitek and Sherlock (2011) all showed a general pattern of diminishing returns epistasis when beneficial mutations were inserted into closely related backgrounds. Similar results are found in double-mutant studies. Trindade et al. (2009) found that when antibiotic resistance mutations in E. coli are combined, 42% of those showing significant epistasis are antagonistic, while only 15% show synergistic epistasis. Rokyta et al. (2011) inserted nine beneficial single mutations in a G4-like bacteriophage to form 18 double mutants and found antagonistic epistasis for all 18. Finally, a synthesis of 21 studies by Sanjuán and Elena (2006) indicated that antagonistic epistasis is more prevalent in viruses and prokaryotes, while synergistic or no epistasis is more common in eukaryotes. Thus studies have tended to show patterns of epistasis broadly consistent with the two stickbreaking models and additivity on λ.
It is important to clarify that the values of ε and hence the patterns of antagonistic vs. synergistic epistasis depend on the null model used to calculate predicted fitness. It is easy to see what the patterns would be under other nulls by noting that the “predicted” and observed labels in Figure 6 are arbitrary. Figure 6 can also be thought of as showing the fitness divergence between different models as mutations of the same intrinsic effect are introduced. For any null and alternative model, the distance between them corresponds to the values of ε.
Conclusion
The stickbreaking model is based on the simple idea that mutational fitness effects should diminish the nearer the background is to the maximum fitness boundary. It solves the bookkeeping problem while also producing patterns of fitness trajectory and epistasis broadly consistent with experimental findings. The next important step is to develop statistical methods for fitting and testing the stickbreaking model on real data. Like the additive and multiplicative models, stickbreaking is too simple to be biologically correct. Rather, our hope is that stickbreaking is mathematically tractable like those models, but also captures a basic biological property and provides an explanatory power that those models seem to miss.
Appendix
Distribution of Total Fitness Effects After m Steps of Adaptation
We show here that there are three limiting distributions for the fitness achieved after m steps in a walk: the normal distribution under additivity, the log normal under the multiplicative model, and negative log normal under stickbreaking.
Denote the “intrinsic” fitness effect of the beneficial mutation Ai by xi. For the additive model xi = Δwi, for the multiplicative model xi = si, and for the stickbreaking model xi = ui. Note that ui and si are just different ways to scale Δwi. That is, ui = Δwi/(d − wwt) and si = Δwi/wwt. Therefore
and similarly
Throughout we assume that the walks evolve according to SSWM conditions. We also assume that for each value of M, x1, x2, … , xM is fixed. That is, we use the same set of intrinsic fitness effects for replicate walks.
Consider an adaptive walk of length m. Let Yi be the intrinsic fitness effect of the mutation arising at step i. The joint distribution of Y1, Y2, … , Ym can be described as
| (A1) |
Note that Y1, Y2, … , Ym are dependent random variables. The dependence comes from the fact that once a mutation is used in a walk it will not be used again, thus reducing the number of available mutations at each step. However, if M is large enough, we show below that Y1, Y2, … , Ym are approximately independent and identically distributed. Let x(1) = max{x1, x2, … , xM}. Note that . Therefore,
| (A2) |
Here is where the relationship between M and m becomes important. We assume that m is an order of magnitude smaller than M. More precisely, we assume that as M → ∞, then m ln(M)/M → 0 and m → ∞. It follows from extreme value theory that for large M, x(1) ≈ c ln M. [More precisely x(1)/ln(M) converges to a constant c as M → ∞.] Taking the limit as M → ∞ in inequality (A2) reveals that Thus for large M, , for k = 1, 2, … , m. If we replace the denominators in Equation A1 with , then this leads to the assumption that Y1, Y2, … , Ym are approximately independent and identically distributed with Note that and . Both converge as M → ∞.
Normal, Log Normal, and Negative Log Normal
Below we review the three central distributions associated with m steps of an adaptive walk. The normal distribution is given by
| (A3) |
If X follows the normal distribution, we say that V = eX follows the log-normal distribution with probability density function given by
| (A4) |
and if V follows the log normal, we say that W = 1 − V follows the negative log-normal distribution with probability density function given by
| (A5) |
Note that the parameters μ and σ appear in all three probability densities, but must be interpreted differently in each. While μ represents the mean and σ2 represents the variance of a normal, the mean of the log-normal distribution is and the variance of a log normal is . If W is negative log normal, then the mean is and the variance of a negative log normal is the same as that of a log normal.
Now if Yi represents the fitness differences, then the fitness after m steps is given by Under the additive model, the central limit theorem applies and the distribution of w1,2,…,m will be approximately normal with mean and . However, if the multiplicative model applies, then . This implies The central limit theorem now applies to Thus and
So w1,2,…,m/wwt is distributed log normal.
Under the stickbreaking model, is approximately negative log normal, where μ = mE(1 − Y) and σ2 = mVar(1 − Y) and the formulas are analogous to those of the log normal.
The assumption that M is large enough so that m is an order of magnitude smaller yet m is still large enough for the central limit theorem to apply is not always going to be achieved. Simulations can help in determining the degree to which violation of assumptions matters.
Number of Steps vs. Time to Adaptation
Under SSWM conditions the time it takes a mutation with selection coefficient s to arise and fix in the population is exponentially distributed with mean 1/Nμs, where μ is the beneficial mutation rate and N is the population size. Now if there are a total of M beneficial mutations available, the time in generations to fixation of the first beneficial mutation is on average where is the average selection coefficient among the M available mutations. All of our theory is based on the asymptotic results formed by taking the limit as M goes to infinity. As M goes to infinity the time to fixation converges to zero. So a timescale change is required. If we assume that 1 unit of time is equivalent to NμM generations, then the mean time for the first beneficial mutation to fix using this timescale will be exponentially distributed with mean . In the limit as M goes to infinity, converges to the mean of the distribution of beneficial mutations, which we denote by γ. We now use an extension of the central limit theorem that states converges to the normal distribution as t → ∞. This shows that the time limit prediction of the additive model is normal. Applying the analogous central limit theorem result to shows that the multiplicative model leads to a log-normal distribution. Applying the analogous central limit theorem result to shows that the stickbreaking model leads to a negative log-normal distribution.
Footnotes
Communicating editor: M. K. Uyenoyama
Literature Cited
- Barrett R. D. H., M’Gonigle L. K., Otto S. P., 2006. The distribution of beneficial mutant effects under strong selection. Genetics 174: 2071–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247 [DOI] [PubMed] [Google Scholar]
- Betancourt A. J., 2009. Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage MS2. Genetics 181: 1535–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer S., Chappey C., Parkin N. T., Whitcomb J. M., Petropoulos C. J., 2004. Evidence for positive epistasis in HIV-1. Science 306: 1547–1550 [DOI] [PubMed] [Google Scholar]
- Bull J. J., Badgett M. R., Wichman H. A., Huelsenbeck J. P., Hillis D. M., et al. , 1997. Exceptional convergent evolution in a virus. Genetics 147: 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., Badgett M. R., Wichman H. A., 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17: 942–950 [DOI] [PubMed] [Google Scholar]
- Burch C. L., Chao L., 1999. Evolution by small steps and rugged landscapes in the RNA virus ϕ6. Genetics 151: 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch C. L., Turner P. E., Hanley K. A., 2003. Patterns of epistasis in RNA viruses: a review of the evidence from vaccine design. J. Evol. Biol. 16: 1223–1235 [DOI] [PubMed] [Google Scholar]
- Chevin L., 2010. On measuring selection in experimental evolution. Biol. Lett. 7: 210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H., Chiu H., Delaney N., Segrè D., Marx C., 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper V. S., Lenski R. E., 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736–739 [DOI] [PubMed] [Google Scholar]
- Cowperthwaite M. C., Bull J. J., Ancel Myers L., 2005. Distributions of beneficial fitness effects in RNA. Genetics 170: 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., Murray A. W., 2007. The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406 [DOI] [PubMed] [Google Scholar]
- Donnelly P., Joyce P., 1989. Continuity and weak convergence of ranked and size-biased permutations on an infinite simplex. Stoch. Proc. Appl. 31: 89–103 [Google Scholar]
- Elena S. F., Lenski R. E., 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–398 [DOI] [PubMed] [Google Scholar]
- Elena S. F., Davila M., Novella I. S., Holland J. J., Domingo E., et al. , 1998. Evolutionary dynamics of fitness recovery from the debilitating effects of Muller’s ratchet. Evolution 52: 309–314 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618 [DOI] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103: 127–144 [PubMed] [Google Scholar]
- Gillespie J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, New York [Google Scholar]
- Hartl D. L., Clark A. G., 1997. Principles of Population Genetics, Ed. 3 Sinauer Associates, Sunderland, MA [Google Scholar]
- Holder K., Bull J., 2001. Profiles of adaptation in two similar viruses. Genetics 159: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce P., Rokyta D. R., Beisel C. J., Orr H. A., 2008. A general extreme value theory model for the adaptation of DNA sequences under strong selection and weak mutation. Genetics 180: 1627–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H., Burns J., 1981. The molecular basis of dominance. Genetics 97: 639–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman S. A., 1993. The Origins of Order. Oxford University Press, New York [Google Scholar]
- Khan A., Dinh D., Schneider D., Lenski R., Cooper T., 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Kim Y., Orr H. A., 2005. Adaptation in sexuals vs. asexuals: clonal interference and the Fisher–Muller model. Genetics 171: 1377–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., Tkačik G., Plotkin J., 2009. The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. USA 106: 18638–18643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D., Sherlock G., 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7: e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Travisano M., 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur R., 1957. On the relative abundance of bird species. Proc. Natl. Acad. Sci. USA 43: 293–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C., Joyce P., Wichman H., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella I. S., Duarte E. A., Elena S. F., Moya A., Domingo E., et al. , 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA 92: 5841–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2002. The population genetics of adaptation: the adaptation of DNA sequences. Evolution 56: 1317–1330 [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2003. The distribution of fitness effects among beneficial mutations. Genetics 163: 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil C., Taillie C., 1977. Diversity as a concept and its implications for random communities. Bull. Int. Stat. Inst. 47: 497–515 [Google Scholar]
- R Development Core Team, 2009. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rokyta D. R., Beisel C. J., Joyce P., Ferris M. T., Burch C. L., et al. , 2008. Beneficial fitness effects are not exponential for two viruses. J. Mol. Evol. 67: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Abdo Z., Wichman H. A., 2009. The genetics of adaptation for eight microvirid bacteriophages. J. Mol. Evol. 69: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Joyce P., Caudle S., Miller C., Beisel C., et al. , 2011. Epistasis between beneficial mutations and the phenotype-to-fitness map for a ssdna virus. PLoS Genet. 7: e1002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., de Visser J. A. G. M., Gerrish P. J., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045 [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Elena S. F., 2006. Epistasis correlates to genomic complexity. Proc. Natl. Acad. Sci. USA 103: 14402–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R., Moya A., Elena S. F., 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101: 8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra S. E., Bataillon T., Gifford D. R., Kassen R., 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol. 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander O. K., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dymanics of mutational effects. PLoS Biol. 5: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade S., Sousa A., Xavier K., Dionisio F., Ferreira M., et al. , 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5: e1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D. M., Delaney N. F., DePristo M. A., Hartl D. L., 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114 [DOI] [PubMed] [Google Scholar]
- Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J., 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424 [DOI] [PubMed] [Google Scholar]
- Wichman H. A., Millstein J., Bull J. J., 2005. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R., Schneider D., Winkworth C., Riley M., Lenski R., 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 103: 9107–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]