Abstract
The Saccharopolyspora erythraea mutB knockout strain, FL2281, having a block in the methylmalonyl-CoA mutase reaction, was found to carry a diethyl methylmalonate-responsive (Dmr) phenotype in an oil-based fermentation medium. The Dmr phenotype confers the ability to increase erythromycin A (erythromycin) production from 250 – 300% when the oil-based medium is supplemented with 15 mM levels of this solvent. Lower concentrations of the solvent stimulated proportionately less erythromycin production, while higher concentrations had no additional benefit. Although the mutB strain is phenotypically a low-level erythromycin producer, diethyl methylmalonate supplementation allowed it to produce up to 30% more erythromycin than the wild type (control) strain--a strain that does not show the Dmr phenotype. The Dmr phenotype represents a new class of strain improvement phenotype. A theory to explain the biochemical mechanism for the Dmr phenotype is proposed. Other phenotypes found to be associated with the mutB knockout were a growth defect and hyper-pigmentation, both of which were restored to normal by exposure to diethyl methylmalonate. Furthermore, mutB fermentations did not significantly metabolize soybean oil in the presence of diethyl methylmalonate. Finally, a novel method is proposed for the isolation of additional mutants with the Dmr phenotype.
Keywords: Diethyl methylmalonate, erythromycin, mutB, methylmalonyl-CoA mutase, Dmr, Saccharopolyspora erythraea
Introduction
The polyketide antibiotic erythromycin A (erythromycin) is produced by Saccharopolyspora erythraea, a gram-positive, mycelial, soil-dwelling actinomycete. This bacterium is widely used for studies of polyketide biosynthesis (Rawlings 2001; Kwan et al. 2011) and strain improvement (Wu et al. 2011; Chen et al 2010; Wang et al 2007); and its genome has been completely sequenced and annotated (Oliynyk et al. 2007). Although erythromycin has been in use as an antibiotic since the 1950’s, it is now primarily used as an active pharmaceutical ingredient (API) for the production of several semi-synthetic erythromycin derivatives, including azithromycin, clarithromycin, dirithromycin and roxithromycin. There is an ongoing need by manufacturers to reduce the cost of erythromycin production through strain or process improvements, or as is frequently the case, a combination of the two. This is particularly relevant now since the semi-synthetic erythromycins will soon be entering the generic market where lowering the cost of production is critical to commercial success.
With erythromycin being one of the antibiotics from the golden-age of antibiotic discovery, most of the best commercial strains available today were improved by classical strain improvement technology (Queener and Lively 1986). The classical method involves the trial and error generation of random mutations to find the one strain, usually out of tens of thousands, that makes more erythromycin. The improvements that can be found by the classical method diminish in frequency and magnitude over time. Therefore newer metabolic engineering strain improvement methods are being investigated (Keasling 2010). Some of the newer processes under development are heterologous systems using Escherichia coli (Pfeifer et al. 2002; Boghigian et al. 2011; Zhang et al. 2010; Weber 2010) and Streptomyces coelicolor (Lombo et al. 2001); or native systems using the unicellular Aeromicrobium erythreum (Brikun et al. 2004; Miller 1991) or the mycelial S. erythraea (Minas 2005; Reeves et al. 2007). Meanwhile, current large-scale fermentations still use the classically improved S. erythraea strain as the producing organism, and a fermentation medium consisting of a mixture of agricultural ingredients such as soybean meal, soybean oil, and starch.
The approach used in this study was to work with S. erythraea to develop a process improvement to complement the mutB genetic strain improvement target (Fig. 1). Previously, genetic experiments showed that erythromycin production was limited by the polyketide precursor methylmalonyl-coenzyme A (CoA). Since then, similar findings for other polyketide natural products have been reported (Mo et al. 2009). Our results were inferred from the mutB knockout strain, which is blocked in the propionate pathway at the methylmalonyl-CoA mutase (MCM) reaction between methylmalonyl-CoA and succinyl-CoA (Fig. 1; Reeves et al. 2006). This mutation, therefore, blocks the flow of polyketide precursors into the polyketide synthase (PKS) from the tricarboxylic acid (TCA) cycle. This block causes a significant reduction (40 - 65 %) in the amount of erythromycin produced in an oil-based medium (this study, and Reeves et al. 2006). Since in the past erythromycin yield improvements were made possible by genetic manipulations that resulted in increased flow of methylmalonyl-CoA to the polyketide synthase (Reeves et al. 2006, 2007), the study here investigated the prediction that the addition of a methylmalonyl-CoA-related compound directly to the fermentation medium could also increase yields.
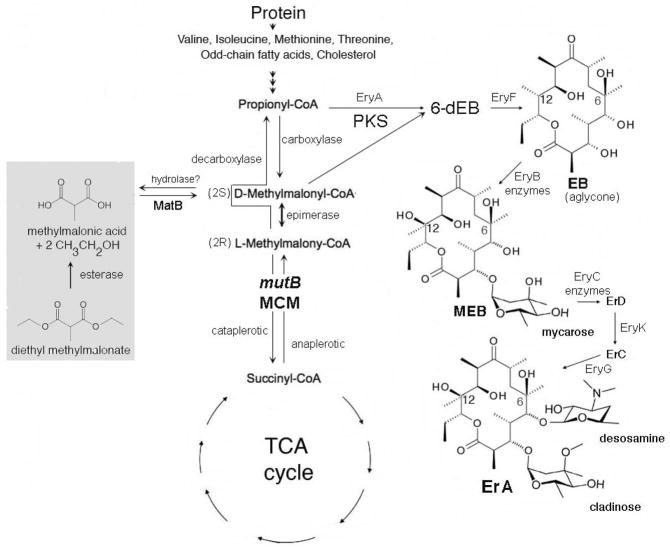
Fig. 1.
Metabolic map of the propionate pathway and erythromycin biosynthetic pathway. Hypothetical pathway (gray box) shows diethyl methylmalonate assimilation into erythromycin (Lombo et al. 2001). A hydrolase, converting methylmalonyl-CoA to methylmalonic acid is also postulated. Abbreviations: MCM, methylmalonyl-CoA mutase; EB, erythronolide B; 6-dEB, 6-deoxyerythronolide B; MEB, 3-O-α-mycarosylerythronolide B; ErA, C, and D, erythromycin A, C and D. EryA, polyketide synthase; EryF, C-6 hydroxylase; EryB, dTDP-mycarose biosynthetic and transferase enzymes; EryC, dTDP-desosamine biosynthetic and transferase enzymes; EryK, C-12 hydroxylase; EryG, S-adenosylmethionine methylase acting on the attached mycarose to convert it to cladinose.
Diethyl methylmalonate was chosen as the methylmalonyl-CoA-related compound because it was predicted, due to the ubiquity of cellular esterases, to feed directly into the propionate pathway (Fig. 1). It was also chosen because it is used in other industrial processes and is readily available in bulk quantities. Another advantage of diethyl methylmalonate is that it is soluble in the oil component of the erythromycin fermentation medium, and therefore does not lower the pH of the aqueous culture medium as does methylmalonic acid. Diethyl methylmalonate is also 2 – 3 times less expensive than methylmalonate, so it is plausible that it could be cost effective to use in large scale fermentations, particularly since low concentrations would be effective.
The benefits of using diethyl methylmalonate with the S. erythraea mutB strain are reported here. Also discussed is a new strategy for modifying the classical strain improvement method to take S. erythraea commercial strains beyond their current production plateau.
Materials and methods
Bacterial strains, culture conditions, chemicals and biochemicals
The two bacterial strains of Saccharopolyspora erythraea used in this study were the wild type, FL2267, and the mutB polar knockout strain, FL2281 (Reeves et al. 2006). Both strains are derivatives of the S. erythraea ATCC 11635 strain. The knockout in methylmalonyl-CoA mutase in FL2281 is caused by the insertion of the kanamycin resistance gene in mutB (Reeves et al. 2006). Erythromycin A, B and C were obtained from the U.S. Pharmacopeia. Erythronolide B (EB, 99% pure) and 3-α-O-mycarosyl erythronolide B (MEB, 99% pure) were produced by Fermalogic, Inc. Fermentations were performed in OFM1-2x starch broth (Oil-based Fermentation Medium with 2x Starch) per liter: toasted nutrisoy flour (ADM, Decatur, IL), 22 g; DifcoTM soluble starch, 30 g; CaCO3 powder (JT Baker, Phillipsburg, NJ), 3 g; MgSO4.7H2O (JT Baker), 0.5 g; FeSO4 7H2O (JT Baker), 15 mg; Soy oil, 50 ml (ADM). Diethyl methylmalonate 99% (a solvent) was obtained from Sigma-Aldrich (St. Louis, MO) and was added to cultures after autoclaving the medium. To achieve the 15 mM supplementation level, 64 μl of diethyl methylmalonate was added to 25 ml of broth and 1.25 ml of soy oil in a 250 ml shake flask.
Fermentations, bioassays, growth, oil utilization, and thin layer chromatography (TLC)
The fermentation, bioassay and TLC procedures used in this study were described previously by (Reeves et al. 2006). Shake flask fermentations were performed in triplicate for erythromycin production (Fig. 2) and duplicate (Fig. 3)The. Two hundred fifty ml shake flasks were used with 25 ml of fermentation broth per flask. For preparation of the thin layer chromatograms, two of the three fermentation extracts were spotted. For pH, samples were taken and measured on a Beckman model Φ34 pH meter and an average value was calculated from three fermentations. Culture viscosity was based on a visual assessment: a score of 1 for low (watery), 2 for intermediate, and 3 for high viscosity (Figs. 2 and 3). Relative cell pellet size was used for estimating growth (Fig. 3), and oil layer thickness was used for estimating oil utilization. The respective pellet and oil layers were measured following centrifugation of a sample of the culture broth in a 15-ml conical test tube.
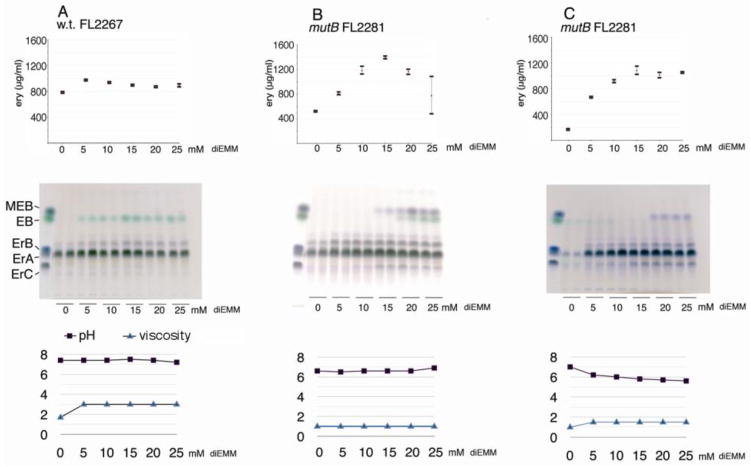
Fig. 2.
A comparison of the wild type (FL2267) and mutB (FL2281) strains showing how they respond differently to diethyl methylmalonate supplementation. A Effects of single batch diethyl methylmalonate supplementation on the wild type strain, FL2267, (experiment SFF05B-09) and B effects on FL2281, the mutB knockout strain (experiment SFF01A-09). Also shown is C, effects of daily 5 mM supplementation of diethyl methylmalonate on the mutB knockout strain FL2281 (experiment SFF11A-09). Top panel, bioassay results (error bars represent standard deviation of the mean); middle panel, TLC results; bottom panel, pH and viscosity at day 5. Abbreviations: ErA or ery, erythromycin A; ErB, ErC, erythromycins B and C; EB, erythronolide B; MEB, 3-O-α-mycarosylerythronolide B; diEMM, diethyl methylmalonate.
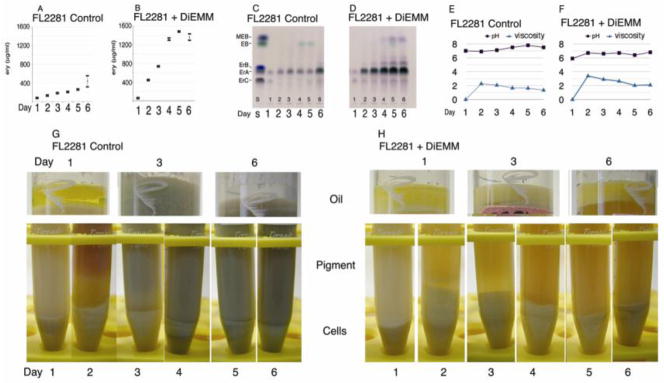
Fig. 3.
A comparison between diethyl methylmalonate unsupplemented and supplemented cultures of the mutB strain FL2281. A and B, erythromycin production, (error bars represent standard deviation from the mean, experiment SFF10B-09) C and D, thin layer chromatography of broth extracts; E and F, viscosity and pH; G and H, growth, pigmentation, and oil utilization.
Calculation of bioconversion efficiency for diethyl methylmalonate to erythromycin
The calculation for the bioconversion of diethyl methylmalonate into erythromycin A at a 15 mM concentration level was determined by taking the amount of erythromycin produced due to the addition of diethyl methylmalonate, which was 1.4 g/L, (Fig. 2B, 15 mM) minus the amount of erythromycin produced with no diethyl methylmalonate, which was 0.52 g/L (Fig 2B, 0 mM DiEMM control), with the difference being 0.88 g/L. Converting to millimolar units, 0.88 g/L is divided by 734 g/M (molecular weight of erythromycin A) to give 1.2 mM. Assuming that seven moles of diethyl methylmalonate are used for each mole of erythromycin A, 15mM was divided by 7, to give 2.14 mM. Since 2.14 moles of the “7-mer” diethyl methylmalonate went into making 1.2 mM of erythromycin A; this represents a molar bioconversion rate of 1.2 mM divided by 2.14 mM, or 56%.
Results
Diethyl methylmalonate supplementation of wild-type S. erythraea (FL2267)
In order to observe the effect of diethyl methylmalonate on the wild-type strain, shake flask fermentations were performed in triplicate in oil-based broth (OFM1-2x starch) supplemented with diethyl methylmalonate in concentrations ranging from 0 to 25 mM. After five days the cultures were analyzed for erythromycin concentration, pH, viscosity, and presence of non- or mono-glycosylated erythromycin precursors (EB or MEB, respectively).
The results show that without supplementation, the wild-type fermentations produced a mean of 790 μg/ml (SD = 32 μg/ml) of erythromycin. With supplementation, the yields went up slightly (Fig. 2A, top). Qualitative TLC analysis (Fig. 2A middle) showed that diethyl methylmalonate caused accumulation of the non-bioactive, non-glycosylated erythromycin precursor, EB. However, it is presumed that not all the diethyl methylmalonate was converted to EB because the intensity of the EB spots did not rise beyond the 15 mM level. The pH of the cultures were not affected by supplementation, but viscosity (measured at day five) was improved with supplementation at all levels.
Diethyl methylmalonate supplementation of S. erythraea mutB (FL2281)
As with the wild type strain, the FL2281 mutB strain was grown in triplicate fermentations in OFM1-2x starch broth supplemented at the beginning of the fermentation with diethyl methylmalonate in concentrations ranging from 0 to 25 mM. The same analysis was performed as above.
The results (Fig. 2B) show that diethyl methylmalonate supplementation caused a strong positive effect on erythromycin production at the 5, 10 and 15 mM levels. At the 15 mM level, the mutB fermentations rose to a mean of 1395 μg/ml (SD = 58 μg/ml) from a baseline of 520 μg/ml (SD = 38 μg/ml) at 0 mM, representing an increase in erythromycin production of over 250%.
When the mutB and wild type strains were compared, with both strains growing under their most productive conditions, (15 mM for the mutB strain, and 5 mM for the wild type) the mutB strain produced over 30% more erythromycin than the wild type strain. At the 15 mM concentration, only 64 μl of diethyl methylmalonate is required per 25 ml of broth (0.25% v/v), to cause this large increase. This represented a highly efficient molar conversion rate of diethyl methylmalonate to erythromycin of over 55% (Materials and Methods).
The mutB knockout strain did not accumulate non- or mono-glycosylated erythromycin precursors (MEB or EB) until the 15 mM supplementation level was reached, at which point the mono-glycosylated precursor MEB began to accumulate. At the 20 and 25 mM concentrations both EB and MEB accumulated.
Also seen was a strong negative effect on erythromycin production by diethyl methylmalonate at the 20 and 25 mM levels of supplementation (Fig. 2B), a result that indicated that higher concentrations of diethyl methylmalonate may be toxic to the mutB strain, or inhibitory to erythromycin production, or both. To circumvent this problem in the mutB strain, an experiment was set up to feed diethyl methylmalonate at smaller (5mM) doses on a daily basis instead of at one large dose at the beginning of the fermentation. The results (Fig. 2C) showed, however, that multiple daily dosing of lower levels of diethyl methylmalonate only improved the yield of the fermentation at the 25 mM level--at lower concentrations of diethyl methylmalonate there was no benefit.
Dmr and associated phenotypes of the mutB knockout strain FL2281
OFM1-2x starch fermentations of the mutB strain, with and without DiEMM supplementation, were compared at 24 hr intervals over a period of six days (Fig. 3) to study the effects of diethyl methylmalonate supplementation on erythromycin production and other phenotypes, including growth, viscosity, pigment production, and oil utilization. The defective growth and viscosity of the mutB strain were restored back to normal when diethyl methylmalonate was added to the medium (Fig. 3E and F). This restoration of normal growth is particularly visible at days 2 and 3, and 4 (Fig. 3G and H).
Culture pigmentation was also significantly affected by diethyl methylmalonate supplementation. FL2281 fermentations were pigmented red by day 2 and turned a dark grayish-green color at later time points (Fig. 3G). However, FL2281 cultures with diethyl methylmalonate supplementation showed no signs of pigmentation, that is, besides their normal amber color which is characteristic of the healthy wild type strain (Fig. 3H).
Finally, diethyl methylmalonate supplementation reduced the consumption of soybean oil in the medium. The soybean oil layer in the unsupplemented culture was completely depleted by the end of the five day fermentation. The oil layer of the supplemented culture became cloudier, but showed no visible reduction in thickness, indicating that it was not highly metabolized unchanged throughout the fermentation (Fig. 3G and H).
Erythromycin production was strongly improved by supplementation, as seen previously. The peak levels of production for the mutB strain were at day 5 (M = 1480 μg/ml, SD = 36 μg/ml) which were over 300% above the maximum production level of the mutB strain without supplementation at its peak level at day 6 (430 μg/ml, SD = 235 μg/ml). (Fig. 3A and B).
Discussion
Erythromycin strain and process improvement
In previous studies, genetic experiments indicated that methylmalonyl-CoA was a limiting factor for erythromycin biosynthesis (Reeves et al. 2004, 2006, 2007); therefore, in this study, experiments were performed to see whether direct supplementation of the medium with a di-ester of methylmalonate could improve the erythromycin fermentation.
The results of figures 2 and 3 show that the mutB strain FL2281 did respond to diethyl methylmalonate supplementation by producing significantly more erythromycin. When the mutB strain’s diethyl methylmalonate-responsive (Dmr) phenotype was measured in two separate experiments at the 15 mM level it resulted in a 250% increase (Fig. 2) and a 300% increase (Fig. 3) in erythromycin production. The 250% increase represented a 55% mole/mole conversion efficiency of diethyl methylmalonate into erythromycin (Materials and Methods). In striking contrast, the wild type strain FL2267 did not make significantly more erythromycin in response to diethyl methylmalonate. Since the mutB strain naturally produces approximately 40% less erythromycin than the wild type strain, diethyl methylmalonate not only brought the mutB strain up to the wild type level of production, but it took its production level up 30% beyond that. A discussion of how the mutB strain could use diethyl methylmalonate to create this improvement follows.
Starting with the wild type strain, in unsupplemented OFM1-2x starch medium, the results (Fig. 2A) show that diethyl methylmalonate is not converted into erythromycin, indicating that the polyketide precursor pool is not the (sole) limiting factor. Therefore, the model for the wild type strain shows a molar-balanced set of precursor pools leading to erythromycin production (Fig. 4A). In this case, no one precursor is limiting and therefore all precursor pools need to be raised together to cause a strain improvement. Global mutations that raise all precursor pools simultaneously are the type of mutation that is needed, and are most likely already incorporated into classically-produced commercial strains. When diethyl methylmalonate is added to the wild type strain, it simply feeds into the polyketide synthase (PKS) and causes the strain to produce more of the polyketide intermediate EB (Fig. 4B). A spot for EB can be seen on the TLC of the wild type strain as soon as even the lowest level of diethyl methylmalonate is added to the culture broth (Fig. 2A).
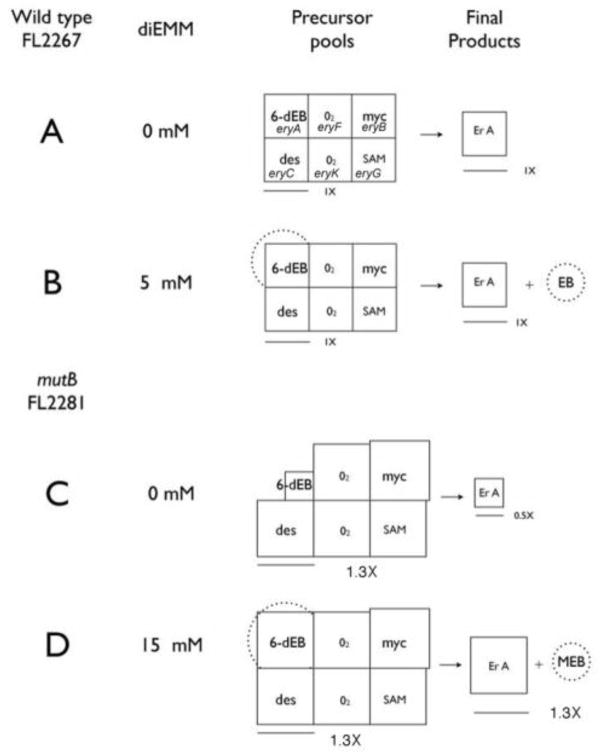
Fig. 4.
“Precursor pool” maps of erythromycin biosynthesis for the wild type (FL2267) and mutB knockout (FL2281) strains to explain data in Fig. 2. One mole of erythromycin (ErA, box under “final products”) is constructed by combining one mole of each of the six precursors (six boxes under “precursor pools”) going from the 6-dEB pool to the SAM pool. Boxes with size markers represent relative molar size of the precursor pools and panel A contains the label for the corresponding biosynthetic gene designation (Fig. 1). The assumption from the diagram is that the size of the ErA pool can never be bigger than the size of the smallest precursor pool (that is, the limiting factor pool). A and B: addition of diethyl methylmalonate to wild type (FL2267) strain. C and D: addition of diethyl methylmalonate to mutB (FL2281) strain. The diagram includes the explanation for why the supplemented mutB strain can make more erythromycin A than the wild type strain, because it is postulated to have larger non-polyketide precursor pool sizes (1.3x) than the wild type strain (1x). The diagram also provides an explanation for why the mutB strain accumulates MEB instead of EB at the 15 mM supplementation level (Fig. 2B), because the dTDP-mycarose precursor pool is predicted to be slightly larger than the dTDP-desosamine precursor pool. Abbreviations: 6-dEB, 6-deoxyerythronolide B; myc, dTDP-mycarose; des, desosamine; SAM, S-adenosylmethionine, DiEMM, diethyl methylmalonate.
The model for the mutB strain on the other hand shows a strain with an imbalance in the precursor pools, with the polyketide precursor pool being significantly smaller than the other five pools (Fig. 4C), which in turn, causes the strain to make less erythromycin. The five non-polyketide-related pools are also shown 30% larger than in the wild type strain, a condition whose origin is unknown, but is another postulated indirect phenotype of the mutB knockout. When diethyl methylmalonate is added to the mutB strain it is able to convert the diethyl methylmalonate to erythromycin because the other precursor pools are available in excess. Also, because these other pools are shown 30% larger in the mutB strain, the supplemented mutB strain can make up to 30% more erythromycin than the wild type strain, as reflected in the experimental data.
This model requires that diethyl methylmalonate first be converted to methylmalonate, which we propose would be performed by esterases native to Actinomycetes (Anthonsen et al. 1995; Fig. 1). The methylmalonate would then be ligated to coenzyme A using a native ATP-dependent ligase, homologous to the well-studied MatB from S. coelicolor (Hughes and Keatinge-Clay, 2011). A homolog for MatB exists in S. erythraea (SACE_ 3857) and has 65% identity to the amino acid sequence of the homologous protein from S. coelicolor.
The Dmr phenotype allows the strain to make more erythromycin through the addition of the diethyl methylmalonate to the medium. The Dmr phenotype has significant analogies to the previously described EryA phenotype (Weber et al. 1985, 1990). Although a mutant with the EryA phenotype cannot make EB due to a mutation in the PKS, it can absorb EB from an exogenous source and can convert that EB to erythromycin A, such as described in cooperative biosynthesis (cosynthesis) experiments (Weber et al. 1990; Fig. 5). The similarity between the EryA and Dmr phenotypes is that the polyketide precursor pathways are not fully functional in either strain, and yet both strains still produce and accumulate the non-polyketide precursors.
Fig. 5.
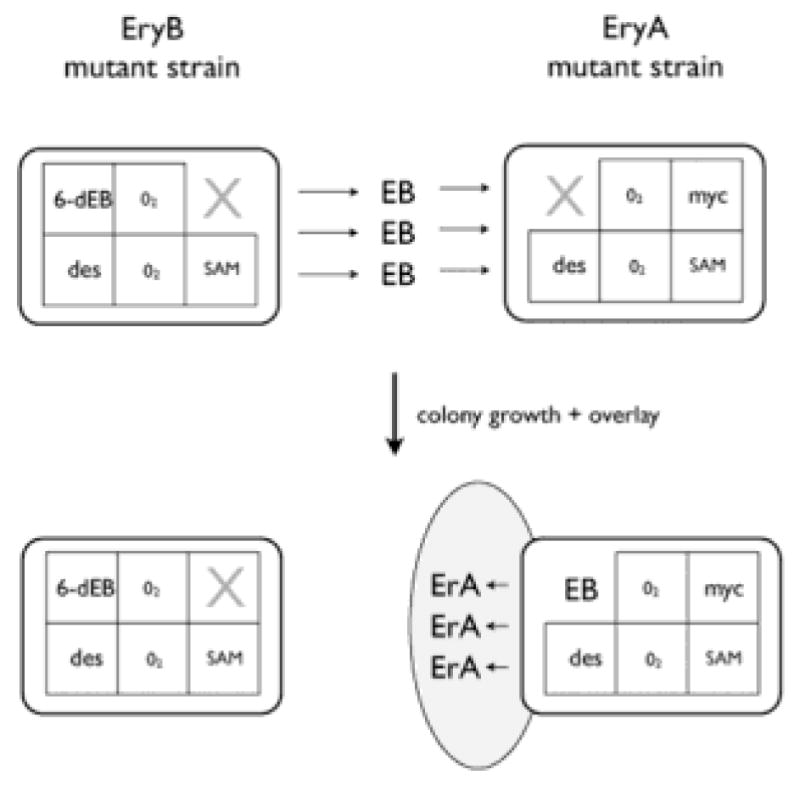
Cooperative biosynthesis of erythromycin A (ErA) by an EryA mutant (eryA PKS-knockout) growing next to an EryB mutant (blocked in one step of mycarose biosynthesis) on an agar plate surface (Weber et al., 1990). Neither the EryB or EryA mutants can make ErA by themselves; however, the EryA mutant can make erythromycin A using the EB that it obtains cooperatively from the EryB mutant growing next to it. The grey eliptical zone represents the zone of growth inhibition produced by the EryA mutant on the agar plate after overlay with a sensitive assay organism. The EryB mutant does not secrete a similar zone of erythromycin, and we assume this is because the deoxysugars are not secreted or exported from the EryA mutant.
If this model is correct, and S. erythraea does not actively balance or coordinately regulate erythromycin polyketide and non-polyketide precursor levels, then it would be useful to actively seek other mutations with the Dmr phenotype that specifically cause an increased accumulation of the non-polyketide precursor pools, in particular the pools for the two deoxythymidine diphosphate (dTDP)-deoxysugars, dTDP-L-mycarose and dTDP-D-desosamine (Fig. 6). Current commercial strains could theoretically harbor a silent Dmr phenotype, and they could be tested for this phenotype in an oil-based medium by simply adding diethyl methylmalonate to the fermentation broth to see if erythromycin levels increase.
Fig. 6.
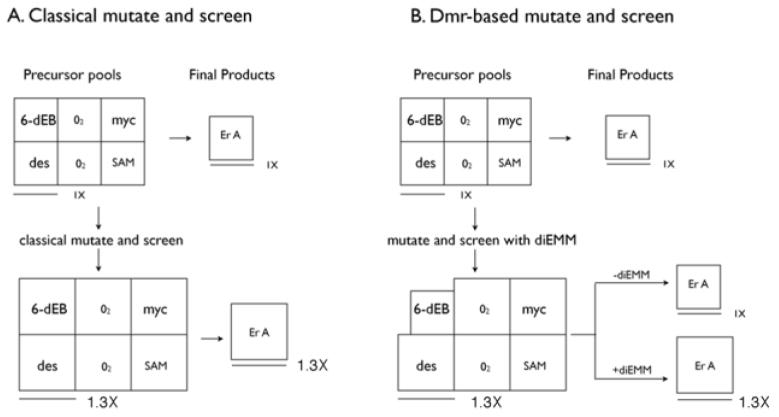
Comparison of the results obtained from classical mutate and screen program and the Dmr-based mutate and screen program. A. Classical mutate and screen produces global mutations that increase levels of all six precursor pools. No supplementation is needed to produce the strain improvement effect. B. Dmr-based screening produces Dmr-dependent mutations require that diethyl methylmalonate is added to the growth medium to see the strain improvement effect. These may be mutations, for example, that specifically increase the deoxysugar precursor pool levels. Abbreviations: ErA, erythromycin A; 6-dEB, 6-deoxyerythronolide B; myc, dTDP-mycarose; des, desosamine; SAM, S-adenosylmethionine; diEMM, diethyl methylmalonate.
It is more likely that mutations conferring Dmr phenotypes will have to be incorporated into existing commercial strains by directed screening for such a mutation (Fig. 6). Fortunately the Dmr phenotype does not require that broths be screened for dTDP-L-mycarose or dTDP-D-desosamine levels. Traditional screening for erythromycin titers can be used as long as diethyl methylmalonate is added to the fermentation broth used in the screen (Fig. 6B).
Soy bean oil utilization
Another finding from this study was that diethyl methylmalonate supplementation (0.13% v/v) in the mutB fermentation strongly inhibited or stopped soybean oil (5% v/v) consumption during the fermentation, most likely because diethyl methylmalonate is metabolized preferentially and is converted more efficiently into erythromycin than is soybean oil by the strain.
The Dmr phenotype suggests a new class of mutations could lead to better strains
A diethyl methylmalonate-responsive (Dmr) phenotype is described for the mutB knockout strain which causes it to respond to this solvent with a significant (250 – 300%) increase in erythromycin production. Previously mutations that caused overproduced the erythromycin deoxysugars were not useful for strain improvement unless they concurrently led to overproduction of the polyketide precursors. The results of this study, however, suggest that mutations that affect deoxysugar production alone could also have significant value. First, they have value because this type of mutation would be absent from current commercial strains, and secondly they could be combined with with the diethyl methylmalonate process improvement, and known tailoring improvements (Chen et al 2008; Zou et al 2010), to create a commercial fermentation that is better than those currently available.
Acknowledgments
The authors acknowledge Andrey Fedashtchin, Noelle Kwan, and Eric Miller for helpful discussions. This work was supported by the National Institute for General Medical Sciences GM079945 through a Small Business Innovation Research award to Fermalogic.
References
- Anthonsen HW, Baptista A, Drabløs F, Martel P, Petersen SB, Sebastião M, Vaz L. Lipases and esterases: a review of their sequences, structure and evolution. Biotechnology Annual Review. 1995;1:315–371. doi: 10.1016/s1387-2656(08)70056-5. [DOI] [PubMed] [Google Scholar]
- Boghigian BA, Zhang H, Pfeifer BA. Multi-factorial engineering of heterologous polyketide production in Escherichia coli reveals complex pathway interactions. Biotechnol Bioeng. 2011;108:1360–1371. doi: 10.1002/bit.23069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brikun IA, Reeves AR, Cernota WH, Luu MB, Weber JM. The erythromycin biosynthetic gene cluster of Aeromicrobium erythreum. J Ind Microbiol Biotechnol. 2004;31:335–344. doi: 10.1007/s10295-004-0154-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Deng W, Wu JQ, Qian JC, Chu J, Zhuang YP, Zhang SL, Liu W. Genetic modulation of the overexpression of tailoring genes eryK and eryG leading to the improvement of erythromycin a purity and production in Saccharopolyspora erythraea fermentation. Appl Environ Microb. 2008;74:1820–1828. doi: 10.1128/AEM.02770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AJ, Keatinge-Clay A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB. Chemistry & Biology. 2011;18:165–76. doi: 10.1016/j.chembiol.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- Kwan DH, Tosin M, Schläger N, Schulz F, Leadlay PF. Insights into the stereospecificity of ketoreduction in a modular polyketide synthase. Org Biomol Chem. 2011 Apr 7;9(7):2053–6. doi: 10.1039/c1ob00022e. Epub 2011 Feb 21. [DOI] [PubMed] [Google Scholar]
- Lombo F, Pfeifer B, Leaf T, Ou S, Kim YS, Cane DE, Licari P, Khosla C. Enhancing the atom economy of polyketide biosynthetic processes through metabolic engineering. Biotechnol Prog. 2001;17:612–617. doi: 10.1021/bp010045j. [DOI] [PubMed] [Google Scholar]
- Miller ES. Cloning vectors, mutagenesis, and gene disruption (ermR) for the erythromycin-producing bacterium Aeromicrobium erythreum. Appl Environ Microbiol. 1991;57 :2758–61. doi: 10.1128/aem.57.9.2758-2761.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas W. Production of erythromycin with Saccharopolyspora erythraea. In: Barredo JL, editor. Methods in Biotechnology, Vol. 18: Microbial Processes and Products. Humana Press; Totowa, NJ: 2005. pp. 65–89. [Google Scholar]
- Mo SJ, Ban Y-H, Park JW, Yoo YJ, Yoon YJ. Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. J Ind Microbiol Biotechnol. 2009;36:1473–1482. doi: 10.1007/s10295-009-0635-7. [DOI] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Pfeifer B, Hu Z, Licari P, Khosla C. Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl Environ Microbiol. 2002;68 :3287–3292. doi: 10.1128/AEM.68.7.3287-3292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queener SW, Lively DH. Screening and selection for strain improvement. In: Demain AL, Soloman NA, editors. Manual of Industrial Microbiology and Biotechnology. Amer. Society for Microbiology; Washington DC: 1986. pp. 155–169. [Google Scholar]
- Rawlings B. Type I polyketide biosynthesis in bacteria (Part A--erythromycin biosynthesis) Nat Prod Rep. 2001;18:190–227. doi: 10.1039/b009329g. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Cernota WH, Brikun IA, Wesley RK, Weber JM. Engineering precursor flow for increased erythromycin production in Aeromicrobium erythreum. Metab Eng. 2004;6:300–312. doi: 10.1016/j.ymben.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM. Effects of methylmalonyl CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. J Ind Microbiol Biotechnol. 2006;33:600–609. doi: 10.1007/s10295-006-0094-3. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Weber JM. Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab Eng. 2007;9:293–303. doi: 10.1016/j.ymben.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Chu J, Zhuang Y, Zhang L, Zhang S. Improved production of erythromycin A by expression of a heterologous gene encoding S-adenosylmethionine synthetase. Appl Microbiol Biotechnol. 2007;75:837–842. doi: 10.1007/s00253-007-0894-z. [DOI] [PubMed] [Google Scholar]
- Weber JM, Wierman CK, Hutchinson CR. Genetic analysis of erythromycin production in Streptomyces erythreus. J Bacteriol. 1985;164:425–433. doi: 10.1128/jb.164.1.425-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JM, Leung JO, Maine GT, Potenz RHB, Paulus TJ, DeWitt JP. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol. 1990;172:2372–2383. doi: 10.1128/jb.172.5.2372-2383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T. Making E. coli an Erythromycin Production Plant. Chemistry & Biology. 2010;17:1168–9. doi: 10.1016/j.chembiol.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang Q, Deng W, Qian J, Zhang S, Liu W. An artificial attB site for specific recombination facilitates genetic manipulations towards improving the erythromycin A production in an industrial Saccharopolyspora erythraea strain. Appl Environ Microbiol. 2011 Aug 12; doi: 10.1128/AEM.06034-11. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Wu J, Skalina K, Pfeifer BA. Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chemistry & Biology. 2010;17:1232–40. doi: 10.1016/j.chembiol.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Zou X, Zeng W, Chen C, Qi X, Qian J, Chu J, Zhuang Y, Zhang S, Li W. Fermentation optimization and industrialization of recombinant Saccharopolyspora erythraea strains for improved erythromycin a production. Biotechnol Bioproc E. 2010;15:959–968. [Google Scholar]