Abstract
Although mapping quantitative traits in inbred strains is simpler than mapping the analogous traits in humans, classical inbred crosses suffer from reduced genetic diversity compared to experimental designs involving outbred animal populations. Multiple crosses, for example the Complex Trait Consortium's eight-way cross, circumvent these difficulties. However, complex mating schemes and systematic inbreeding raise substantial computational difficulties. Here we present a method for locally imputing the strain origins of each genotyped animal along its genome. Imputed origins then serve as mean effects in a multivariate Gaussian model for testing association between trait levels and local genomic variation. Imputation is a combinatorial process that assigns the maternal and paternal strain origin of each animal on the basis of observed genotypes and prior pedigree information. Without smoothing, imputation is likely to be ill-defined or jump erratically from one strain to another as an animal's genome is traversed. In practice, one expects to see long stretches where strain origins are invariant. Smoothing can be achieved by penalizing strain changes from one marker to the next. A dynamic programming algorithm then solves the strain imputation process in one quick pass through the genome of an animal. Imputation accuracy exceeds 99% in practical examples and leads to high-resolution mapping in simulated and real data. The previous fastest quantitative trait loci (QTL) mapping software for dense genome scans reduced compute times to hours. Our implementation further reduces compute times from hours to minutes with no loss in statistical power. Indeed, power is enhanced for full pedigree data.
THERE are trade-offs in mapping quantitative trait loci (QTL) in humans vs. model organisms. The primary advantage of human data is that any mapped gene is guaranteed to be relevant. In addition, traits such as psychometric measures are limited to humans. On the other hand, gene mapping in model organisms is considerably easier. For many model organisms, generation times are short and environmental effects can be rigidly controlled. Any genes mapped can be quickly located in humans by synteny. Murine mapping exploits inbred strains where all mice are completely homozygous and genetically identical. Diversity is regained by crossing the strains. It seems obvious that the more strains involved in a cross, the greater the chance of mapping a relevant gene. For this reason geneticists are contemplating more ambitious crosses with more contributing strains. Unfortunately, these complex crosses are harder to analyze statistically, particularly when pedigree structures are poorly documented. In this article we tackle some challenges of analyzing data from arbitrarily complex crosses. The Complex Trait Consortium's eight-way cross (Churchill et al. 2004; Aylor et al. 2011) is just one of many conceptual possibilities. Heterogeneous stocks (HS) also find wide application in mapping mouse QTL (Valdar et al. 2009). In addition to murine mapping, ongoing efforts in Drosophila (Macdonald and Long 2007) and Arabidopsis (Kover et al. 2009) mapping are upping the ante in the analysis of complex-cross data. Even though we focus on mice, readers should keep in mind the broader implications of our statistical and algorithmic agenda.
In humans the dominant mapping strategies are linkage and association mapping. The former is more robust; the latter has better resolution. The shift from linkage analysis to association mapping has been accompanied by the replacement of pedigree data by random sample and case–control data. Although one can imagine random sampling of wild mice, the opportunities for strict environmental and dietary control are lost. Association mapping is certainly possible by sampling all available strains, but traditionally the number and availability of rare strains have imposed limits on mapping resolution and power (Chesler et al. 2001; Grupe et al. 2001; Cervino et al. 2007; Scudellari 2010). Thus, pedigree data retain some real advantages in mapping mouse genes. Linkage mapping operates by tracking recombination events. These accumulate more readily in deep pedigrees and allow a trait to be mapped to the smallest region of overlap defined by conserved strain blocks. The polymorphisms defining the blocks usually do not drive trait variation. Of course, as single-nucleotide polymorphism (SNP) panels in mice become more dense (Frazer et al. 2007; Saar et al. 2008), the chances of a panel including causative variants increases.
It seems to us that the best route to success in association mapping with inbred strains is to use the local strain origins of each mouse as fixed effects in a mixed-effects statistical model. Although confined to quantitative traits, this strategy has several advantages. First, it mimics what linkage mapping is seeking to accomplish in tracking recombination events and strain blocks. Second, in contrast to standard association mapping, it does not rely on a single SNP at a time to distinguish local strain origins. Third, the random effects part of a mixed-effects model readily captures polygenic background. Our recent model of polygenic inheritance in inbred strains (Bauman et al. 2008) makes it possible to calculate trait variances and covariances across a pedigree, regardless of the number of founding strains and the internal complexity of the pedigree.
The literature on QTL mapping strategies for inbred strains is longstanding and too large to review here. Recent articles touting random effects models in inbred strains include Xie et al. (1998), Liu and Zeng (2000), and Bauman et al. (2008). Bennett et al. (2010) argue that association mapping with large SNP mouse panels has the potential for much higher mapping resolution. Early results from the Collaborative Cross support this contention (Aylor et al. 2011).
The new polygenic models for inbred strain data derived by Bauman et al. (2008) involve certain combinatorial (strain) coefficients that bear a strong resemblance to standard global (theoretical) kinship coefficients appropriate to outbred populations. Both kinds of coefficients can be quickly computed by simple recurrence relations. Calculating the local (conditional) analogs of these global coefficients is much more challenging. These depend on all observed marker genotypes in the vicinity of a putative QTL. On small pedigrees it is possible to compute local strain coefficient matrices exactly by generating all possible descent graphs (gene flow patterns) at the QTL and neighboring markers (Kruglyak et al. 1996). In practice, inbred strain pedigrees are so large that the number of possible descent graphs is astronomical, and current computation is limited to slow Monte Carlo sampling (Sobel and Lange 1996). In this article, we dispense with computation of local strain coefficients and propose as a substitute direct imputation of strain origins locally along each animal's genome. Once imputation is done by a very fast dynamic programming algorithm, local strain origins serve as mean effects in a multivariate Gaussian model for association testing.
Our imputation approach is based on minimizing animal by animal an objective function incorporating both loss and penalty terms. The loss function cumulates the negative log-likelihood of the observed data from each marker given the local strain origins at the marker. The penalty terms suppress switches in strain origin and encourage origin constancy over long stretches of the animal's genome. When a switch occurs, the jump to another strain is biased by the global fraction of the animal's genome attributable to that strain. Here the global strain coefficients supply prior information. In effect, the penalty terms serve to smooth and guide origin imputation. Our dynamic programming algorithm for minimizing the objective function requires a single pass through the data and operates with linear time and storage. The algorithm is also crafted to accommodate missing strain genotypes, which are filled in by application of a majorization–minimization (MM) algorithm (Hunter and Lange 2004). The entire process is very fast and acceptably accurate. The few errors made in imputation occur at strain origin boundaries. Day-Williams et al. (2011) introduce an analogous approach to accurately imputing local kinship coefficients in human data when pedigree origins are unknown.
It is worth emphasizing our modeling choices and how they compare with traditional choices. First, our QTL effects are mean effects rather than variance effects. In QTL mapping in humans, the opposite is true. Variance effects are preferred to mean effects in statistical modeling when the underlying predictors are unobserved or too numerous for parsimonious parameterization. Neither of these conditions holds for complex crosses between inbred strains. Strain origins succinctly capture the underlying genetics without committing to the information provided by a single SNP. Second, in reconstructing strain origins most statisticians turn to hidden Markov models (Mott et al. 2000; Liu et al. 2010). In our opinion, penalized likelihoods achieve the same goal at a fraction of the computational cost. Reasonable penalties introduce prior information into frequentist inference in basically the same way that priors do in Bayesian inference. Of course, penalties have to be tuned. Fortunately, we show that statistical inference in the current setting is relatively insensitive to the value of the penalty tuning constant.
The remainder of this article is organized as a progression from theory and algorithms to data analysis. Ordered strain coefficients and fractions are first introduced along with simple algorithms for their computation. These global combinatorial indexes summarize prior pedigree information. The dynamic programming algorithm for imputing strain origins and missing genotypes in the various founding strains is then sketched. This is followed by a summary of our mixed-effects model and how it plays out in QTL association testing. Both simulated and real data demonstrate the accuracy of strain imputation and its effectiveness in QTL association mapping. Finally, the broader implications of the model and its limitations are discussed.
Methods
Ordered strain coefficients and fractions
To pave the way for our imputation method, we generalize the notion of strain coefficients (Bauman et al. 2008). Imagine a pedigree generated by a set of complicated crosses involving a certain number of inbred strains. Each founder of the pedigree is assigned to a definite strain; different founders are allowed to belong to the same strain. The pedigree is then filled in with descendants of the original crosses, who are bred according to an experimental protocol. The ordered strain coefficient is decorated by several indexes. The subscripts i and j denote two animals in the pedigree; the possibility i = j is permitted. The superscripts m and p stress that a maternal gene is sampled from animal i and a paternal gene is sampled from animal j. Finally, the ordered pair (a, b) refers to two strains. In this notation represents the joint probability that the maternal gene of animal i at a random locus is drawn from strain a and the paternal gene at the same locus of j is drawn from strain b. Similarly, we can define the coefficients and . In our previous article, we defined (unordered) strain coefficients related to the current coefficients by the equation
The coefficient ψij(a, b) corresponds to random sampling from the combined pool of maternal and paternal genes. When i = j, sampling is done with replacement. Neither the ordered nor the unordered strain coefficients take into account observed genotypes.
The marginal probabilities
are referred to as ordered strain fractions. The corresponding unordered strain fractions
were introduced by Bauman et al. (2008). Unordered strain coefficients are analogous to global kinship coefficients in outbred populations. Thus, it is not too surprising that one can derive simple recurrences for computing unordered strain coefficients and unordered strain fractions.
Recurrence relations
The various recurrences presuppose that parents are numbered before children in a pedigree. For a founder i belonging to strain a, it is obvious that γi(a) = 1; all other entries of γi are 0. If a nonfounder i has parents k and l, then the averaging law holds. If i is a founder belonging to strain a and j is a founder belonging to strain b, then ψij(a, b) = 1; all other entries of ψij are 0. Finally, if i is a nonfounder with parents k and l and j is an animal previously considered, then
These recurrences are logical consequences of simple sampling arguments as noted by Bauman et al. (2008).
In computing the ordered versions of strain coefficients and strain fractions, it is again convenient to begin with the founders. If the founders i and j belong to strains a and b, respectively, then we set
All other strain coefficients and fractions involving founders i and j are set to 0. The symmetries
apply to nonfounders as well as to founders.
The remainder of the strain coefficients is computed recursively on the basis of the founder values. If we number the animals so that parents precede children, then we can compute all coefficients in one pass through the pedigree. Consider an animal i whose mother k and father l have already been visited. Taking into account the maternal and paternal origins of i’s two genes at an arbitrary locus gives the averaging laws
The analogous recurrences for ordered strain coefficients are
| (1) |
| (2) |
Finally for any previously visited animal j ≠ i, we set
and employ the symmetry relations noted earlier to facilitate switching the order of i and j.
Imputation of strain origins
As the Introduction suggests, we approach imputation of local strain origins through loss functions, penalty functions, and dynamic programming. Our discrete optimization strategy has the virtues of speed, simplicity, and accuracy. With less dense genotyping, soft probabilistic imputation might be preferable, but the information content of modern genome scans is so great that hard imputation errors are confined to the borders of recombination blocks. Although competing methods of imputation such as hidden Markov chains have proved their worth in haplotyping (Mott et al. 2000; Liu et al. 2010), we see no compelling reason to commit to models with more than the minimal number of parameters. Furthermore, hidden Markov chains involve their own sometimes dubious assumptions, such as the left to right flow of the underlying probabilistic process. In our experience with haplotyping, penalized likelihood estimation is competitive with hidden Markov modeling in accuracy and computationally faster (Ayers and Lange 2008).
Strain origins can be imputed with or without defined pedigrees. If pedigree status is available, then it furnishes prior information that should improve imputation accuracy. In practice, meticulous records are often lacking, and empirically derived strain coefficients and fractions are helpful. If strains are typed on different marker sets, then missing strain genotypes (founder genotypes) also become an issue. Before dealing with these complications, we first turn to the case of full pedigree and strain genotype data. Genotypes on individual pedigree members may be missing.
Imputation with full data:
Consider the ordered strain origin pair uk = (ak, bk) for animal i with observed genotype rk/sk at marker k. Our imputation process incorporates the log-penetrance (conditional log-likelihood)
as the negative loss at marker k. At the first marker the log-likelihood should also take into account the prior probabilities determined by the strain coefficients; accordingly, we set
For the sake of simplicity, we calculate the underlying penetrances on the basis of a simple genotyping error model that assigns probability 1 – ε to a match between a strain and an allele and probability ε to a mismatch. Typically ε > 0 is small, say ≤0.01. Table 1 specifies penetrances under this SNP model with alleles labeled 1 and 2. In Table 1 ta is the allele carried by strain a. The ordered genotype (ta, tb) displays its maternal allele on the left and its paternal allele on the right.
Table 1.
Penetrances Pr[r/s | (a, b)] for a SNP
| Phenotype r/s |
||||
| Genotype (ta, tb) | 1/1 | 1/2 | 2/2 | Missing |
| (1, 1) | (1 – ε)2 | 2ε(1 – ε) | ε2 | 1 |
| (1, 2) | ε(1 – ε) | (1 – ε)2 + ε2 | ε(1 – ε) | 1 |
| (2, 1) | ε(1 – ε) | (1 – ε)2 + ε2 | ε(1 – ε) | 1 |
| (2, 2) | ε2 | 2ε(1 – ε) | (1 – ε)2 | 1 |
The objective function for animal i also includes a penalty Pk(uk, uk+1) for each pair of adjacent markers. Here the state of the system at marker k is an element uk = (ak, bk) from the Cartesian product set {1, … , s} × {1, … , s} of strain origin pairs possible for s strains. As the genome of animal i is traversed, the penalty is designed to suppress jumps between strains and guide jumps, when they do occur, toward more likely states. With uk = (ak, bk) and uk+1 = (ak+1, bk+1), one term of our penalty can be written as
For n consecutive markers and u = (u1, … , un), the overall objective function becomes
| (3) |
Dynamic programming algorithm:
One can find the optimal sequence of states by a one-pass dynamic programming algorithm. Dynamic programming proceeds by solving the sequence of intermediate problems
for m taking the successive values 1, … , n, starting with O1(u1) = –L1(u1). When we reach m = n, the value equals the minimum of the objective function. If we keep track of one solution sequence u1(um), … , um–1(um) for each partial objective Om(um), then we can construct a best overall sequence by taking the best un and appending to it u1(un), … , un–1(un). To better understand the recursive phase of the algorithm, note that the partial solution Om(um) is found by minimizing
over all um–1.
The astute reader will note the analogy between our optimal strain origin sequence and the most probable sequence delivered by the Viterbi algorithm in hidden Markov modeling. The Viterbi algorithm is a special case of dynamic programming. In general, the Viterbi algorithm is preceded by maximum-likelihood estimation of the underlying parameters and is therefore not fully Bayesian despite its reliance on Bayes’ rule.
Imputation with missing data:
We now extend our imputation method to handle missing pedigree information and missing strain genotypes. The obvious tactic is to substitute empiric estimates of strain coefficients and fractions for their theoretical counterparts in the imputation process. It is important to keep in mind that imputation of strain origins requires only the diagonal strain coefficients, where the two underlying animals i and j coincide. Besides estimating these quantities, we must also impute missing strain genotypes. The latter goal is achieved by estimation as well. Let πak be the unknown frequency of allele 1 in strain a at marker k. Assuming strain a has a fixed allele at this marker, the estimate of πak should obviously hover around either 0 or 1.
Our overall strategy is to put all of the mentioned ingredients into one large pot and estimate global coefficients and fractions and missing strain allele frequencies simultaneously with imputing strain origins. To succeed, the process should be performed iteratively until successive refinements stabilize. In fact, we simplify matters by alternating two steps. The first is dynamic programming imputation of strain origins given current strain coefficients and fractions and current frequencies for the missing strain alleles. The second is reestimation of all parameters given imputed strain origins. The second step is iterative and depends on an MM algorithm discussed in the Appendix. This two-step strategy sounds complicated, possibly slow, and potentially error prone. However, the amount of data delivered by modern genotyping chips is so overwhelming that these fears are unwarranted. Observe that the data from all animals inform estimation of missing allele frequencies. Thus, we iterate over all animals simultaneously. The MM algorithm is fast enough in this setting to cope with iterations within iterations. Convergence is declared in the outer iterations when all imputations stabilize. In practice this happy state of affairs is achieved after only five or six rounds of the two-step process.
QTL mapping
In this section, we briefly review the QTL association model introduced by Bauman et al. (2008) and show how imputation can be incorporated. The basic model involves s strains and t traits. These traits follow a multivariate Gaussian distribution over a pedigree, so it suffices to specify means, variances, and covariances.
Let Xik denote the polygenic contribution to trait k of animal i. Bauman et al. (2008) derive the means and covariances
| (4) |
where μk(a) is the polygenic mean effect of trait k for strain a, Cij is an s × s combinatorial matrix with entries Cij(a, b) = ψij(a, b) – γi(a)γj(b), and Ωkl is an s × s matrix of covariance effects for traits k and l. The st × st matrix Ω with blocks Ωkl is positive semidefinite. Note that Cij is defined by unordered strain coefficients and fractions. Although the parameter matrix Ω is not identifiable, one can subtract its nonidentifiable part and estimate the residue. Readers are referred to Bauman et al. (2008) for complete details.
The full null model adds random error/environment and various fixed effects. In this setting, the means and covariances for the trait values Yik are
| (5) |
| (6) |
where η is an intercept vector, zim is the mth of p predictors measured on animal i, and βmk is the corresponding regression coefficient for trait k. The matrix ϒ captures the environmental covariation of the traits within a single animal. It is noteworthy that the polygenic effects appear in both the mean and the variance levels in the null model. To avoid confounding polygenic mean effects with the intercept η, we set for each trait k.
Under the alternative hypothesis in association mapping, the QTL mean effects are tied to the trait location along the chromosome under consideration. This location is viewed as containing a candidate gene whose alleles shift trait mean values. These alleles are not directly observable, so we take imputed strain origins as surrogates for alleles. Think of the strain origin pair (a, b) (maternal strain a and paternal strain b) as a kind of genotype. For an additive model, strain a has impact εk(a) on trait k, and the strain origin pair (a, b) has overall impact εk(a) + εk(b), with the constraint understood. Therefore, the means under the alternative are
| (7) |
The covariances displayed in Equation 6 remain the same. This is the most parsimonious QTL model possible. For crosses with just a few strains, one can contrast this model with a nonadditive model with strain effects δk(a, b) on trait k. Here the constraint is relevant. In Equation 7 we exchange the single term δk(a, b) for the sum εk(a) + εk(b).
If we stack the observed values of the random traits Yik in a vector y, the corresponding means in a vector ν, and the corresponding covariances in a matrix Σ, then the Gaussian log-likelihood of the given pedigree can be written as
Association testing against the alternative hypothesis reduces to computing a likelihood-ratio statistic that asymptotically follows a chi-square distribution with (s – 1)t d.f. To implement likelihood-ratio testing (LRT), iterative maximum-likelihood estimation must be undertaken over the entire parameter vector for each marker.
Results
We now evaluate strain origin imputation and its impact on association testing in both simulated and real data. The next section records strain imputation results for simulated data mimicking the Collaborative Cross (Churchill et al. 2004; Aylor et al. 2011). We pay particular heed to the consequences of missing pedigree information and missing strain genotypes. Given the reassuring outcomes of imputation, we examine QTL association mapping for simulated data under random mating and for real expression QTL (eQTL) data with MF1 mice, an outbred population constructed from eight founding strains.
Imputation performance
To evaluate imputation accuracy, we employed the Gene Dropping option of the genetic analysis program MENDEL (Lange et al. 2001) and simulated the outcomes of various mating designs assuming linkage equilibrium and a postulated marker map. One of the virtues of simulated data is that true strain origins are known. Imputation accuracy is computed as the percentage of sites where the estimated founder ancestry matches the truth. Our matching criterion takes into account that many inbred strains are related (Flint 2010) and have common chromosome blocks identical by descent (IBD). If two or more founder strains’ genotypes are identical across an entire window of consecutive markers, then we lump strains identical within the window and assess matches accordingly. Our reported averages cover all markers and assume a window 51 markers long with the current marker at the center. Imputation accuracy is relatively insensitive to the choice of the penalty tuning constant λ, which we take as 1 unless otherwise mentioned.
Collaborative cross example:
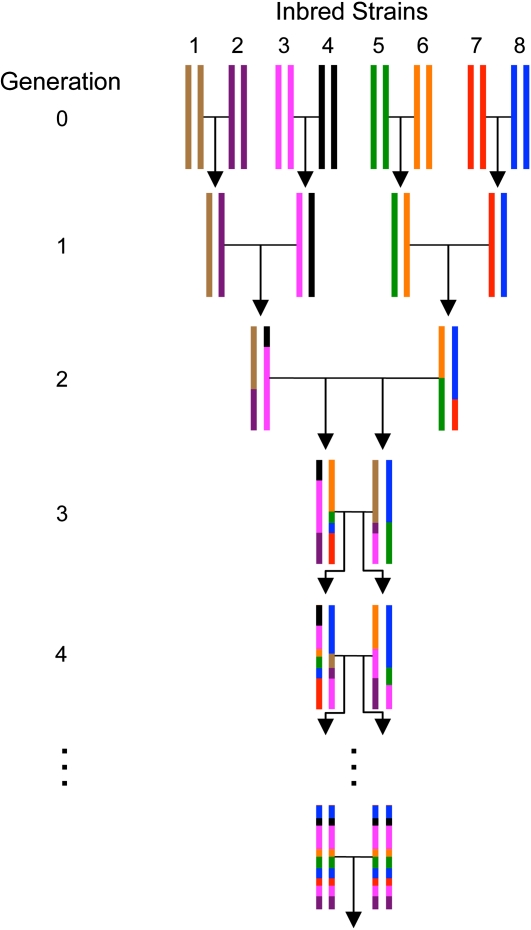
As an example of data that researchers may encounter in practice, we turn to the Collaborative Cross (CC), a large panel of recombinant inbred (RI) strains derived from eight genetically diverse founder strains. The founding strains include five classical inbred strains (C57BL/6J, 129S1/SvImJ, A/J, NOD/LtJ, and NZO/H1LtJ) and three wild-derived strains (CAST/EiJ, PWK/PhJ, and WSB/EiJ). The CC is specifically designed for complex trait analysis (Churchill et al. 2004; Aylor et al. 2011). Similar study designs are being implemented with other model organisms (Macdonald and Long 2007; Kover et al. 2009). As depicted in Figure 1, three generations of rigid mating are followed by ≥20 rounds of brother–sister mating. In each mating design the founder strains are permuted to randomize and balance the genomes of the resulting RI lines. Each permutation of the founders is called a funnel. With no loss of generality, we analyze a data set on the basis of only one funnel.
Figure 1 .
The eight-way funnel breeding scheme for generating recombinant inbred (RI) strains.
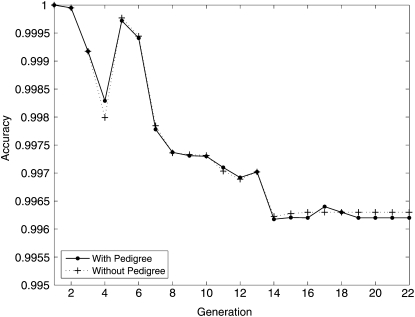
Based loosely on the Collaborative Cross mating scheme, we simulated a 23-generation pedigree with 414 mice, 20 generations of inbreeding, and 20 mice per inbred generation. Note that we simulated random mating rather than brother–sister mating after the first few generations. The genotypes of the founder strains were downloaded from The Jackson Laboratory mouse phenome database at http://phenome.jax.org/SNP. We randomly chose 10,000 contiguous SNPs on chromosome 19 from among the 221,798 SNPs in the database. Our SNPs span the chromosome 19 map from 3.2 Mb to 61.3 Mb. The distances between adjacent markers range from 2 bp to 545.9 kb, with an average of 5.8 kb. All markers are informative. Data across the entire mouse genome can be handled in exactly the same manner. Figure 2 plots imputation accuracy as a function of generational depth for a single random replicate of the pedigree. Each point on the solid curve represents an average across 20 mice × 10,000 SNPs = 200,000 data points. Imputation accuracy ranges from 99.6 to 100%, with a mean of 99.7%. The maximum standard deviation of these estimates is 0.87%. When we compare accuracy for each generation across 20 simulation replicates, the standard errors range from 0.0 to 0.31%.
Figure 2 .
Imputation performance for Collaborative Cross (CC) mice with and without pedigree information.
The CC example assumes full pedigree information and gives high imputation accuracy. Across a pedigree, accuracy drops as we descend to lower generations. This phenomenon simply reflects the gradual accumulation of recombination events and the number of strain origin switches that must be explained in imputation.
Imputation without pedigree information:
In this section, we comment on imputation performance in the simulated data ignoring prior pedigree information. The dashed curve in Figure 2 plots imputation accuracy against generation number ignoring pedigree information in the simulated pedigree. Accuracy suffers no discernible degradation. It may seem odd that imputation accuracy is equally good with and without pedigree information, but there is no guarantee that the average strain fractions and coefficients across a mouse genome conform to theoretical strain fractions and coefficients, which are valid only in an expected sense across many replicates of the same pedigree. In any case, the comparison in Figure 2 makes it clear that detailed pedigree records are unnecessary to achieve high imputation accuracy.
Imputation with missing founders’ genotypes:
Many strains are only incompletely typed on existing chips. For a test of imputation in the partial absence of strain genotypes, we again used our simulated pedigree with CC founder strains. We randomly deleted 20% of the genotypes from the markers of each founder strain. Average imputation accuracy is now 98.1%, ranging from 90 to 100%. Across all selected markers and strains, the average absolute difference between the true allele frequency and the estimated allele frequency for the minor allele is 7.05 × 10−5. In fact, only four of these allele frequency differences, 6 × 10−5, 0.02, 0.03, and 0.93, fall outside the interval [0, 10−6]. At the marker with the most egregious difference, very few descendants carry the allele in question, and a single putative genotyping error exerts enormous influence. Imputation errors at the beginning of the iterative process of imputation and allele frequency estimation can also occasionally steer frequencies in the wrong direction.
Specification of the penalty constant λ:
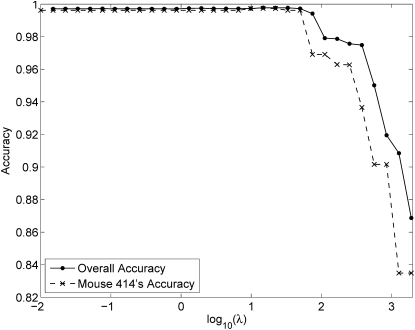
As an illustration of the relative insensitivity of imputation to the choice of the penalty constant λ, we consider again the 414 mice of the simulated CC pedigree. Figure 3 plots imputation accuracy as a function of the logarithm of λ for one randomly chosen mouse from the last generation of the pedigree and for the average over all mice. Imputation accuracy stays >99.8% over a broad range of λ-values, including our recommended value λ = 1.
Figure 3 .
Overall imputation accuracy and mouse 414’s accuracy as a function of the logarithm of the tuning constant λ. Mouse 414 appears in the last generation of the pedigree and gives the poorest imputation results.
QTL association testing
Simulated univariate trait example:
For mapping purposes, we simulated a cross involving a univariate trait, four inbred strains, and six pedigrees of 15 generations each. Given the short life span of mice, we used only the 600 mice from the last 5 generations for imputation and association testing. The four founding strains 129S1/SvImJ, A/J, PWK/PhJ, and CAST/EiJ from the CC contributed equally to the pedigrees. From the second generation onward, 10 mice were randomly mated in each generation to form the next generation. We employed the Gene Dropping option of MENDEL to generate genotypes at 19,000 random SNPs evenly distributed across the 19 mouse chromosomes. From these 19,000 SNPs, we singled out SNP 5408 on chromosome 6 as the QTL and omitted its genotypes from association testing. We then generated univariate trait values independently for each pedigree by sampling from a multivariate Gaussian distribution with means and covariances prescribed by the model. Table 2 displays the parameter values used in the simulations. These values were chosen randomly subject to the constraints
In total we tested for association at 372 evenly spaced locations, each location corresponding to the center of a window of 51 SNPs. Founder strains that were IBD across the window were lumped. The extended spacing between window centers adjusts for linkage disequilibrium and reduces the number of tests performed. Center-to-center spacing is a user option in MENDEL. To examine whether pedigree information is essential for association testing, we analyzed the data with and without pedigree structure specified.
Table 2.
Simulation parameters for the univariate QTL simulation example
| Inbred strain | μstrain | εstrain |
| 129S1 | −2.81 | −1.53 |
| A | 2.13 | −0.66 |
| PWK | 4.62 | 1.14 |
| CAST | −3.94 | 1.05 |
| Intercept | 6.75 |
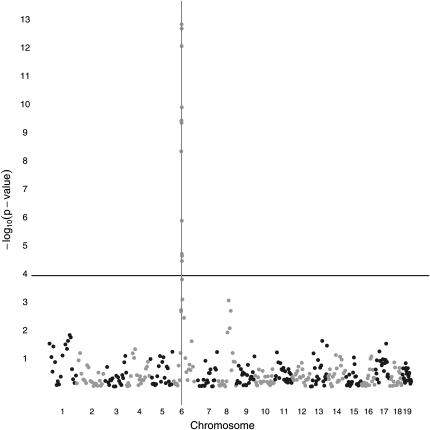
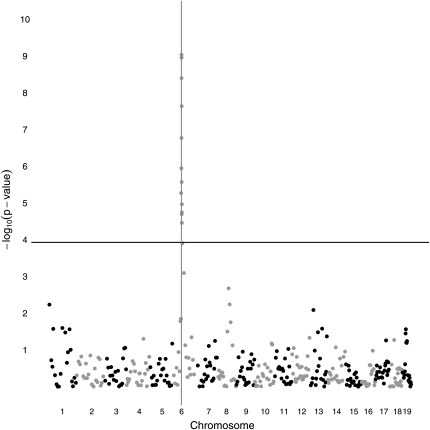
Imputation was performed under the tuning constant λ = 1. Regardless of whether pedigree structure is specified, imputation accuracy for all mice exceeded 98%. The per site accuracy ranged from 64.7 to 100%. There are 10 sites with accuracy <80%, of which 8 are the first site of a chromosome. There are 335 sites out of 19,000 with accuracy <90%. Most of these are also near the 5′ end of a chromosome. Figures 4 and 5 plot –log10(P-value) from the LRT as a function of map position in base pairs. Polygenic background is taken into account in both plots. In Figure 4, where pedigree structure is exploited, 41 SNPs rise above the Bonferroni threshold specified by the horizontal line. The SNP at location 61,209,472 bp immediately adjacent to the QTL (61,222,084 bp) gives the highest –log10(P-value). In Figure 5, where pedigree structure is ignored, 42 SNPs rise above the Bonferroni correction threshold. The overlap between the two sets of SNPs is almost complete. Ignoring pedigree structure causes the most significant P-value to increase from ∼10−13 to ∼10−9.
Figure 4 .
Four-way cross QTL association mapping of a univariate trait using windows 51 SNPs long and pedigree structure information. The vertical line represents the QTL location. The horizontal line represents the genome-wide significance threshold after Bonferroni correction.
Figure 5 .
Four-way cross QTL association mapping of a univariate trait using windows 51 SNPs long and excluding pedigree structure. The vertical line represents the QTL location. The horizontal line represents the genome-wide significance threshold after Bonferroni correction.
In fact, the plots are more complicated than meet the eye. For one thing, they were constructed in two stages. The first stage involved the 372 SNPs defined by the subsampling procedure. These stage-one SNPs were then supplemented by 50 stage-two SNPs drawn from the window centered around the best SNP discovered in stage one. Graphed P-values are also adjusted by the conservative method of genomic control (Devlin and Roeder 1999; Devlin et al. 2004). In genomic control, one multiplies all LRT statistics by the ratio of the theoretical median of the relevant asymptotic chi-square distribution to the sample median of the LRT statistic across the genome. This reduces the largest computed –log10(P-value) from ∼9.6 to the 9.0 value seen in Figure 5. The method of genomic control is a crude attempt to compensate for model failures and the large sample approximations inherent in the LRT. Only the stage-one SNPs were used to compute the genomic control adjustment.
Comparison with competing software is subtle. The program EMMA (Efficient Mixed-Model Association) (Kang et al. 2008) is certainly the fastest of the competing programs and arguably the most sophisticated in how it handles background polygenic inheritance. On the basis of computational speed, MENDEL easily bests EMMA. On a standard personal computer, stage one of the MENDEL run took about 30 min to impute strain origins and test for association on these data when pedigree structure is included. Total computational time increased to ∼1 hr when pedigree structure was ignored. In contrast, EMMA took ∼1 day to analyze these data. The differences between MENDEL and EMMA are entirely attributable to the smaller number of locations MENDEL tests.
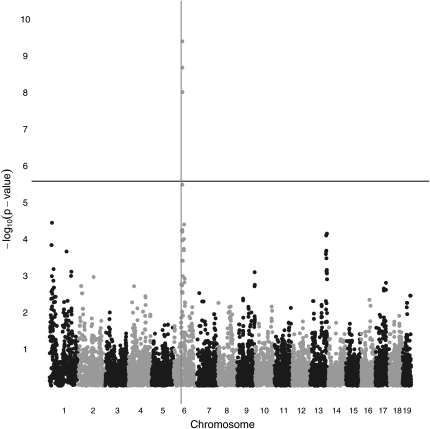
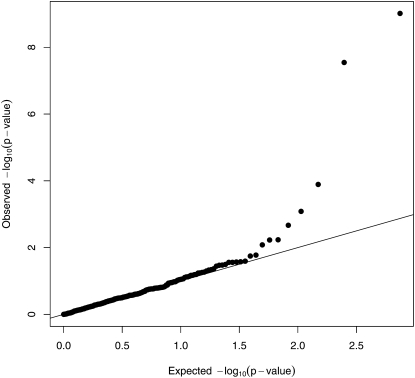
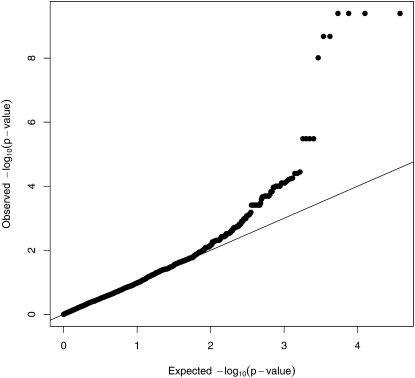
EMMA also correctly localizes the QTL in these simulated data. See Figure 6, where seven SNPs rise above the Bonferroni correction level. Four of these SNPs share the lowest P-value. Probably the most relevant statistical comparison between the programs is the increment of the maximum –log10(P-value) over the Bonferroni threshold. By this measure EMMA’s power is slightly worse than the power of our strain origin test without pedigree structure. EMMA’s power is notably worse than the power of the strain origin test with pedigree data. Note that EMMA’s P-values have also been adjusted by the method of genomic control. In our view this adjustment is less successful for EMMA than it is for MENDEL. EMMA’s test statistics undergo more radical adjustment, suggesting a poorer match between the model and the data (Price et al. 2010). (The peak value of 9.4 in Figure 6 was 10.6 before recalibration.) Furthermore, q-q plots of the adjusted statistic suggests that further adjustment of EMMA’s P-values is probably needed. See Figures 7 and 8.
Figure 6 .
Four-way cross QTL association mapping of a univariate trait using the program EMMA. The vertical line represents the QTL location. The horizontal line represents the genome-wide significance threshold after Bonferroni correction.
Figure 7 .
q-q plot of the adjusted MENDEL P-values for the simulated data assuming no pedigree structure information.
Figure 8 .
q-q plot of the adjusted EMMA P-values for the simulated data.
Bivariate analysis of pleiotropic traits:
An attractive feature of the MENDEL software is its ability to analyze multiple traits simultaneously. This capacity can increase the power to detect associations (Bauman et al. 2005). To illustrate this, we simulated a single replicate of a CC funnel cross with measured bivariate traits. The second column of Table 3 records the parameter values used during the simulation. The data involve four inbred strains (129S1, A, PWK, and CAST) and four pedigrees. Each pedigree had four founders, 15 generations, and 154 mice. To avoid confounding and permit estimation of all four global strain coefficients, each pedigree omits a different strain from its founder list. Specifically, in pedigree 1 the founder crosses involved strains 129S1 × A and 129S1 × CAST; in pedigree 2, A × CAST and A × PWK; in pedigree 3, CAST × PWK and CAST × 129S1; and in pedigree 4, PWK × 129S1 and PWK × A. Again to maintain realism, we use only the trait values for the 104 mice in the bottom 5 generations of each pedigree, 416 mice in total. As in the previous example, we simulated 1000 SNPs per mouse chromosome. We also introduced a QTL at SNP 555 of chromosome 1 (rs30642162). This major gene accounted for ∼5% of the variability in each of the two traits. The strains CAST and PWK carry genotype 2/2 at this locus and the strains 129S1 and A carry genotype 1/1. This locus was omitted from subsequent imputation and association analyses.
Table 3.
Univariate and bivariate analyses of simulated pleiotropic traits
| Values used in simulation | Estimates (SE) from univariate analysis of trait 1 | Estimates (SE) from univariate analysis of trait 2 | Estimates (SE) from bivariate analysis of traits 1 and 2 | |
| QTL Location (Mb) | 118.74 | 118.99 | 118.99 | 118.99 |
| CI (Mb) | 2.98 | 7.03 | 2.98 | |
| Trait 1 values | ||||
| μ | 5.00 | 4.47 (0.33) | 4.53 (0.44) | |
| βmale = –βfemale | 1.00 | 1.00 (0.11) | 1.00 (0.11) | |
| μ129S1 | −0.20 | 0.29 (0.57) | 0.31 (0.58) | |
| μA | 0.10 | 0.25 (0.60) | 0.32 (0.58) | |
| μCAST | 0.30 | −0.68 (0.54) | −0.74 (0.59) | |
| μPWK | −0.20 | 0.14 (0.55) | 0.11 (0.57) | |
| ε129S1 | −1.25 | −0.86 (0.31) | −0.82 (0.32) | |
| εA | −1.25 | −0.61 (0.33) | −0.68 (0.36) | |
| εCAST | 1.25 | 0.90 (0.27) | 0.97 (0.31) | |
| εPWK | 1.25 | 0.57 (0.30) | 0.53 (0.31) | |
| Trait 2 values | ||||
| μ | 6.00 | 6.21 (0.42) | 6.48 (0.43) | |
| βmale = –βfemale | 0.00 | −0.14 (0.12) | −0.13 (0.12) | |
| μ129S1 | −0.10 | 0.34 (0.59) | 0.48 (0.58) | |
| μA | 0.10 | 0.45 (0.61) | 0.44 (0.62) | |
| μCAST | 0.10 | −0.03 (0.58) | −0.18 (0.61) | |
| μPWK | −0.10 | −0.75 (0.59) | −0.74 (0.58) | |
| ε129S1 | −1.25 | −0.48 (0.32) | −0.53 (0.31) | |
| εA | −1.25 | −0.70 (0.36) | −0.70 (0.37) | |
| εCAST | 1.25 | 1.08 (0.32) | 1.09 (0.33) | |
| εPWK | 1.25 | 0.10 (0.30) | 0.14 (0.31) | |
| 5.0 | 5.04 (0.42) | 4.97 (0.42) | ||
| σtrait1,trait2 | 0.5 | 0.52 (0.33) | ||
| 5.0 | 5.06 (0.49) | 4.89 (0.48) | ||
| LRT | 46.98 | 24.94 | 54.88 | |
| DF | 3 | 3 | 6 | |
| P-value | 4.05 × 10−9 | 1.60 × 10−5 | 4.91 × 10−10 | |
We simulated a single replicate of a Collaborative Cross funnel with measured bivariate traits. The data involve four inbred strains (129S1, A, PWK, and CAST) and four pedigrees. Each pedigree had four founders, 15 generations, and 154 mice. See the text for more details on pedigree structure and simulation procedure. The second column displays the parameter values used to simulate the data, including intercept, μ; sex effect, βmale = –βfemale; polygenic mean effects, μstrain; major gene mean effects, εstrain; environmental variances, σ2trait; and environmental covariance, σtrait1,trait2. Simulation values for the polygenic variance effects in Equation 4 were Ω1,1(i, i) = Ω2,2(i, i) = 5.0, Ω1,2(i, i) = 0.50, and Ω1,1(i, j) = Ω2,2(i, j) = Ω1,2(i, j) = 0.0 for strains i ≠ j. Ω is not identifiable in the model, and thus no estimates are provided. The parameter estimates displayed pertain to the most likely SNP found by the MENDEL package. SE is the standard error. CI is the width of the one-LOD credible interval. LRT is the likelihood-ratio test statistic. DF is the degrees of freedom.
Our statistical analysis pinpoints the region around the QTL; indeed, no other region reaches genome-wide significance in association testing. Table 3 provides the parameter estimates and their standard errors, likelihood-ratio statistics, P-values, and 1-LOD credible intervals (CI) at the most likely positions. Two of the data columns of Table 3 tabulate these values for each trait analyzed separately. The rightmost column lists the values for the two traits analyzed jointly. In all three analyses, the most likely position for the QTL occurs at the SNP nearest to the simulated QTL, only 0.25 Mb distant. All of the 1-LOD credible intervals cover the true position of the QTL. Trait 1 is more strongly associated than trait 2 in the univariate analyses and has a smaller credible interval. Joint analysis leads to little change in parameter estimates. Almost all estimates are within two standard errors of their simulation values. These results are reasonable for a single simulation replicate. It is also noteworthy that the P-value for the bivariate analysis is more significant than for either univariate analysis, even though the degrees of freedom increase to 6. The bivariate analysis also maintains the tight credible interval seen in the trait 1 univariate analysis despite the larger interval seen in the trait 2 univariate analysis. These results reflect the extra information exploited in a joint analysis.
Real MF1 mice expression data:
The MF1 outbred mouse lineage was created in the early 1970s by crossing the LACA line, a standard prolific outbred mouse line, with another outbred albino line called CF. It is thought that the MF1 mouse genome represents a complex mosaic of the genomes of the inbred lines C3H, BALB/cJ, RIII, AKR, DBA/2, I, A/J, and C57BL/6J (Yalcin et al. 2004). Because MF1 mice lack good pedigree records, we used empiric strain fractions and coefficients in strain origin imputation. The average genetic contributions from strains C3H and BALB/cJ are only 5.9% and 2.7%, respectively, so we assumed for the sake of simplicity that the last six strains are the founding strains.
Ghazalpour et al. (2008) studied a total of 110 MF1 mice, measuring their gene transcript levels in liver and genotyping them at 5024 SNPs on the Affymetrix 5K Mouse Chip. Their motivation was to replicate earlier QTL mapping results from an F2 intercross between the parental strains C57BL/6J.ApoE2/2 and C3H/HeJ.ApoE2/2 (Wang et al. 2006). Mapping these eQTL in the MF1 mice appears to give better resolution and partially vindicates the use of outbred lines. Some of the eQTL are cis-eQTL and consequently involve variants in a gene influencing expression levels of that gene.
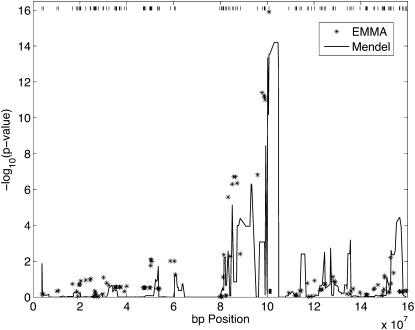
The Ttf2 gene is the most conspicuous eQTL in the study. Its expression levels provide an opportunity for eQTL association mapping based on imputed strain origins. The Ttf2 gene is located on chromosome 3: 100,742,783–100,773,586 bp on the minus (−) strand. Figure 9 compares MENDEL’s mapping results with the results output by the program EMMA (Ghazalpour et al. 2008). Both programs map the QTL to the correct interval but differ in their peak P-values. EMMA’s slightly better performance likely stems from five reasons. First, the QTL may appear among the genotyped SNPs. Second, EMMA’s test involves fewer degrees of freedom and thus is at an advantage when genotypes at the mapped SNP are highly correlated with genotypes at the underlying causative mutation. Third, this example features a sparse marker map. Bonferroni corrections of EMMA’s and MENDEL’s P-values are therefore comparable, and imputation of strain origin is more problematic. Fourth, the lack of decent pedigree records also makes strain origin imputation more challenging for MENDEL in these deep pedigrees. Fifth, analyzing a small region of the genome is inconsistent with genomic control. Despite these handicaps, MENDEL performs well.
Figure 9 .
eQTL mapping of the Ttf2 gene on chromosome 3 using results from EMMA and MENDEL. Six founder strains were used: C57Bl6, DBA2, A, AKR, ILn, and RIII. The solid curve displays the –log10(P-values) from MENDEL’s association test. EMMA’s results are displayed as asterisks. The physical location of the Ttf2 gene is shown by the small solid rectangle near the x-axis directly under the dominant peak. The tick marks at the top of the graph are the locations of the SNPs used by EMMA.
Discussion
Several recent innovations have improved the prospects for mapping mouse genes influencing complex traits. First, geneticists are now undertaking more ambitious crosses with multiple strains and sophisticated mating schemes. Second, it is now possible to incorporate polygenic background correctly in a mixed-effects model. Mixed-effects models accommodate large pedigrees, arbitrary numbers of contributing strains, and multivariate traits. Third, high-density SNP mapping panels provide unprecedented mapping resolution. Fourth, recently introduced inbred lines from wild mice capture more genetic diversity and reveal the blind spots in the mouse genome where traditional laboratory strains show little variation. Fifth, using strain origins as predictors is arguably superior to using SNP-by-SNP allele counts as predictors. The recent article by Solberg-Woods et al. (2010) confirms the value of strain-origin predictors.
Although mixed-effects models are ideal vehicles for association testing, they carry considerable computational baggage. The rate-limiting step is the imputation of local strain origins in each animal. In this article we propose an accurate and efficient imputation method that takes advantage of dense SNP maps and prior pedigree information when available. Alternation of dynamic programming and the MM algorithm quickly solves the imputation problem. Our examples demonstrate that it is possible to impute missing genotypes for founding strains and to estimate global strain fractions and coefficients in the absence of full pedigree information. In highly symmetric pedigrees, empirically derived global fractions and coefficients are nearly as accurate as the corresponding theoretical fractions and coefficients.
Imputation accuracy in a given pedigree is affected by the mating scheme, the number of generations of crossing, the diversity of the founder strains, and the density of the markers. Under the Collaborative Cross design, we attain an imputation accuracy of >99.6% even at generation 22, regardless of whether we include or ignore pedigree information. Local lumping of strains substantially improves imputation accuracy as anticipated by Yalcin et al. (2005). It also increases statistical power in subsequent QTL association testing by reducing the degrees of freedom of the likelihood-ratio test. When founder strains are closely related and several founder strains make approximately equal contributions to subsequent generations, programs such as GAIN (Liu et al. 2010) and HAPPY (Mott et al. 2000) give probability distributions of strain origins that are likely preferable to hard imputation.
The imputation methods in MENDEL scale linearly in computational complexity and storage. For a pedigree with s founding strains, n animals, and m markers, computational complexity is proportional to s4mn. Other programs such as MERLIN (Abecasis et al. 2001), GAIN (Liu et al. 2010), and HAPPY (Mott et al. 2000) scale less well. On a computer with 2 GB of memory, we had trouble running MERLIN on a pedigree with five generations of inbreeding, 19 animals, and 10,000 markers. Although GAIN incorporates prior pedigree information in imputation and enjoys high imputation accuracy, it relies on slow MCMC sampling. HAPPY has the ability to include imputed strain origins in QTL analysis, but its posterior distributions are less sharp than GAIN’s and lead to less efficient mapping inference (Liu et al. 2010). Our combination of methods performs well in both local strain imputation and subsequent QTL association mapping.
Our simulation example suggests that accurate pedigree records can improve the quality of gene mapping. However, good records do not appear to help much in strain imputation. Experimentalists might well object to the added burden of pedigree record keeping, so it is reassuring that considerable signal survives even when pedigree structure is ignored.
It is worth stressing again the advantages of strain association testing over single SNP association testing. In a modern genome scan, the former strategy mitigates the severity of Bonferroni corrections because the number of locations tested is much smaller than the number of SNPs genotyped. A ratio of 1:1000 is realistic. Unless most SNPs are genotyped, it is also likely that the causative SNP will be omitted. A correlated SNP can substitute, but if its correlation with the primary SNP is too weak, then origin attribution is apt to lead to more accurate prediction of strain vulnerability to extreme trait values. Of course, there will be exceptions where correlated SNPs align perfectly with the primary SNP. Thus, our confidence in strain origin predictors is tempered by a wait-and-see attitude. It is worth noting that in simulating the Collaborative Cross, Valdar et al. (2006) reach the general conclusion that single SNP analysis is inferior to strain-origin analysis.
Our previous article (Bauman et al. 2008) was written before mouse high-density genotyping attained its present status. The current release of MENDEL incorporates all methods discussed here. It relies on dense SNP scans, handles multivariate traits, salvages missing data whenever possible, reports outlier pedigrees and outlier animals, performs maximum-likelihood estimation under both Gaussian and multivariate t models, and accommodates arbitrarily complex crosses. Readers can download a free copy of MENDEL from http://www.genetics.ucla.edu/software. Versions of MENDEL are available for several different computing platforms. Extensive documentation and sample problems are provided. Mendel input files including the raw genotypes and MF1 gene expression are provided in Supporting Information, File S1. We encourage the use of MENDEL and further refinement of the techniques discussed here.
Acknowledgments
We thank Eleazar Eskin and Jae-Hoon Sul for their help in using the program EMMA. We also thank Jake Lusis for sharing the MF1 mice expression data with us. This research was supported by U.S. Public Health Service grants GM53275 and MH59490 and a University of California, Los Angeles, dissertation year fellowship to Jin Zhou.
Appendix: Application of the MM Algorithm
In the dynamic programming algorithm, penetrances depend on the alleles that the strains possess at the different markers. If the allele for strain a is unknown at marker k, then a penetrance at that marker depends on the postulated frequency πak of allele 1. Let ρl denote the probability that a gene contributed by strain a at marker k is interpreted as allele l. Clearly, we have
Table A1 specifies penetrances under this naive model. A question mark in Table A1 indicates a missing strain allele. Penetrances for the partially observed genotypes (1, ?) and (2, ?) follow the same rules as those governing the partially observed genotypes (?, 1) and (?, 2). A fully missing phenotype still has penetrance 1 as in Table 1.
Table A1.
Penetrances Pr[r/s | (a, b)] for SNP k with missing data
| Phenotype r/s |
|||
| Genotype (ta, tb) | 1/1 | 1/2 | 2/2 |
| (?, 1) | (1 – ε)ρ1(πak) | ερ1(πak) + (1 – ε)ρ2(πak) | ερ2(πak) |
| (?, 2) | ερ1(πak) | (1 – ε)ρ1(πak) + ερ2(πak) | (1 – ε)ρ2(πak) |
| (?, ?) | ρ1(πak)ρ1(πbk) | ρ1(πak)ρ2(πbk) + ρ2(πak)ρ1(πbk) | ρ2(πak)ρ2(πbk) |
ta is the allele carried by strain a and tb is the allele carried by strain b.
Initialization of parameters is required. The allele frequencies πak are set to . The strain fractions bear a strong resemblance to ethnic ancestry fractions and can be roughly estimated for each animal by the well-known EM algorithm (Tang et al. 2005) implemented in the Ethnic Admixture option of MENDEL (Lange et al. 2001). Finally, strain coefficients are initialized by product rules such as
assuming independent transmission of maternal and paternal gametes. For the sake of simplicity, let θ denote the parameter vector corresponding to the unknowns
An MM algorithm for minimization operates by majorizing an objective function f(θ) by a surrogate function g(θ | θr) anchored at the current iterate θr of a search (Hunter and Lange 2004). Majorization is defined by the two properties
In other words, the surface lies above the surface and is tangent to it at the point . Construction of the majorizing function constitutes the first M of the MM algorithm. The second M of the algorithm minimizes the surrogate rather than If denotes the minimum point of then the descent property is true. The proof of this claim follows from the inequalities
determined by the definitions of majorization and the next iterate The fact that majorization is preserved under sums permits one to work piecemeal on a complex objective function. The EM algorithm (Dempster et al. 1977) is a special case of the maximization version of the MM algorithm. In this case the first M refers to minorization and the second M to maximization.
In the present application of the MM algorithm, the argument of the objective function (3) is the parameter vector θ rather than the hidden state u, which is fixed throughout the MM iterations. Majorization is driven entirely by the concavity of a logarithm function as manifested in Jensen's inequality
Here cr is a constant that depends on xr and yr but not on x or y. Exploiting the property ln ab = ln a + ln b, this majorization yields, for example,
where dr is another irrelevant constant. For some terms in the objective function such as the penetrance ερ1(πak) + (1 – ε)ρ2(πak), Jensen's inequality must be applied first to separate ερ1(πak) from (1 – ε)ρ2(πak) and then to separate the terms hidden in ρ1(πak) and ρ2(πak).
The purpose of these maneuvers is to construct a surrogate function in which all parameters θj are separated and appear in the form ej ln θj or ej ln(1 – θj) for appropriate constants ej. If the term ej ln(1 – θj) appears, then θj is a binomial parameter; otherwise, θj is a multinomial parameter. In either case we consider the nonnegative constant ej to be a pseudocount of successes and update θj by the ratio of its pseudosuccesses to its pseudotrials. The ultimate formulas are messy and omitted here, but the basic idea is simple. One can derive the same updates by setting up an appropriate complete data structure and constructing an EM algorithm. In our view, direct majorization has some didactic advantages over calculating the confusing conditional expectations specifying the EM surrogate function.
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Abecasis G., Cherny S., Cookson W., Cardon L., 2001. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30: 97–101 [DOI] [PubMed] [Google Scholar]
- Ayers K., Lange K., 2008. Penalized estimation of haplotype frequencies. Bioinformatics 24: 1596–1602 [DOI] [PubMed] [Google Scholar]
- Aylor D., Valdar W., Foulds-Mathes W., Buus R., Verdugo R., et al. , 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman L., Almasy L., Blangero J., Duggirala R., Sinsheimer J., et al. , 2005. Fishing for pleiotropic qtls in a polygenic sea. Ann. Hum. Genet. 69: 590–611 [DOI] [PubMed] [Google Scholar]
- Bauman L., Sinsheimer J., Sobel E., Lange K., 2008. Mixed effects models for quantitative trait loci mapping with inbred strains. Genetics 180: 1743–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B., Farber C., Orozco L., Min Kang H., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervino A., Darvasi A., Fallahi M., Mader C., Tsinoremas N., 2007. An integrated in silico gene mapping strategy in inbred mice. Genetics 175: 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E., Rodriguez-Zas S., Mogil J., Darvasi A., Usuka J., et al. , 2001. In silico mapping of mouse quantitative trait loci. Science 294: 2423. [DOI] [PubMed] [Google Scholar]
- Churchill G., Airey D., Allayee H., Angel J., Attie A., et al. , 2004. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Day-Williams A., Blangero J., Dyer T., Lange K., Sobel E., 2011. Linkage analysis without defined pedigrees. Genet. Epidemiol. 35: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster A., Laird N., Rubin D., 1977. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B 39: 1–38 [Google Scholar]
- Devlin B., Roeder K., 1999. Genomic control for association studies. Biometrics 55: 997–1004 [DOI] [PubMed] [Google Scholar]
- Devlin B., Bacanu S., Roeder K., 2004. Genomic control to the extreme. Nat. Genet. 36: 1129–1130 [DOI] [PubMed] [Google Scholar]
- Flint J., 2011. Mapping quantitative traits and strategies to find quantitative trait genes. Methods 53: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K., Eskin E., Kang H., Bogue M., Hinds D., et al. , 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Ghazalpour A., Doss S., Kang H., Farber C., Wen P.-Z., et al. , 2008. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet. 4: e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A., Germer S., Usuka J., Aud D., Belknap J., et al. , 2001. In silico mapping of complex disease-related traits in mice. Science 292: 1915–1918 [DOI] [PubMed] [Google Scholar]
- Hunter D., Lange K., 2004. A tutorial on MM algorithms. Am. Stat. 58: 30–37 [Google Scholar]
- Kang H., Zaitlen N., Wade C., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L., Daly M., Reeve-Daly M., Lander E., 1996. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 58: 1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lange K., Cantor R., Horvath S., Perola M., Sabatti C., et al. , 2001. mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am. J. Hum. Genet. 69: A1886 [Google Scholar]
- Liu E., Zhang Q., McMillan L., de Villena F., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zeng Z., 2000. A general mixture model approach for mapping quantitative trait loci from diverse cross designs involving multiple inbred lines. Genet. Res. 75: 345–355 [DOI] [PubMed] [Google Scholar]
- Macdonald S. J., Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Talbot C., Turri M., Collins A., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A., Zaitlen N., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saar K., Beck A., Bihoreau M., Birney E., Brocklebank D., et al. , 2008. SNP and haplotype mapping for genetic analysis in the rat. Nat. Genet. 40: 560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudellari M., 2010. Mouse mash-up. Sci. Am. 302: 20–21 [DOI] [PubMed] [Google Scholar]
- Sobel E., Lange K., 1996. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 58: 1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Solberg-Woods L., Holl K., Tschannen M., Valdar W., 2010. Fine-mapping a locus for glucose tolerance using heterogeneous stock rats. Physiol. Genomics. 41: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Peng J., Wang P., Risch N., 2005. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 28: 289–301 [DOI] [PubMed] [Google Scholar]
- Valdar W., Mott R., Flint J., 2006. Simulating the collaborative cross: power of QTL detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Holmes C., Mott R., Flint J., 2009. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yehya N., Schadt E., Wang H., Drake T., et al. , 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Gessler D. D. G., Xu S., 1998. Combining different line crosses for mapping quantitative trait loci using the identical by descent-based variance component method. Genetics. 149: 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Willis-Owen S. A. G., Fullerton J., Meesaq A., Deacon R. M., et al. , 2004. Genetic dissection of a behavioral quantitative trait locus shows that rgs2 modulates anxiety in mice. Nat. Genet. 36: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Flint J., Mott R., 2005. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 171: 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]