Abstract
Gene transposition puts a new gene copy in a novel genomic environment. Moreover, genes moving between the autosomes and the X chromosome experience change in several evolutionary parameters. Previous studies of gene transposition have not utilized the phylogenetic framework that becomes possible with the availability of whole genomes from multiple species. Here we used parsimonious reconstruction on the genomic distribution of gene families to analyze interchromosomal gene transposition in Drosophila. We identified 782 genes that have moved chromosomes within the phylogeny of 10 Drosophila species, including 87 gene families with multiple independent movements on different branches of the phylogeny. Using this large catalog of transposed genes, we detected accelerated sequence evolution in duplicated genes that transposed when compared to the parental copy at the original locus. We also observed a more refined picture of the biased movement of genes from the X chromosome to the autosomes. The bias of X-to-autosome movement was significantly stronger for RNA-based movements than for DNA-based movements, and among DNA-based movements there was an excess of genes moving onto the X chromosome as well. Genes involved in female-specific functions moved onto the X chromosome while genes with male-specific functions moved off the X. There was a significant overrepresentation of proteins involving chromosomal function among transposed genes, suggesting that genetic conflict between sexes and among chromosomes may be a driving force behind gene transposition in Drosophila.
INTERCHROMOSOMAL gene transposition, the movement of genes between chromosome arms, has historically been regarded as relatively rare in Drosophila, on the basis of the observation that homology of the chromosome arms (referred to as “Muller elements”) is generally maintained among the species within the genus (Muller 1940). This observation has been upheld by the mapping of molecular markers between different species (Ranz et al. 2003) and borne out again by a comparison of orthologs across the 12 completed genomes (Bhutkar et al. 2007). Nonetheless, specific instances of genes apparently moving between chromosomes have been reported since the 1970s (Ranz et al. 2003), including the recent report of a gene movement that has contributed to reproductive isolation between Drosophila melanogaster and D. simulans (Masly et al. 2006).
Gene transpositions occur through gene duplication, either by a DNA-based mechanism (ectopic recombination) or by an RNA-based mechanism (retrotransposition by reverse transcription of an mRNA). Once a duplicate has arisen in a new location, the original copy can be maintained or the original copy can be lost, resulting in an apparent map change of the locus. Hereafter, we refer to the former case as duplicative transpositions and the latter as relocations (Meisel et al. 2009).
With the sequencing of the genomes of 12 Drosophila species (Clark et al. 2007), transpositions could finally be systematically identified at gene-by-gene resolution. Several studies that looked at gene transpositions at a genome-wide scale have since been published. Bhutkar et al. (2007) found ∼500 positionally relocated genes, although these amounted to <5% of all orthologs. Bai et al. (2007) focused on duplicated retrogenes that changed chromosome arms and found ∼0.5 retrogenes transposed per million years. More recently, Meisel et al. (2009) studied gene duplicates created by all mechanisms and found 368 duplicative transpositions and 195 relocations. Although these studies have expanded the knowledge of gene transpositions considerably, they are limited to gene families with simple lineage-specific transposition events. This is because they considered only the movement of single-copy orthologs (relocations) or unambiguous gains along only a single lineage (e.g., changes from one to two copies, where all other species have one copy). One result of this limited set of movements is that many gene families, especially the larger ones, have been overlooked. This oversight is significant because most of the genes identified as transposed in early studies of Drosophila were members of large gene families dispersed across many arms, e.g., rRNAs (Alonso and Berendes 1975), actins (Fyrberg et al. 1980), tubulins (Sánchez et al. 1980), and histones (Felger and Pinsker 1987). Thus, we might predict that a large proportion of transposed genes will have been missed. Studies of lineage-specific transpositions will become even more limited in the future, due to the fact that phylogenetic patterns of gain and loss will become more complicated as more genomes are added to the phylogeny. In this article, we attempt to expand the set of families examined for evidence of transposition.
There are many interesting evolutionary dynamics introduced by duplicated genes transposing to new locations. For instance, there are sufficient reasons to presume that the evolution of the transposed (daughter) gene should be different from that of the original (parent) gene. The movement puts the transposed gene into a new genomic context, and the change in spatial environment can result in changes to many variables that affect the fate of the new gene: e.g., mutation rate (Marques-Bonet et al. 2007), recombination rate (Zhang and Kishino 2004), and regulatory environment (Vinckenbosch et al. 2006). The transposed copy can also have immediate changes in expression pattern on the basis of the different regulatory elements in the vicinity and changed chromatin environments. Although a change in the local and temporal expression pattern of a gene is more likely to be detrimental, it is also a source for accidental novelties that could promote the retention of the gene. Dispersed duplications are also less likely to experience homogenizing gene conversions that could hinder the divergence process (Ohta and Dover 1983; Casola et al. 2010). For all of these reasons, previous studies have found that transposed gene duplicates evolve faster than their original counterparts in rodents and primates (Cusack and Wolfe 2007; Han et al. 2009), and initial studies in Drosophila seemed to bear this pattern out (Clark et al. 2007).
In addition to having an effect on the transposed genes, the process of gene transposition is itself subject to multiple evolutionary forces. Betrán et al. (2002) found an excess of retrogenes transposing from the X chromosome to the autosomes in D. melanogaster, but not the reverse. More recent studies have confirmed this excess of retrogenes moving off the X and neo-X chromosomes in multiple Drosophila species (Dai et al. 2006; Bai et al. 2007; Meisel et al. 2009; Vibranovski et al. 2009). Unlike retrogenes, DNA-based transpositions do not appear to consistently move in excess from the X to autosomes: while there is an excess found for DNA-based relocations in Drosophila (Bhutkar et al. 2007; Meisel et al. 2009; Vibranovski et al. 2009; Moyle et al. 2010), there is not an excess of DNA-based duplicative transpositions (Meisel et al. 2009). However, this last study has been criticized on the grounds that too few instances of DNA-based transpositions were found for conclusive inference (Zhang et al. 2010). The same patterns of movement are repeated in mammals, with an excess of X-to-autosome gene movements for RNA-based duplicative transpositions (Emerson et al. 2004; Potrzebowski et al. 2008) and for DNA-based relocations (Moyle et al. 2010), but not for DNA-based duplications (Jiang et al. 2007; Han and Hahn 2009). There are several hypotheses that attempt to explain the excess of X-to-autosome gene traffic, including sexually antagonistic selection (Rice 1984; Wu and Xu 2003; Connallon and Clark 2011), escape from X inactivation (Betrán et al. 2002), meiotic drive (Meiklejohn and Tao 2010), and dosage compensation (Bachtrog et al. 2010). All of these hypotheses invoke natural selection, differing only in the particular selective agent responsible for driving movement. All of these hypotheses also predict that the pattern of X-to-autosome gene traffic should be consistent regardless of the mechanism of duplication, although they may differ in which types of duplication events are most able to respond to selection. Having a larger set of transposed genes would enable us to address whether X-to-autosome movement is truly limited to retrogenes or whether it is a general pattern found across all classes of transposed genes.
In this article, we show that by using well-studied phylogenetic inference methods we can better utilize the wealth of information provided by whole genomes to more completely identify the set of gene transpositions, including gene families with multiple parallel transpositions across a phylogeny. We first introduce our parsimony-based method and demonstrate its accuracy and then apply it to the whole genomes of 10 Drosophila species.
Methods
Data
We used the GLEANR gene annotations from the whole genomes of D. simulans, D. erecta, D. yakuba, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis, D. virilis, D. grimshawi, D. sechellia, and D. persimilis (Clark et al. 2007) and the D. melanogaster annotations from FlyBase release 4.3. The homologous gene families were defined by a clustering scheme called fuzzy reciprocal blast (FRB) that uses pairwise sequence similarities within and across the 12 genomes (Clark et al. 2007; Hahn et al. 2007). Another independent set of gene family definitions was produced using the Markov clustering (MCL) algorithm (Enright et al. 2002) for comparison. The mapping of each gene to the Muller elements was done using the gene-scaffold-chromosome mapping from Schaeffer et al. (2008). Only five Muller elements (A–E) were used; we excluded genes on the small fourth and the repeat-rich/gene-poor Y chromosome. The sequences of each gene in each gene family were aligned using MAFFT (Katoh et al. 2005). The gene trees were constructed as in Hahn et al. (2007), using the neighbor-joining method. Although the initial data set included the genes of D. sechellia and D. persimilis, these genomes were excluded from our downstream analyses because we suspected the higher number of gene duplications and losses in these lineages (Hahn et al. 2007) were due to the lower-coverage genome assemblies that could affect our results.
Parsimonious ancestral reconstruction
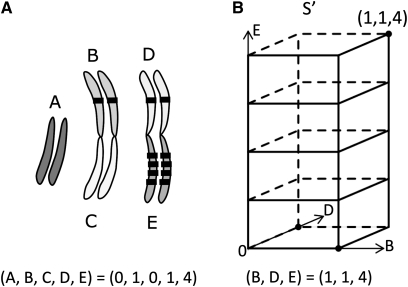
The parsimony approach uses a set of simplifying assumptions and finds the smallest number of changes that can explain the variance we see among extant taxa, while at the same time inferring the ancestral states of a character. In our problem, we can think of the locations of the gene members of a family as a character and formulate the problem as finding the ancestral gene location distribution that minimizes the total change across the whole tree. This can give us at least a lower bound on the number of changes, in our case the number of transpositions. As Sankoff and Rousseau (1975) elegantly showed, the problem of parsimonious ancestral reconstruction is a special case of a Steiner tree problem that can be solved generally with dynamic programming. Our contribution in this study is to find a way to encode the location distribution of multiple genes and define the state space, S, and the costs, d, for moving between states. Once we have a state space, the mapping between the inner nodes and the states that minimize the total distance across the tree is found easily by applying the Sankoff algorithm. In the case of Drosophila, there is a straightforward homology between the chromosome arms across the species we are comparing (i.e., the Muller elements). So we divided the genome by the chromosome arms into m different nonoverlapping partitions, x1 . . . xm ∈ X, where X is the set of all partitions that correspond to the total genome. We encoded the distribution of a gene family as a vector u = (u1, u2, … , um) with each element specifying the number of copies on each of the arms. Let ui (i = 1, . . . , m) be the number of genes that reside on each partition xi for a certain gene family f (i.e., the number of genes on each chromosome arm) (Figure 1). Then we define the state space S as the Manhattan metric space S = Nm, where
This type of distance is biologically realistic, as a relocation into a different partition involves a gain in a new location and a loss in the original location.
Figure 1 .
Character encoding and state space. (A) The genomic distribution of a gene family is encoded as a vector with each element corresponding to the number of genes in each partition (chromosome arm or Muller element). Shown is an example of a gene family with six genes distributed across three arms in D. melanogaster. (B) The total space, S, is an n-dimensional space when there are n partitions, but we need to consider only the subspace, S′, up to the maximum number of genes in each partition. In this case, we consider only a three-dimensional space, where each axis represents each arm containing at least one gene, and up to point (1, 1, 4). Moving in the increasing direction on any axis represents a gene gain on the corresponding chromosome arm.
Our operations for moving between states are gains and losses, and each operation entails a cost. We experimented with an equal additive cost of 1 for each gain and loss and a slightly bigger cost of 1.1 for gain compared to 1 for loss. We did not have a separate operation for transpositions and thus did not have an explicit cost for transpositions. Instead, we inferred transpositions after the reconstruction, by identifying specific gains as transpositions. Whenever there was a gain from 0 to 1 in any arm, we inferred the gain to be a transposition. The origin of the transposition was determined to be the arms containing homologs in the parental node. This method underestimates the number of transpositions because only the first gain on an arm is counted as a transposition and any subsequent transposition that lands on the same arm is not counted as a transposition, only as an ordinary gain. Since recurrent births of a whole gene family are not biologically reasonable, we also added a special penalty for the transitions originating from the zero vector (i.e., [0, 0, 0, 0, 0]). We used a penalty of 5 to ensure that there are not any recurrent births of a family within the Drosophila tree.
Filtering based on gene trees
As an additional way to remove errors in the ancestral reconstruction, we used the topologies of the gene trees from each reconstructed family. The only scenario that could lead to an overestimate of transpositions is when the event we identified is not actually a transposition, but a loss of a gene that predated the most recent common ancestor (MRCA) of the 10 species we studied. Because we count only the first occurrence of a gene on a new chromosome arm as a transposition, as long as the gene first appears on the new arm after the MRCA, the count of the transposition should be correct even if the branch it is mapped to may be inaccurate. Fortunately, we can identify these cases by checking the gene tree. If the genes on different chromosomes both existed before the MRCA, the genes on different chromosomes should be genetically distant from each other, and the reconciled gene tree should show a duplication node before the MRCA leading to two different clades corresponding to two chromosomal positions (Supporting Information, Figure S1A). We reconciled gene trees with the species tree using NOTUNG (Durand et al. 2006) with three different thresholds of bootstrap support (90, 60, and 0). If the reconciled topology showed the arrangement described above (Figure S1A), we considered the transposition to be older than the MRCA and removed these transpositions from further analyses. We report here the results based on the bootstrap threshold of 60; there were no qualitative differences in the results when using different bootstrap thresholds.
Simulation and accuracy
We ran a total of 20 types of simulations, with a combination of five categories of rates described in Table S1 and each starting from four different root states (1, 0, 0, 0, 0), (1, 1, 0, 0, 0), (1, 1, 1, 0, 0), and (1, 1, 1, 1, 0). The state space was defined as a five-dimensional space limited by the zero vector (0, 0, 0, 0, 0) and the arbitrary maximum of (10, 10, 10, 10, 10). For each set we ran 200 simulations on the Drosophila phylogeny of 10 species (Clark et al. 2007). We compare the rates either by varying duplication, loss, and transposition at the same time—to preserve the ratio of events—or by varying only the transposition rate to see the effect of transpositions on the accuracy. The infinitesimal transition matrix (Q) used to simulate data was defined on the basis of the transition rates (Table S1), so that all state transitions by one gene loss had a transition rate of μ and all state transitions by one gene increase had a transition rate of λ, except that any transition involving a gain on a new chromosome (going from 0 to 1 in any arm) had a transition rate of ν. The diagonal entries were one minus the row sums, and the rest of the entries in the substitution matrix were filled with zero. We set the “medium” transition rate matrix to have a birth rate of 0.0012/genes per MY and a loss rate of 0.0015/genes per MY; these gain and loss rates are of a similar order of magnitude with the rates estimated previously using a likelihood-based method (Hahn et al. 2007). Starting from the root along each branch, we randomly sampled the time lag for staying in the same state from the exponential distribution with the rate corresponding to the correct diagonal entry in the matrix Q. After the time lag we randomly sampled the new state according to the probability corresponding to the correct row of the matrix Q. We repeated this procedure for the time equal to the branch length or until the absorbing state was reached and continued to the next branch until the leaves of the tree. We also recorded the true number of transpositions for each branch while we simulated the families.
To assess the accuracy of our reconstruction algorithm we compared the true inner-node states with the reconstructed states and reported the percentage of correct reconstructions from 200 families for each inner node. We also compared the true count of events for each branch and compared them with the count of events inferred by the reconstruction. Again the percentage of correct counts for gain, loss, and transpositions from 200 families was reported for each branch.
Inferring the mechanism of transposition
We inferred the molecular mechanism of duplications by comparing the exon numbers of the original (parental) locus and the transposed locus. For duplicative transpositions, we used the parental genes within the same species, and for relocations we used the parental genes in the closest sister species. We inferred a DNA-based duplication when there was at least one parental gene with more than one exon and at least one transposed gene with more than one exon. We inferred retrotransposed duplicates when all parental genes had more than one exon and all transposed genes had only one exon. For all other cases we classified the mechanism as ambiguous. If the alignments of the genes were <60% of the total length of the genes, they were also classified as ambiguous.
Sequence analysis
Because our goal was to compare the evolution of the daughter gene sequence with that of the parent gene sequence, we examined only duplicative transpositions that retained the original sequence. The branch of the transposition event was mapped to the reconciled gene tree and we tested the two branches right after the transposition event, the transposed branch leading to the duplicated gene and the sister branch leading to the original gene (Figure S2). The test for higher dN/dS ratios on the transposed branch was done using the likelihood-ratio tests in PAML (Yang 1998) with five different models (Figure S2). There are two ways for the transposed branch to have higher dN/dS than the background using this approach—it can be significantly higher under model B compared to model A or it can be significantly higher under model E compared to model C. Likewise, there are two ways for the sister (parent) branch to have higher dN/dS (“model C vs. model A” or “model E vs. model D” in Figure S2). We denoted a branch as accelerated only if the branch had significantly higher dN/dS in both of the tests.
Testing for direction in movements
Large transpositions can move multiple linked genes at the same time, so the number of events can be different from the number of genes identified. This can be a problem when testing for trends in the data since we are counting multiple genes as independent samples when they may not be. To avoid this potential problem we scanned the transposed genes that we had identified to find linked genes. When two genes transposed on the same branch and were adjacent to each other, or at most three genes apart, we merged the transpositions into one event. Testing for direction of movements between arms was done using the counts of movements and not the counts of genes. Pericentric movements were defined as gene movements between Muller elements that correspond to two fused arms of a metacentric chromosome, on the basis of the karyotype of the species. These movements were excluded from the analyses because they could be confounded with pericentric inversions.
There were two kinds of uncertainties when inferring the direction of movements.
-
i.
There is one parsimonious ancestral state but the ancestral state has genes on more than two chromosome arms. We excluded these cases because we cannot distinguish which of the arms the movement originates from.
-
ii.
There is more than one parsimonious ancestral state. This happens mostly in relocations where the two children nodes have genes on reciprocal arms and the ancestral state can be either one of them. In this case if we choose the ancestral state to be the same as child 1, the movement is automatically assigned to the branch leading to the other child 2, and the direction of the movement is determined to be from child 1’s state to child 2’s state. But the choice of the ancestral state is arbitrary and the direction of the movement may just as likely be the opposite. Therefore, we exclude these cases as well.
The expected proportions of X → autosome (A) movements and A → X movements were calculated using the formula presented in Betrán et al. (2002), but with the number of genes and length of arms corresponding to weighted averages among the species considered here. The weights were proportional to the number of transpositions found on each branch (Table S2).
Results and Discussion
Thousands of gene duplications among Drosophila genomes
We applied our parsimonious ancestral reconstruction method to the gene families of 10 Drosophila species. To ensure that the gene family annotations were well supported, we considered only gene families found in at least 5 species. Under these conditions, we were able to study 11,108 gene families containing 121,466 genes in total. The parsimony method described above allows us to infer the minimum number of duplications and losses in total, regardless of the chromosomal location of genes (3 gene families had to be excluded from the analysis because they were too large; see below for details). Among these families we identified 2696 gene duplications and 5751 gene losses across the phylogeny. Since the phylogeny comprises a total branch length of ∼393 MY (Clark et al. 2007), the rate of duplication is ∼6.9 genes per MY, while the rate of loss is ∼14.6 genes per MY. This result is based on unweighted parsimonious reconstruction with a cost scheme that penalizes gains more than losses to minimize the number of transpositions inferred (see below). When we used a scheme of equal costs for gain and loss, it still resulted in more gene losses than gains, with 2716 genes duplicated and 5775 genes lost. Previously, the gene duplication and loss rate was estimated using a likelihood framework to be ∼17 genes per genome per million years (Hahn et al. 2007). The rates from our parsimony method are somewhat smaller, as expected from a parsimony method relative to a likelihood method.
Fourteen percent of all gene duplicates in Drosophila are transpositions onto a different chromosome arm
We found a total of 782 genes transposed between chromosome arms across the 10 species (Table S3); 142 gene movements were filtered out on the basis of the gene-tree topology. Of the total number of gene duplicates we observed, 14% (311/2279) were duplicates between chromosome arms (we exclude relocations from this count to make a fair comparison between intra- and interchromosomal duplication events). In addition to the 311 gene duplicates that retained both copies (duplicative transposition), there were 471 genes where the new duplicate survived on a different chromosome while the original copy was lost (relocation). Finding more relocations than duplicative transpositions may seem unexpected, but we can interpret this pattern as revealing the higher rates of losses compared to retentions after gene duplication, as is expected. In total, the rate of gene movement between chromosomal arms in Drosophila is ∼2 genes per million years, with slightly less than one gene gained by duplicative transposition every million years.
Because of our cost scheme, if there is a Muller element difference that precisely splits the Drosophila and Sophophora subgenera, it is more parsimonious to infer two independent losses on the two branches leading to each subgenus, rather than inferring one gain (transposition) and one loss on each Muller element on each branch, respectively. As a result, no relocations were identified on the two branches right below the root. We think this is conservative since without more information from an outgroup species we cannot confidently infer the state at the root. Although we have missed these and possibly some other transpositions on specific branches, we were able to identify a number of transpositions that were overlooked in previous studies, especially several occurrences of multiple gene transpositions within a family that resulted in complicated phylogenetic patterns (see below). In total, our data set contains 421 new gene transpositions that were not identified in previous studies (Bhutkar et al. 2007; Meisel et al. 2009; Vibranovski et al. 2009). While we have identified more moved genes than previous studies, we have also missed some movements. Among the 782 moved genes in our study, 361 of them overlapped with the high-confidence relocations in Bhutkar et al. (2007), but there were also 176 high-confidence relocations in this previous article that were not identified by our methods (Table S4). A total of 148 of the 176 were cases that we excluded due to the event being at the root of the tree (82/148), a problem in the assembly (33/148), or an invalidating gene-tree topology (27/148). Twenty-eight were movements that we missed because of some fault in our analysis (17/28, explained in Table S4) or because we excluded the D. sechellia and D. persimilis lineages from our study (11/28).
More than half of the movements, 461 (59%) in total, were DNA-based duplications, 111 (14%) were RNA-based duplications, and 210 (27%) were ambiguous. These data indicate that there were four times as many DNA-based transpositions as RNA-based. Previously, Bhutkar et al. (2007) estimated that 24% of the relocated genes identified were due to retrotransposition events. We observed that retrogenes are more likely to keep the parental copy compared to DNA-based duplicates (P = 6.57e-08; Figure S3); i.e., they are less likely to be relocated. This is expected if we consider the fact that DNA-based duplications often bring along the flanking regions around the gene, while retrotransposed duplicates lose the introns and the flanking noncoding regions of the original gene. Since retrotranspositions are less likely to be able to recreate the whole range of expression patterns of the original gene, they are less likely to replace the original gene altogether compared to DNA-based duplicates. This explanation is also consistent with the broader spatial and temporal expression pattern found in relocated genes compared to transposed genes that have the original copy retained (Meisel et al. 2009). Alternatively, there may also be a mechanistic reason retrogenes are less likely to be relocated: DNA- and RNA-based mechanisms differ not only in the precise molecular steps that produce new gene duplicates but also with respect to whether the initial mutation is actually duplicative. In the case of RNA-based mechanisms, the duplication does not result in the loss of copies on any chromosomes and is therefore truly duplicative. Because the original parental locus is not lost during the initial duplication event that creates a retrogene, a relocation can occur only when a subsequent mutation causing the loss of the parental gene arises and fixes. On the other hand, for DNA-based transpositions that arise through nonallelic homologous recombination (NAHR), it is almost always the case that ectopic recombination results in two meiotic products: one with an additional copy and one missing a copy. In this mechanism, the duplication event is not truly duplicative; it merely involves the movement of a locus from one haploid genome to another. Therefore, at least in male Drosophila (where both meiotic products can be present in gametes), both the duplication and loss alleles may be segregating in the next generation, increasing the likelihood of relocation.
Assessing the accuracy of the n-dimensional parsimony method
To estimate the accuracy of our reconstruction, we tested the algorithm against simulated gene families. We assessed the accuracy first by changing the rate of all events (gain, loss, and transposition), while maintaining the relative ratio of events, and then by changing only the rate of transposition events while gain and loss rates were held constant. The results show that the accuracy decreases as the rate of transposition events increases, as would be expected. However, with realistic rates the accuracy is at least 95% for the total count of transpositions on each branch (Figure 2), across all rate categories. The accuracy of the ancestral states at each node was at least 60%, with the lowest values for the ancestral states of nodes near the root (Figure S4). When we compare the inferred count of transpositions to the true count of transpositions, we see that the inferred counts are smaller than the true counts on most branches (Figure S5).
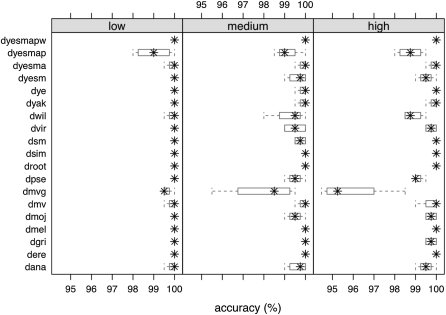
Figure 2 .
Accuracy of the count of transposition events on each branch measured by the percentage of correct counts of 200 runs on each branch of the phylogeny. Branch labels are explained in Figure S7. Simulations are shown under low (0.00004), medium (0.0002), and high (0.0004) transposition rates. Box plots are based on the four sets of runs starting from different root states.
We observed a trade-off between the number of losses and the number of transpositions. This is because one can explain the same state by inferring either a transposition to the new location in one branch or losses of the corresponding location on the neighboring branches of the phylogenetic tree. In general, we found that the inferred counts of events were always lower than the true counts, but for the simulations with the lowest rate there were a few cases where we overestimated the number of transpositions by inaccurately inferring transpositions instead of the true case of multiple losses. By assigning slightly higher costs for gains compared to losses (1.1 vs. 1), we found more losses and fewer transpositions. Because we are most interested in accurately identifying gene transpositions, we used the model with the higher cost for gains in all the results reported in this article.
We also compared the accuracy between parsimony that ignores branch length (unweighted) and parsimony that takes into account the branch length by weighting the costs accordingly (weighted). The accuracy between weighted and unweighted parsimony was comparable, but weighted parsimony tended to infer more events than unweighted parsimony by splitting events into longer branches instead of inferring one event on a short branch. Again, because we wanted to be conservative on the count of transpositions, we decided to use unweighted parsimony for downstream analyses.
To evaluate the effect of gene family definition on the inferred transpositions, we ran the analyses on a different data set of gene families prepared with an independent method of clustering. The FRB clustering that we use in our main results produced 11,433 gene families with a median size of 12 (approximately one gene in each species) and a mean size of 12.93. In contrast, the MCL clustering used for comparison produced 8777 gene families with a median size of 13 and a mean size of 19.34. The variance between the gene family sizes was larger for the MCL (1502.83) compared to the FRB (66.93). The MCL clustering inferred 1728 transpositions while FRB inferred 936 transpositions before filtering; after filtering out duplicates older than the MRCA, the difference between the data sets decreased. The remaining MCL clusters resulted in 1094 transpositions after filtering. MCL clusters have larger families because they tend to merge families that are split apart in FRB clusters. There can be an overcounting of transpositions in MCL clusters if large families include many duplication events that predate the MRCA and coupled with several losses they appear as several independent transpositions. On the other hand, there can be an undercounting in FRB clusters if valid transpositions are split into new families because of accelerated sequence evolution in the transposed gene. Currently, we cannot determine what proportions of the differences are underestimates in the FRB clusters or overestimates in the MCL clusters. Gene family definition is an important source of uncertainty that is not captured in the simulation accuracy and warrants further investigation. In either data set, transposition between chromosome arms is common.
Sequence divergence after transposition
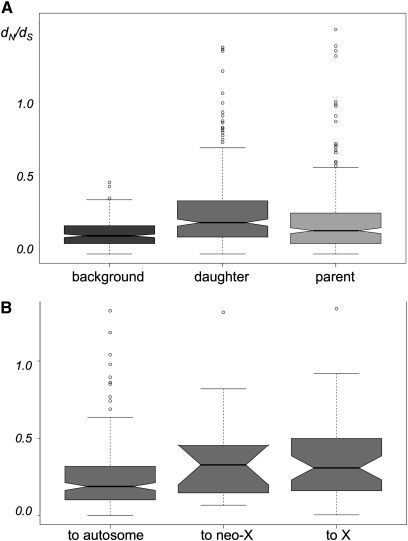
We hypothesized that the new genomic location of a transposed gene will facilitate the gain of new function. To test this hypothesis we compared the transposed (daughter) copy and the original (parent) copy in terms of their sequence divergence (cf. Cusack and Wolfe 2007; Han et al. 2009). The above results allow us to identify these pairs and to polarize them as parents and daughters. Naturally, our comparison was restricted to the 311 duplicative transpositions only and did not take into account the relocations because no such sequence comparisons can be made in these cases. To estimate the sequence divergence on the branches leading to the parent and daughter copies, we used the branch model of PAML to test whether either of the two branches just after the duplication event had experienced higher levels of nonsynonymous substitutions compared to synonymous substitutions (i.e., dN/dS). In 165 of 311 cases, there was a significantly increased dN/dS on at least one of the branches after the transposition. Fifty of these 165 had elevated dN/dS on both branches compared to the background and the remaining 115 had elevated values of dN/dS on only one branch. When we examined families where only one branch had experienced accelerated evolution, in 89 of 115 transpositions (77.4%) the daughter branch had an elevated dN/dS ratio compared to the 26 of 115 (22.6%) cases where the parent branch did. We also compared the distribution of dN/dS values between the daughter branch and the parent branch and found that the median value of dN/dS is higher on the daughter branch (Figure 3A). This result is consistent with previous findings in D. pseudoobscura, where there was an overall excess of accelerated evolution of derived transposed copies (Meisel et al. 2010), and is consistent with the comparison among single-copy orthologs (Clark et al. 2007). If we divide the movements into whether they go to an autosome, the X chromosome, or the neo-X chromosome, we found that genes landing on the X chromosome show elevated values of dN/dS (Figure 3B). Again, this agrees with the previous results among single-copy orthologs (see Supplementary Figure 7 in Clark et al. 2007). We did not test for positive selection explicitly, so we cannot tell whether the elevated values of dN/dS are due to adaptive evolution or relaxed selection. But the results suggest that the transposed copy is more likely to be functionally diverged than the original copy.
Figure 3 .
dN/dS estimates for branches following the transposition event. (A) The distribution of dN/dS estimates for the background branches, daughter (transposed) branches, and parent (original) branches. (B) The distribution of dN/dS estimates of the daughter branches with the daughter gene landing on the autosomes, the neo-X chromosome, or the X chromosome. The bottom and top of the box mark the lower and upper quantiles, while the band in the middle of the box marks the median. The ends of the whiskers extend to 1.5 × interquartile range (IQR). Outliers not included within the range of 1.5 × IQR are plotted as open circles.
Multiple independent transpositions within a gene family
Our study allowed us to identify new gene transpositions that were previously discarded due to complicated genomic distributions. An example is the family containing D. melanogaster gene CG32625, which is distributed along all five Muller elements. We infer that the gene has moved from the X chromosome to three different chromosome arms independently on the branches leading to D. simulans, D. willistoni, and the ancestral branch of D. mojavensis and D. virilis through both DNA-based and RNA-based duplications. We know little about this gene family other than that some members have weak sequence similarity to the gametocyte-specific factor 1 (GTSF1) protein and show enriched expression in the ovary of D. melanogaster. In total, we discovered 87 gene families with multiple movements on different branches of the phylogeny, comprising 193 gene transpositions (Table S5). There have been previous reports describing parallel transpositions from the X chromosome to the autosomes. In particular, cervantes, Ntf-2, and ran all gave rise to multiple retrogenes in independent lineages (Bai et al. 2007), and Meisel et al. (2009) found homologous genes independently transposing out of the independently evolved neo-X chromosomes of D. pseudoobscura and D. willistoni. These genes also show up in our data set (cerv, Ntf-2, and Prosβ2R2) and our set of gene families with multiple movements also shows an excess of genes moving out of the X and neo-X chromosomes (and see below). More than half of these gene families were uncharacterized, so we were not able to find any functional category significantly associated with these genes, but examples include odorant receptors, chemosensory receptors, actin-related proteins, and genes involved in RNA silencing (armi, mael), oogenesis (gus), chromosome segregation (CAP-D2, Smc5, and SA-2), and meiosis (fwd).
Chromosome segregation functions are enriched among transposed genes
Previous studies of retrogenes have found several gene families that appear to be recurrently retrotransposed (Bai et al. 2007; Tracy et al. 2010). The most prominent examples are the collection of nuclear-encoded mitochondrial genes with functions in energy production. Gallach and Betrán (2011) have argued that these genes are under sexually antagonistic selection due to high-energy production being beneficial to males but detrimental to females. Although several functional categories have been repeatedly found among retrotransposed genes, there was not any noticeable functional overrepresentation among DNA-based duplicates other than the few historical studies mentioned above. Finding similar functional annotations among genes with multiple parallel movements suggests the possibility that genes with particular functions could be transposed more often than others and could provide clues to the possible selective forces driving these movements.
We used GOrilla (Eden et al. 2009) and the DAVID annotation server (Huang et al. 2008) to find functionally enriched categories among the transposed genes. Both analyses gave similar results. With the GOrilla analyses, we found 15 gene ontology (GO) terms enriched among the transposed genes (Table S6). Among them, 6 terms are related to chromosomal activity. Some of these terms are in accordance with previous studies. For example, most of the genes under the term “structural constituent of cytoskeleton” are actins and tubulins, and studies of these gene families were among the earliest works that discovered homologous genes dispersed across several chromosome arms in Drosophila (Fyrberg et al. 1980; Sánchez et al. 1980).
The overrepresentation of transposed genes with functions related to chromosomes is unexpected and has not been reported before. This enrichment is more striking when we look at the results from DAVID. DAVID clusters the functional annotations that are closely related to each other—measured by the degree of shared gene members—so the results are reported in clusters of annotations (Huang et al. 2008). The cluster with the highest score includes 58 genes that function in the M phase, meiosis, and chromosome segregation. The next three clusters also involved chromosome part (a parent term that covers many structural components of a chromosome including centromere, telomere, kinetochore, chromatin, nucleosome, condensin, cohesion, etc.), mitosis, or chromosome condensation, so the top four clusters contain a total of 97 genes (Table S7). Although not included in this list by the GO term database, we note that the cervantes/quijote gene family first identified by Betrán et al. (2006) as having transposed multiple times is also likely to have a function in chromosome maintenance: the constituent genes of the family show sequence similarity to the sumo ligase Nse2 proteins (non-SMC element 2) in other species.
We found that movements in genomes with neo-X chromosomes have higher representation in these clusters (54/107) than in the whole set (321/782), so it is possible that the enrichment in chromosome function is specific to the lineages with the neo-X fusion. When we excluded the D. pseudoobscura and D. willistoni lineages from tests for enrichment, we found mixed results depending on the tool we used; similar categories were significant in DAVID but no terms were significant in GOrilla. If indeed the excess were specific to the two lineages, a natural hypothesis would be that the movement of these genes is a response to or is related to the X/neo-X fusion event.
Since many of the genes on the list are known to evolve rapidly at the protein level (Anderson et al. 2009), movement onto different chromosomes may be a by-product of the rapid turnover of genes under an evolutionary arms race. The developmental stage of germ-line cell division is vulnerable to the intrusion of selfish elements, e.g., transposable elements and meiotic drive alleles. The genes involved in chromosome replication, condensation, and segregation may be undergoing constant conflict between selfish elements that invade the genome and alleles that counter these elements. Previous hypotheses even single out sex-ratio drive as a force underlying biased patterns of retrotransposition (Meiklejohn and Tao 2010). In Drosophila, the process of meiosis is also different between males and females, so there is potential for sexual conflict during this stage as well. Recently, Meisel et al. (2010) found two genes that are involved in chromosome segregation that moved out of the neo-X chromosome of D. pseudoobscura and hypothesized that the duplication may be a resolution of the sexual conflict the gene was under to specialize in male-specific vs. female-specific meiosis.
Bias in the direction of the movements
We discovered 20 movements that involved ≥2 linked genes, covering 52 genes in total (Table S8). Merging the linked movements trimmed down the 782 transposed genes to 750 independent movements. Removing uncertainties in the direction of movement reduced the number to a total of 665 transpositions. Since pericentric movements can be confounded with inversions, we excluded those as well, and ended up with 584 independent movements between chromosomes that we can confidently polarize. Across all types of transpositions, we found that there was an overall excess of genes moving off the X chromosome (Table S9), consistent with previous findings (Betrán et al. 2002; Bai et al. 2007; Meisel et al. 2009; Vibranovski et al. 2009). However, there has been some disagreement about whether this pattern applies to all duplicates or just those formed by retrotransposition and how it varies between duplicative transpositions and relocations (Han and Hahn 2009; Meisel et al. 2009; Vibranovski et al. 2009; Moyle et al. 2010; Zhang et al. 2010). Our results contain the largest set of gene transpositions to date and should be able to provide a definitive answer.
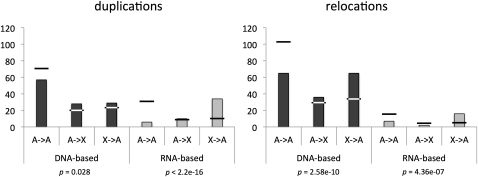
When we compared the movements by their mechanism, we did find excess movement off the X across both DNA- and RNA-based movements—consistent with previous reports—but we also observed a clear and significant quantitative difference in the excess between DNA-based movements and RNA-based movements (Figure S6). This difference in the extent of bias was present even when we divided the whole data set into four subsets [(duplicative transposition, relocation) × (DNA-based, RNA-based)] (Figure 4). We found that, especially among DNA-based duplicative transpositions, there were as many genes moving onto the X as genes moving off of the X. This pattern was present regardless of whether we included D. willistoni and D. pseudoobscura, the lineages with neo-X chromosomes. Among the genes moving onto the X were several genes involved in female meiosis, such as mei-41 and ballchen.
Figure 4 .
Movements between autosomes and the X chromosome. Columns show the frequency of movements between autosomes and the X chromosome separated by the mechanism of transposition and whether the original gene is retained (duplications) or not (relocations). Horizontal bars represent the expected frequencies. All categories show significant deviation from the expectation calculated on the basis of the number of genes on and the length of each chromosome arm, although the degree and pattern of the deviation are different for DNA-based vs. RNA-based transpositions.
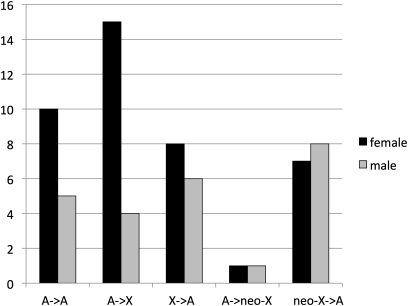
Despite the deficit of male-biased expression among genes on the X (Sturgill et al. 2007), to our knowledge there has not been any report of female-biased genes moving onto the X. We attempted to contrast the movements of genes involved in female-specific functions and male-specific functions. We used the controlled vocabulary in FlyBase to find gene families involved in female meiosis, female gamete generation, and female sex differentiation and contrasted these with families involved in male meiosis, male gamete generation, and male sex differentiation. Although both male- and female-associated genes show transpositions off the neo-X chromosome, only female genes show a pattern of excess genes moving onto the established X chromosome (Figure 5, Table 1). In addition, there is an overrepresentation of DNA-based duplications among the genes with female-specific functions (18/46 compared to 172/782 overall) and this association partially explains the different pattern of movements we see for the DNA-based duplications relative to RNA-based duplications (Figure 4).
Figure 5 .
Movements between autosomes and the X chromosome for genes involved in sex-specific functions. Shown is the frequency of movements between autosomes and the X chromosome for genes involved in female meiosis, female gamete generation, and female sex differentiation compared to those involved in male meiosis, male gamete generation, and male sex differentiation. Genes involved in female-specific functions show an excess of movement onto the established X chromosome. The genes used for comparison are listed in Table 1.
Table 1 . Transposed genes involved in female function vs. male function.
| Female function | Male function | ||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Branch | From | To | Gene symbol | Branch | From | To |
| mael | dana | D | F | Chc | dana | A | E |
| vir | dana | C | F | mael | dana | D | F |
| gus | dyesm | BC | BC | Grip84 | dgri | A | D |
| armi | dgri | D | A | mia | dgri | E | A |
| Hira | dgri | A | C | Kap3 | dmoj | A | F |
| tkv | dmv | B | C | Dhod | dmv | E | A |
| fzy | dpse | B | A | fwd | dmv | D | B |
| cuff | dpse | C | A | fwd | dmv | D | E |
| JIL-1 | dpse | D | A | Grip84 | dpse | A | D |
| RpS2 | dpse | B | A | polo | dpse | D | B |
| mus304 | dyesmap | B | D | mfr | dpse | D | B |
| polo | dpse | D | B | DnaJ-60 | dpse | C | B |
| mfr | dpse | D | B | can | dpse | D | B |
| baf | dpse | B | A | nes | dpse | D | B |
| mei-41 | dyesmap | C | A | BG4 | dyesmap | D | E |
| c(3)G | dyesmap | A | E | mael | dpse | D | E |
| Rala | dpse | A | E | PpY-55A | dpse | C | A |
| tej | dpse | C | E | fan | dpse | D | B |
| mael | dpse | D | E | r-cup | dsim | A | E |
| nonA | dyesmap | A | E | gdl | dwil | D | B |
| Bj1 | dpse | D | E | Hsp83 | dwil | D | C |
| Pxt | dpse | E | A | uri | dwil | C | D |
| kuz | dsim | B | A | MED20 | dyesm | A | B |
| CycB | dsim | C | A | otu | dyesma | A | B |
| gus | dsim | C | A | sub | dwil | C | A |
| ball | dsim | E | A | ||||
| stc | dvir | B | E | ||||
| mud | dvir | A | E | ||||
| shu | dwil | C | A | ||||
| fsd | dwil | C | E | ||||
| spn-D | dwil | EF | EF | ||||
| del | dwil | B | D | ||||
| gdl | dwil | D | B | ||||
| sca | dwil | C | B | ||||
| Hsp83 | dwil | D | C | ||||
| sub | dwil | C | A | ||||
| Fs(2)Ket | dwil | B | A | ||||
| pav | dwil | D | C | ||||
| armi | dwil | BD | C | ||||
| Top1 | dwil | A | B | ||||
| Hlc | dwil | A | B | ||||
| mirr | dwil | D | A | ||||
| otu | dyesma | A | B | ||||
| rhi | dyesma | C | A | ||||
| wek | dyesma | E | B | ||||
| alpha-Cat | dyesmap | E | D | ||||
Shown are gene transpositions in gene families involved in female meiosis, female gamete generation, and female sex differentiation compared to gene transpositions in gene families involved in male meiosis, male gamete generation, and male sex differentiation. Branch labels follow the names defined in Figure S7. More details on the transpositions are listed in Table S11.
If duplication off the X is driven by selection—as multiple studies have demonstrated (Emerson et al. 2004; Schrider et al. 2011)—then why do we see not only a difference in the degree of bias in movements but also a difference in the representation of sex-specific functions among different types of mutations? One possibility is that there may be a sex bias in the types of mutations that lead to transpositions. Sex-biased mutation rates can influence the relative rate of substitution on the sex chromosome and the autosomes. Kirkpatrick and Hall (2004) showed that the ratio of the rate of adaptive substitution between autosomes and the X shifts to be faster on autosomes if there is a higher mutation rate in males. Extending the same logic, we expect the rate difference to be much smaller if there is higher mutation rate in females. This result is contingent on the dominance of the mutation, so one has to assume that duplicative mutations are dominant. Drosophila has different mechanisms of meiosis between males and females, with no recombination in males, and this may lead to higher rates of DNA-based duplications in females. There also appears to be more reverse transcriptase present in the germ line of males (due to retroelements on the Y chromosome), which could allow the rate of retrogene mutation to be higher in males. Indeed, certain retrotransposons, such as copia elements, are expressed at much higher levels in males and show higher transposition activity in males (Pasyukova et al. 1997). This sex difference in mechanism of transposition could lead to a much greater autosomal fixation rate for retrogenes and an only marginally faster autosomal fixation rate for DNA-based duplicates when compared to the X chromosomes (M. V. Han and M. W. Hahn, unpublished results). Further analyses will be needed to distinguish among mechanistic and selective explanations for these patterns.
Limitations and possible extensions
Our approach considers the distribution of a gene family across chromosomes as a single multivariate trait. This view has its advantages and disadvantages compared to considering the number of gene copies on each chromosome arm as independent univariate traits. One advantage is that we can take into account the biological reality that loss of a gene family from the whole genome followed by a whole resurrection of a gene family is not likely. This is difficult to take into account when you consider each chromosome separately because you have no information on the state of other chromosomes. By keeping the state multidimensional, one can distinguish between a creation of a family from a zero state (0 → 1 when all other chromosomes have 0 genes) and a gain of a gene on a chromosome through transposition (0 → 1 when there is at least 1 gene on another chromosome), and costs can be assigned accordingly. In our method, we added a special penalty for the transitions originating from the zero vector (i.e., [0, 0, 0, 0, 0]), similar to the Dollo parsimony cost (Farris 1977). The disadvantage of the multivariate encoding is that the search space is exponential in the number of chromosomes. However, although the space is multidimensional, the total space we need to search is limited. The observation that makes our algorithm feasible for the problem considered here is that in general the genes of a gene family are clustered onto a small number of chromosome arms; therefore we do not have to explore the whole space of S, only the subset, S′. For example, for a gene family that resides on three different chromosome arms, we need to explore only the three-dimensional subspace of S and only up to the maximum number of genes observed among the leaves (Figure 1B). So even though the complexity of the algorithm is O(|S′|2), S′ ⊂ S, it is possible to reconstruct the states of most gene families. For our data set, there were only three families that we had to exclude because they were too large to analyze in reasonable running time. These three families were a histone family, a serine protease family, and a zinc ion-binding family, with each family containing ≥294 genes across 10 Drosophila species (Table S10). The largest single family included in our analyses had a total of 184 genes.
For species that have more chromosomes than Drosophila, larger gene families, or extensive genome rearrangements, our method may not work as well. A larger number of chromosomes means that a larger state space must be considered for each gene family. Likewise, larger gene families are likely to be spread across more chromosomes, and the size of the state space will grow with the total number of genes on any single chromosome. For extensively rearranged genomes—i.e., those without almost perfect correspondences between arms across species—we could instead segment the genome into syntenic blocks that are conserved across the species we are interested in. However, once again this will result in an extremely large state space: one that is equivalent to the number of syntenic blocks that contain paralogs for any single family. One solution to all of these problems would be to consider each chromosome independently, but as discussed above, this results in a loss of information from different chromosomes, which could lead to unrealistic inferences. Finally, the cost scheme we use is arbitrary; although this is a limitation of all parsimony methods, we used a higher cost for gains relative to losses to make our model more realistic. One future extension of this work would be to allow estimation of the transition rates as parameters in a likelihood framework, which could make dealing with a larger state space more manageable. Although there are also problems inherent to likelihood models, this is an approach that should be pursued in the future.
Conclusions
We have used a novel implementation of parsimony to analyze the location and size of gene families among 10 Drosophila species. We found many transpositions that were previously overlooked, including multiple parallel movements within single gene families. In total, our data set contains 782 interchromosomal movements, which include 421 transposed genes that we have newly identified. Using this set of transposed genes, we confirmed several previous hypotheses, including a link between gene transposition and increased rates of sequence evolution, as well as the excess of gene movement off Drosophila X chromosomes. We also detected new patterns among gene transpositions that could not be detected using previous data sets. We observed an excess of female-associated genes moving onto the established X chromosome. We also found that genes with chromosome segregation- and meiosis-related functions are not only evolving rapidly in their sequence but also frequently transposing across chromosomes through duplication. These results suggest that gene movement between chromosomes can have an important role in resolving intragenomic conflicts, both between the sexes and among chromosomes.
Acknowledgments
We thank Rich Meisel, Nitin Phadnis, and two anonymous reviewers for providing valuable comments on the manuscript. This work was supported by National Science Foundation grant DBI-0845494 (to M.W.H.).
Footnotes
Communicating editor: N. Perrimon
Literature Cited
- Alonso C., Berendes H. D., 1975. The location of 5S (ribosomal) RNA genes in Drosophila hydei. Chromosoma 51: 347–356 [DOI] [PubMed] [Google Scholar]
- Anderson J., Gilliland W., Langley C., 2009. Molecular population genetics and evolution of Drosophila meiosis genes. Genetics 178: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Toda N. R. T., Lockton S., 2010. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr. Biol. 20: 1476–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Casola C., Feschotte C., Betrán E., 2007. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 8: R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E., Thornton K., Long M., 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betrán E., Bai Y., Motiwale M., 2006. Fast protein evolution and germ line expression of a Drosophila parental gene and its young retroposed paralog. Mol. Biol. Evol. 23: 2191–2202 [DOI] [PubMed] [Google Scholar]
- Bhutkar A., Russo S., Smith T., Gelbart W., 2007. Genome-scale analysis of positionally relocated genes. Genome Res. 17: 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C., Ganote C. L., Hahn M. W., 2010. Nonallelic gene conversion in the genus Drosophila. Genetics 185: 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Connallon T., Clark A. G., 2011. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack B., Wolfe K. H., 2007. Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol. Biol. Evol. 24: 679–686 [DOI] [PubMed] [Google Scholar]
- Dai H., Yoshimatsu T., Long M., 2006. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 385: 96–102 [DOI] [PubMed] [Google Scholar]
- Durand D., Halldorsson B., Vernot B., 2006. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol. 13: 320–335 [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Kaessmann H., Betrán E., Long M., 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540 [DOI] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S., Ouzounis C. A., 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris J. S., 1977. Phylogenetic analysis under Dollo’s law. Syst. Zool. 26: 77–88 [Google Scholar]
- Felger I., Pinsker W., 1987. Histone gene transposition in the phylogeny of the Drosophila obscura group. J. Zool. Syst. Evol. Res. 25: 127–140 [Google Scholar]
- Fyrberg E., Kindle K., Davidson N., Sodja A., 1980. The actin genes of Drosophila: a dispersed multigene family. Cell 19: 365–378 [DOI] [PubMed] [Google Scholar]
- Gallach M., Betrán E., 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol. Evol. 26: 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Han M. V., Han S.-G., 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3: e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M. V., and M. W. Hahn, 2009 Identifying parent-daughter relationships among duplicated genes. Pac. Symp. Biocomput. 2009: 114–125. [PubMed]
- Han M. V., Demuth J., McGrath C., Casola C., Hahn M. W., 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res. 19: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Tang H., Ventura M., Cardone M. F., Marques-Bonet T., et al. , 2007. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39: 1361–1368 [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K.-i., Toh H., Miyata T., 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Hall D. W., 2004. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution 58: 437–440 [PubMed] [Google Scholar]
- Marques-Bonet T., Sànchez-Ruiz J., Armengol L., Khaja R., Bertranpetit J., et al. , 2007. On the association between chromosomal rearrangements and genic evolution in humans and chimpanzees. Genome Biol. 8: R230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly J. P., Jones C., Noor M., Locke J., Orr H. A., 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313: 1448–1450 [DOI] [PubMed] [Google Scholar]
- Meiklejohn C., Tao Y., 2010. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 25: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R. P., Han M. V., Hahn M. W., 2009. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 1: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel R., Hilldorfer B., Koch J., Lockton S., Schaeffer S., 2010. Adaptive evolution of genes duplicated from the Drosophila pseudoobscura neo-X chromosome. Mol. Biol. Evol. 27: 1963–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Muir C. D., Han M. V., Hahn M. W., 2010. The contribution of gene movement to the “two rules of speciation”. Evolution 64: 1541–1557 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1940. Bearings of the ‘Drosophila’ work on systematics, pp. 185–268 The New Systematics, edited by Huxley J. Clarendon Press, Oxford [Google Scholar]
- Ohta T., Dover G., 1983. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc. Natl. Acad. Sci. USA 80: 4079–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova E., Nuzhdin S., Li W., Flavell A. J., 1997. Germ line transposition of the copia retrotransposon in Drosophila melanogaster is restricted to males by tissue-specific control of copia RNA levels. Mol. Gen. Genet. 255: 115–124 [DOI] [PubMed] [Google Scholar]
- Potrzebowski L., Vinckenbosch N., Marques A. C., Chalmel F., Jégou B., et al. , 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J., Gonzalez J., Casals F., Ruiz A., 2003. Low occurrence of gene transposition events during the evolution of the genus Drosophila. Evolution 57: 1325–1335 [DOI] [PubMed] [Google Scholar]
- Rice W., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742 [DOI] [PubMed] [Google Scholar]
- Sánchez F., Natzle J., Cleveland D., Kirschner M., McCarthy B., 1980. A dispersed multigene family encoding tubulin in Drosophila melanogaster. Cell 22: 845–854 [DOI] [PubMed] [Google Scholar]
- Sankoff D., Rousseau P., 1975. Locating the vertices of a Steiner tree in an arbitrary metric space. Math. Program. 9: 240–246 [Google Scholar]
- Schaeffer S., Bhutkar A., McAllister B., Matsuda M., Matzkin L., et al. , 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179: 1601–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider, D. R., K. A. Stevens, C. M. Cardeno, C. H. Langley, and M. W. Hahn, 2011 Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Res 21: 2087–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill D., Zhang Y., Parisi M., Oliver B., 2007Demasculinization of X chromosomes in the Drosophila genus. Nature 450: 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy C., Rio J., Motiwale M., Christensen S., Betrán E., 2010. Convergently recruited nuclear transport retrogenes are male biased in expression and evolving under positive selection in Drosophila. Genetics 184: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski M. D., Zhang Y., Long M., 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N., Dupanloup I., Kaessmann H., 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 103: 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-I., Xu Y. E., 2003. Sexual antagonism and X inactivation-the SAXI hypothesis. Trends Genet. 19: 243–247 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Vibranovski M., Krinsky B., Long M., 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20: 1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Kishino H., 2004. Genomic background predicts the fate of duplicated genes: evidence from the yeast genome. Genetics 166: 1995–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]