Abstract
Non-alcoholic fatty liver disease (NAFLD) is the hepatic manifestation of metabolic syndrome and the leading cause of chronic liver disease in the Western world. Twenty percent of NAFLD individuals develop chronic hepatic inflammation (non-alcoholic steatohepatitis, NASH) associated with cirrhosis, portal hypertension and hepatocellular carcinoma, yet causes of progression from NAFLD to NASH remain obscure. Here, we show that the NLRP6 and NLRP3 inflammasomes and the effector protein IL-18 negatively regulate NAFLD/NASH progression, as well as multiple aspects of metabolic syndrome via modulation of the gut microbiota. Different animal models reveal that inflammasome deficiency-associated changes in the configuration of the gut microbiota are associated with exacerbated hepatic steatosis and inflammation through influx of TLR4 and TLR9 agonists into the portal circulation, leading to enhanced hepatic TNF-α expression that drives NASH progression. Furthermore, co-housing of inflammasome-deficient animals to wild type mice results in exacerbation of hepatic steatosis, glucose intolerance, and obesity. Thus, altered interactions between the gut microbiota and the host, produced by defective NLRP3 and NLRP6 inflammasome sensing, may govern the rate of progression of multiple metabolic syndrome-associated abnormalities, highlighting the central role of the microbiota in the pathogenesis of heretofore seemingly unrelated systemic auto-inflammatory and metabolic disorders.
Title words: Inflammasome, non-alcoholic fatty liver disease, gut microbiota
Introduction
The prevalence of Non-alcoholic fatty liver disease (NAFLD) ranges from 20–30% in the general population and up to 75–100% in obese individuals1,2. NAFLD is considered one of the manifestations of metabolic syndrome3. While most patients with NAFLD remain asymptomatic, 20% progress to develop chronic hepatic inflammation (non-alcoholic steatohepatitis, NASH), which in turn can lead to cirrhosis, portal hypertension, hepatocellular carcinoma and increased mortality4,5,6. Despite its high prevalence, factors leading to progression from NAFLD to NASH remain poorly understood and no treatment has proven effective7,8.
A “two hit” mechanism is proposed to drive NAFLD/NASH pathogenesis9. The first hit, hepatic steatosis, is closely associated with lipotoxicity-induced mitochondrial abnormalities that sensitize the liver to additional pro-inflammatory insults. These second hits include enhanced lipid peroxidation and increased generation of reactive oxygen species (ROS)10. Inflammasomes are cytoplasmic multi-protein complexes composed of one of several NLR and PYHIN proteins, including NLRP1, NLRP3, NLRC4, and AIM2. Inflammasomes are sensors of endogenous or exogenous pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs)11 that govern cleavage of effector pro-inflammatory cytokines such as pro-IL-1β and pro-IL-1812,13. Most DAMPs trigger the generation of ROS, which are known to activate the NLRP3 inflammasome14. Therefore, we hypothesized that inflammasome-dependent processing of IL-1β and IL-18 may play an important role in progression of NAFLD.
Results
Feeding adult mice a methionine-choline deficient diet (MCDD) for 4 weeks beginning at 8 weeks of age induces several features of human NASH, including hepatic steatosis, inflammatory cell infiltration and ultimately fibrosis15. To investigate the role of inflammasomes in NASH progression, we fed MCDD to C57Bl/6 wt (NCI), Asc−/−, and caspase-1−/− mice to induce early liver damage in the absence of fibrosis (Fig. 1a–d, Supplementary fig. 1c). Compared to wt animals, age- and gender matched Asc−/− and caspase-1−/− mice that were fed MCDD were characterized by significantly higher serum ALT and AST activity, by enhanced microvesicular and macrovesicular hepatic steatosis, and by accumulation of multiple immune subsets in the liver from the innate and adaptive arms of the immune system (as defined by pathological examination and flow cytometry; n=7–11 mice/group; Fig. 1a–d; Supplementary fig. 1c; Supplementary fig. 2a). Remarkably, the hepatic accumulation of T and B cells seems to be dispensable for this phenotype since Asc−/− mice lacking adaptive immune cells (Asc−/−; Rag−/−) also exhibited more severe NASH compared to wt animals, and comparable degrees of pathology to Asc−/− animals (Supplementary fig. 2b–d).
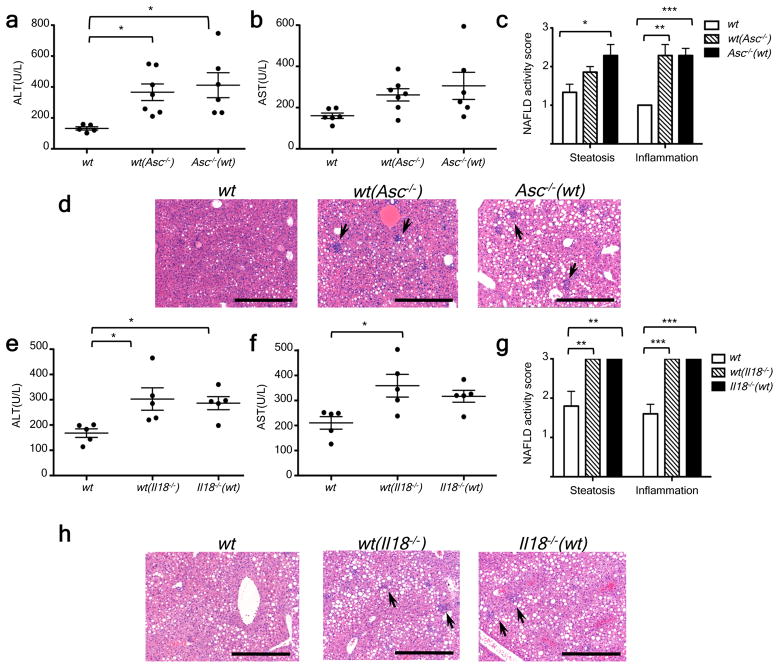
Figure 1. Increased severity of NASH in inflammasome-deficient mice.
To induce NASH, mice were fed with MCDD for 24 d. Their serum ALT and AST activities measured and NAFLD histological activity scores were determined. (a–h) Comparison of ALT, AST, and NAFLD activity, plus histological scores for steatosis and inflammation between singly-housed wild-type (wt) mice and caspase-1−/− (a,b), Asc−/− (c,d), Nlrp3−/−(e,f), or Il18−/−(g,h). Data represent two independent experiments (n=7–19 mice/treatment group). Error bars represent the SEM of samples within a group. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001 (Student’s t test).
To test whether the increased NASH observed in Asc- and caspase-1-deficient mice was mediated by IL-1β or IL-18, we performed similar experiments using mice deficient in either the IL-1 receptor (Il1r−/−), or IL-18 (Il18−/−). Il1r−/− mice did not show any changes in the severity of NASH when compared to wt mice when fed MCDD (Supplementary fig. 1a–b). In contrast, but similar to Asc−/− and caspase-1−/− mice, MCDD-fed Il18−/− animals featured a significant exacerbation of NASH severity (Fig. 1g–h; Supplementary fig. 1c).
To assess the role of the NLRP3 inflammasome in NASH progression, we fed singly-housed Nlrp3−/− and wt animals MCDD for 24 days and evaluated disease progression. Nlrp3−/− mice developed exacerbated NASH compared to wt mice as judged by increased levels of serum ALT and AST, plus NAFLD activity inflammation scores (Fig. 1e–f; Supplementary fig. 1c). Remarkably, bone marrow (BM) chimeric mice in which NLRP3 and ASC deficiency was limited to the hematopoietic compartment did not show any increase in the severity of NASH when compared to wt mice reconstituted with wt BM (Supplementary fig. 3a–f). Likewise, knock-in mice that specifically express a constitutively active NLRP3 inflammasome in CD11c+ myeloid cells (Nlrp3KI;CD11c+-Cre) or hepatocytes (Nlrp3KI;Albumin-Cre)16 did not feature any significant differences in MCDD-induced NASH severity as compared to wt mice (Supplementary figure 3g–l). These results suggest that aberrations in inflammasome function in cells other than hepatocytes or myeloid cells are key determinants of the enhanced disease progression in inflammasome-deficient mice.
We recently discovered that inflammasomes act as steady-state sensors and regulators of the colonic microbiota, and that a deficiency in components of two inflammasomes, NLRP617 and NLRP3 (unpublished), both of which include ASC, caspase-1, and involve IL-18 but not IL-1R, results in the development of an altered transmissible, colitogenic intestinal microbial community17. This microbiota is associated with increased representation of members of Bacteroidetes (Prevotellaceae) and the bacterial phylum TM7, and reductions in representation of members of the genus Lactobacillus in the Firmicutes phylum17. Moreover, electron microscopy (EM) studies disclosed aberrant colonization of crypts of Lieberkuhn with bacteria with morphologic features of Prevotellaceae17. Therefore, we sought to investigate whether enhanced NASH severity in inflammasome-deficient mice is driven by their altered microbiota. Strikingly, co-housing of Asc−/− and Il18−/− mice with wt animals for 4 weeks (beginning at 4–6 weeks of age), prior to induction of NASH with MCDD resulted in significant exacerbation of NASH in the wt cagemates [which we will refer to as wt(Asc−/−) and wt(Il18−/−), respectively, in the following text], as compared to singly-housed, age- and gender-matched wt controls (n=5–7 mice/genotype/housing condition). In co-housed wt mice, disease severity reached comparable levels to that of co-housed Asc−/− and Il18−/− mice (Fig. 2a–h). Moreover, significantly increased numbers of multiple inflammatory cell types were present in the liver of wt(Asc−/−) compared to wt mice (Supplementary fig. 2a). Similar findings were observed in wt mice co-housed with caspase-1−/−, Nlrp3−/−, and Nlrp6−/− mice (Supplementary fig. 4a–f). To exclude the possibility that aberrant microbiota represented in all mice maintained in our vivarium, we co-housed wt mice with other strains of NLR-deficient mice that were either obtained from the same source as Asc−/− and Nlrp3−/− mice (Nlrc4−/−, Nlrp12−/−), or generated in our laboratory (Nlrp4c−/−). None of these strains featured a similar phenotype (Supplementary fig. 4g–l). These results indicate that the transmissible colitogenic microbiota present in inflammasome-deficient mice is a major contributor to their enhanced NASH. In agreement with this, combined antibiotic treatment with Ciprofloxacin and Metronidazole, previously shown to abrogate the colitogenic activity of the microbiota associated with inflammasome-deficient mice associated microbiota17, significantly reduced the severity of NASH in Asc−/− mice, and abolished transmission of the phenotype to wt(Asc−/−) animals (Supplementary fig. 5).
Figure 2. Increased severity of NASH in Asc and Il18-deficient mice is transmissible to co-housed wild-type animals.
Asc−/− or Il18−/− mice and wt mice were co-housed for 4 weeks and then fed MCDD. (a–d) ALT (a), AST (b), NAFLD activity scores (c), and H&E-stained sections of livers (d) of singly-housed wt mice (wt), wt mice co-housed with Asc−/− (wt (Asc−/−)), and Asc−/− mice co-housed with wt (Asc−/−(wt)). (e–h) ALT (e), AST (f), NAFLD activity histological scores (g), and H&E-stained sections of livers (h) of wt, wt(Il18−/)−, and Il18−/−(wt). Data are representative of two independent experiments. Error bars represent SEM. Scale bars = 200 μm (d,h). *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
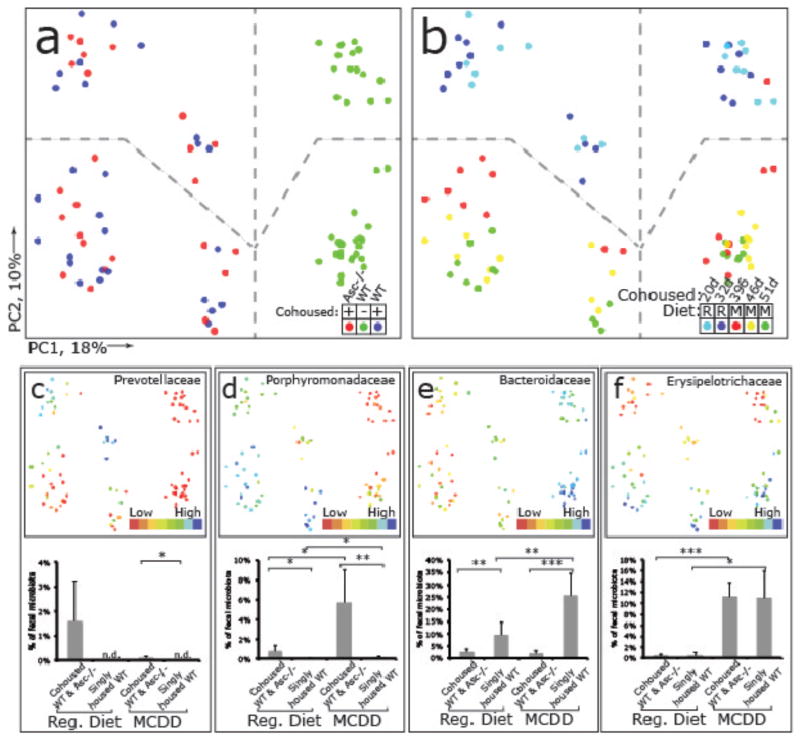
To ascertain the effects of MCDD on the gut microbiota, we performed a culture-independent analysis of amplicons generated by primers directed against variable region 2 of bacterial 16S rRNA genes of fecal samples collected from wt mice co-housed with Asc−/− animals [wt(Asc−/−)], their Asc−/− cagemates [Asc−/−(wt)] as well as singly-housed wt controls 1d and 12d prior to, and 7, 14 and 19 days after initiation of this diet (n=20 animals; 8 singly-housed wt, 6 co-housed wt and 6 Asc−/− mice). The structures of bacterial communities were compared based on their phylogenetic content using unweighted UniFrac. The results are illustrated in Figure 3. Table 1 provides a list of all phylotypes that, based on criteria outlined in Methods, discriminate co-housed wt(Asc−/−) from their singly-housed wt counterparts. Prior to MCDD, and consistent with our previous findings17, the fecal microbiota of wt(Asc−/−) mice adopted a configuration similar to Asc−/− cagemates, including the appearance of Prevotellaceae (Table 1 and Fig. 3a–c). There was also a significant increase in proportional representation of members of the family Porphyromonadaceae (primarily in the genus Parabacteroides) in wt(Asc−/−) mice compared to their singly-housed wt counterparts (Fig. 3d,e). The representation of Porphyromonadaceae was greatly increased in both the co-housed wt and Asc−/− mice (but not in singly-housed wt) when they were switched to a MCDD diet (p<0.01; t-test; Fig. 3d). A dramatic increase in the family Erysipelotrichaceae (phylum Firmicutes) also occurred with MCDD in co-housed animals, to a level that was >10% of the community (Fig. 3f). Although the Prevotellacaeae decreased when co-housed wt(Asc−/−) mice were placed on MCDD, their relative abundance remained significantly higher than in singly-housed wt animals (Fig. 3c).
Figure 3. 16S rRNA sequencing demonstrates diet and co-housing associated changes in gut microbial ecology.
(a) Principal Coordinates Analysis (PCoA) of Unweighted UniFrac distances of 16S rRNA sequences demonstrating clustering according to co-housing status on principal coordinate 1 (PC1). (b) PCoA of same plot as in (a) colored for experimental day. Mice were co-housed and fed a regular diet for the first 32 days of the experiment (two timepoints taken at day 20 and 32) before being switched to MCDD (sampled at days 39, 46 and 51 of the experiment). (c–f) PCoA and bargraphs of family level taxa Prevotellaceae, Porphyromonadaceae, Bacteroidaceae and Erysipelotrichaceae demonstrating diet-and microbiota-dependent differences in taxonomic representation. PCoA plots contain spheres represented a single fecal community colored according relative representation of the taxon (Blue represents relatively higher levels; red indicates lower levels). Bar graphs represent averaged taxonomic representation for singly or co-housed mouse while on either regular or MCD diet (n=8 for singly-housed wt, n=12 co-housed Asc−/−(wt) and wt(Asc−/−) animals;) * p < 0.05, ** p < 0.01, *** p < 0.001 by t-test after Bonferonni correction for multiple hypotheses. n.d. = not detected.
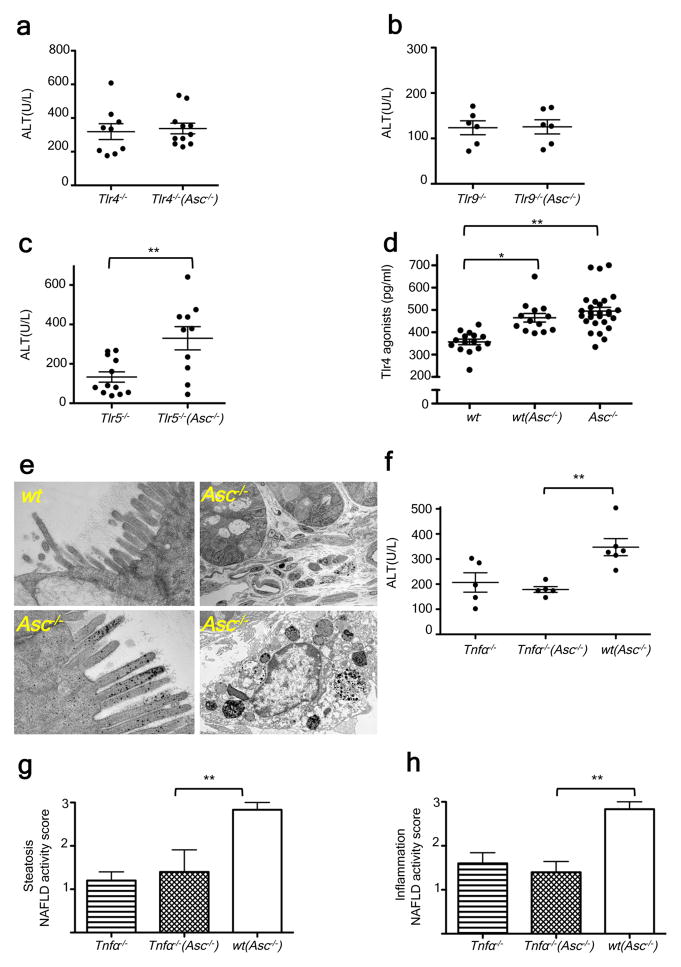
Together, these results pointed to the possibility that members of the altered intestinal microbiota in inflammasome-deficient MCDD-treated mice may promote a signaling cascade in the liver upon translocation, resulting in progression to NASH in susceptible animals. Toll Like Receptors (TLR) play a major role in NAFLD pathophysiology due to the liver’s exposure to relatively large amounts of PAMPs derived from the intestine and delivered via the portal circulation18–20. Therefore, we hypothesized that TLR signaling mediates the increased susceptibility to progression to NASH in mice exposed to the gut microbiota of Asc−/− animals. Myd88−/−;Trif−/−(Asc−/−) mice are devoid of all TLR signaling pathways. When co-housed with Asc−/− mice between 5 and 9 weeks of age, they exhibited decreased severity of NASH after exposure to MCDD for 24 days, compared to wt(Asc−/−) mice (Supplementary fig. 6a–b). To define which specific TLRs was responsible for the inflammatory response, we co-housed Tlr4-, Tlr9-, or Tlr5-deficient mice with Asc−/− animals and induced NASH with MCDD as previously described. Similar to wt mice, Tlr5−/− mice co-housed with Asc−/− mice ((Tlr5−/−(Asc−/−)) featured a statistically significant exacerbation of hepatic injury, steatosis, and inflammation, when compared to singly-housed Tlr5−/− controls (Fig. 3c; supplementary fig. 6g–h), indicating that TLR5 does not mediate the microbiota-mediated exacerbation in disease severity. In contrast, Tlr4−/−(Asc−/−) and Tlr9−/−(Asc−/−) mice did not show the customary increase in disease severity when compared to their singly-housed Tlr4−/− and Tlr9−/− counterparts (Fig. 3a–b; supplementary fig. 6c–f).
These observations suggest that intact bacteria or bacterial products derived from the intestine trigger TLR4 and TLR9 activation, which results in an increased rate of disease progression in mice that house a colitogenic gut microbiota associated with inflammasome deficiency (i.e. Asc−/−, wt(Asc−/−) mice). Efforts to sequence 16S rRNA genes that might be present in total liver DNA, microbial qPCR assays of portal vein blood DNA, histologic analysis of intact liver, and aerobic and anaerobic cultures of liver homogenates did not reveal any evidence of intact bacteria in wt or Asc−/− mice fed MCDD (data not shown). Notably, transmission electron microscopy studies of colon harvested from wt and Asc−/− mice revealed an abundance of electron dense material, suggestive of some black-pigmented bacterial species, in colonic epithelial cells and macrophages located in the lamina propria of Asc−/− mice but not in wt animals (Fig. 3e; supplementary fig. 7c). In agreement with previous results, we did not detect any translocation of intact bacteria (Fig. 3e; supplementary fig. 7c).
These observations provide evidence for the uptake of bacterial products from locally invasive gut microbes in Asc−/− mice (Fig. 3e; supplementary fig. 7c). If microbial components, rather than whole organisms were transmitted to the liver then they should be detectable in the portal circulation. Indeed, levels of TLR4 and TLR9 agonists but not TLR2 agonists (assayed by their ability to activate TLR reporter cell lines), were markedly increased in the portal circulation of MCDD-fed wt(Asc−/−), and Asc−/− mice as compared to wt controls (n= 13–28 mice/group; fig. 3d; supplementary figure 7a–b). Altogether, these results indicate a mechanism whereby TLR4 and TLR9 agonist efflux from the intestines of inflammasome-deficient mice or their co-housed partners, through the portal circulation, to the liver where they trigger TLR4 and TLR9 activation that in turn results in enhanced progression of NASH.
We next explored the downstream mechanism whereby microbiota-induced TLR signaling enhances NASH progression. Pro-inflammatory cytokines, and in particular tumor necrosis factor (TNF)-α, a downstream cytokine of TLR signaling, are known to contribute to progression of hepatic steatosis to steatohepatitis and eventually hepatic fibrosis in a number of animal models and in human patients21,22. Following induction of NASH by the MCDD, hepatic Tnfα mRNA expression was significantly upregulated in Asc−/− and Il18−/− mice, which exhibit exacerbated disease, but not in Il1r−/− mice, which do not (Supplementary fig. 8a–c). Moreover, Tnfα mRNA levels were significantly increased in wt mice that had been previously co-housed with Asc−/− or Il18−/− mice and then fed MCDD (Supplementary fig. 8d–e), suggesting that its enhanced expression was mediated by elements of the microbiota responsible for NASH exacerbation. In contrast, we did not observe any changes in Il6 or Il1β mRNA levels in the livers of Asc−/−, Il18−/−, or Il1r−/− mice compared to wt controls (Supplementary fig. 8a–c). Furthermore, while MCDD-administered singly-housed Tnfα−/− mice had comparable NASH severity to singly-housed wt animals (Fig. 3f–h; supplementary fig. 8f) co-housing with Asc-deficient mice prior to MCDD induction of NASH resulted in increased liver injury, hepatic steatosis, and inflammation in wt mice but not in Tnfα−/− mice (Fig. 3f–h; supplementary fig. 8f). These results indicate that TNF-α mediates the hepatotoxic effects downstream of the transmissible gut microbiota present in Asc−/− mice.
The aberrant gut microbiota in NLRP3 and NLRP6 inflammasome-deficient mice induces colonic inflammation through epithelial induction of CCL5 secretion17. To test whether this colon inflammation influences TLR agonist influx into the portal circulation and NASH progression, we induced NASH in wt and Ccl5−/− mice that had been either singly-housed or co-housed. MCDD-fed, singly-housed wt and Ccl5−/− mice exhibited equivalent levels of NASH severity (Supplementary fig. 9a–c), suggesting that CCL5 does not play a role in the early stages of NAFLD/NASH in the absence of the inflammasome-associated colitogenic microbiota. However, we documented significantly increased levels of liver injury, inflammation, and steatosis in wt(Asc−/−) but not Ccl5−/−(Asc−/−) mice (Fig. 5a–c), which led us to conclude that CCL5 is required for the exacerbation of disease through cohousing with inflammasome-deficient mice. Moreover, Ccl5−/−(Asc−/−) animals exhibited significantly reduced levels of TLR4 and TLR9 agonists in their portal vein blood than wt(Asc−/−) mice (Supplementary fig. 9d–f). Together, these results suggest that microbiota-induced subclinical colon inflammation is a determining factor in the rate of TLR agonist influx from the gut, and in NAFLD/NASH progression.
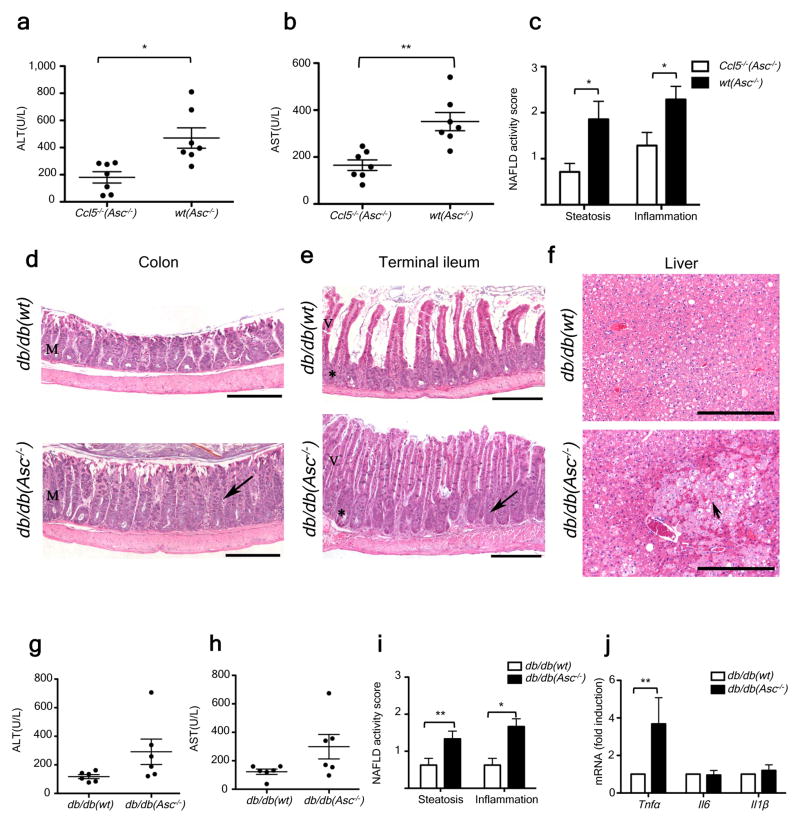
Figure 5. Increased severity of NASH in Asc-deficient mice is transmissible to db/db by co-housing and is mediated by CCL5-induced intestinal inflammation.
(a–c) ALT (a), AST (b), and NAFLD activity histological scores (c) of wt(Asc−/−) and Ccl5−/−(Asc−/−) mice. Data represents two independent experiments. (d–j) db/db mice were co-housed with wt or Asc−/− mice for 12 weeks. (d–f) Representative H&E-stained sections of colon (d), terminal ileum (e), and liver (f) from db/db(wt) and db/db(Asc−/−) mice fed a standard chow diet. Mucosal and crypt hyperplasia (arrow). Hepatocyte degeneration (arrowhead). Scale bars = 500 μm (d–e), Scale bars = 200 μm (f). (g–i) ALT (g), AST (h), and NAFLD activity scores (i) of db/db(wt), and db/db(Asc−/−) mice. (j) Hepatic Tnfα, il6, and il1β mRNA levels. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
The MCDD system is a common model for studying inflammatory processes associated with progression from NAFLD to NASH, yet it lacks many of the associated metabolic phenotypes of NAFLD, such as obesity and insulin resistance23. As such, our results in this model might conceivably be limited to the way dysbiosis can influence NASH progression in patients with enhanced intestinal permeability, such as those with IBD24, but not for the majority of patients who suffer from NASH in the context of metabolic syndrome. To test whether alterations in the gut microbiota of inflammasome-deficient mice may affect the rate of progression of NAFLD and other features associated with metabolic syndrome, we extended our studies to genetically obese mice and mice fed with high fat diet (HFD).
Leptin-receptor deficient (db/db) animals develop multiple metabolic abnormalities, including NAFLD and impaired intestinal barrier function25, that closely resemble the human disease26. However, significant hepatocyte injury, inflammation, and fibrosis are not observed in the absence of a “second hit”27. Upon cohousing of db/db mice with Asc−/− (db/db(Asc−/−)) or WT mice (db/db(wt)) for a period of twelve weeks, and as previously shown for Asc−/− mice17, the colon and ileum of all db/db(Asc−/−) mice exhibited mild to moderate mucosal and crypt hyperplasia (Fig. 5d–f), that was not seen in db/db(wt) mice.
Strikingly, co-housed db/db(Asc−/−) mice also showed increased levels of hepatocyte injury as evidenced by higher levels of ALT and AST in their sera, and significantly exacerbated steatosis and hepatic inflammation scores when compared with db/db(wt) mice (Fig. 5g–i). In addition to a parenchymal inflammatory exudate, patchy areas of markedly degenerated hepatocytes and hepatocytes undergoing necrosis were observed, but only in db/db(Asc−/−) animals (Fig. 5f). Furthermore, some areas of congestion were seen in the centro-lobular zone as well as in the hepatic parenchyma – features that resemble Peliosis Hepatis, a condition observed in a variety of pathological settings including infection (data not shown). In accord with our MCDD results, hepatic Tnfα mRNA levels were significantly higher in co-housed db/db(Asc−/−) mice than in db/db(wt) animals (Fig. 5j). Again, no significant differences were observed in hepatic Il6 or Il1β mRNA levels (Fig. 5j).
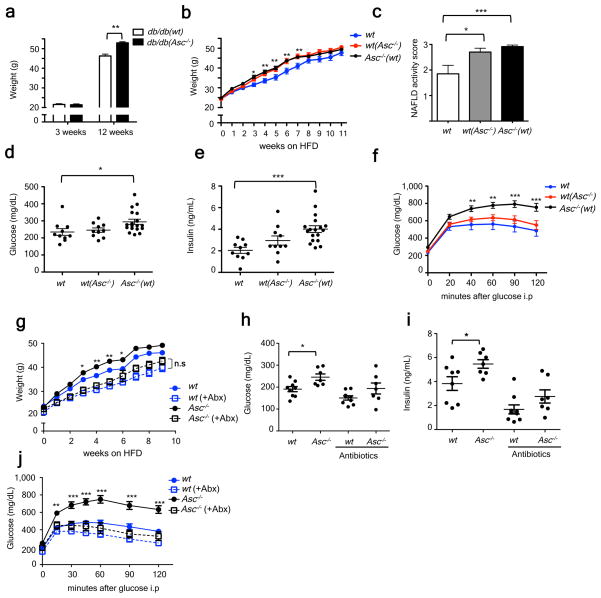
Interestingly, db/db(Asc−/−) mice developed significantly more weight gain as compared to db/db(wt) after 12 weeks of co-housing (Figure 6a), suggesting that the inflammasome-associated gut microbiota could exacerbate additional processes associated with the metabolic syndrome, such as obesity. To address this possibility, we monitored multiple metabolic parameters in wt, wt(Asc−/−), and Asc−/− mice fed a high fat diet (HFD) for 12 weeks. Strikingly, Asc−/− mice gained body mass more rapidly and featured enhanced hepatic steatosis (Fig. 6b–c; supplementary fig. 11f). Asc−/− mice also showed elevated fasting plasma glucose and insulin levels, and decreased glucose tolerance as compared to singly-housed weight matched wt mice (Fig. 6d–f). Interestingly, wt(Asc−/−) mice recapitulated the same increased rate of body mass gain and steatosis when compared to singly-housed wt controls, although they did not show significant alterations in glucose homeostasis (Fig. 6d–f). Nevertheless, antibiotic treatment (Ciprofloxacin and Metronidazole) abrogated all these effects, including altered rate of gain in body mass, glucose intolerance, and fasting plasma insulin levels in Asc−/− mice as compared to wt mice (Fig. 6g–j). Alterations of these metabolic parameters were not caused by changes in feeding behavior between the antibiotic-treated and untreated groups (data not shown). These results suggest different levels of microbiota-mediated regulation of the various manifestations of the metabolic syndrome: i.e., some features (obesity, steatosis) are pronounced and transmissible by co-housing, while others (glycemic control) are affected by alterations in the microbiota but not readily transferable by co-housing. Additionally, we performed a 16S rRNA-based analysis of the fecal microbiota of Asc−/− and wt animals that were treated with or without Ciprofloxacin and Metronidazole (4 weeks) prior to switching to HFD for 4 additional weeks. We also observed an overrepresentation of Porphyromonadaceae in Asc−/− as compared to wt mice (p=0.052) at week eight of HFD in the absence of antibiotics. Importantly, the analysis demonstrated that Prevotellaceae and Porphyromonadaceae, two family level taxa, were undetectable 8 weeks after antibiotic treatment (Supplementary fig. 12a–c; Table 2).
Figure 6. Asc-deficient mice develop increased obesity and loss of glycemic control on HFD.
(a) Weight of db/db(wt) or (db/db(Asc−/−) at 3 weeks of age and at 12 weeks of co-housing. (b–f) Asc−/− and wt mice were co-housed for 4 weeks and then fed HFD. (b) Body weights. (c) NAFLD histological activity score. (d–e) Fasting plasma glucose and insulin after 11 weeks of HFD. (f) IPGTT after 12 weeks of HFD. (g–j) Mice were untreated, or treated orally with antibiotics, for 3 weeks prior to HFD feeding for 12 weeks. (g) Body weights. (h–i) Fasting plasma glucose and insulin levels after 8 weeks on a HFD. (j) IPGTT after 10 weeks of HFD. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
To assess whether these metabolic abnormalities are specific to Asc−/− mice, we performed similar experiments with Nlrc4−/− mice. These mice showed an equal rate of body mass gain, and similar glucose tolerance phenotypes as singly-housed wt mice, confirming the specificity of the phenotype (Supplementary fig. 10a–d). 16S rRNA analysis revealed that there was an increased representation of Porphyromonadaceae in Nlrc4−/− mice when compared to wt mice (Table 3). These results indicate that (i) some metabolic aberrations associated with the dysbiosis of inflammasome-deficient mice can be horizontally transferred from one mouse to another,(ii) the gut microbiota of inflammasome-deficient mice negatively impacts NAFLD progression and glucose homeostasis, and (iii) configurational changes in the microbiota, which involve overrepresentation Porphyromonadaceae in combination with alterations in additional taxa, are likely required to produce these host phenotypes.
Discussion
The results presented here provide evidence that modulation of the intestinal microbiota through multiple inflammasome components is a critical determinant of NAFLD/NASH progression as well as multiple other aspects of metabolic syndrome such as weight gain and glucose homeostasis. Our results demonstrate a complex and cooperative effect of two sensing protein families, namely NLR’s and TLR’s, in shaping metabolic events. In the gut, the combination of host related factors such as genetic inflammasome deficiency-associated dysbiosis result in abnormal accumulation of bacterial products in the portal circulation. The liver, being a ‘first pass’ organ and thus exposed to the highest concentration of portal system products such as PAMPs, is expected to be most vulnerable to their effects, particularly when pre-conditioned by subclinical pathology such as lipid accumulation in hepatocytes. Indeed in our models, accumulation of TLR agonists was sufficient to drive progression of NAFLD/NASH even in genetically intact animals.
This ‘gut-liver axis’, driven by alterations in gut microbial ecology, may offer an explanation for a number of long-standing albeit poorly understood clinical associations. One example is the occurrence of primary sclerosing cholangitis (PSC) in patients with inflammatory bowel disease, particularly those with inflammation along the length of the colon. Celiac disease, another inflammatory disorder with increased intestinal permeability, is associated with a variety of liver disorders, ranging from asymptomatic transaminasemia, NAFLD, to primary biliary cirrhosis (PBC). In fully developed cirrhosis, complications associated with high mortality such as portal hypertension, variceal bleeding, spontaneous bacterial peritonitis, and encephalopathy are triggered by translocation of bacteria or bacterial components, providing another important example of the importance of the interplay between the microbiome, the immune response and liver pathology28.
Recent reports suggest a complex role of inflammasome function in multiple manifestations of the metabolic syndrome. Activation of IL-1β, mainly through cleavage by the NLRP3 inflammasome, promotes insulin resistance 29,30, atherosclesotic plaque formation 31, and β cell death 32,33. Moreover, caspase-1 activation seems to direct adipocytes toward a more insulin-resistant phenotype34. Conversely, Il18-deficient mice are prone to develop obesity, hyperphagia and insulin resistance35. These discrepancies most likely reflect a hierarchical contribution of multiple inflammasome components in different metabolic processes, tissues, and mouse models. In agreement with previous studies, we found increased obesity and insulin resistance in Il18-deficient mice fed with a HFD (data not shown). However, and in contrast to two previous reports30,34, we showed that Asc−/− mice are prone to obesity induction, hepatosteatosis, as well as impaired glucose homeostasis when fed a HFD. We propose that alterations in intestinal microbiota communities associated with multiple inflammasome deficiencies could account for these discrepancies and it should be added to the list of major environmental/host factors affecting manifestations and progression of metabolic syndrome in susceptible populations.
A significant expansion of Porphyromonadaceae was found in Asc−/− and wt(Asc−/−) following administration of MCDD and HFD, which was abolished by antibiotic treatment. Interestingly, one member of the family, Porphyromonas, has been associated with several components of the metabolic syndrome in both mice and humans, including atherosclerosis and diabetes mellitus 36,37. Moreover, expansion of this taxa is strongly associated with complications of chronic liver disease 36. More work is needed to further delineate the relevance of the suggested taxa discovered in our work to the pathogenesis and progression of human NAFLD/NASH and other features of the metabolic syndrome. Elucidation of similar or distinct mechanisms to the ones presented here, possibly linking Porphyromonadaceae expansion to a propensity for development of the metabolic syndrome, would be of importance to the field.
Methods summary
Six to eight week-old male mice were fed a methionine-choline-deficient diet for 24 days. Eight to ten week-old male mice were fed a high fat diet (HFD) ad libitum. This diet consists of 60% calories from fat and was administered for 10–12 weeks. Standard histology of liver, terminal ileum, and colon were described previously17. The presence of immune cells in liver tissue was analyzed by flow cytometry on livers digested with 0.5mg/ml collagenase IV. Glucose tolerance test were performed after 10–12 weeks of consuming the HFD and mice were fasted overnight (~14 h), and injected intraperitoneally with D-glucose. Transmission electron microscopy was performed as previously described17. Data are expressed as mean ± SEM. Differences were analyzed by Student’s t test or ANOVA and post hoc analysis for multiple group comparison. p values ≤ 0.05 were considered significant.
Methods
Mice
Caspase-1−/− mice (Casp1tm1Flv) and NLRP4c were generated in our laboratory38. Production of ASC−/− (Pycardtm1Flv), Nlrp3−/−, Nlrp6−/−, Nlrc4−/−, and Nlrp12−/− mice is described elsewhere17. Il18−/− (Il18tm1Aki), Il1r−/− (Il1r1tm1Imx), Tnfα −/−(Tnftm1Gkl), Tlr4−/− (Tlr4lps−del), Tlr5−/− (Tlr5tm1Flv), Myd88−/− (Myd88tm1Defr), Ccl5−/−(Ccl5tm1Hso), Rag1−/− (Rag1tm1Mom), CD11c-Cre (Itgax-cre), albumin-Cre (Alb-cre), Trif−/− (Ticam1Lps2), and db/db(Leprdb) mice were obtained from Jackson Laboratories. Tlr9−/− mice have been described in another report39. Production of Nlrp3KI (A350V) mice is described elsewhere16. wt C57Bl/6 mice were purchased from the NCI. For co-housing experiments, age-matched wt and KO mice at the age of 4–6 weeks were co-housed in sterilized cages for 4 or 12 weeks at a ratio of 1:1 (wt:KO), with unrestricted access to food and water. No more than 6 mice in total were housed per cage. For antibiotic treatment, mice were given a combination of ciprofloxacin (0.2 g/L) and metronidazole (1 g/L) for 4 weeks in the drinking water. All antibiotics were obtained from Sigma Aldrich. All experimental procedures were approved by the local IACUC.
NASH model
6–8 week-old male mice were fed a methionine-choline-deficient diet (MP Biomedicals, Solon, OH) for 24 days. Methionine-choline-sufficient control diet was the same but supplemented with choline chloride (2g/kg diet) and DL-methionine (3g/kg diet). Mice had unrestricted access to food and water.
High fat diet model
8–10 week-old male mice were fed a high fat diet (HFD) ad libitum. This diet consists of 60% calories from fat (D12492i; Research Diets) and was administered for 10–12 weeks.
Histology
The intact liver was excised immediately after mice were euthanized by asphyxiation, fixed in 10% neutral buffered formalin and embedded in paraffin. Liver sections were stained with hematoxylin and eosin, or trichrome. Histological examination was performed in a blinded fashion by a experienced gastrointestinal pathologist with the histological scoring system for NAFLD40. Briefly, steatosis and inflammation scores ranged from 0 to 3 with 0 being within normal limits and 3 being most severe. Individual scores were assigned for each parameter. The most severe area of hepatic inflammation of representative histology sections were photographed using an Olimpus microscope.
Colons were fixed in Bouin’s medium and embedded in paraffin. Blocks were serially sectioned along the cephalocaudal axis of the gut to the level of the lumen; 5 μm-thick sections was stained with hematoxylin and eosin. Digital light microscopic images were recorded with a Zeiss Axio Imager.A1 microscope (Thornwood, NY), AxioCam MRc5 camera and AxioVision 4.7.1 imaging software (Carl Zeiss Microimaging).
Gene expression analysis
Tissues were preserved in RNAlater solution (Ambion), and subsequently homogenized in TRIZOL reagent (Invitrogen). 1μg of RNA was used for generate cDNA using the HighCapacity cDNA Reverse Transcription kit (Applied Biosystems). RealTime-PCR was performed using gene-specific primer/probe sets (Applied Biosystems) and Kapa Probe Fast qPCR kit (Kapa Biosystems) on a 7500 Fast Real Time PCR instrument (Applied Biosystems). The reaction conditions were 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. Data was analyzed using the Sequence Detection Software according to the ΔCt method with hprt1 serving as the reference housekeeping gene.
Glucose Tolerance Test (GTT)
GTT were performed after 10–12 weeks of consuming the HFD. Mice were fasted overnight (~14 h), and injected intraperitoneally with 10% dextrose at a dose of 1 g/kg body weight. Blood was collected from tail vein and plasma glucose levels measured at indicated times using a YSI 2700 Select Glucose Analyzer (YSI Life Sciences). Plasma insulin levels were determined by radioimmunoassay (Linco).
FACS analysis
Livers were harvested, digested with 0.5mg/ml collagenase IV (Sigma) for 45 min at 37°C, homogenized and repeatedly centrifuged at 400 × g for 5 min to enrich for hematopoietic cells. Cells were stained for flow cytometry using antibodies against CD45.2, CD11b, CD11c, NK1.1, B220, CD4, CD8, TCRβ, F4/80, Gr-1, MHC class II (Biolegend) and analyzed on a BD LDR II.
Portal vein blood collection
Mice were anesthetized with ketamine 100mg/kg and xylazine 10mg/kg. Mice were placed on a clean surgical field, and the abdominal fur was clipped and cleaned with a two stage surgical scrub consisting of Betadine and 70% ethanol. A 1 to 1.5 cm midline incision was made in the skin and abdominal wall. The peritoneum was moved to the left and the portal vein was punctured with a 30G needle. Between 0.2 and 0.3 ml of blood were collected per mouse. Serum was recovered by centrifugation at 4000 rpm for 15 min at room temperature and then stored at −80°C in endotoxin-free tubes until assayed.
Measurement of PAMPs
TLR2, TLR4, and TLR9 agonists were assayed in portal vein serum using HEK-blue mTLR2, HEK-blue mTLR4, and HEK-blue mTLR9 reporter cell lines (Invivogen, SanDiego, CA) and the manufacturer’s protocol with modifications. In brief, 2.2 × 105 HEK-blue mTLR2, 1.0 × 105 HEK-blue mTLR4, and 2.0 × 105 HEK-blue mTLR9 cells were plated in 96-well plates containing 10 μl of heat inactivated (45 min at 56°C) portal vein serum. Cells were then incubated for 21 h at 37°C under an atmosphere of 5% CO2/95% air. Twenty microliters of the cell culture supernatants were collected and added to 180 μl of the QUANTI-Blue substrate in a 96-well plate. The mixtures were then incubated at 37°C in 5% CO2/95% air for 3 h and SEAP levels were determined using an spectrophotometer at 655 nm.
Transmission electron microscopy
Mice were perfused via their left ventricles using 4% PFA in PBS. Selected tissues were fixed in 2.5% gluteraldehyde in 0.1M sodium cacodylate buffer pH7.4 for 1–2 h. Samples were rinsed 3 times in sodium cacodylate buffer and post-fixed in 1% osmium tetroxide for 1 h, en bloc stained in 2% uranyl acetate in maleate buffer pH 5.2 for a further hour then rinsed, dehydrated, infiltrated with Epon812 resin, and baked over night at 60°C. Hardened blocks were cut using a Leica UltraCut UCT. 60nm-thick sections were collected and stained using 2% uranyl acetate and lead citrate. Samples were all viewed FEI Tencai Biotwin TEM at 80Kv. Images were taken using Morada CCD and iTEM (Olympus) software.
Bone Marrow Chimeras
Bone marrow was flushed from femurs with DMEM with 10% FBS, red cells were lysed, and the material filtered through a 70um filter. 1×106 cells in 100 ul PBS were delivered by retro-orbital injection into lethally irradiated (1000 rad) mice. For two weeks post-engraftment, mice were maintained on antibiotics (Sulfatrim). Six weeks after transplantation animals were switched to MCDD. A wt non-irradiated mouse was co-housed with the engrafted mice for four weeks prior to NASH induction. Under our standardized protocol, bone marrow chimeras routinely show a level of engraftment of ≥93%.
Bacterial 16S rRNA amplicon sequencing
Total DNA was isolated from the livers of mice fed a MCDD diet and used for attempted PCR amplification of variable region 2 of bacterial16S rRNA genes20 that may be present in the tissue. Thirty cycles of amplification of liver DNA prepared from 7 wt, and 7 ASC−/− mice yielded detectable product (>60ng/reaction) in 3 samples from the wt group and 3 samples from the Asc−/− group. All amplicons were then subjected to multiplex pyrosequencing with a 454 instrument using FLX Titanium chemistry (137–1510 reads/sample, average read length, 360 nt). Reads were analyzed using the QIIME software package. Operational taxonomic unit (OTU) picking was performed using uclust and taxonomic assignments made with RDP41. This analysis demonstrated inconsistent representation of taxa between animals and taxa that largely represented organisms not associated with the gut microbiota. G-test indicated that there was no significant correlation between any of these taxa and the presence of NASH.
For analysis of the fecal microbiota of MCDD-fed Asc−/−(wt), wt(Asc−/−) and singly-housed wt mice, fecal pellets were collected at the time points indicated in Figure 3.. The protocols that we used to extract fecal DNA and to perform multiplex pyrosqequencing of amplicons generated by PCR from the V2 regions of bacterial 16S rRNA genes, have been previously described20. A total of 366,283 sequences were generated from 181 fecal samples (average 2023±685 reads/sample; average read length, 360 nt). Sequences were de-multiplexed and binned into species-level operational taxonomic units (OTUs; 97% nucleotide sequence identity; %ID) using QIIME 1.2.146. Taxonomy was assigned within QIIME using RDP. Chimeric sequences were removed using ChimeraSlayer and OTUs were filtered to a minimum of 10 sequences per OTU and 1000 OTUs per sample. PCoA plots were generated by averaging the unweighted UniFrac distances of 100 subsampled OTU tables. Statistical analysis was performed on the proportional representation of taxa (summarized to Phyla, Class, Order, Family and Genus levels), using paired (where possible) and unpaired t tests. Taxa that were significantly different after multiple hypothesis testing were included in Supplemental Tables X–Z.
Statistical Analysis
Data are expressed as mean ± SEM. Differences were analyzed by Student’s t test or ANOVA and post hoc analysis for multiple group comparison. p values ≤ 0.05 were considered significant.
Supplementary Material
Figure 4. Increased severity of NASH in Asc-deficient and co-housed wild-type animals is mediated by TLR4, TLR9, and TNF-α.
Asc−/− mice were co-housed with wt, Tnfα −/−, Tlr4−/−, Tlr9−/−, or Tlr5−/− mice for 4 weeks and then fed MCDD. (a–c) ALT levels of Tlr4−/−(Asc−/−) (a), Tlr9−/−(Asc−/−) (b), and Tlr5−/−(Asc−/−) (c) and their singly-housed counterparts. (d) TLR4 agonists in portal vein sera from MCDD-fed wt, wt(Asc−/−), and Asc−/− animals. (e) Transmission electron microscopy images of colon from wt and Asc−/−. (f–h) ALT (f), and NAFLD activity histological scores (g–h) of Tnfα −/−, wt(Asc−/−), and Tnfα −/− (Asc−/−). Data are representative of two independent experiments. Error bars represent SEM. *p≤ 0.05, **p≤ 0.01, ***p≤ 0.001.
Acknowledgments
We would like to thank Elisabeth Eynon, Jon Alderman, Adam Williams, Frances Manzo, and Hila Elinav for technical assistance and discussions; Morven Graham and Christoph Rahner for performing electron microscopy; David R. Peaper for assistance in microbiological culture procedures. EE is supported by Cancer Research Institute (2010–2012) and by a supplemental grant from the Israel-US educational foundation (2009) and is a recipient of the Claire and Emmanuel G. Rosenblatt award from the American Physicians for Medicine in Israel Foundation (2010–2011). J.H.M. and T.S are supported by Leukemia and Lymphoma Society Postdoctoral Fellowships. S.C.E is supported by T32HL007974 and K08A1085038. W.Z.M is supported by R01DK076674-01 and the VA Merit award. This work was supported in part by the Howard Hughes Medical Institute (G.I.S., R.A.F.), the United States-Israel binational Foundation grant (E.E. and R.A.F.), the Crohn’s and Colitis Foundation of America (A.K. and J.I.G) and R01 DK-40936, R24 DK-085638, P30 DK-45735 and U24 DK-059635
Footnotes
The authors report no conflict of interest.
Author contributions
J.H-M, E.E., and R.A.F. designed the study and wrote the manuscript. J.H-M., E.E., C.J., L.H., W.Z.M., M.J.J., J.C., G.I.S., and C.A.T. performed the in-vitro and in-vivo experimental work and edited the manuscript. T.S. and S.C.E. supported the work with key suggestions and editing of the manuscript. H.M.H. provided the Nlrp3 knock-in mice and provided valuable feedback on the manuscript. A.L.K. and J.I.G performed the stool processing and metagenomic analysis of the microbiota and provided key suggestions to the manuscript and participated in its editing. R.A.F. directed the project.
References
- 1.Sheth SG, Gordon FD, Chopra S. Nonalcoholic steatohepatitis. Ann Intern Med. 1997;126:137–145. doi: 10.7326/0003-4819-126-2-199701150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig J, Viggiano TR, McGill DB, Oh BJ. Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin Proc. 1980;55:434–438. [PubMed] [Google Scholar]
- 3.Marchesini G, et al. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell SH, et al. Cryptogenic cirrhosis: clinical characterization and risk factors for underlying disease. Hepatology. 1999;29:664–669. doi: 10.1002/hep.510290347. [DOI] [PubMed] [Google Scholar]
- 5.Shimada M, et al. Hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J Hepatol. 2002;37:154–160. doi: 10.1016/s0168-8278(02)00099-5. [DOI] [PubMed] [Google Scholar]
- 6.Propst A, Propst T, Judmaier G, Vogel W. Prognosis in nonalcoholic steatohepatitis. Gastroenterology. 1995;108:1607. doi: 10.1016/0016-5085(95)90724-6. [DOI] [PubMed] [Google Scholar]
- 7.Charlton M. Cirrhosis and liver failure in nonalcoholic fatty liver disease: Molehill or mountain? Hepatology. 2008;47:1431–1433. doi: 10.1002/hep.22246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hjelkrem MC, Torres DM, Harrison SA. Nonalcoholic fatty liver disease. Minerva Med. 2008;99:583–593. [PubMed] [Google Scholar]
- 9.Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 10.Sanyal AJ, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 11.Sutterwala FS, Ogura Y, Flavell RA. The inflammasome in pathogen recognition and inflammation. J Leukoc Biol. 2007;82:259–264. doi: 10.1189/jlb.1206755. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 13.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 15.Varela-Rey M, et al. Non-alcoholic steatohepatitis and animal models: understanding the human disease. Int J Biochem Cell Biol. 2009;41:969–976. doi: 10.1016/j.biocel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 16.Brydges SD, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivera CA, et al. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47:571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, et al. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology. 2010;139:323–334. e327. doi: 10.1053/j.gastro.2010.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki E, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 21.Crespo J, et al. Gene expression of tumor necrosis factor alpha and TNF-receptors, p55 and p75, in nonalcoholic steatohepatitis patients. Hepatology. 2001;34:1158–1163. doi: 10.1053/jhep.2001.29628. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, et al. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37:343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- 23.Diehl AM. Lessons from animal models of NASH. Hepatol Res. 2005;33:138–144. doi: 10.1016/j.hepres.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Broome U, Glaumann H, Hultcrantz R. Liver histology and follow up of 68 patients with ulcerative colitis and normal liver function tests. Gut. 1990;31:468–472. doi: 10.1136/gut.31.4.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, et al. Leptin signaling in intestinal epithelium mediates resistance to enteric infection by Entamoeba histolytica. Mucosal Immunol. 2011;4:294–303. doi: 10.1038/mi.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikejima K, et al. The role of leptin in progression of non-alcoholic fatty liver disease. Hepatol Res. 2005;33:151–154. doi: 10.1016/j.hepres.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Guebre-Xabier M, et al. Altered hepatic lymphocyte subpopulations in obesity-related murine fatty livers: potential mechanism for sensitization to liver damage. Hepatology. 2000;31:633–640. doi: 10.1002/hep.510310313. [DOI] [PubMed] [Google Scholar]
- 28.Almeida J, Galhenage S, Yu J, Kurtovic J, Riordan SM. Gut flora and bacterial translocation in chronic liver disease. World J Gastroenterol. 2006;12:1493–1502. doi: 10.3748/wjg.v12.i10.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandanmagsar B, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen H, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 33.Masters SL, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1beta in type 2 diabetes. Nat Immunol. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stienstra R, et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc Natl Acad Sci U S A. 2011;108:15324–15329. doi: 10.1073/pnas.1100255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Netea MG, et al. Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat Med. 2006;12:650–656. doi: 10.1038/nm1415. [DOI] [PubMed] [Google Scholar]
- 36.Bajaj JS, et al. Linkage of Gut Microbiome with Cognition in Hepatic Encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2011 doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makiura N, et al. Relationship of Porphyromonas gingivalis with glycemic level in patients with type 2 diabetes following periodontal treatment. Oral Microbiol Immunol. 2008;23:348–351. doi: 10.1111/j.1399-302X.2007.00426.x. [DOI] [PubMed] [Google Scholar]
- 38.Sutterwala FS, et al. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 40.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.