Abstract
The Altai region of southern Siberia has played a critical role in the peopling of northern Asia as an entry point into Siberia and a possible homeland for ancestral Native Americans. It has an old and rich history because humans have inhabited this area since the Paleolithic. Today, the Altai region is home to numerous Turkic-speaking ethnic groups, which have been divided into northern and southern clusters based on linguistic, cultural, and anthropological traits. To untangle Altaian genetic histories, we analyzed mtDNA and Y chromosome variation in northern and southern Altaian populations. All mtDNAs were assayed by PCR-RFLP analysis and control region sequencing, and the nonrecombining portion of the Y chromosome was scored for more than 100 biallelic markers and 17 Y-STRs. Based on these data, we noted differences in the origin and population history of Altaian ethnic groups, with northern Altaians appearing more like Yeniseian, Ugric, and Samoyedic speakers to the north, and southern Altaians having greater affinities to other Turkic speaking populations of southern Siberia and Central Asia. Moreover, high-resolution analysis of Y chromosome haplogroup Q has allowed us to reshape the phylogeny of this branch, making connections between populations of the New World and Old World more apparent and demonstrating that southern Altaians and Native Americans share a recent common ancestor. These results greatly enhance our understanding of the peopling of Siberia and the Americas.
Introduction
The Altai Republic is located in south-central Russia, situated at the borders of Mongolia, China, and Kazakhstan. It sits at a crossroads where the Eurasian steppe meets the Siberian taiga and serves as an entry point into northern Asia. Having been habitable throughout the last glacial maximum (LGM), the Altai region has had a human presence for some 45,000 years.1 The archaeology of the region shows that, during this time, a number of different cultures and peoples lived in and migrated through the area.2–4 The confirmation of Neanderthals and the recent discovery of a new hominin at the Denisova cave in the Altai region indicates that this area has long hosted extremely diverse populations.5–7 It is also the area from which the ancestors of Native American populations are thought to have arisen prior to their expansion into the New World.8–11 In addition, archaeological evidence suggests that a few of the later cultural horizons (Afanasievo and Andronovo) arose in western Eurasia and spread eastward to the Altai region during the Eneolithic and Bronze Ages, respectively.12,13 Such interactions increased during the Iron Age, as evidenced by the frozen Pazyryk kurgans in the southern Altai Mountains,14 which contained examples of the typical “Scytho-Siberian animal style” observed throughout the entire Eurasian steppe.3,15 These populations further intermingled with expanding Altaic speaking groups, and specifically the movements involving the Xiongnu, Xianbei, and Yuezhi, as recorded by ancient Chinese historians in the second century BCE.16,17
Ethnographic studies of Turkic-speaking tribes indigenous to the Altai region of southern Siberia noted cultural differences among ethnic groups such that they could be classified into northern or southern Altaians.18,19 Northern Altaian ethnic groups include the Chelkan, Kumandin, and Tubalar. The Altai-kizhi, Teleut, and Telengit were grouped together as southern Altaians, along with a few other smaller populations. Similarly, linguistic studies have shown that languages from northern and southern populations are mutually unintelligible, despite their having similar Turkic roots. The northern Altai languages also showed greater influences from Samoyedic, Yeniseian, and Ugric languages, possibly reflecting their origin among the ancestors of these present-day peoples. By contrast, southern Altaian languages belong to the Kipchak branch of Turkic language family and have been greatly influenced by Mongolian, especially after the expansion of the Mongol Empire.16,20 These linguistic differences are further mirrored by differences in anthropometric traits, traditional subsistence strategies, religious traditions, and clan names for northern and southern Altaians.18,19,21
Genetic analysis of Altaian populations initially focused on protein polymorphisms to assess levels of diversity and the relationships between them and other Siberian populations by comparing relative proportions of West and East Eurasian genotypes.22–24 The role that the Altai region played in the dispersal of humans into northern Eurasia and subsequently into the Americas gained increasing importance with the search for the founding mitochondrial DNAs (mtDNAs) and Y chromosomes for the New World.8,25,26 As a result, the issue of where Native American progenitors originated became a hotly debated topic, with suggested source areas being Central Asia, Mongolia, and different parts of Siberia.8–10,27–46 However, much of the previous genetic research into this issue focused mainly on southern Altaian populations, leaving our understanding of the genetic diversity of northern Altaian groups incomplete.
Given the ethnographic and historical background of Altaian peoples, we characterized the mtDNA and Y chromosome variation in these populations to elucidate their genetic history. Our first objective was to determine whether the ethnographic classifications of northern and southern Altaians reflected their patterns of genetic variation, and specifically whether they shared a common ancestry. If differences were observed, we then wanted to know whether they were attributable to demographic factors, social organization, or some combination of the two. The second goal was to examine whether northern Altaians' genetic variation is structured by tribe and clan identity. The third goal was to use these data to investigate larger questions concerning the peopling of Siberia (and the Americas). In particular, we were interested in learning whether these genetic data would reveal the effects of ancient and/or recent migrations into or out of the Altai region, including that giving rise to the ancestors of indigenous populations from America. Overall, this paper attempts to understand the population history of Altaians by placing them into a Siberian genetic context and uses a phylogeographic approach to dissect the layers of history, uncovering the formation of these ethnic groups and their importance for understanding the peopling of Northern Asia and the Americas.
Subjects and Methods
Sample Collection
Between 1991 and 2002, we conducted ethnographic fieldwork and sample collection in a number of settlements within the southern part of the Altai Republic (Figure 1). During this period, a total of 267 self-identified Altai-kizhi individuals living in the villages of Mendur-Sokkon, Cherny Anuy, Turata, and Kosh-Agach participated in the study. In addition, another nine Altai-kizhi individuals from villages in the northern Altai Republic participated in the study (see below), bringing the total number of Altai-kizhi participants to 276, of whom 120 were men.
Figure 1.
Map of the Altai Republic and Locations of Sample Collection
In 2003, we worked with 214 Northern Altaians living in the Turochak District of the Altai Republic. These persons included 91 Chelkans, 52 Kumandins, and 71 Tubalars living in nine different villages in the Biya and Lebed' River basins and along Teletskoe Lake (Figure 1). The villages included Artybash, Biika, Dmitrievka, Kebezen, Kurmach-Baigol, Sank-Ino, Shunarak, Tandoshka, and Yugach. Of the northern Altaian participants, 69 were men.
Blood samples were drawn from all participants with informed consent written in Russian and approved by the University of Pennsylvania IRB and the Institute of Cytology and Genetics in Novosibirsk, Russia. Genealogical data were also obtained from each person at the time of sample collection to ensure that the individuals were unrelated through at least three generations and to assess the level of admixture in these communities. Individuals were categorized by self-identified ethnicity for this study.
Molecular Genetic Analysis
Sample Preparation
Bloods were fractionated through low-speed centrifugation to obtain plasma and red cell fractions. Total genomic DNAs were isolated from buffy coats with a lysis buffer and standard phenol-chloroform extraction protocol modified from earlier studies.27,47
mtDNA Analysis
The mtDNA of each sample was characterized by high-resolution SNP analysis and control region sequencing. PCR-RFLP analysis was employed to assign individuals to West48–52 and East30,53–56 Eurasian mtDNA haplogroups by screening them for known diagnostic markers, as per previous studies57,58 (Table S1 available online), with the nomenclature used to classify the mitochondrial haplotype according to PhyloTree.org.59
The hypervariable segment 1 (HVS1) of the control region was directly sequenced for each sample by published methods,58 and hypervariable segment 2 (HVS2) was sequenced with the primers indicated in Table S2. Sequences were read on ABI 3130xl Gene Analyzers located in the Laboratory of Molecular Anthropology and the Department of Genetics Sequencing Core Facility at the University of Pennsylvania and aligned and edited with the Sequencher 4.8 (Gene Codes Corporation). All polymorphic nucleotides were reckoned relative to the revised Cambridge reference sequence (rCRS).60,61 The combination of SNP data and control region sequences defined maternal haplotypes in these individuals.
Y Chromosome Analysis
The nonrecombining portion of the Y chromosome (NRY) from each male participant was characterized by assaying phylogenetically informative biallelic markers in a hierarchical fashion according to published information62,63 and previously published methods.64 A total of 116 biallelic markers were tested to define sample membership in respective NRY haplogroups. Most of the SNPs and fragment length polymorphisms were characterized by custom TaqMan assays read on an ABI Prism 7900 HT Real-Time PCR System (Applied Biosystems). These polymorphisms included L53, L54, L55, L56, L57, L213, L329, L330, L331, L332, L333, L365, L400, L456, L472, L474, L475, L476, L528, LLY22g, M3, M9, M12, M15, M18, M20, M25, M35, M45, M55, M56, M69, M70, M73, M81, M86, M89, M93, M96, M102, M117, M119, M120, M122, M123, M124, M128, M130, M134, M143, M147, M157, M162, M170, M172, M173, M174, M178, M186, M201, M204, M207, M214, M217, M223, M230, M242, M253, M265, M267, M269, M285, M304, M323, M335, M346, M410, M417, M434, M458, P15, P25, P31, P36.2, P37.2, P47, P60, P63, P105, P215, P256, P261, P297, and PK2. Additional markers were detected through direct sequencing (L191, L334, L401, L527, L529, M17, M46 [Tat], M343, M407, MEH2, P39, P43, P48, P53.1, P62, P89, P98, P101, PageS000104, and PK5) and by PCR-RFLP analysis (M175).65 Seventeen short tandem repeats (STRs) were characterized with the AmpFlSTR Yfiler PCR Amplification Kit (ABI) and read on an ABI 3130xl Genetic Analyzer with GeneMapper ID v3.2 software. Each paternal haplotype was designated by its 17-STR profile. Y chromosome lineages were defined as the unique combinations of SNP and STR data present in the samples. DYS389b was calculated by subtracting DYS389I from DYS389II, which was used for all statistical and network analyses.64
Comparative Data
To place their genetic histories in a broader contextual framework, we compared Altaian mtDNA and NRY data with those from populations in southern Siberia, Central Asia, Mongolia, and East Asia. For the mtDNA analysis, the populations included Telengits, Teleuts, Shors, Khakass, Tuvinians, Todzhans, Tofalars, Soyots, Buryats, Khanty, Mansi, Ket, Nganasan, Western Evenks, Uyghurs, Kazakhs, Kyrgyz, Uzbeks, and Mongolians.41,43,44,66–71 For the NRY analysis, only populations that were represented by full Y-STR data sets (not just Y-STRs for specific haplogroups) were used for comparative purposes. These populations included Teleuts, Khakass, Mansi, Khanty, Kalmyks, Mongolians, and Uyghurs.68,72–75 The STR haplotypes were reduced to ten loci (DYS19, DYS389I, DYS398b, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, and DYS439) to allow for as broad a comparison as possible. In the coalescence analysis, we used the 15 Y-STR loci Q-M3 haplotypes from Geppert et al.76
Data Analysis
Summary statistics, including gene diversity and pairwise differences, were calculated with Arlequin v3.1177 for mtDNA HVS1 (np 16024-16400) and NRY Y-STRs. FST and RST values between populations were also calculated with Arlequin v3.11 for the HVS1 sequences and Y-STRs, respectively. FST values were estimated with the Tamura and Nei model of sequence evolution.78 Pairwise genetic distances were visualized by multidimensional scaling (MDS) with SPSS 11.0.0.79 In addition, nucleotide diversity, Tajima's D, and Fu's FS were calculated with mtDNA HVS1 sequences.
We analyzed the phylogenetic relationships among Y-STR haplotypes and complete mtDNA genomes by using Network 4.6.0.0 (Fluxus Technology Ltd). These networks employed a reduced median-median joining approach and MP post-processing.80–82 The NRY haplotypes used to generate the networks consisted of 15 Y-STRs. DYS385 was excluded from the network analysis because differentiation between DYS385a and DYS385b is not possible with the Y-Filer kit.83 The Y-STR loci were weighted based on the inverse of their variances. Mitogenomes used in this analysis came from the published literature and GenBank.
The time to the most recent common ancestor (TMRCA) for mitogenomes was estimated with the methods of Soares et al.84 The Y-STR diversity within each haplogroup was assessed by two methods.64 The first involved calculation of rho statistics with Network 4.6.0.0, where the founder haplotype was inferred as in Sengupta et al.85 The second used Batwing,86 a Bayesian analysis where the TMRCA and expansion time of each population (or haplogroup) were calculated by previously published methods.64,72,87 Both the evolutionary and the pedigree-based mutation rates were used to estimate coalescence dates with generation times of 25 and 30 years, respectively.88–90 Because a definitive consensus does not yet exist as to which rate should be used, the validity of the resulting estimates are discussed. In addition, Batwing was used to estimate the split or divergence times of several haplogroups. This method assumes that, after populations split, no further migration occurs between them. In this case, the haplogroups investigated were not shared between populations but derive from a common source, thereby justifying this approach. Duplicated loci and new STR variants detected in this study were excluded from statistical analysis.
Results
Mitochondrial DNA and Y Chromosome Diversity
The maternal genetic ancestry of northern and southern Altaian populations was explored by characterizing coding region SNPs and control region sequences from 490 inhabitants of the Altai Republic, which yielded 99 distinct mtDNA haplotypes defined by SNP and HVS1 mutations (Table S3). The majority of mtDNAs were of East Eurasian origin, although the relative proportion of these haplotypes was greater in Chelkans (91.5%) compared to other Altaian populations (75.2% in Tubalars, 75.6% in Kumandins, and 76.4% in Altai-kizhi) (Table 1). Despite exhibiting a lower overall frequency of West Eurasian haplogroups, Altaians (specifically, the Altai-kizhi, Tubalar, and Kumandins) had a higher proportion of them as compared to other southern Siberians.41,43 Differences in mtDNA haplogroup profiles were observed among northern Altaian ethnic groups and between northern Altaians and Altai-kizhi, with the Chelkans being extraordinarily distinct. Nevertheless, comparisons among other Altaian ethnic groups revealed some consistent patterns. mtDNA haplogroups B, C, D, and U4 were found in all Altaian populations, but at varying frequencies, whereas southern Altaians (Altai-kizhi, Telengits, and Teleuts) tended to have a greater variety of West Eurasian haplogroups at low frequencies. Shors, who have sometimes been categorized as northern Altaians,18 exhibited a similar haplogroup profile to other northern Altaian ethnic groups, including moderate frequencies of C, D, and F1, although they lacked others (N9a and U).41
Table 1.
mtDNA Haplogroup Frequencies of Altaian Populations
| Hg | Chelkan | Kumandin | Tubalar1 | Tubalar2 | Shor | Altai-kizhi1 | Altai-kizhi2 | Telengit | Teleut |
|---|---|---|---|---|---|---|---|---|---|
| # | 91 | 52 | 71 | 72 | 28 | 276 | 48 | 55 | 33 |
| C | 15.1 | 41.5 | 35.6 | 20.8 | 17.9 | 31.4 | 25.0 | 14.6 | 24.2 |
| Z | 2.7 | 3.6 | 4.3 | 4.2 | 3.0 | ||||
| M8 | 3.6 | 4.2 | |||||||
| D4 | 13.9 | 15.1 | 24.7 | 15.3 | 25.0 | 13.0 | 6.3 | 18.2 | 24.2 |
| D5 | 8.6 | 3.8 | 4.1 | 5.6 | 3.6 | 0.7 | 3.0 | ||
| G | 3.2 | 4.0 | 4.2 | 3.6 | |||||
| M7 | 1.8 | ||||||||
| M9 | 1.4 | ||||||||
| M10 | 1.1 | 3.6 | 0.4 | 2.1 | |||||
| M11 | 2.1 | 1.8 | 3.0 | ||||||
| M∗ | 1.8 | ||||||||
| A | 1.9 | 11.1 | 3.6 | 2.9 | 4.7 | 7.3 | |||
| I | 3.6 | 1.4 | 2.1 | 1.8 | |||||
| N1a | 1.8 | ||||||||
| N1b | 0.4 | ||||||||
| W | 1.1 | ||||||||
| X | 3.8 | 1.4 | 2.2 | 2.1 | 3.0 | ||||
| N9a | 19.4 | 1.9 | 2.7 | 6.9 | 1.8 | ||||
| B | 3.2 | 3.8 | 2.7 | 4.2 | 3.6 | 1.4 | 6.3 | 14.6 | 6.1 |
| F1 | 10.8 | 3.8 | 1.4 | 14.3 | 8.3 | 4.2 | 1.8 | 3.0 | |
| F2 | 15.1 | 2.7 | 3.6 | 2.5 | 2.1 | ||||
| H | 1.1 | 2.7 | 1.4 | 3.6 | 2.5 | 8.3 | 9.1 | 9.1 | |
| H2 | 3.3 | 2.1 | |||||||
| H8 | 5.7 | 2.7 | 4.2 | 3.6 | 1.4 | ||||
| HV | 1.8 | ||||||||
| V | 6.1 | ||||||||
| J | 3.6 | 4.0 | 6.3 | 1.8 | |||||
| T | 1.9 | 0.4 | 3.6 | 6.1 | |||||
| U2 | 2.8 | 0.7 | 1.8 | 3.0 | |||||
| U3 | 2.1 | ||||||||
| U4 | 4.3 | 3.8 | 15.1 | 18.1 | 3.6 | 0.7 | 2.1 | 1.8 | 3.0 |
| U5 | 2.2 | 9.4 | 4.1 | 5.6 | 3.3 | 2.1 | 1.8 | ||
| U8 | 1.8 | ||||||||
| K | 3.6 | 3.3 | 6.3 | 3.0 | |||||
| R9 | 1.1 | 3.8 | 1.4 | 2.2 | 5.5 | ||||
| R11 | 2.1 |
Haplogroups C and D were the most frequent mtDNA lineages in the Altaians, consistent with the overall picture of the Siberian mtDNA gene pool. However, phylogeographic analysis of these lineages showed a greater diversity of haplotypes in the southern Altaians compared to northern Altaians. Although haplotypes were shared between regions, northern Altaians largely had C4 with the root HVS1 motif (16223-16298-16327) and C5c, whereas the southern Altaians had C4a1 and C4a2. Although C5c is largely confined to Altaians, it has been suggested that an early migration from Siberia to Europe brought haplogroup C west, where the branch differentiated during the Neolithic and then was taken back into southern Siberia.83 Also noteworthy, D4j7 appears to be specific to Altaians and Shors.41,91 In addition, a D5a haplotype was shared by Tubalars and Altai-kizhi, and a rare D5c2 haplotype was shared by the Chelkans and Kumandins. Interestingly, complete mtDNA genome sequencing of a subset of our D5c2 samples showed few differences from those present in Japan,55 suggesting a possible connection resulting from the dispersal of Altaic speaking populations.92 The remainder of the D haplotypes were found in other southern Siberian and Central Asian populations.
To explore the NRY variation in Altaian populations, 116 biallelic polymorphisms were characterized in 189 male individuals, resulting in 106 Y chromosome lineages (Table 2). Northern Altaian populations were composed largely of haplogroups Q and N-P43, whereas southern Altaians had a higher proportion of R-M417, C-M217/PK2, C-M86, and D-P47. Haplogroups typical of south Asia, western Europe, and East Asia were not found in appreciable frequencies.72,93–99 The haplogroup frequency differences between northern and southern Altaians were statistically significant (χ2 = 66.03, df = 9, p = 9.09 e−11).
Table 2.
High-Resolution NRY Haplogroup Frequencies in Altaian Populations
| Haplogroup | Chelkan | Kumandin | Tubalar | Altai-kizhi |
|---|---|---|---|---|
| C3∗ | 19 (0.158) | |||
| C3c1 | 5 (0.042) | |||
| D3a | 6 (0.050) | |||
| E1b1b1c | 1 (0.037) | |||
| I2a | 1 (0.037) | |||
| J2a | 3 (0.025) | |||
| L | 1 (0.040) | |||
| N1∗ | 1 (0.059) | 3 (0.111) | ||
| N1b∗ | 5 (0.200) | 8 (0.471) | 2 (0.017) | |
| N1c∗ | 1 (0.008) | |||
| N1c1 | 2 (0.017) | |||
| O3a3c∗ | 1 (0.008) | |||
| O3a3c1 | 1 (0.037) | 1 (0.008) | ||
| Q1a2 | 1 (0.037) | |||
| Q1a3a∗ | 15 (0.600) | 10 (0.370) | ||
| Q1a3a1c∗ | 20 (0.167) | |||
| R1a1a1∗ | 4 (0.160) | 2 (0.118) | 10 (0.370) | 60 (0.500) |
| R1b1a1 | 6 (0.353) | |||
| T | ||||
| Total | 25 | 17 | 27 | 120 |
As with the mtDNA data set, we also observed differences in NRY haplogroup composition among northern Altaian populations, where each ethnic group shared haplogroups with the other two, yet had distinct haplogroup profiles. Overall, Kumandins had the most disparate haplogroup frequencies of the northern Altaians, exhibiting similar number of N-P43 chromosomes as the Chelkans, which were quite similar to those found in Khanty and Mansi populations in northwestern Siberia.68,100 In addition, a large proportion of Kumandin Y chromosomes belonged to R-M73. This haplogroup is largely restricted to Central Asia101 but has also been found in Altaian Kazakhs and other southern Siberians.64,102 In fact, Myres et al.101 noted two distinct clusters of R-M73 STR haplotypes, with one of them containing Y chromosomes bearing a 19 repeat allele for DYS390, which appears to be unique to R-M73. Interestingly, the majority of Kumandin R-M73 haplotypes fell into this category, although haplotypes from both clusters are found in southern Siberia.102
In all cases, the haplotypes present in Altaians fit into known modern human phylogenies. None of the Altaians had a mitochondrial lineage similar to those of Neanderthals or the Denisovan hominin. Although there are no ancient Denisovan or Neanderthal Y chromosome data to compare with the Altaian data set, the Altaian Y chromosomes clearly derived from more recent expansions of modern humans out of Africa.
Altaian Genetic Relationships
Summary statistics were calculated to assess the relative amounts of genetic diversity in Altaian populations (Table 3). Gene diversities based on HVS1 of the mtDNA showed that, overall, the Altai-kizhi were more diverse than the northern Altaians. The average pairwise differences for the Altai-kizhi were also smaller. In fact, the estimates for the Altai-kizhi and Tubalars were comparable to other southern Siberians.43 By contrast, those for the Chelkans and Kumandins were lower and more similar to Soyots, but not as low as that of Tofalars. Mismatch distributions were smooth and bell-shaped for all populations except the Chelkans, which had a significant raggedness index. This statistic indicated that Tubalars, Kumandins, and Altai-kizhi had experienced sudden expansions or expansions from population bottlenecks.103 Tests of neutrality confirmed these findings in yielding significantly negative Tajima's D and Fu's FS estimates for all populations, except the Chelkans, indicating that this particular population probably experienced a reduction in population size or was subdivided.
Table 3.
HVS1 Summary Statistics for Altaian Populations
| Population |
Northern Altaian |
Southern Altaian |
||
|---|---|---|---|---|
| Chelkan | Kumandin | Tubalar1 | Altai-kizhi1 | |
| # of samples | 91 | 52 | 71 | 276 |
| # of haplotypes | 22 | 18 | 26 | 75 |
| Haplotype diversity | 0.923 ± 0.013 | 0.914 ± 0.021 | 0.953 ± 0.010 | 0.976 ± 0.003 |
| Nucleotide diversity | 0.020 ± 0.011 | 0.022 ± 0.011 | 0.019 ± 0.010 | 0.018 ± 0.009 |
| Pairwise differences | 7.68 ± 3.61 | 8.22 ± 3.87 | 7.03 ± 3.34 | 6.84 ± 3.23 |
| Raggedness index | 0.032 | 0.022 | 0.010 | 0.011 |
| Raggedness p value | 0.000 | 0.149 | 0.635 | 0.388 |
| Tajima D | 1.201 | −0.644 | −0.701 | −1.180 |
| Tajima D p value | 0.000 | 0.000 | 0.000 | 0.000 |
| Fu's FS | 3.417 | −0.497 | −3.877 | −24.416 |
| Fu's FS p value | 0.002 | 0.000 | 0.000 | 0.000 |
To understand Altaian maternal genetic background, we compared our data with those from other North Asian and Central Asian populations. FST values between populations were calculated with HVS1 sequences and viewed through multidimensional scaling (Figure 2). In this analysis, southern Siberians formed a rather diffuse cluster, with most Central Asian and Mongolian populations being separated from them. Altaian populations also did not constitute a distinct cluster unto themselves. Based on the FST values, the Chelkans were distinctive from all other ethnic groups. Although falling closest to the Khakassians in the MDS plot, they shared a smaller genetic distance with the Tubalars2, which was expected because of the inclusion of some Chelkans in that sample set.44 Kumandins and Tubalars1 were not significantly different, and appeared close to Tuvinians and southern Altaians. In fact, both populations had smaller FST values with southern Altaians than they did with the Chelkans, although the genetic distances between Tubalars1 and Tubalars2, Altai-kizhi, and Teleuts were also nonsignificant. Unlike northern Altaians, most of the southern Altaian populations clustered together. The Altai-kizhi, Teleuts, and Tubalars1 formed one small cluster with Kyrgyz, whereas the Telengits showed greater affinities with Central Asian populations. The southern Altaian cluster sat near a cluster of Tuvinian populations, suggesting a similar population history and likely gene flow between these groups.
Figure 2.
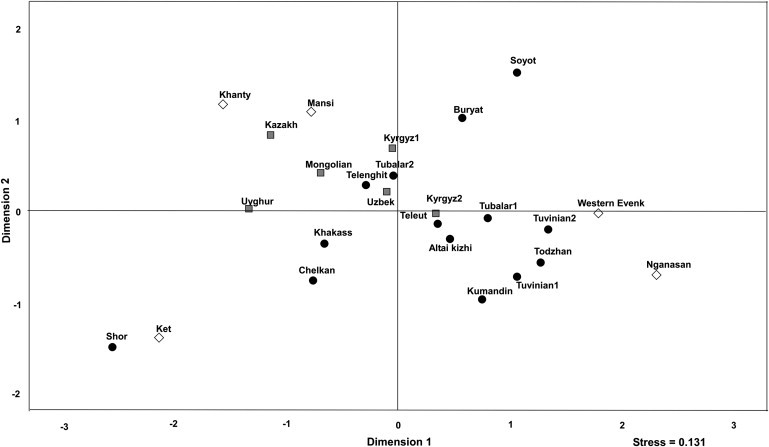
MDS Plot of FST Genetic Distances Generated from mtDNA HVS1 Sequences in Siberian and Central Asian Populations
Circle, southern Siberian; diamond, northwestern Siberian; square, Central Asian.
Summary statistics were calculated to assess the genetic diversity of paternal lineages in Altaian populations (Table 4). Gene diversities based on Y-STR haplotypes (15-loci Y-STR haplotypes; Table S4) showed that the Altai-kizhi were more diverse than the northern Altaians. Unlike the mtDNA data, within group pairwise differences were greater in the southern Altaian and Tubalar Y-STR haplotypes than in the Chelkans and Kumandins.
Table 4.
Y-STR Summary Statistics for Altaian Populations
| Population |
Northern Altaian |
Southern Altaian |
||
|---|---|---|---|---|
| Chelkan | Kumandin | Tubalar | Altai-kizhi | |
| # of samples | 25 | 17 | 27 | 120 |
| # of haplotypes | 14 | 9 | 18 | 62 |
| Haplotype diversity | 0.910 ± 0.043 | 0.912 ± 0.042 | 0.954 ± 0.025 | 0.978 ± 0.005 |
| Pairwise differences | 6.59 ± 3.22 | 6.39 ± 3.19 | 7.40 ± 3.57 | 7.58 ± 3.56 |
Y-chromosomal variation in the four populations in our data set provided a slightly different picture than the mitochondrial data. In this analysis, RST values were calculated with 15-loci Y-STR haplotypes (Table S6). These estimates indicated that only the Chelkans and Tubalars were not significantly different from each other. The Kumandins were quite distant from all populations, although these distances were slightly smaller among northern Altaians than with the Altai-kizhi. The Altai-kizhi were again closest to the Tubalars.
These relationships were affirmed by the haplotype sharing between the four populations. The Chelkans and Tubalars shared a large proportion of their haplotypes, mostly those from haplogroups Q and R-M417, whereas the Kumandins shared only one haplotype with Tubalars (a rare N-LLY22g haplotype). In addition, the northern and southern Altaians shared only a single haplotype, belonging to haplogroup O-M117, which is more commonly found in southern China.104 In fact, these two Y chromosomes were the only occurrences of haplogroup O in our data set.
The Y-STR profiles were reduced to 10-loci STR haplotypes in order to compare Y chromosome diversity in several Siberian and Central Asian populations (Table 5; Figure 3). The genetic distances in our sample set remained high despite the greater haplotype sharing that resulted from this reduction. Overall, the genetic distances were much greater with the Y-STR haplotypes compared to mtDNA haplotypes, indicating greater genetic differentiation in paternal lineages compared to maternal lineages. In addition to the Chelkans and Tubalars, two other groups of populations exhibited nonsignificant RST values. One group included Uyghur (from Urumqi and Yili) and Mongolian (Kalmyks and Mongolians) populations, and the other included the Mansi and a Sagai population identified as part of the Khakass ethnic group. In contrast with their position in the mtDNA MDS plot, northern Altaians were separated from all other populations, including other southern Siberians. The three groups of Khakass (Sagai, Sagai/Shor, and Kachin) fell much closer to the Khanty and Mansi, which probably indicates a common ancestry for these populations. Unfortunately, more complete Y-STR data sets were not available for other southern Siberian populations. Nonetheless, these results indicated a different history for northern Altaians compared to Central Asians and even other southern Siberians. A specific reason for this difference is that Mongolians had a much greater genetic impact on southern Altaians, which is expected given the historical and linguistic evidence.18,19,105
Table 5.
Low-Resolution NRY Haplogroup Frequency Comparison of Altaians
| Hg | Chelkan | Kumandin | Tubalar | Altai-kizhi1 | Altai-kizhi2 | Teleut1 | Teleut2 | Shor |
|---|---|---|---|---|---|---|---|---|
| C | 20.0 | 13.0 | 8.5 | 5.7 | 2.0 | |||
| D | 5.0 | 3.3 | ||||||
| E | 3.7 | |||||||
| F (xJ,K) | 3.7 | 3.3 | 10.7 | 2.0 | ||||
| J | 2.5 | 2.2 | 2.1 | |||||
| K (xN1c,O,P) | 24.0 | 52.9 | 11.1 | 1.7 | 2.2 | 13.7 | ||
| N1c | 2.5 | 5.4 | 10.6 | 28.6 | 2.0 | |||
| O | 3.7 | 1.7 | ||||||
| P (xR1a1a) | 60.0 | 35.3 | 40.7 | 16.7 | 28.3 | 34.3 | 2.0 | |
| R1a1a | 16.0 | 11.8 | 37.0 | 50.0 | 42.4 | 68.1 | 31.4 | 78.4 |
| Total | 25 | 17 | 27 | 120 | 92 | 47 | 35 | 51 |
Figure 3.
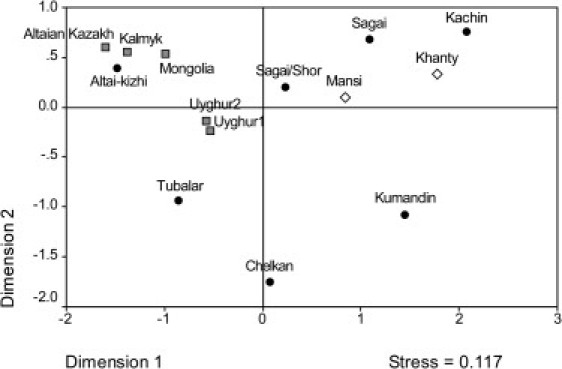
MDS Plot of RST Genetic Distances Generated from Y Chromosome STR Haplotypes in Siberian and Central Asian Populations
Circle, southern Siberian; diamond, northwestern Siberian; square, Central Asian.
Altaian and Native American Connections
To test the hypothesis that Native Americans share a more recent common ancestor with Altaians relative to other Siberian and East Asian populations, we specifically examined the mtDNA and NRY haplogroups that appeared in both locations. For the mtDNA, it is well known that haplogroups A–D and X largely make up the maternal genetic heritage of indigenous peoples in the Americas.27,29,39,47,106 Complete mtDNA genome sequencing has led to a greater comprehension of the phylogeny of Native American mtDNAs and, consequently, a better understanding of their origins.107–110 Although Altaians possess the five primary mtDNA haplogroups found in the Americas, these lineages are not exactly the same as those appearing in Native Americans at the subhaplogroup level. This is also true for other Siberian populations except in those few instances where gene flow across the Bering Strait brought some low frequency types back to northeastern Siberians.
An example of this pattern is haplogroup C1a. Southern Altaians possessed C1a, which is an exclusively Asian branch of the predominately American C1 haplogroup.107,108 To date, only four complete C1a genomes have been published. These sequences produced a more recent TMRCA than other genetic evidence had previously suggested for the peopling of the Americas. Although Tamm et al.107 viewed this haplogroup as representing a back migration into Siberia, it does not occur in Siberian populations that are geographically closest to the Americas, but rather those living in southern and southeastern Siberia.41,89 However, given the small effective population sizes from the northeastern Siberian groups that have been studied thus far, this haplogroup could have been lost because of drift.
The other mtDNA haplogroup found in northern and southern Altaians that is a close relative of a Native American lineage is D4b1a2a1a. This haplogroup has been found in Altaians, Shors, and Uzbeks from northwestern China.41,44,70 Analysis of complete mtDNA genomes identified a sister branch (D4b1a2a1a1), which is found only in northeastern Siberian populations and Inuit from Canada and Greenland.42,45,54,91,111 TMRCAs were calculated from the complete mtDNA genomes of this branch and those from Native American D4b1a2a1a1. By analyzing only synonymous mutations from these sequences with the method of Soares et al.,84 we estimated the TMRCAs of these two branches at 11.8 kya and 15.8 kya, respectively.
For the Y chromosome, indigenous American lineages are derived mostly from haplogroups C and Q, and, as such, are crucial for understanding of the genetic histories of peoples from the Americas and how they relate to populations of Central Asia and Siberia.9,39,93,98,112,113 Just as Seielstad et al.114 and Bortolini et al.38 used M242 to clarify the genetic relationship between Asian and American Y chromosomes, the characterization of this haplogroup at an even higher level of resolution has led to a much greater understanding of the origins of Native American Y chromosomes and their connections to Asian types. In this regard, it was recently shown that the American Q-M3 SNP is located on an M346-positive background.63 The presence of M346 in Central Asia and Siberia has strengthened the argument for a southern Siberian or Central Asian origin for many American Y chromosomes.85,99,102,115
Given the importance of haplogroup Q for Native American origins, we subjected samples from this lineage to high-resolution SNP analysis involving 37 biallelic markers to better understand the relationship between Old and New World populations and the migration(s) that connect them. All Y chromosomes in this study that belonged to haplogroup Q (as indicated by the presence of M242) were also found to have the P36.2, MEH2, L472, and L528 markers (Figure S1). Thus, these haplotypes fell into the Q1a branch of the Y chromosome phylogeny. Because Q1b Y chromosomes were not found in Altaian samples, we were not able to definitively place the L472 and L528 SNPs at the same phylogenetic position as MEH2. For this reason, their placement is tentative until L275/L314/M378 Y chromosomes are screened for these markers. Furthermore, M120/M265-positive, P48-positive, and P89-positive samples were not found in the Altai region. Therefore, the placement of these branches at the same phylogenetic level as M25/M143 and M346/L56/L57 should also be considered as provisional (although see Karafet et al.63).
The M346, L56, and L57 SNPs were positioned as ancestral to three derived branches in the Family Tree DNA phylogeny. We found that the L474, L475, and L476 SNPs were present in all of our M346-positive samples. However, because M323- and L527/L529-positive samples were not found in the Altaians, we could not confirm the exact position of these markers at either the Q1a3 or Q1a3a level. On the other hand, all Altaians that possessed the M346, L56, L57, L474, L475, and L476 SNPs also had L53, L55, L213, and L331.
Interestingly, northern and southern Altaian Q Y chromosomes differed by three markers. L54, L330, and L333 were found in Q haplotypes in the southern Altaians and one Altaian Kazakh, whereas the northern Altaians Q haplotypes lacked these derived SNPs. Thus, according to the standard nomenclature set by the Y Chromosome Consortium62 and followed by others, the northern Altaian Q haplotypes belonged to Q1a3a∗ and the southern Altaians belonged to Q1a3a1c∗. We have further confirmed that M3 haplotypes belong to L54-derived Y chromosomes (unpublished data). These alterations in the phylogeny change the haplogroup name of the Native American Q-M3 Y chromosomes from Q1a3a to Q1a3a1a. Moreover, the position of M3 and L330/L333 in the phylogeny indisputably showed that the MRCA of most Native American Y chromosomes was shared with southern Altaians.
The differences between the northern and southern Altaian Q Y chromosomes were also reflected in the analysis of high-resolution Y-STR haplotypes (Figure S2).116 Comparisons of Altaian Q-M346 Y chromosomes with those from southern Siberian, Central Asian, and East Asian populations revealed affinities between southern Altaian and these other groups. However, the northern Altaians remained distinctive, even in networks constructed from fewer Y-STR loci (Figure S3).
The time required to evolve the extent of haplotypic diversity observed in each of the subhaplogroups can aid in determining when particular mutations arose and possibly when these mutations were carried to other locations. The TMRCA for the northern Altaian Q1a3a∗ Y chromosomes indicated a relatively recent origin for them, one dating to either the Bronze Age or recent historical period, depending on the Y-STR mutation rate being used (Table 6). The southern Altaian/Altaian Kazakh Q1a3a1c∗ Y chromosomes had a slightly older TMRCA that dated them to either the late Neolithic or early Bronze Age. By using Bayesian analysis, we further estimated the divergence time of the two Q haplogroups at about 1,000 years after the TMRCA of all Altaian Q lineages (∼20 kya), indicating an ancient separation of northern and southern Altaian Q Y chromosomes (Table 7).
Table 6.
TMRCAs and Expansion Times for Altaian and Native American NRA Haplogroup Q Lineages
| Hg | N |
Network |
Batwing - TMRCA |
Batwing - Expansion |
||
|---|---|---|---|---|---|---|
| ρ ± σ | Median | 95% C.I. | Median | 95% C.I. | ||
| Pedigree-Based Mutation Rate | ||||||
| All Q1a3a | 97 | 5,390 ± 1,000 | 8,420 | [5,620–14,290] | 7,230 | [1,220–20,510] |
| Q1a3a∗ | 25 | 1,410 ± 580 | 1,480 | [680–3,060] | 2,100 | [380–6,830] |
| Q1a3a1a∗ | 52 | 5,820 ± 1,280 | 7,630 | [4,870–12,920] | 4,680 | [480–14,940] |
| Q1a3a1c∗ | 20 | 2,420 ± 700 | 2,970 | [1,500–5,960] | 2,680 | [450–8,610] |
| Evolutionary-Based Mutation Rate | ||||||
| All Q1a3a | 97 | 14,970 ± 2,760 | 25,580 | [14,230–51,140] | 17,220 | [1,380–54,950] |
| Q1a3a∗ | 25 | 3,910 ± 1,610 | 5,320 | [2,300–12,160] | 4,340 | [1,000–13,080] |
| Q1a3a1a∗ | 52 | 16,170 ± 3,550 | 22,160 | [11,960–44,340] | 9,800 | [620–39,543] |
| Q1a3a1c∗ | 20 | 6,750 ± 1,950 | 8,720 | [3,960–20,010] | 5,600 | [1,030–17,910] |
Note: ρ, rho statistic; σ, standard error; Q1a3a∗, Northern Altaians (this study); Q1a3a1a, Native Americans (Geppert et al.76); Q1a3a1c, Southern Altaians (this study).
Table 7.
Divergence Times between Haplogroups/Populations
|
TMRCA |
Split Time |
|||
|---|---|---|---|---|
| Median | 95% Confidence Interval | Median | 95% Confidence Interval | |
| Pedigree-Based Mutation Rate | ||||
| Northern and Southern Altaians | 5,490 | [3,000–11,100] | 4,490 | [1,730–10,070] |
| Southern Altaians and Native Americans | 7,740 | [5,170–12,760] | 4,950 | [2,360–9,490] |
| Evolutionary-Based Mutation Rate | ||||
| Northern and Southern Altaians | 21,890 | [9,900–57,440] | 19,260 | [7,060–54,600] |
| Southern Altaians and Native Americans | 21,960 | [12,260–42,690] | 13,420 | [5,220–30,430] |
A similar analysis was conducted to determine when the L54 haplogroup arose and gave rise to M3 and L330/L334 subbranches. The indigenous American Y chromosomes used in this analysis were more diverse than those of southern Altaians. The resulting TMRCA for the South American Q1a3a1a∗ samples was 22.2 kya or 7.6 kya, depending on the mutation rate used. The divergence between the M3 and L330/L334 Y chromosomes was ∼13.4 kya, with a TMRCA of 22.0 kya, via the evolutionary rate. By contrast, the TMRCA and divergence time via a pedigree-based mutation rate were 7.7 kya and 4.9 kya, respectively.
The time required to generate the haplotypic diversity in the L54-positive Y chromosomes clearly showed that the evolutionary rate provided a more reasonable estimate. The Americas were inhabited well before 5–8 kya, based on various lines of evidence, making the use of the pedigree-based mutation rate questionable. The estimates generated with the evolutionary-based mutation rate provided times that are more congruent with the known prehistory of the Americas.117 They are also similar to the TMRCAs calculated for Native American mtDNA haplogroups.107,108
Discussion
Origins of Northern and Southern Altaians
In this paper, we characterized mtDNA and NRY variation in northern and southern Altaians to better understand their population histories and elucidate the genetic relationship between Altaians and Native American populations. The evidence from the mtDNA and NRY data supports the hypothesis that northern and southern Altaians generally formed out of separate gene pools. This complex genetic history involves repeated migrations into (and probably out of) the Altai-Sayan region. In addition, the histories as revealed by these data added nuances that could not be attained through low-resolution characterization alone.
The NRY data provided the clearest evidence for a significant genetic difference between the two sets of Altaian ethnic groups. Although sharing certain NRY haplogroups, the two population groups differed in the frequencies of these lineages, and, more importantly, shared few haplotypes with them. By contrast, northern and southern populations shared considerably more mtDNA haplotypes, indicating that some degree of gene flow had occurred between them, albeit in a sex-specific manner. As seen in other populations from Siberia and Central Asia, the patrilineality of these groups probably helped to shape this difference in patterns of mtDNA and Y-chromosomal variation.64,118
In addition, each northern Altaian ethnic group showed different genetic relationships with the Altai-kizhi. The Tubalars consistently grouped closer to the Altai-kizhi than the other two northern Altaians based on both mtDNA and NRY data. Thus, the higher genetic diversity of mtDNA and NRY haplotypes in the Tubalars is probably the result of admixture with other groups, such as southern Altaians. The Chelkans, on the other hand, have the most divergent set of mtDNAs of the three populations. Mismatch analysis and tests of neutrality indicated that the Chelkans show signs of decreasing population size or population structure. Long-term endogamy has probably also played a role in shifting the pattern of mtDNA diversity in Chelkans from that seen in other northern Altaians. Because of this endogamy (and genetic drift), only a few lineages attained high frequencies, resulting in reduced mtDNA diversity. Based on the NRY data, the Kumandins were distinct from both the Chelkans and Tubalars, who were composed of mostly the same set of lineages. Thus, the genetic diversity in northern Altaians is structured by ethnic group membership, and, therefore, can be viewed as reflecting distinctive histories for each population.
Not much is known about the ethnogenesis of northern Altaians. However, it has been suggested that they descended from groups that historically lived around the Yenisei River and spoke either southern Samoyedic, Ugric, or Yeniseian languages.18,19 These populations are the same ones that later contributed to the formation of the Kets, Selk'ups, Shors, and Khakass in northwestern Siberia and the western Sayans of southern Siberia.4,105 Furthermore, the Chelkans and Tubalars possess a large number of Q1a3a∗ Y chromosomes with dramatically different STR profiles compared to other southern Siberians (Altai-kizhi and Tuvinians) and Mongolians. Thus, it is possible that similar lineages will be found in the Kets and/or Sel'kups, where high frequencies of Q1-P36 have already been noted.119 Should this be the case, it would provide additional evidence for northern Altaians having common ancestry with Samoyedic, Yeniseian, and Ugric speakers. In fact, Chelkans and Kumandins also have N-P43 Y chromosomes very similar to ones found in the Ugric-speaking Khanty. Regardless, there is notable genetic discontinuity between northern Altaians and other Turkic-speaking people of southern Siberia.
Southern Altaians share greater affinities with Mongolians and Central Asians than they do with northern Altaians. This is partly because of the high frequencies of Y chromosome haplogroup C in these groups. In fact, present-day Kyrgyz are nearly indistinguishable from the Altai-kizhi based on their NRY haplogroup profile.120,121 They share similar C-M217 and R-M417 lineages with the Altai-kizhi, suggesting a recent common ancestry for the two groups, which further supports the theory of a recent common ancestry among southern Siberians and Kyrgyz.122
As evident in the disparities in genetic history between northern and southern Altaians, the Altai has served as a long-term genetic boundary zone. These disparities reflect the different sources of genetic lineages and spheres of interaction for both groups. The northern Altaians share clan names, similar languages, subsistence strategies, and other cultural elements with populations that today live farther to the north.4 By contrast, southern Altaians share these same features with populations in Central Asia, mostly with Turkic- (Kipchak) but also Mongolic-speaking peoples. Thus, the geography of the Altai (taiga versus steppe) has helped to maintain these cultural and biological (mtDNA, Y chromosome, and cranial-morphological) differences.
Furthermore, no evidence of Denisovan or Neanderthal ancestry was found in the Altaian mtDNA and Y chromosome data. However, this does not preclude such admixture in the autosomes of Altaian populations. Greater numbers of derived Denisovan SNPs were found in some southeastern Asian and Oceanian populations, although native Siberians were not included in that study.123 Therefore, this issue requires further investigation.
Native American Origins
Many earlier genetic studies looked for the origins of Native Americans among the indigenous peoples of Siberia, Mongolia, and East Asia. Often, the identification of source populations conflicted between studies, depending largely on the loci or samples being studied. Cranial morphology has been used to demonstrate a connection between the Native Americans and Siberian populations.124,125 Various researchers have suggested sources such as the Baikal region of southern Siberia, the Amur region of southeastern Siberia, and more generally Eurasia and East Asia.126–128 A study of autosomal loci also showed an affinity between populations in the New World and Siberian regions but did not attempt to pinpoint a particular area of Siberia as the source area.129 In addition, mtDNA studies have suggested New World origins from a number of different locations including different parts of Siberia, Mongolia, and northern China.34,41–45,47,71,130
Our own analysis of Altaian mtDNAs showed that the five primary haplogroups (A–D, X) were present among these populations. However, Altaian populations (and generally all Siberian populations outside of Chukotka) lack mtDNA haplotypes that are identical to those appearing in the Americas. The only exceptions are the Selk'ups and Evenks who bear A2 haplotypes, with their presence in those groups being explained as a result of a back migration to northeast Asia.107
Despite the general absence of Native American haplotypes in southern Siberia, there are sister branches whose MRCAs are shared with those in Native Americans. One such lineage is C1a, which was found in two Altai-kizhi individuals and has also been observed at low frequencies in Mongolia, southeastern Siberia, and Japan.44,46,55,71 Tamm et al.107 attribute its presence in northeast Asia to a back migration from the New World, where haplogroups C1b–d are prevalent, whereas Starikovskaya et al.44 argue that C1a and C1b arose in the Amur region, with C1b migrating to the Americas later. A similar lineage is D4b1a2a1a, a sister branch to D4b1a2a1a1, which is found in northern North America. Although both of these lineages date to around 15,000 years ago, additional mitogenome sequences from these haplogroups are needed to estimate more precise TMRCAs for them and thereby delineate their putative Asian and American origins.
Results obtained from the Y chromosome analysis support the view that southern Siberians and Native Americans share a common source.8,9,11,38,131 This connection was initially suggested by a low-level Y-SNP resolution and an alphoid heteroduplex system by Santos et al.8 Subsequently, Zegura et al.11 showed a similarity in NRY Q and C types among southern Altaians and Native Americans by using only fast evolving Y-STR loci and, again, low-level Y-SNP resolution. We focused on haplogroup Q in this study because of the greater number of new mutations published for this branch and corresponding levels of Y-STR resolution (15–17 loci), which are currently lacking for published Native American haplogroup C Y chromosomes. This high-resolution characterization is critical because it allows for a more accurate dating of TMRCAs and estimates of divergence between the ancestors of Native Americans and indigenous Siberians. For example, with this approach, Seielstad et al.114 dated the origin of the M242, which defines the NRY haplogroup Q, and, in turn, provided a more accurate upper bound to the timing of the initial peopling of the Western Hemisphere.
Several studies have shown that the American-specific Q-M3 arose on an M346-positive Y chromosome.63,115,132 The M346 marker was also discovered in Altaians and other Siberian populations.102,116 However, it has a broad geographic distribution, being found in Siberia, Central Asia, East Asia, India, and Pakistan, albeit at lower frequencies.85,99 We have shown that southern Altaians M346 Y chromosomes also possess L54, a SNP marker that also is shared by Native Americans who have the M3 marker and which is more derived than M346. Because L54 is found in both Siberia and the Americas, it most probably defines the initial founder haplogroup from which M3 later developed.
Our coalescence analysis suggests that the two derived branches of L54 (M3 and L330/L334) diverged soon after this mutation arose. Estimates using the evolutionary Y-STR mutation rate place the origin of this marker at around 22,000 years ago, with the two branches diverging at roughly 13,400 years ago. Although the 95% confidence intervals for the Bayesian analyses are broad, the median values of the TMCRAs estimated with this method closely match those obtained through the analysis with rho statistics. In addition, the coalescence estimates of northern and southern Altaian Q Y chromosomes show that they, too, are similar to the overall TMRCA estimates. This concordance suggests that a rapid expansion probably occurred for this particular Y chromosome branch around 15,000–20,000 years ago. Given previous estimates for the timing of the initial peopling of the Americas, this scenario seems plausible, because these estimates fall in line with recent estimates of indigenous American mitogenomes.107,133
As in any study, there are limitations to this analysis. The primary issues are the accuracy and precision of using microsatellites for dating origins and dispersals of haplotypes. The stochastic nature of mutational accumulation will continue to be a source of some uncertainty in any attempt at dating TMRCAs. For this reason, the question of which Y-STR mutation rate to use for coalescence estimates has been debated.88,134,135 In this study, the evolutionary rate seems the most realistic, because estimates generated with the pedigree rate provided times that are much too recent, given what is known about the peopling of the New World from nongenetic studies.117 There is no evidence that the majority of Native Americans (men with Q-M3 Y chromosomes) derived from a migration less than 8 kya, as would be suggested from the TMRCAs calculated with the pedigree rate. However, other studies have used the pedigree mutation rate to explore historical events with great effect—the most-well-known case being the Genghis Khan star cluster.136 It is possible that such rates are, like that of the mtDNA, time dependent or that the Y chromosomes to which the Y-STRs are linked have been affected by purifying selection.84,133,137,138 In this regard, the pedigree-based mutation rate would be more appropriately used with lower diversity estimates, reflecting recent historical events, while the evolutionary rate would be used in scenarios with higher diversity estimates, reflecting more ancient phenomena. Although beyond the scope of this paper, it is likely that the Y-STR mutation rate follows a similarly shaped curve as that of the mitochondrial genome.
Furthermore, haplogroup divergence dates need not (and mostly do not) equate with population divergence dates. In this case, however, the mutations defining the southern Altaian and Native American branches of the Q-L54 lineage most probably arose after their ancestral populations split, given the geographic exclusivity of each derived marker. Yet, sample sets that are not entirely representative of a derived branch could potentially skew the coalescent results. In all likelihood, the L54 marker will be found in other southern Siberian populations, because southern Altaians show some genetic affinities with Tuvinians and other populations from the eastern Sayan region. Even so, the consistency of TMRCA estimates and the divergence dates for the different Q branches examined here suggest that our data sets are sufficiently representative. Moreover, even though the M3 haplotypes used in this analysis came exclusively from indigenous Ecuadorian populations, the diversity found within this data set is similar to previous estimates of the age of the Q-M3 haplogroup.11
Although different lines of evidence point to different source populations for Native Americans, the alternatives need not be exclusive. The effects of historical and demographic events and evolutionary processes, particularly recent gene flow, have shaped modern-day populations such that we should not expect that any one population in the Old World would show the same genetic composition as populations in the New World. That (an) ancestral population(s) probably differentiated into the numerous populations of Siberia and Central Asia, which have interacted over the past 15,000 years, is not lost on us. Historical expansions of people and the effects of animal and plant domestication have played critical roles in shaping the genetics of both Old and New World populations, particularly in the past several thousand years. Modern populations have complex, local histories that need to be understood if these are to be used in larger interregional (or biomedical) analyses. Through the use of phylogeographic methods, we can attain a better understanding of these populations for such purposes. It is through this type of approach that it becomes quite clear that southern Altaians and Native Americans share a recent common paternal ancestor.
Acknowledgments
The authors would like to thank all of the indigenous Altaian participants for their involvement in this study. We also thank Fabricio Santos for his careful review of and helpful suggestions for the manuscript, and two anonymous reviewers for their constructive comments. In addition, we would like to acknowledge the people who facilitated and provided assistance with our field research in the Altai Republic. They include Vasiliy Semënovich Palchikov, the staff of the Biochemistry Lab at the Turochak Hospital, Dr. Maria Nikolaevna Trishina, Vitaliy Trishin, Alexander A. Guryanov, the staff of the Native Affairs office in Gorniy Altaiask, Galina Nikolaevna Makhalina, and Tatiana Kunduchinovna Babrasheva. In addition, we received help from a number of people living in local villages around the Turochakskiy Raion, particularly Alexander Adonyov. This project was supported by funds from the University of Pennsylvania (T.G.S.), the National Science Foundation (BCS-0726623) (T.G.S., M.C.D.), the Social Sciences and Humanities Research Council of Canada (MCRI 412-2005-1004) (T.G.S.), and the Russian Basic Fund for Research (L.P.O.). T.G.S. would also like to acknowledge the infrastructural support provided by the National Geographic Society.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Arlequin, version 3.11, http://cmpg.unibe.ch/software/arlequin3/
Batwing, http://www.mas.ncl.ac.uk/∼nijw/
Network, version 4.6.0.0, http://www.fluxus-engineering.com/sharenet.htm
Network Publisher, version 1.3.0.0, http://www.fluxus-engineering.com/nwpub.htm
Y-DNA Haplogroup Tree 2011, version 6.46, http://www.isogg.org/tree
References
- 1.Goebel T. Pleistocene human colonization of Siberia and peopling of the Americas: An ecological approach. Evol. Anthropol. 1999;8:208–227. [Google Scholar]
- 2.Gryaznov M.P. Cowles Book Company, Inc.; New York: 1969. The Ancient Civilization of Southern Siberia. [Google Scholar]
- 3.Okladnikov A.P. Ancient population of Siberia and its culture. In: Levin M.G., Potapov L.P., editors. The Peoples of Siberia. The University of Chicago Press; Chicago: 1964. pp. 13–98. [Google Scholar]
- 4.Levin M.G., Potapov L.P. University of Chicago Press; Chicago: 1964. The Peoples of Siberia. [Google Scholar]
- 5.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L.F. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause J., Fu Q., Good J.M., Viola B., Shunkov M.V., Derevianko A.P., Pääbo S. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krause J., Orlando L., Serre D., Viola B., Prüfer K., Richards M.P., Hublin J.J., Hänni C., Derevianko A.P., Pääbo S. Neanderthals in central Asia and Siberia. Nature. 2007;449:902–904. doi: 10.1038/nature06193. [DOI] [PubMed] [Google Scholar]
- 8.Santos F.R., Pandya A., Tyler-Smith C., Pena S.D., Schanfield M., Leonard W.R., Osipova L., Crawford M.H., Mitchell R.J. The central Siberian origin for native American Y chromosomes. Am. J. Hum. Genet. 1999;64:619–628. doi: 10.1086/302242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karafet T.M., Zegura S.L., Posukh O., Osipova L., Bergen A., Long J., Goldman D., Klitz W., Harihara S., de Knijff P. Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am. J. Hum. Genet. 1999;64:817–831. doi: 10.1086/302282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lell J.T., Sukernik R.I., Starikovskaya Y.B., Su B., Jin L., Schurr T.G., Underhill P.A., Wallace D.C. The dual origin and Siberian affinities of Native American Y chromosomes. Am. J. Hum. Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zegura S.L., Karafet T.M., Zhivotovsky L.A., Hammer M.F. High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas. Mol. Biol. Evol. 2004;21:164–175. doi: 10.1093/molbev/msh009. [DOI] [PubMed] [Google Scholar]
- 12.Anthony D.W. Princeton University Press; Princeton, N.J.: 2007. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World. [Google Scholar]
- 13.Kuzmina E.E., Mair V.H. University of Pennsylvania Press; Philadelphia: 2008. The Prehistory of the Silk Road. [Google Scholar]
- 14.Rudenko S.I. University of California Press; Berkeley: 1970. Frozen Tombs of Siberia, the Pazyryk Burials of Iron Age Horsemen. [Google Scholar]
- 15.David-Kimball J., Bashilov V.A., Yablonsky L.T., editors. Nomads of the Eurasian Steppes in the Early Iron Age. Zinat Press; Berkeley, CA: 1995. [Google Scholar]
- 16.Golden P.B. Otto Harrassowitz; Wiesbaden: 1992. An Introduction to the History of the Turkic Peoples: Ethnogenesis and State-Formation in Medieval and Early Modern Eurasia and the Middle East. [Google Scholar]
- 17.Grousset R. Rutgers University Press; New Brunswick, N.J.: 1970. The Empire of the Steppes: A History of Central Asia. [Google Scholar]
- 18.Potapov L.P. The origins of the Altayans. In: Michael H.N., editor. Studies in Siberian Ethnogenesis. University of Toronto Press; Toronto: 1962. pp. 169–196. [Google Scholar]
- 19.Potapov L.P. The Altays. In: Levin M.G., Potapov L.P., editors. The Peoples of Siberia. University of Chicago Press; Chicago: 1964. pp. 305–341. [Google Scholar]
- 20.Menges K.H. Otto Harrassowitz; Wiesbaden: 1968. The Turkic Languages and Peoples: An Introduction to Turkic Studies. [Google Scholar]
- 21.Levin M.G. The anthropological types of Siberia. In: Levin M.G., Potapov L.P., editors. The Peoples of Siberia. The University of Chicago Press; Chicago: 1964. pp. 99–104. [Google Scholar]
- 22.Osipova L.P., Sukernik R.I. [Polymorphism of immunoglobulin Gm- and Km-allotypes in northern Altaians (western Sibiria)] Genetika. 1978;14:1272–1275. [PubMed] [Google Scholar]
- 23.Posukh O.L., Osipova L.P., Kashinskaia IuO., Ivakin E.A., Kriukov IuA., Karafet T.M., Kazakovtseva M.A., Skobel'tsina L.M., Crawford M.G., Lefranc M.P., Lefranc G. [Genetic analysis of the South Altaian population of the Mendur-Sokkon village, Altai Republic] Genetika. 1998;34:106–113. [PubMed] [Google Scholar]
- 24.Sukernik R.I., Karafet T.M., Abanina T.A., Korostyshevskiĭ M.A., Bashlaĭ A.G. [Genetic structure of 2 isolated populations of native inhabitants of Sibiria (Northern Altaics) according to the results of a study of blood groups and isoenzymes] Genetika. 1977;13:911–918. [PubMed] [Google Scholar]
- 25.Sukernik R.I., Shur T.G., Starikovskaia E.B., Uolles D.K. [Mitochondrial DNA variation in native inhabitants of Siberia with reconstructions of the evolutional history of the American Indians. Restriction polymorphism] Genetika. 1996;32:432–439. [PubMed] [Google Scholar]
- 26.Shields G.F., Schmiechen A.M., Frazier B.L., Redd A., Voevoda M.I., Reed J.K., Ward R.H. mtDNA sequences suggest a recent evolutionary divergence for Beringian and northern North American populations. Am. J. Hum. Genet. 1993;53:549–562. [PMC free article] [PubMed] [Google Scholar]
- 27.Torroni A., Schurr T.G., Yang C.C., Szathmary E.J., Williams R.C., Schanfield M.S., Troup G.A., Knowler W.C., Lawrence D.N., Weiss K.M. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992;130:153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace D.C., Torroni A. American Indian prehistory as written in the mitochondrial DNA: a review. Hum. Biol. 1992;64:403–416. [PubMed] [Google Scholar]
- 29.Torroni A., Schurr T.G., Cabell M.F., Brown M.D., Neel J.V., Larsen M., Smith D.G., Vullo C.M., Wallace D.C. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 30.Torroni A., Sukernik R.I., Schurr T.G., Starikorskaya Y.B., Cabell M.F., Crawford M.H., Comuzzie A.G., Wallace D.C. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am. J. Hum. Genet. 1993;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- 31.Forster P., Harding R., Torroni A., Bandelt H.J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 32.Merriwether D.A., Ferrell R.E. The four founding lineage hypothesis for the New World: a critical reevaluation. Mol. Phylogenet. Evol. 1996;5:241–246. doi: 10.1006/mpev.1996.0017. [DOI] [PubMed] [Google Scholar]
- 33.Bonatto S.L., Salzano F.M. Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am. J. Hum. Genet. 1997;61:1413–1423. doi: 10.1086/301629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merriwether D.A., Hall W.W., Vahlne A., Ferrell R.E. mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am. J. Hum. Genet. 1996;59:204–212. [PMC free article] [PubMed] [Google Scholar]
- 35.Neel J.V., Biggar R.J., Sukernik R.I. Virologic and genetic studies relate Amerind origins to the indigenous people of the Mongolia/Manchuria/southeastern Siberia region. Proc. Natl. Acad. Sci. USA. 1994;91:10737–10741. doi: 10.1073/pnas.91.22.10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karafet T.M., Zegura S.L., Vuturo-Brady J., Posukh O., Osipova L., Wiebe V., Romero F., Long J.C., Harihara S., Jin F. Y chromosome markers and Trans-Bering Strait dispersals. Am. J. Phys. Anthropol. 1997;102:301–314. doi: 10.1002/(SICI)1096-8644(199703)102:3<301::AID-AJPA1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 37.Lell J.T., Brown M.D., Schurr T.G., Sukernik R.I., Starikovskaya Y.B., Torroni A., Moore L.G., Troup G.M., Wallace D.C. Y chromosome polymorphisms in native American and Siberian populations: identification of native American Y chromosome haplotypes. Hum. Genet. 1997;100:536–543. doi: 10.1007/s004390050548. [DOI] [PubMed] [Google Scholar]
- 38.Bortolini M.C., Salzano F.M., Thomas M.G., Stuart S., Nasanen S.P., Bau C.H., Hutz M.H., Layrisse Z., Petzl-Erler M.L., Tsuneto L.T. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am. J. Hum. Genet. 2003;73:524–539. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurr T.G., Sherry S.T. Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: evolutionary and demographic evidence. Am. J. Hum. Biol. 2004;16:420–439. doi: 10.1002/ajhb.20041. [DOI] [PubMed] [Google Scholar]
- 40.Derenko M.V., Malyarchuk B., Denisova G.A., Wozniak M., Dambueva I., Dorzhu C., Luzina F., Miścicka-Sliwka D., Zakharov I. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum. Genet. 2006;118:591–604. doi: 10.1007/s00439-005-0076-y. [DOI] [PubMed] [Google Scholar]
- 41.Derenko M.V., Malyarchuk B., Grzybowski T., Denisova G., Dambueva I., Perkova M., Dorzhu C., Luzina F., Lee H.K., Vanecek T. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am. J. Hum. Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volodko N.V., Starikovskaya E.B., Mazunin I.O., Eltsov N.P., Naidenko P.V., Wallace D.C., Sukernik R.I. Mitochondrial genome diversity in arctic Siberians, with particular reference to the evolutionary history of Beringia and Pleistocenic peopling of the Americas. Am. J. Hum. Genet. 2008;82:1084–1100. doi: 10.1016/j.ajhg.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Derenko M.V., Grzybowski T., Malyarchuk B.A., Dambueva I.K., Denisova G.A., Czarny J., Dorzhu C.M., Kakpakov V.T., Miścicka-Sliwka D., Woźniak M., Zakharov I.A. Diversity of mitochondrial DNA lineages in South Siberia. Ann. Hum. Genet. 2003;67:391–411. doi: 10.1046/j.1469-1809.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 44.Starikovskaya E.B., Sukernik R.I., Derbeneva O.A., Volodko N.V., Ruiz-Pesini E., Torroni A., Brown M.D., Lott M.T., Hosseini S.H., Huoponen K., Wallace D.C. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann. Hum. Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starikovskaya Y.B., Sukernik R.I., Schurr T.G., Kogelnik A.M., Wallace D.C. mtDNA diversity in Chukchi and Siberian Eskimos: implications for the genetic history of Ancient Beringia and the peopling of the New World. Am. J. Hum. Genet. 1998;63:1473–1491. doi: 10.1086/302087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schurr T.G., Wallace D.C. Genetic prehistory of Paleoasiatic-speaking populations of northeastern Siberia and their relationships to Native Americans. In: Kendall L., Krupnik I., editors. Constructing cultures then and now: celebrating Franz Boas and the Jesup North Pacific Expedition. Arctic Studies Center, National Museum of Natural History, Smithsonian Institution; Washington, D.C.: 2003. pp. 239–258. [Google Scholar]
- 47.Schurr T.G., Ballinger S.W., Gan Y.Y., Hodge J.A., Merriwether D.A., Lawrence D.N., Knowler W.C., Weiss K.M., Wallace D.C. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am. J. Hum. Genet. 1990;46:613–623. [PMC free article] [PubMed] [Google Scholar]
- 48.Macaulay V., Richards M., Hickey E., Vega E., Cruciani F., Guida V., Scozzari R., Bonné-Tamir B., Sykes B., Torroni A. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am. J. Hum. Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richards M., Macaulay V., Hickey E., Vega E., Sykes B., Guida V., Rengo C., Sellitto D., Cruciani F., Kivisild T. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 50.Torroni A., Bandelt H.J., D'Urbano L., Lahermo P., Moral P., Sellitto D., Rengo C., Forster P., Savontaus M.L., Bonné-Tamir B., Scozzari R. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am. J. Hum. Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torroni A., Huoponen K., Francalacci P., Petrozzi M., Morelli L., Scozzari R., Obinu D., Savontaus M.L., Wallace D.C. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torroni A., Lott M.T., Cabell M.F., Chen Y.S., Lavergne L., Wallace D.C. mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am. J. Hum. Genet. 1994;55:760–776. [PMC free article] [PubMed] [Google Scholar]
- 53.Kivisild T., Tolk H.V., Parik J., Wang Y., Papiha S.S., Bandelt H.J., Villems R. The emerging limbs and twigs of the East Asian mtDNA tree. Mol. Biol. Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- 54.Schurr T.G., Sukernik R.I., Starikovskaya Y.B., Wallace D.C. Mitochondrial DNA variation in Koryaks and Itel'men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am. J. Phys. Anthropol. 1999;108:1–39. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka M., Cabrera V.M., González A.M., Larruga J.M., Takeyasu T., Fuku N., Guo L.J., Hirose R., Fujita Y., Kurata M. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14(10A):1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao Y.G., Kong Q.P., Bandelt H.J., Kivisild T., Zhang Y.P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gokcumen O., Dulik M.C., Pai A.A., Zhadanov S.I., Rubinstein S., Osipova L.P., Andreenkov O.V., Tabikhanova L.E., Gubina M.A., Labuda D., Schurr T.G. Genetic variation in the enigmatic Altaian Kazakhs of South-Central Russia: insights into Turkic population history. Am. J. Phys. Anthropol. 2008;136:278–293. doi: 10.1002/ajpa.20802. [DOI] [PubMed] [Google Scholar]
- 58.Rubinstein S., Dulik M.C., Gokcumen O., Zhadanov S., Osipova L., Cocca M., Mehta N., Gubina M., Posukh O., Schurr T.G. Russian Old Believers: genetic consequences of their persecution and exile, as shown by mitochondrial DNA evidence. Hum. Biol. 2008;80:203–237. doi: 10.3378/1534-6617-80.3.203. [DOI] [PubMed] [Google Scholar]
- 59.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 60.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 61.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 62.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karafet T.M., Mendez F.L., Meilerman M.B., Underhill P.A., Zegura S.L., Hammer M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dulik M.C., Osipova L.P., Schurr T.G. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS ONE. 2011;6:e17548. doi: 10.1371/journal.pone.0017548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox M.P. Minimal hierarchical analysis of global human Y-chromosome SNP diversity by PCR-RFLP. Anthropol. Sci. 2006;114:69–74. [Google Scholar]
- 66.Derbeneva O.A., Starikovskaia E.B., Volod'ko N.V., Wallace D.C., Sukernik R.I. [Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia] Genetika. 2002;38:1554–1560. [PubMed] [Google Scholar]
- 67.Derbeneva O.A., Starikovskaya E.B., Wallace D.C., Sukernik R.I. Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am. J. Hum. Genet. 2002;70:1009–1014. doi: 10.1086/339524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pimenoff V.N., Comas D., Palo J.U., Vershubsky G., Kozlov A., Sajantila A. Northwest Siberian Khanty and Mansi in the junction of West and East Eurasian gene pools as revealed by uniparental markers. Eur. J. Hum. Genet. 2008;16:1254–1264. doi: 10.1038/ejhg.2008.101. [DOI] [PubMed] [Google Scholar]
- 69.Comas D., Calafell F., Mateu E., Pérez-Lezaun A., Bosch E., Martínez-Arias R., Clarimon J., Facchini F., Fiori G., Luiselli D. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am. J. Hum. Genet. 1998;63:1824–1838. doi: 10.1086/302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao Y.G., Kong Q.P., Wang C.Y., Zhu C.L., Zhang Y.P. Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china. Mol. Biol. Evol. 2004;21:2265–2280. doi: 10.1093/molbev/msh238. [DOI] [PubMed] [Google Scholar]
- 71.Kolman C.J., Sambuughin N., Bermingham E. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics. 1996;142:1321–1334. doi: 10.1093/genetics/142.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xue Y., Zerjal T., Bao W., Zhu S., Shu Q., Xu J., Du R., Fu S., Li P., Hurles M.E. Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khar'kov V.N., Medvedeva O.F., Luzina F.A., Kolbasko A.V., Gafarov N.I., Puzyrev V.P., Stepanov V.A. [Comparative characteristics of the gene pool of Teleuts inferred from Y-chromosomal marker data] Genetika. 2009;45:1132–1142. [PubMed] [Google Scholar]
- 74.Khar'kov V., Khamina K., Medvedeva O., Shtygasheva O., Stepanov V. Genetic diversity of the Khakass gene pool: Subethnic differentiation and the structure of Y-chromosome haplogroups. Mol. Biol. (Mosk.) 2011;45:446–458. [PubMed] [Google Scholar]
- 75.Roewer L., Krüger C., Willuweit S., Nagy M., Rodig H., Kokshunova L., Rothämel T., Kravchenko S., Jobling M.A., Stoneking M., Nasidze I. Y-chromosomal STR haplotypes in Kalmyk population samples. Forensic Sci. Int. 2007;173:204–209. doi: 10.1016/j.forsciint.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 76.Geppert M., Baeta M., Núñez C., Martínez-Jarreta B., Zweynert S., Cruz O.W., González-Andrade F., González-Solorzano J., Nagy M., Roewer L. Hierarchical Y-SNP assay to study the hidden diversity and phylogenetic relationship of native populations in South America. Forensic Sci. Int. Genet. 2011;5:100–104. doi: 10.1016/j.fsigen.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 77.Excoffier L., Laval G., Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 79.SPSS Inc . SPSS Inc.; Chicago, IL: 2001. SPSS for Windows Release 11.0.0. [Google Scholar]
- 80.Polzin T., Daneschmand S.V. On Steiner trees and minimum spanning trees in hypergraphs. Oper. Res. Lett. 2003;31:12–20. [Google Scholar]
- 81.Bandelt H.J., Forster P., Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 82.Bandelt H.J., Forster P., Sykes B.C., Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gusmão L., Butler J.M., Carracedo A., Gill P., Kayser M., Mayr W.R., Morling N., Prinz M., Roewer L., Tyler-Smith C., Schneider P.M., DNA Commission of the International Society of Forensic Genetics DNA Commission of the International Society of Forensic Genetics (ISFG): an update of the recommendations on the use of Y-STRs in forensic analysis. Forensic Sci. Int. 2006;157:187–197. doi: 10.1016/j.forsciint.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 84.Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sengupta S., Zhivotovsky L.A., King R., Mehdi S.Q., Edmonds C.A., Chow C.E., Lin A.A., Mitra M., Sil S.K., Ramesh A. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson I., Balding D., Weale M. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. [Ser A] 2003;166:155–188. [Google Scholar]
- 87.Xue Y., Zerjal T., Bao W., Zhu S., Shu Q., Xu J., Du R., Fu S., Li P., Hurles M.E. Modelling male prehistory in east Asia using BATWING. In: Matsumura S., Forster P., Renfrew C., editors. Simulations, Genetics and Human Prehistory. McDonald Institute for Archaeological Research; Cambridge: 2008. pp. 79–88. [Google Scholar]
- 88.Zhivotovsky L.A., Underhill P.A., Cinnioğlu C., Kayser M., Morar B., Kivisild T., Scozzari R., Cruciani F., Destro-Bisol G., Spedini G. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dupuy B.M., Stenersen M., Egeland T., Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum. Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- 90.Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 91.Derenko M., Malyarchuk B., Grzybowski T., Denisova G., Rogalla U., Perkova M., Dambueva I., Zakharov I. Origin and post-glacial dispersal of mitochondrial DNA haplogroups C and D in northern Asia. PLoS ONE. 2010;5:e15214. doi: 10.1371/journal.pone.0015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhadanov S.I., Dulik M.C., Markley M., Jennings G.W., Gaieski J.B., Elias G., Schurr T.G., Genographic Project Consortium Genetic heritage and native identity of the Seaconke Wampanoag tribe of Massachusetts. Am. J. Phys. Anthropol. 2010;142:579–589. doi: 10.1002/ajpa.21281. [DOI] [PubMed] [Google Scholar]
- 93.Hammer M.F., Karafet T.M., Redd A.J., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L. Hierarchical patterns of global human Y-chromosome diversity. Mol. Biol. Evol. 2001;18:1189–1203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- 94.Kivisild T., Rootsi S., Metspalu M., Mastana S., Kaldma K., Parik J., Metspalu E., Adojaan M., Tolk H.V., Stepanov V. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am. J. Hum. Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wells R.S., Yuldasheva N., Ruzibakiev R., Underhill P.A., Evseeva I., Blue-Smith J., Jin L., Su B., Pitchappan R., Shanmugalakshmi S. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosser Z.H., Zerjal T., Hurles M.E., Adojaan M., Alavantic D., Amorim A., Amos W., Armenteros M., Arroyo E., Barbujani G. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintana-Murci L., Krausz C., Zerjal T., Sayar S.H., Hammer M.F., Mehdi S.Q., Ayub Q., Qamar R., Mohyuddin A., Radhakrishna U. Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am. J. Hum. Genet. 2001;68:537–542. doi: 10.1086/318200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Underhill P.A., Passarino G., Lin A.A., Shen P., Mirazón Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 99.Zhong H., Shi H., Qi X.-B., Duan Z.-Y., Tan P.-P., Jin L., Su B., Ma R.Z. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol. Biol. Evol. 2011;28:717–727. doi: 10.1093/molbev/msq247. [DOI] [PubMed] [Google Scholar]
- 100.Mirabal S., Regueiro M., Cadenas A.M., Cavalli-Sforza L.L., Underhill P.A., Verbenko D.A., Limborska S.A., Herrera R.J. Y-chromosome distribution within the geo-linguistic landscape of northwestern Russia. Eur. J. Hum. Genet. 2009;17:1260–1273. doi: 10.1038/ejhg.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Myres N.M., Rootsi S., Lin A.A., Järve M., King R.J., Kutuev I., Cabrera V.M., Khusnutdinova E.K., Pshenichnov A., Yunusbayev B. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Malyarchuk B., Derenko M., Denisova G., Maksimov A., Wozniak M., Grzybowski T., Dambueva I., Zakharov I. Ancient links between Siberians and Native Americans revealed by subtyping the Y chromosome haplogroup Q1a. J. Hum. Genet. 2011;56:583–588. doi: 10.1038/jhg.2011.64. [DOI] [PubMed] [Google Scholar]
- 103.Rogers A.R., Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 104.Shi H., Dong Y.L., Wen B., Xiao C.J., Underhill P.A., Shen P.D., Chakraborty R., Jin L., Su B. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am. J. Hum. Genet. 2005;77:408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Forsyth J. Cambridge University Press; Cambridge, England: 1992. A History of the Peoples of Siberia: Russia's North Asian Colony, 1581–1990. [Google Scholar]
- 106.Brown M.D., Hosseini S.H., Torroni A., Bandelt H.J., Allen J.C., Schurr T.G., Scozzari R., Cruciani F., Wallace D.C. mtDNA haplogroup X: An ancient link between Europe/Western Asia and North America? Am. J. Hum. Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tamm E., Kivisild T., Reidla M., Metspalu M., Smith D.G., Mulligan C.J., Bravi C.M., Rickards O., Martinez-Labarga C., Khusnutdinova E.K. Beringian standstill and spread of Native American founders. PLoS ONE. 2007;2:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Achilli A., Perego U.A., Bravi C.M., Coble M.D., Kong Q.P., Woodward S.R., Salas A., Torroni A., Bandelt H.J. The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies. PLoS ONE. 2008;3:e1764. doi: 10.1371/journal.pone.0001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perego U.A., Achilli A., Angerhofer N., Accetturo M., Pala M., Olivieri A., Kashani B.H., Ritchie K.H., Scozzari R., Kong Q.P. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr. Biol. 2009;19:1–8. doi: 10.1016/j.cub.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 110.Perego U.A., Angerhofer N., Pala M., Olivieri A., Lancioni H., Kashani B.H., Carossa V., Ekins J.E., Gómez-Carballa A., Huber G. The initial peopling of the Americas: a growing number of founding mitochondrial genomes from Beringia. Genome Res. 2010;20:1174–1179. doi: 10.1101/gr.109231.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Helgason A., Pálsson G., Pedersen H.S., Angulalik E., Gunnarsdóttir E.D., Yngvadóttir B., Stefánsson K. mtDNA variation in Inuit populations of Greenland and Canada: migration history and population structure. Am. J. Phys. Anthropol. 2006;130:123–134. doi: 10.1002/ajpa.20313. [DOI] [PubMed] [Google Scholar]
- 112.Bortolini M.C., Salzano F.M., Bau C.H., Layrisse Z., Petzl-Erler M.L., Tsuneto L.T., Hill K., Hurtado A.M., Castro-De-Guerra D., Bedoya G., Ruiz-Linares A. Y-chromosome biallelic polymorphisms and Native American population structure. Ann. Hum. Genet. 2002;66:255–259. doi: 10.1017/S0003480002001148. [DOI] [PubMed] [Google Scholar]
- 113.Underhill P.A., Shen P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonné-Tamir B., Bertranpetit J., Francalacci P. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 114.Seielstad M., Yuldasheva N., Singh N., Underhill P., Oefner P., Shen P., Wells R.S. A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am. J. Hum. Genet. 2003;73:700–705. doi: 10.1086/377589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schurr T.G., Osipova L.P., Zhadanov S.I., Dulik M.C. Genetic diversity in Native Siberians: Implications for the prehistoric settlement of te Cis-Baikal region. In: Weber A.W., Katzenberg M.A., Schurr T.G., editors. Prehistoric Hunter-Gatherers of the Baikal Region, Siberia. University of Pennsylvania Press; Philadelphia: 2010. pp. 121–134. [Google Scholar]
- 116.Dulik, M.C. (2011). A molecular anthropological study of Altaian histories utilizing population genetics and phylogeography. PhD thesis, University of Pennsylvania, Philadelphia, PA.
- 117.Fiedel S.J. The peopling of the New World: present evidence, new theories, and future directions. J. Archaeol. Res. 2000;8:39–103. [Google Scholar]
- 118.Martínez-Cruz B., Vitalis R., Ségurel L., Austerlitz F., Georges M., Théry S., Quintana-Murci L., Hegay T., Aldashev A., Nasyrova F., Heyer E. In the heartland of Eurasia: the multilocus genetic landscape of Central Asian populations. Eur. J. Hum. Genet. 2011;19:216–223. doi: 10.1038/ejhg.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Karafet T.M., Osipova L.P., Gubina M.A., Posukh O.L., Zegura S.L., Hammer M.F. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum. Biol. 2002;74:761–789. doi: 10.1353/hub.2003.0006. [DOI] [PubMed] [Google Scholar]
- 120.Balaresque P., Parkin E.J., Roewer L., Carvalho-Silva D.R., Mitchell R.J., van Oorschot R.A., Henke J., Stoneking M., Nasidze I., Wetton J. Genomic complexity of the Y-STR DYS19: inversions, deletions and founder lineages carrying duplications. Int. J. Legal Med. 2009;123:15–23. doi: 10.1007/s00414-008-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Underhill P.A., Myres N.M., Rootsi S., Metspalu M., Zhivotovsky L.A., King R.J., Lin A.A., Chow C.E., Semino O., Battaglia V. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Soucek S. Cambridge University Press; Cambridge, New York: 2000. A History of Inner Asia. [Google Scholar]
- 123.Reich D., Patterson N., Kircher M., Delfin F., Nandineni M.R., Pugach I., Ko A.M., Ko Y.C., Jinam T.A., Phipps M.E. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hrdlička A. Crania of Siberia. Am. J. Phys. Anthropol. 1942;29:435–481. [Google Scholar]
- 125.González-José R., Bortolini M.C., Santos F.R., Bonatto S.L. The peopling of America: craniofacial shape variation on a continental scale and its interpretation from an interdisciplinary view. Am. J. Phys. Anthropol. 2008;137:175–187. doi: 10.1002/ajpa.20854. [DOI] [PubMed] [Google Scholar]
- 126.Kozintsev A.G., Gromov A.V., Moiseyev V.G. Collateral relatives of American Indians among the Bronze Age populations of Siberia? Am. J. Phys. Anthropol. 1999;108:193–204. doi: 10.1002/(SICI)1096-8644(199902)108:2<193::AID-AJPA5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 127.Crawford M.H. Cambridge University Press; Cambridge: 1998. The Origins of Native Americans: Evidence from Anthropological Genetics. [Google Scholar]
- 128.Brace C.L., Nelson A.R., Seguchi N., Oe H., Sering L., Qifeng P., Yongyi L., Tumen D. Old World sources of the first New World human inhabitants: a comparative craniofacial view. Proc. Natl. Acad. Sci. USA. 2001;98:10017–10022. doi: 10.1073/pnas.171305898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S., Lewis C.M., Jakobsson M., Ramachandran S., Ray N., Bedoya G., Rojas W., Parra M.V., Molina J.A., Gallo C. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Horai S., Kondo R., Nakagawa-Hattori Y., Hayashi S., Sonoda S., Tajima K. Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol. Biol. Evol. 1993;10:23–47. doi: 10.1093/oxfordjournals.molbev.a039987. [DOI] [PubMed] [Google Scholar]
- 131.Kaessmann H., Zöllner S., Gustafsson A.C., Wiebe V., Laan M., Lundeberg J., Uhlén M., Pääbo S. Extensive linkage disequilibrium in small human populations in Eurasia. Am. J. Hum. Genet. 2002;70:673–685. doi: 10.1086/339258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bailliet G., Ramallo V., Muzzio M., García A., Santos M.R., Alfaro E.L., Dipierri J.E., Salceda S., Carnese F.R., Bravi C.M. Brief communication: Restricted geographic distribution for Y-Q∗ paragroup in South America. Am. J. Phys. Anthropol. 2009;140:578–582. doi: 10.1002/ajpa.21133. [DOI] [PubMed] [Google Scholar]
- 133.Ho S.Y., Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am. J. Hum. Genet. 2008;83:142–146. doi: 10.1016/j.ajhg.2008.06.014. author reply 146–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhivotovsky L.A., Underhill P.A. On the evolutionary mutation rate at Y-chromosome STRs: comments on paper by Di Giacomo et al. (2004) Hum. Genet. 2005;116:529–532. doi: 10.1007/s00439-005-1281-4. [DOI] [PubMed] [Google Scholar]
- 135.Di Giacomo F., Luca F., Popa L.O., Akar N., Anagnou N., Banyko J., Brdicka R., Barbujani G., Papola F., Ciavarella G. Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum. Genet. 2004;115:357–371. doi: 10.1007/s00439-004-1168-9. [DOI] [PubMed] [Google Scholar]
- 136.Zerjal T., Xue Y., Bertorelle G., Wells R.S., Bao W., Zhu S., Qamar R., Ayub Q., Mohyuddin A., Fu S. The genetic legacy of the Mongols. Am. J. Hum. Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhivotovsky L.A., Underhill P.A., Feldman M.W. Difference between evolutionarily effective and germ line mutation rate due to stochastically varying haplogroup size. Mol. Biol. Evol. 2006;23:2268–2270. doi: 10.1093/molbev/msl105. [DOI] [PubMed] [Google Scholar]
- 138.Ho S.Y., Phillips M.J., Cooper A., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


