CONSPECTUS
Enzymes catalyze a particular reaction in cells, but only a few control the rate of this reaction and the metabolic pathway that follows. One specific mechanism for such enzymatic control of a metabolic pathway involves molecular feedback, whereby a metabolite further down the pathway acts at a unique site on the control enzyme to alter its activity allosterically. This regulation may be positive or negative (or both), depending upon the particular system. Another method of enzymatic control involves the cooperative binding of the substrate, which allows a large change in enzyme activity to emanate from only a small change in substrate concentration. Allosteric regulation and homotropic cooperativity are often known to involve significant conformational changes in the structure of the protein.
Escherichia coli aspartate transcarbamoylase (ATCase) is the textbook example of an enzyme that regulates a metabolic pathway—namely, pyrimidine nucleotide biosynthesis—by feedback control and by the cooperative binding of the substrate, l-aspartate. The catalytic and regulatory mechanisms of this enzyme have been extensively studied. A series of X-ray crystal structures of the enzyme in the presence and absence of substrates, products, and analogs have provided details, at the molecular level, of the conformational changes that the enzyme undergoes as it shifts between its low-activity, low-affinity form (T state) to its high-activity, high-affinity form (R state). These structural data provide insights into how this enzyme not only catalyzes the reaction between l-aspartate and carbamoyl phosphate to form N-carbamoyl-l-aspartate and inorganic phosphate, but also how the allosteric effectors modulate this activity.
In this Account, we summarize studies on the structure of the enzyme and describe how these structural data provide insights into the catalytic and regulatory mechanisms of the enzyme. The ATCase-catalyzed reaction is regulated by nucleotide binding some 60 Å from the active site, inducing structural alterations that modulate catalytic activity. The delineation of the structure and function in this particular model system will help in understanding the molecular basis of cooperativity and allosteric regulation in other systems as well.
Early History
Biosynthetic control by a feedback inhibition mechanism was established in 1956 by Yates and Pardee1 for pyrimidine nucleotides, and by Umbarger2 for isoleucine. Specifically for pyrimidine nucleotide biosynthesis, Yates and Pardee1 showed that the formation of N-carbamoyl-L-aspartate was feedback-inhibited by the end product of the pathway, CTP, in a specific and potent manner.
The enzyme responsible for the formation of N-carbamoyl-L-aspartate is aspartate transcarbamoylase (E.C.2.1.3.2, aspartate carbamoyltransferase, ATCase), first purified and crystallized in 1960 by Shepherdson and Pardee.3 Although the catalytic reaction was actually deduced in 1955–19564–6 (Fig. 1). Shepherdson and Pardee3 estimated the molecular weight of the E. coli enzyme to be approximately 220,000 ± 40,000 Da, based upon sedimentation velocity measurements. Heating the enzyme at 60°C for 4 min was sufficient to render the enzyme insensitive to feedback inhibition without loss of catalytic activity.7 Then, in 1964, Gerhart and Pardee8 reported that the catalytic and regulatory sites were distinct. In a 1963 summary, Gerhart and Pardee9 described CTP inhibition and ATP activation, and correctly suggested that these nucleotides affected substrate affinity in an indirect manner.
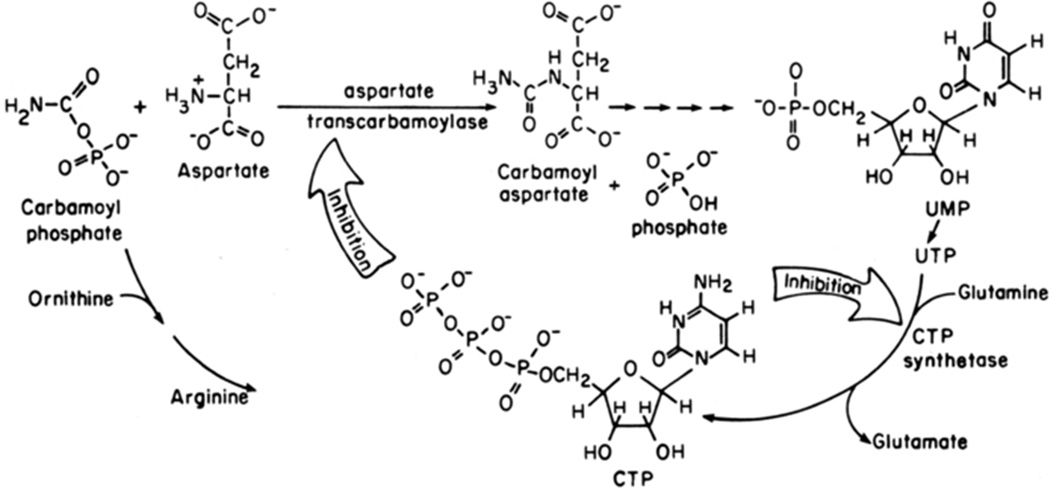
FIGURE 1.
Aspartate transcarbamoylase (ATCase) catalyzes the committed step, the condensation of carbamoyl phosphate and aspartate to form carbamoyl aspartate and inorganic phosphate, in pyrimidine nucleotide biosynthesis for E. coli. Carbamoyl aspartate continues through the pathway leading to the formation of the nitrogenous base in pyrimidine nucleotides. ATCase is feedback inhibited by CTP and the combination of CTP plus UTP. This figure47 has been reproduced here with permission.
Note to editor: I have obtained rights to reuse this figure via Rightslink.
A more exact molecular weight of 310,000 Da was determined in 1965.10,11 Gerhart and Schachman11 demonstrated that the ATCase holoenzyme could be dissociated into two types of subunits, one with a molecular weight of ~96,000 Da and the other with a molecular weight of ~30,000 Da. Furthermore, the larger subunit retained catalytic activity (the catalytic subunit) while the smaller subunit exhibited no activity while it contained the nucleotide binding sites (regulatory subunit). However these same studies11 incorrectly proposed that the holoenzyme was composed of two catalytic subunits and four regulatory subunits.
The aspartate saturation curve for E. coli ATCase is not hyperbolic as expected for an enzyme that obeys Michaelis-Menten kinetics; rather the saturation curve is cooperative7 exhibiting a pH dependent Hill coefficient.8,12 In 1965, the first model for the cooperative kinetic properties of ATCase was proposed by Monod, Wyman and Changeux,13 which required that the enzyme be in a dynamic equilibrium between a low-activity, low-affinity T state and a high-activity, high-affinity R state. The existence of such a dynamic equilibrium was demonstrated for a mutant ATCase in 2007.14
In 1971 Collins and Stark15 synthesized a specific and potent inhibitor of ATCase, N-phosphonacetyl-L-aspartate (PALA). A comparison of the molecular formulas of carbamoyl phosphate (CP) plus L-aspartate (Asp) with the bisubstrate “transition state analogue” PALA is shown in Fig. 2. The ability of PALA to enhance the activity of ATCase at low concentrations of aspartate, in the presence of a saturating concentration of CP, indicates that PALA is able to shift the equilibrium from the T to the R state, a finding that will be important for the determination of the structure of the R state of the enzyme.
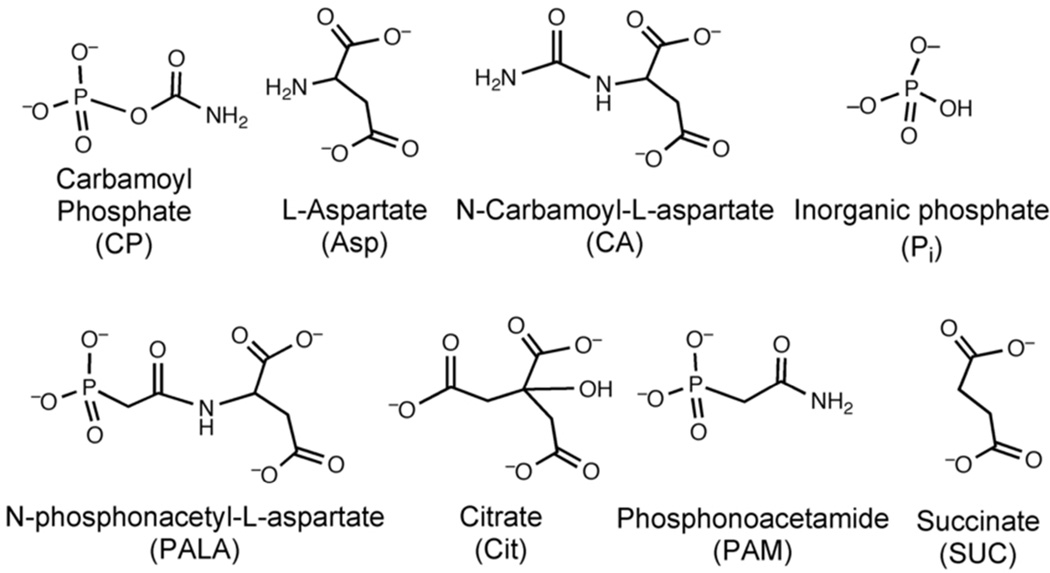
FIGURE 2.
The structures of carbamoyl phosphate (CP), L-aspartate (Asp), N-carbamoyl-L-aspartate (CA), phosphate (Pi), the bisubstrate analogue N-phosphonacetyl- L-aspartate (PALA), the N-carbamoyl-L-aspartate analogue citrate (Cit), the CP analogue phosphonoacetamide (PAM), and the Asp analogue succinate (SUC).
The Quaternary Structure of ATCase
Structural studies in the Lipscomb Laboratory began on the E. coli enzyme in 1967 following a colloquium by J. C. Gerhart, during which the details of a new purification procedure for the holoenzyme as well as the catalytic and regulatory subunits16 was presented. In 1968 K. Weber determined the amino acid sequence of the regulatory subunit17 and used this information to calculate a molecular weight of ~17,000 Da. These data implied that the holoenzyme was composed of six regulatory chains. This fact was combined with the subunit molecular weights,10,11 and crystallographic studies of two crystal forms18 to deduce the quaternary structure of ATCase. One of the crystal forms requires a molecular three-fold axis whereas the other requires at least one non-coincident two-fold axis in the molecule, thus necessitating a c6r6 quaternary structure for the holoenzyme.18 The relative arrangement of the c3 and r2 units in c6r6 was found in the (idealized) R32 T-state crystal form at 5.5Å resolution.19 The symmetry of c6r6 is D3 (Fig. 3, one three-fold axis and three two-fold axes) or the slightly lower C3 (one three-fold axis) in the presence of the allosteric inhibitor CTP or CTP plus UTP.20
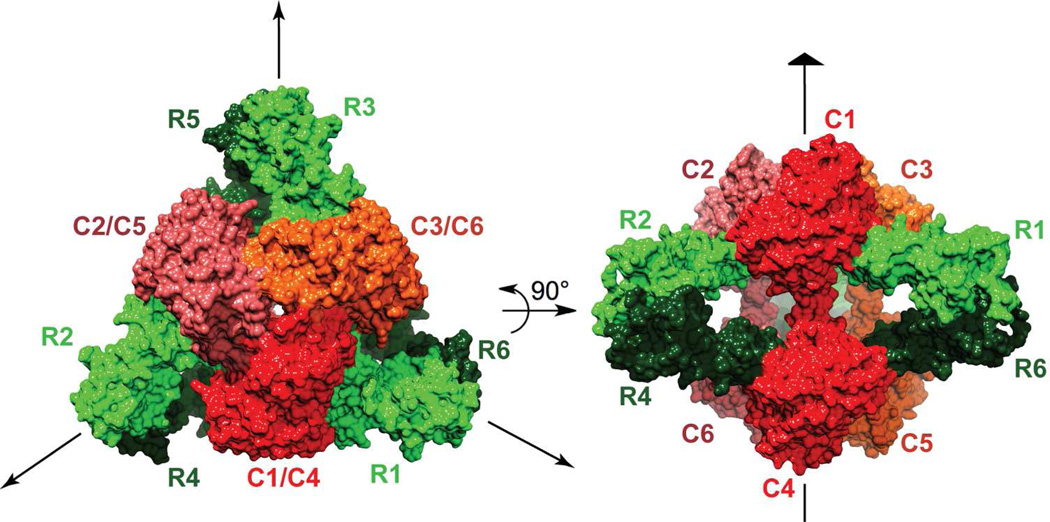
FIGURE 3.
Schematic representation of the quaternary structure of ATCase. Catalytic chains C1-C2-C3 and C4-C5-C6 correspond to the two catalytic trimers while the regulatory chains R1–R6, R2–R4 and R3–R5 correspond to the three regulatory dimers. This figure was drawn with Chimera.48
The Three-dimensional Structure of ATCase
The resolution of the structure of the unliganded E. coli ATCase improved to 2.6Å.21,22 Simultaneously, structural studies were pursued of the T-state enzyme in the presence of ATP or CTP,22 the R-state enzyme in the presence of PALA (Fig. 2) at 2.1 Å resolution,23 and the R-state enzyme with ATP or CTP bound,24 along with additional studies of the T and R states with a variety of substrates and substrate analogues bound.
The secondary structural elements of one catalytic and one regulatory chain are shown in Fig. 4. Each catalytic chain is composed of two folding domains; the Asp domain, primarily involved in the binding of aspartate and the CP domain, primarily involved in the binding of carbamoyl phosphate. Each regulatory chain is also composed of two folding domains: the Zn domain, primarily involved in the binding of the zinc cofactor, and the Al domain, primarily involved in the binding of allosteric effectors. The Asp and CP domains of the catalytic chain, as well as the Al domain of the regulatory chain, are each classified as α/β.25 The Zn domain of the regulatory chain is classified as rubredoxin-like with a metal (zinc or iron) bound that contains usually two CX(n)C motifs.25
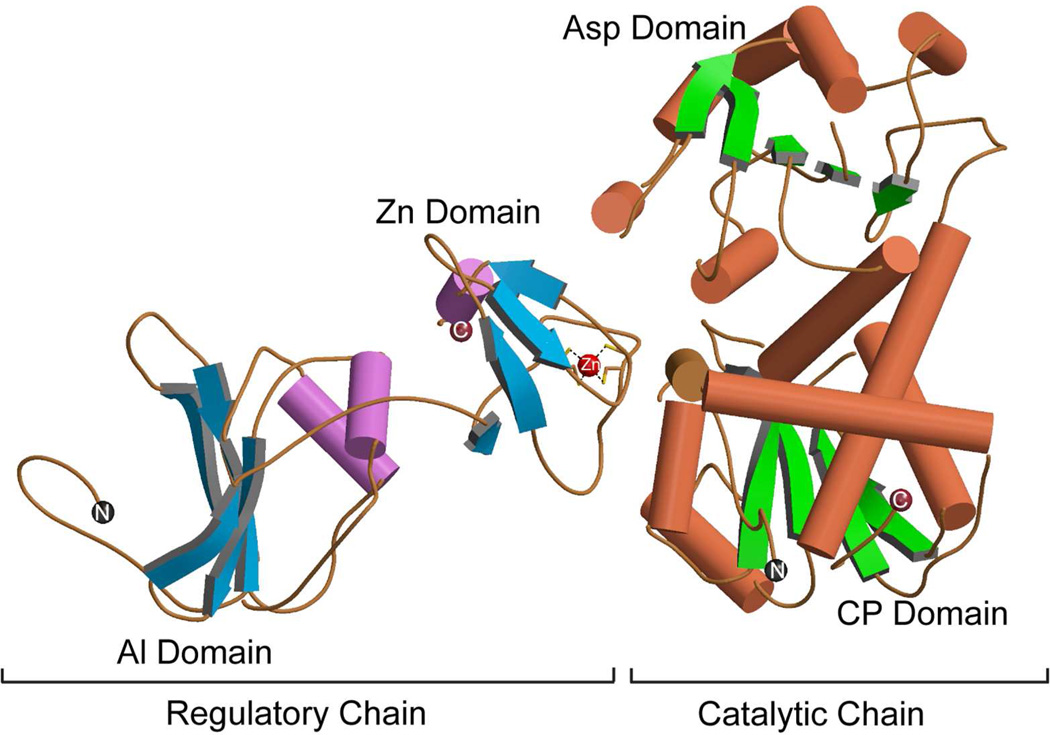
FIGURE 4.
The secondary structural elements of one catalytic and one regulatory chain of E. coli ATCase in the T-state. The N- and C-termini of the catalytic and regulatory chains are indicated, as well as the Zn atom in the regulatory chain that is coordinated tetrahedrally to four cysteine residues. Each catalytic chain is composed of an aspartate (Asp) and a carbamoyl phosphate (CP) domain. Each regulatory chain is composed of an allosteric (Al) and a zinc (Zn) domain. This figure was drawn with MOLSCRIPT49 using data from PDB entry 1ZA1.
The Structure of the ATCase in the T and R States
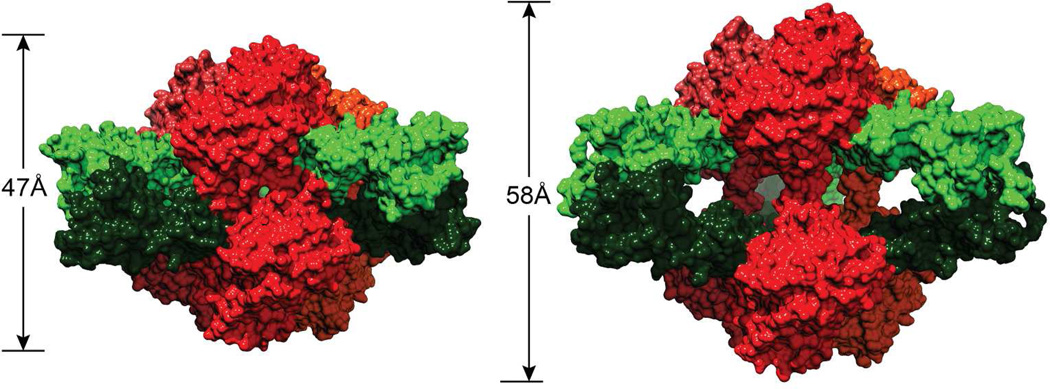
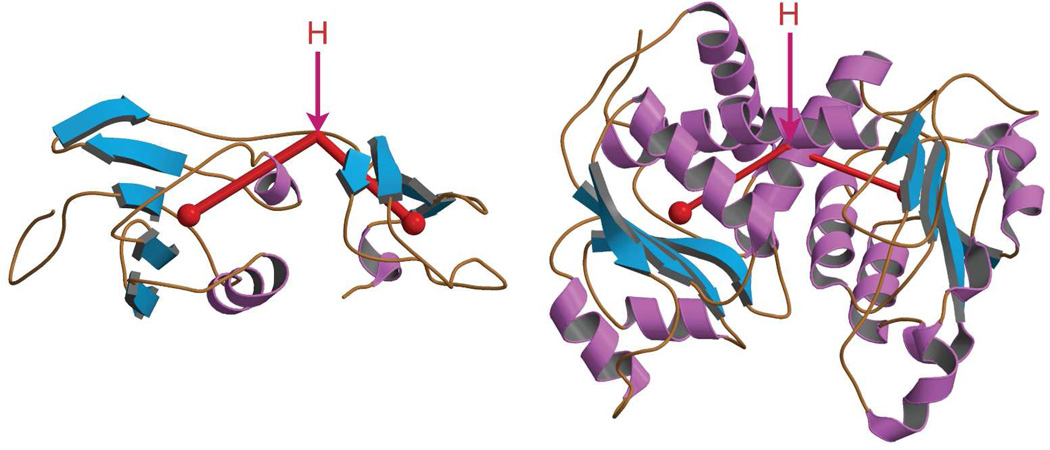
When both substrates are present at physiological concentrations they convert ATCase from the T form to the R form (Fig 5).21,26,27 The same change in quaternary structure can also be induced by the binding of PALA. The changes in the molecular structure between the T and R states include an elongation of 11Å along the molecular three-fold axis. In this transformation there is a rotation of one catalytic trimer c3 relative to the other c3 by 12°, and a rotation of each of the three regulatory dimers about the approximate two-fold axes by 15°. Thus the molecular expansion occurs only along the three-fold axis, with no elongation along the three molecular two-fold directions. The positions of the domains of the catalytic and regulatory chains undergo structural alterations relative to each other. The two domains of the catalytic chain close by 6.8° while the two domains of the regulatory chain open by 1.7° between the T and R states (Fig. 6).
FIGURE 5.
The ATCase holoenzyme in the T (left) and R (right) structures. The catalytic chains are shown in shades of red and the regulatory chains are shown in shades of green. The length of the molecule along the three-fold axis is shorter in the T form than in the R form. This figure was drawn with Chimera.48
FIGURE 6.
Secondary structure of a regulatory chain (left) and a catalytic chain (right) of ATCase. The angle between the domains is measured from the center of mass of each domain (red dot) and a hinge point (H).
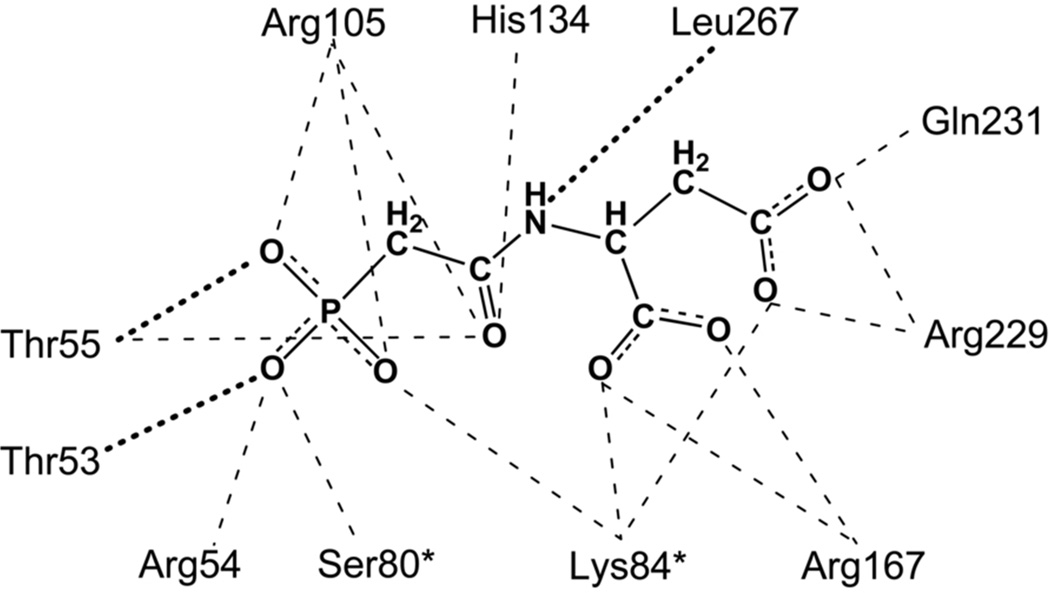
The interactions between the R-state enzyme and PALA are shown in Fig. 7. Ser80 and Lys84 from an adjacent catalytic chain interact with PALA,23 thus each active site is composed of residues from two adjacent chains. Site-specific mutagenesis28 and hybrid experiments29 confirm that the residues from the adjacent catalytic chain are required for catalytic activity.
FIGURE 7.
The interactions between PALA and ATCase in the R-state.27 Dotted lines correspond to interactions with the backbone of the protein. An asterisk after the residue number indicates that it is donated into the active site from the adjacent chain.
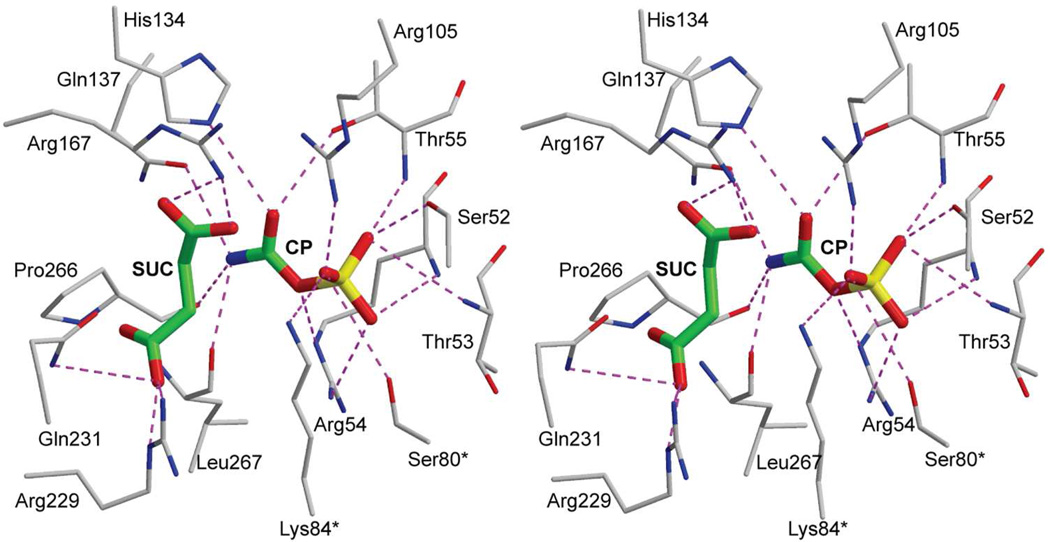
The R-state structure with bound CP and succinate, an analogue of the substrate Asp, is shown in Fig. 8.30 The interactions between the enzyme and the natural substrate CP are virtually identical to those between the enzyme and the phosphonate portion of PALA. However the amide of CP interacts with the side chain of Gln137 and the oxygen between the phosphorus and the carbonyl carbon interacts with Arg54. Using molecular modeling Asp was docked into the enzyme•CP complex. The lowest energy pose has the amino group of Asp within 2.8 Å of the carbonyl carbon of CP, positioned correctly for the nucleophilic attack, which would lead to the formation of the proposed tetrahedral intermediate.31
FIGURE 8.
Stereoview of succinate (SUC) and CP bound in the active site of ATCase in the R state. An asterisk after the residue number indicates that it is donated into the active site from the adjacent chain. This figure was drawn with MOLSCRIPT.49
Structural studies of the enzyme•PALA, enzyme•CP•succinate and enzyme•Pi•citrate complexes were used to model the tetrahedral intermediate and the transition state of the reaction.32 The model of the tetrahedral intermediate bound in the active site of ATCase shows three additional interactions than are observed between the enzyme and PALA (Fig. 9). In addition the model of the tetrahedral intermediate shows significantly more interactions than the summation of interactions between the enzyme and the various combinations of substrates and substrate analogues. Based on the tetrahedral intermediate model the enzyme binds the tetrahedral intermediate, and thus the transition state, significantly more strongly than the substrates or products.
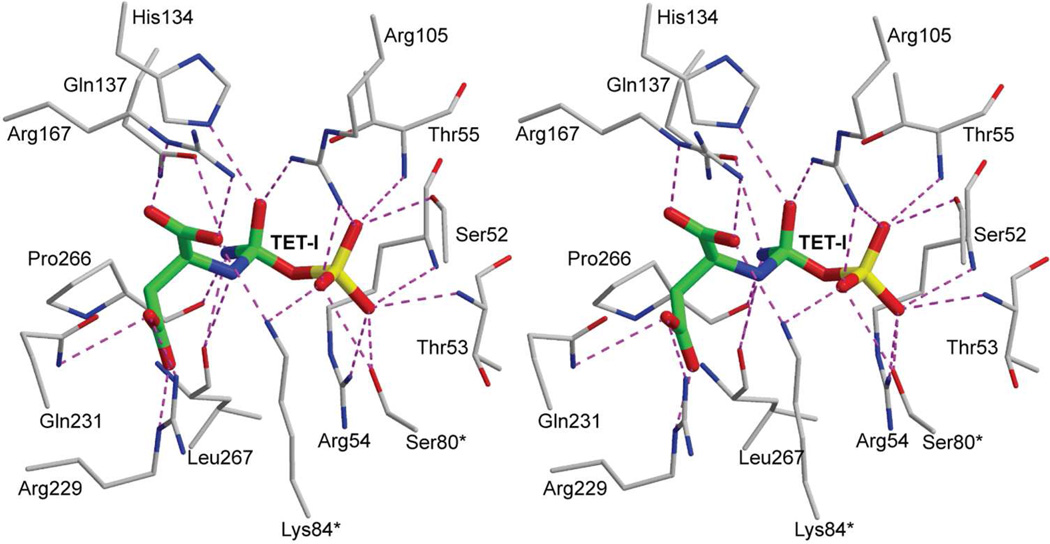
FIGURE 9.
Stereoview of the proposed tetrahedral intermediate (TET-I) bound in the active site of ATCase in the R state. An asterisk after the residue number indicates that it is donated into the active site from the adjacent chain. This figure was drawn with MOLSCRIPT.49
In order to approximate and thereby observe the conformation of the R-state enzyme active site in the presence of products, the structure of the enzyme in the presence of citrate, an analogue of N-carbamoyl-L-aspartate (Fig. 2), plus the product phosphate was determined after displacement of PALA from R-state crystals.33 The carboxylate groups of citrate interact with His134, Arg167, Gln231 and Arg229 (Fig. 10). Lys84, from the adjacent chain, interacts with both citrate and phosphate acting as a bridge between the two molecules. A phosphate oxygen is 3Å away from the carbon in citrate that would correspond to the α-carbon in carbamoyl aspartate. These results imply that this complex represents a reasonable model for the products bound in the active site.33
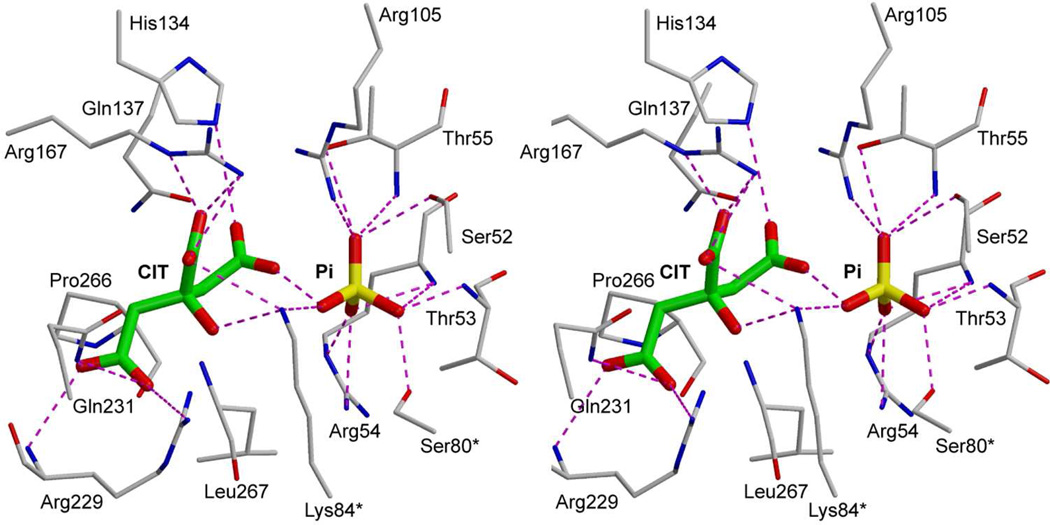
FIGURE 10.
Stereoview of citrate (CIT) and phosphate (Pi) bound in the active site of ATCase in the R state. An asterisk after the residue number indicates that it is donated into the active site from the adjacent chain. This figure was drawn with MOLSCRIPT.49
After the collapse of the oxyanion formed in the transition state, the products carbamoyl aspartate and phosphate become separate as seen in the model structure with citrate and phosphate bound (Fig. 10). The weaker-binding carbamoyl aspartate departs the active site releasing Lys84, which may coincide with the opening of the 240’s loop. The opening of the 240’s loop is necessary to provide a path for the release of carbamoyl aspartate followed by phosphate from the R-state active site, as the active site in the presence of the tetrahedral intermediate shows no opening to the solvent. After phosphate is released the catalytic cycle can begin again with the binding of CP. Time-resolved X-ray scattering studies have shown that as long as substrates are available the enzyme remains in the R state, and only returns to the T state after the substrates are exhausted.34
The Active Site and Mechanistic Aspects
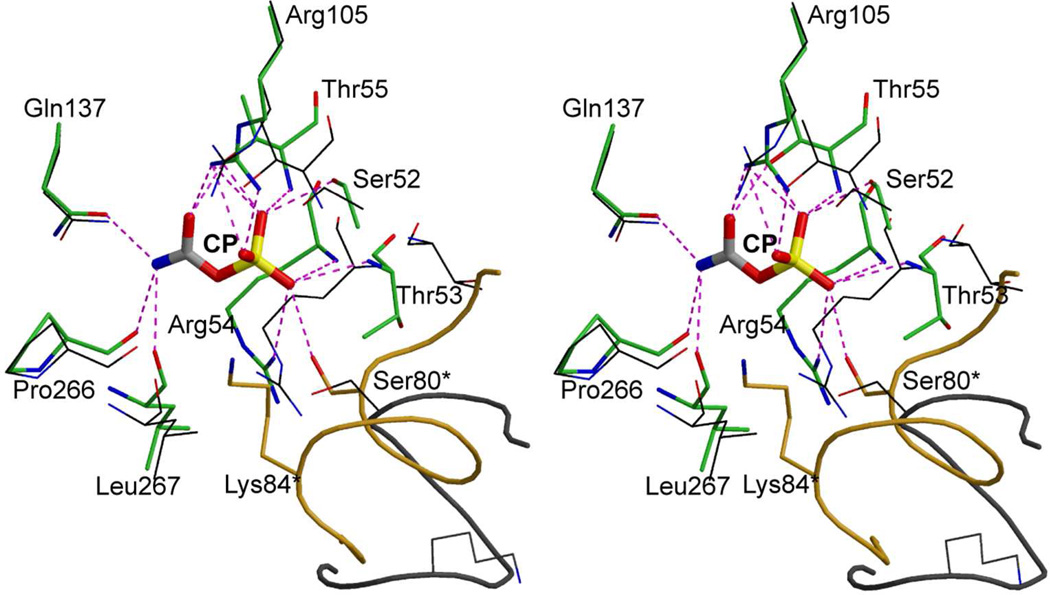
The catalytic mechanism is ordered: CP binds before aspartate; and carbamoyl aspartate leaves before phosphate.35 When CP,31 or CP analogues such as PAM (Fig. 2) bind,36 the enzyme remains in the T state and only tertiary-level conformational changes occur. The largest conformational change that occurs upon the binding of CP involves a reorganization of the 80’s loop that positions Ser80 and Lys84 into the active site. (Fig. 11). In addition, there are some smaller conformational changes involving motions of the 50’s and 240’s loops. The conformational changes induced by the binding of CP create a binding site for aspartate, not only structurally but electrostatically as well (Fig. 12). These conformational changes dramatically alter the electrostatic properties of the active site; they essentially create a positively charged binding pocket for aspartate, the studies of which have provided a mechanism to account for the ordered binding of the substrates.31
FIGURE 11.
Stereoview comparison of the active site of E. coli ATCase in the presence of CP (green carbons, thick) and in the absence of CP (black carbons, thin). The 80's loop from the adjacent chain (residue numbers with asterisks] in the presence of CP (gold carbons, thick) and in the absence of CP (gray carbons, thin) is represented as a coil with Ser80* and Lys84* shown. This figure was drawn with MOLSCRIPT.49
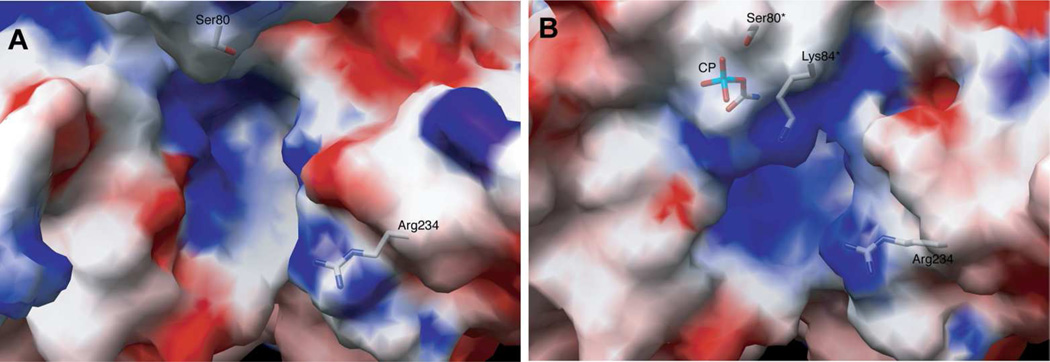
FIGURE 12.
(A) Electrostatic potentials mapped onto the surface of ATCase in the absence (A) and the presence (B) of CP. The positions of Ser80*, Lys84* and Arg234 and CP are overlaid onto the electrostatic map (white carbons, sticks). This figure31 has been reproduced here with permission.
Note to editor [no copyright transfer needed for author to reuse figure PNAS, 102, 8881–8886 (2005)]
The three additional interactions observed in the model of the tetrahedral intermediate bound to the enzyme (Fig. 9) compared to the structure of the enzyme•PALA complex involves interactions between the side chain of Gln137 and the backbone carbonyl oxygen of Pro266 to the amino group on the tetrahedral carbon and the side chain of Arg54 with the ester oxygen between the phosphorus and the tetrahedral carbon. When either Gln137 or Arg54 is replaced by Ala the consequences are extraordinary.37 For the Q137A enzyme, the concentration of CP required to attain one-half of the maximal activity increases by 210-fold. Although Gln137 is not involved directly in binding aspartate the corresponding value for aspartate increases by 76-fold. The extremely reduced affinity for CP of the Q137A enzyme along with the near abolition of aspartate binding points to the critical role that CP plays in preparing the active site for the binding of Asp. The R54A mutation results in a 17,000-fold loss in activity without major influence on the substrate affinity,37 suggesting a role of this residue in transition state stabilization.
The binding of aspartate induces both tertiary and quaternary conformational changes. At the tertiary level, the 240's loop rearranges to position the side chains necessary for catalysis in their high-activity, high-affinity positions (Fig. 13). However, due to steric constraints the movement of the 240's loop cannot occur without the occurrence of a quaternary conformational change (see Fig. 5). The repositioning of the 240's loop and the closure of the two domains of each catalytic chain provide an environment favorable for catalysis. The attack of the amino nitrogen of aspartate on the amide carbon of CP, which forms the tetrahedral intermediate, is enhanced by the binding of Arg105, His134 and Thr5537 to the carbonyl oxygen. The carbonyl oxygen of carbamoyl phosphate, as indicated by the schematic of PALA binding (see Figure 7), allows these three residues to influence substantially the affinity of the enzyme•CP complex for aspartate.
FIGURE 13.
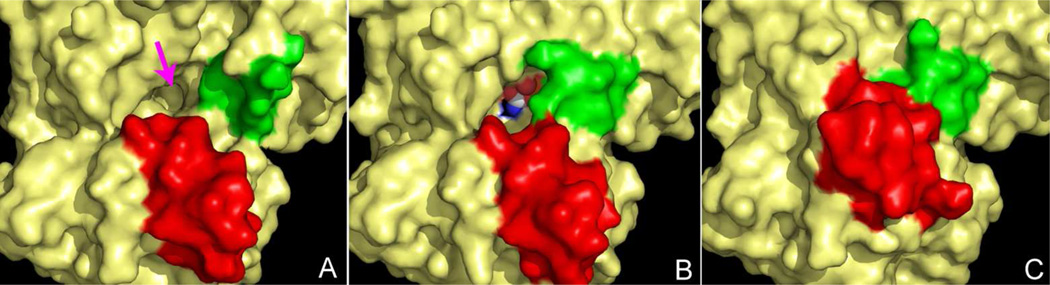
Surface of ATCase near the active site detailing the 80's loop (green) and the 240's loop (red) during catalysis. (A) Conformation of the enzyme before substrates are bound; the approximate position of the active site is indicated by the magenta arrow. (B) Conformation of the enzyme after the binding of CP, which is shown in the active site as a CPK model (white carbon). The 80's loop (green) rearranges to help create the binding site for aspartate. (C) Upon the binding of aspartate the 240's loop (red) undergoes a dramatic conformational change, forcing the substrates toward each other. Solvent is not accessible to the active site in this conformation.
In order for the reaction to occur, the amino group of aspartate must lose two protons starting from –NH3 +. Because of the electropositive field microenvironment encountered by the – NH3 + group, the first proton is lost as aspartate binds. One possibility for the fate of the second proton is that it is captured by the leaving phosphate.32 Another possibility for loss of the second proton involves transfer to either Arg105 or Lys84, the latter being contributed into the active site from the adjacent catalytic chain. It would be expected, based upon the electropositive environment of the active site, that the guanidinium group of Arg105 or the ε-amino group of Lys84 would initially be deprotonated, allowing either one to accept a proton. The roles of Arg105 and Lys84 in the function of the enzyme has been probed via mutagenesis. The R105A and K84N mutations result in a 1,100-fold37 and 1,200-fold23 reduction in activity, respectively.
In models of the tetrahedral intermediate bound in the active site,23,32 Arg54 is bound to the bridging phosphate oxygen, thus partially reducing the negative charge on the phosphate and serving to make the phosphate a better leaving group. In support of the important role of Arg54, when Arg54 is replaced by Ala enzymatic activity is reduced by a factor of about 17,000 fold.37,38
Cooperativity Induced by Aspartate Binding
As indicated above, the binding of CP induces small but significant tertiary conformational changes which not only creates the binding site for aspartate but also weakens those interactions that stabilize the enzyme in the T state.31 The binding of Asp to the enzyme•CP complex, as well, changes the electrostatics of the active site making it more electropositive. The binding of aspartate induces additional major loop motions, particularly of the 80’s and 240’s loops. These conformational changes have two consequences: first, they force the substrates closer together, thereby lowering the activation energy of the reaction, and second, they further weaken the intersubunit interactions that specifically stabilize the T-state structure. The weakening of these interactions triggers the initiation of the global quaternary conformational change. In fact, the 240’s loops cannot attain their final domain-closed conformation without an expansion of the enzyme along the three-fold axis, which allow the 240’s loops from the upper and lower catalytic subunits to slide past each other.31 Thus, the binding of Asp to one or so of the active sites is sufficient to convert the enzyme from the T to the R state.39
Heterotropic Effects and Structural Aspects
The allosteric site of E. coli ATCase is located in the Al domain of the regulatory chain near the N-terminus and the five-stranded β-sheet (Fig. 4). The localization of the N-terminal region of the regulatory chain (residues 1–7) was first modeled in the CTP-bound complex40 near the allosteric site.38 Mutagenesis studies41 support the suggestion that this is the regulatory region of the enzyme. Increased resolution of the X-ray data, 2.1Å23 versus 2.5Å,40 yielded electron density for the independent r1 and r6 chains and allowed tracing of the first seven residues, and remodeling residues 8r–10r. In this new model, the nucleotide site is somewhat more open than that of a mutated T-state structure.42 In addition the trace of residues 1–7 of the regulatory chain shows some inherent asymmetry between the r1 and r6 chains. The structural asymmetry may contribute to the high- and low-affinity binding constants for the nucleotides as determined by equilibrium dialysis43 and fluorescence studies.44 In summary, the N termini of the regulatory chains are involved in both nucleotide binding and heterotropic effects.23,45
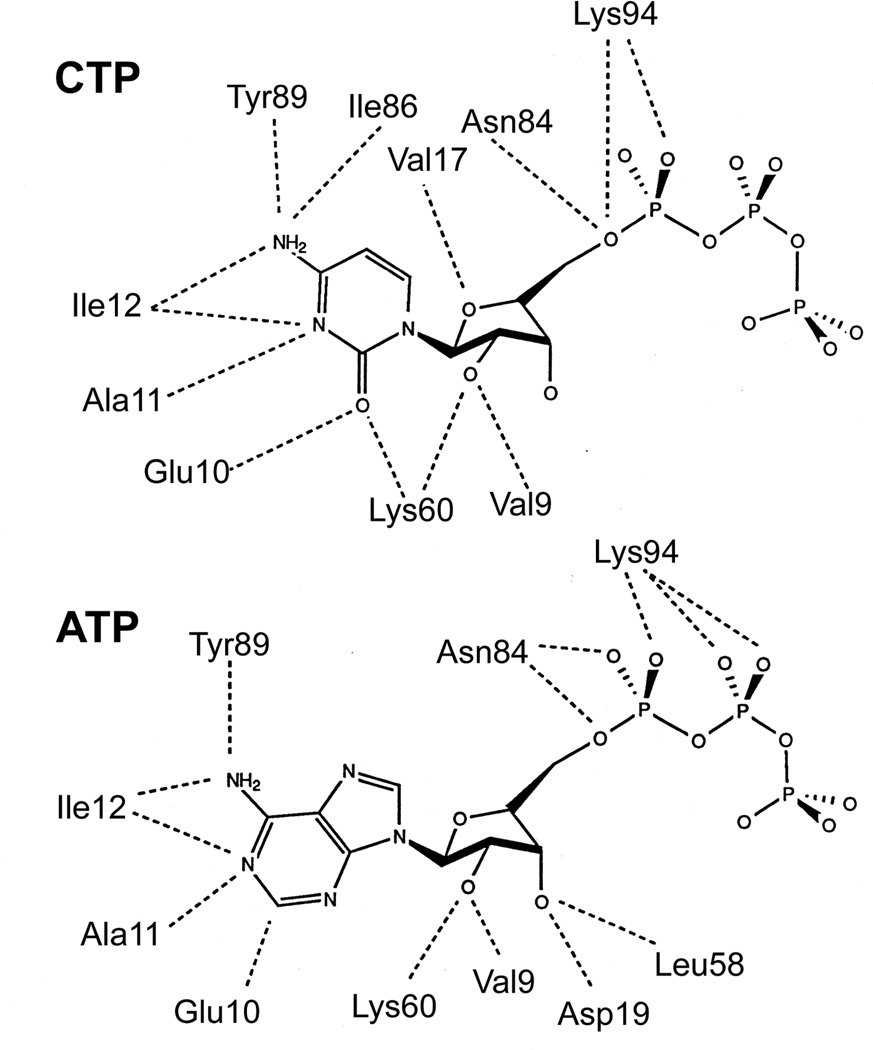
The mode by which CTP and ATP bind to the regulatory site45 of the enzyme in the T state is shown in Fig. 14. This site is about 60Å away from the active site. Although ATP and CTP bind at this regulatory site in both the T state or R state there are differences: for example, CTP binds in a more compact form in which its ribose ring is about 1.5Å deeper in the cleft in the T and R state of the r1 chain and 0.9Å deeper in the T state of the r6 chain than the ribose ring of ATP.45
FIGURE 14.
A comparison of the interactions between CTP (top) and ATP (bottom) and the allosteric site on the regulatory chains of E. coli ATCase. This figure is based on the ATCase•CTP and ATCase•ATP structures in the T state.22
As described above, the domain closure between the CP and the Asp domains of the catalytic chain is caused when aspartate binds to the enzyme•CP complex. A comparison of the active site in the T and R state reveals the change in the position of Arg229 contribute to the affinity of the R state for aspartate compared with the lower affinity for aspartate of the T state. However, the T to R transition is not required for movement of Arg229 into the active site. When unliganded ATCase binds ATP, the guanidinium group of Arg229 moves toward the vacant aspartate cavity, and when CTP binds to the unliganded enzyme, Arg229 moves away from the aspartate cavity. These changes in the T-state structures show that the effects of ATP or CTP binding do not require the quaternary conformational transition. In addition, the binding of CP induces a movement of the side chain of Arg229 toward the aspartate cavity, thus preparing the aspartate region for increased aspartate affinity. The change in the position of Arg229, influenced by the binding of either ATP or CTP some 60Å away, occurs in the C4 chain of the lower trimer, but not in the corresponding C1 chain of the upper trimer. This observation serves as a clear example of the effects of molecular asymmetry in the enzyme mechanism.
Conclusion
The regulation of the rates of metabolic pathways by allosteric enzymes is an extremely important mechanism for cellular control. Allosteric enzymes, such as ATCase, play a pivotal role in metabolism because they have three functions – they catalyze a unique metabolic reaction, alter the rate of catalysis in response to cellular conditions and are responsible for the rate of the larger pathway. Regulation of ATCase involves the binding of signaling molecules to the regulatory sites, and this binding induces an alteration in the rate of catalytic activity. Our studies of ATCase have been directed towards using the three-dimensional structural information to understand, on the molecular level, how ATCase is able to catalyze the reaction between aspartate and carbamoyl phosphate, how the enzyme is able to transition between the T and R conformations, and how the binding of regulatory nucleotides a distance of 60Å from the active site induces alterations to modulate activity. This understanding of the structure and function in this model system will help us understand the molecular basis of cooperativity and allosteric regulation in other systems as well. In 2002 D. Koshland46 emphasized the importance of these type of studies when he said, “this type of cooperativity, which has since been shown in many enzymes, receptors, and ion channels, is of critical importance to both evolution and the field of proteomics because it can serve as a general model for the way in which the networks of interacting enzymes of metabolic pathways are regulated.”
Acknowledgments
The work on aspartate transcarbamoylase was funded by grants GM26237 (E.R.K.) and GM06920 (W.N.L.) from the National Institutes of Health.
Abbreviations used
- ATCase
aspartate transcarbamoylase (EC 2.1.3.2, aspartate carbamoyltransferase)
- c
the catalytic chain of aspartate transcarbamoylase
- r
the regulatory chain of aspartate transcarbamoylase
- c3
the catalytic subunit of aspartate transcarbamoylase composed of three catalytic chains
- r2
the regulatory subunit of aspartate transcarbamoylase composed of two regulatory chains
- CP
carbamoyl phosphate
- Asp
L-aspartate
- PALA
N-phosphonacetyl-L-aspartate
- PAM
phosphonoacetamide
- 80’s loop
a loop in the catalytic chain of aspartate transcarbamoylase comprised of residues 73–88
- 240’s loop
a loop in the catalytic chain of aspartate transcarbamoylase comprised of residues 230–245
Biographies
Evan R. Kantrowitz is Professor of Chemistry at Boston College. His research area focuses on the relationship between the structure and function of allosteric enzymes.
William N. Lipscomb was Emeritus Professor of Chemistry at Harvard University. Professor Lipscomb, or “The Colonel” to those that knew him, investigated a broad array of fields in chemistry, including the inorganic chemistry of boron, crystallography of both small molecules and proteins, the allosteric behavior of enzymes, along with numerous studies in theoretical chemistry. The Colonel was awarded the Chemistry Nobel Prize in 1976 for his work on the structure of boranes that illuminated new aspects of chemical bonding. Before his death on April 15, 2011, at the age of 91, he had finished work on this manuscript.
REFERENCES
- 1.Yates RA, Pardee AB. Control of pyrimidine biosynthesis in Escherichia coli. J. Biol. Chem. 1956;221:757–770. [PubMed] [Google Scholar]
- 2.Umbarger HE. Evidence for a Negative-Feedback Mechanism in the Biosynthesis of Isoleucine. Science. 1956;123:848–848. doi: 10.1126/science.123.3202.848. [DOI] [PubMed] [Google Scholar]
- 3.Shepherdson M, Pardee AB. Production and Crystallization of Aspartate Transcarbamylase. J. Biol. Chem. 1960;235:3233–3237. [Google Scholar]
- 4.Jones ME, Spector L, Lipmann F. Carbamyl phosphate. The carbamyl donor in enzymatic citrulline synthesis. J. Am. Chem. Soc. 1955;77:819–820. [Google Scholar]
- 5.Lowenstein JM, Cohen PP. Studies on the Biosynthesis of carbamylaspartic acid. J. Biol. Chem. 1956;235:57–78. [PubMed] [Google Scholar]
- 6.Reichard P, Hanshoff G. Aspartate carbamyl transferase from Escherichia coli. Acta Chem. Scand. 1956;10:548–560. [Google Scholar]
- 7.Gerhart JC, Pardee AB. Enzymology of control by feedback inhibition. J. Biol. Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 8.Gerhart JC, Pardee AB. Aspartate transcarbamylase, an enzyme designed for feedback inhibition. Fed. Proc. 1964;23:727–735. [PubMed] [Google Scholar]
- 9.Gerhart JC, Pardee AB. The effect of the feedback Inhibitor CTP, on subunit interactions in aspartate transcarbamylase. Cold Spring Harbor Symp. Quant. Biol. 1963;28:491–496. [Google Scholar]
- 10.Gerhart JC. Subunits for control and catalysis in aspartate transcarbamoylase. Brookhaven Symp. Biol. 1964:222–231. [PubMed] [Google Scholar]
- 11.Gerhart JC, Schachman HK. Distinct subunits for the regulation and catalytic activity of aspartate transcarbamylase. Biochemistry. 1965;4:1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- 12.Pastra-Landis SC, Evans DR, Lipscomb WN. The effect of pH on the cooperative behavior of aspartate transcarbamylase from Escherichia coli. J. Biol. Chem. 1978;253:4624–4630. [PubMed] [Google Scholar]
- 13.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 14.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of pre-existing structure equilibrium for a mutant of allosteric aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins KD, Stark GR. Aspartate transcarbamylase: Interaction with the transition state analogue N-(phosphonacetyl)-L-aspartate. J. Biol. Chem. 1971;246:6599–6605. [PubMed] [Google Scholar]
- 16.Gerhart JC, Holoubek H. The purification of aspartate transcarbamylase of Escherichia coli and separation of its protein subunits. J. Biol. Chem. 1967;242:2886–2892. [PubMed] [Google Scholar]
- 17.Weber KK. New Structural Model of E. coli Aspartate Transcarbamylase and the Amino-acid Sequence of the Regulatory Polypeptide Chain. Nature. 1968;218:1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- 18.Wiley DC, Lipscomb WN. Crystallographic Determination of Symmetry of Aspartate Transcarbamylase. Nature. 1968;218:1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]
- 19.Wiley DC, Evans DR, Warren SG, McMurray CH, Edwards BF, Franks WA, Lipscomb WN. The 5.5 Angstrom resolution structure of the regulatory enzyme, asparate transcarbamylase. Cold Spring Harb Symp. Quant. Biol. 1972;36:285–290. doi: 10.1101/sqb.1972.036.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Wild JR, Loughrey-Chen SJ, Corder TS. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamylase. Proc. Natl. Acad. Sci. U. S. A. 1989;86:46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke H-M, Honzatko RB, Lipscomb WN. Structure of unligated aspartate carbamoyltransferase of Escherichia coli at 2.6-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1984;81:4027–4040. doi: 10.1073/pnas.81.13.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevens RC, Gouaux JE, Lipscomb WN. Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: Crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6 Å Resolution. Biochemistry. 1990;29:7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- 23.Jin L, Stec B, Lipscomb WN, Kantrowitz ER. Insights into the mechanism of catalysis and heterotropic regulation of E. coli aspartate transcarbamoylase based upon a structure of enzyme complexed with the bisubstrate analog N-phosphonacetyl-L-aspartate at 2.1 Å. Proteins: Struct. Funct. Genet. 1999;37:729–742. [PubMed] [Google Scholar]
- 24.Gouaux JE, Stevens RC, Lipscomb WN. Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate and CTP or ATP at 2.8 Å resolution and neutral pH. Biochemistry. 1990;29:7702–7715. doi: 10.1021/bi00485a020. [DOI] [PubMed] [Google Scholar]
- 25.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 26.Ke H-M, Lipscomb WN, Cho Y, Honzatko RB. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase: X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J. Mol. Biol. 1988;204:725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- 27.Krause KL, Voltz KW, Lipscomb WN. 2.5 Å structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J. Mol. Biol. 1987;193:527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- 28.Macol C, Dutta M, Stec B, Kantrowitz ER. The 80s loop of the catalytic chain of Escherichia coli aspartate transcarbamoylase is critical for catalysis and homotropic cooperativity. Protein Science. 1999;8:1305–1313. doi: 10.1110/ps.8.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robey EA, Schachman HK. Regeneration of active enzyme by formation of hybrids from inactive derivatives: implications for active sites shared between polypeptide chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U.S.A. 1985;82:361–365. doi: 10.1073/pnas.82.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouaux JE, Lipscomb WN. Three-dimensional structure of carbamyl phosphate and succinate bound to aspartate carbamyltransferase. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4205–4208. doi: 10.1073/pnas.85.12.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Stieglitz KA, Cardia JP, Kantrowitz ER. Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8881–8886. doi: 10.1073/pnas.0503742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouaux JE, Krause KL, Lipscomb WN. The catalytic mechanism of Escherichia coli aspartate carbamoyltransferase: A molecular modeling study. Biochem. Biophys. Res. Commun. 1987;142:893–897. doi: 10.1016/0006-291x(87)91497-5. [DOI] [PubMed] [Google Scholar]
- 33.Gouaux JE, Lipscomb WN. Unpublished study, which can be found in Chapter 5 of the Ph. D. Thesis. 12 Oxford Street, Cambridge, MA 02138: J. E. Gouaux, Harvard University, Department of Chemistry; 1989. [Google Scholar]
- 34.Tsuruta H, Vachette P, Sano T, Moody MF, Tauc P, Amemiya Y, Wakabayashi K, Kihara H. Kinetics of the quaternary structure change of aspartate transcarbamoylase triggered by succinate, a competitive inhibitor. Biochemistry. 1994;33:10007–10012. doi: 10.1021/bi00199a026. [DOI] [PubMed] [Google Scholar]
- 35.Wedler FC. Mechanisms of substrate binding with glutamine synthetase: Equilibrium isotope exchanges with the ovine brain, pea seed, and Escherichia coli enzymes. J. Biol. Chem. 1974;249:5080–5087. [PubMed] [Google Scholar]
- 36.Gouaux JE, Lipscomb WN. Crystal structures of phosphonoacetamide ligated T and phosphonoacetamide and malonate ligated R states of aspartate carbamoyltransferase at 2.8 Å resolution and neutral pH. Biochemistry. 1990;29:389–402. doi: 10.1021/bi00454a013. [DOI] [PubMed] [Google Scholar]
- 37.Stebbins JW, Xu W, Kantrowitz ER. Three residues involved in binding and catalysis in the carbamyl phosphate binding site of Escherichia coli aspartate transcarbamylase. Biochemistry. 1989;28:2592–2600. doi: 10.1021/bi00432a037. [DOI] [PubMed] [Google Scholar]
- 38.Stebbins JW, Robertson DE, Roberts MF, Stevens RC, Lipscomb WN, Kantrowitz ER. Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: A site-specific mutagenesis, NMR and X-ray crystallographic study. Protein Sci. 1992;1:1435–1446. doi: 10.1002/pro.5560011105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macol CP, Tsuruta H, Stec B, Kantrowitz ER. Direct structural evidence for a concerted allosteric transition in Escherichia coli aspartate transcarbamoylase. Nat. Struc. Biol. 2001;8:423–426. doi: 10.1038/87582. [DOI] [PubMed] [Google Scholar]
- 40.Kosman RP, Gouaux JE, Lipscomb WN. Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 Å resolution: Implications for aspartate transcarbamoylase mutants and the mechanism of negative cooperativity. Proteins: Struct. Funct. Genet. 1993;15:147–176. doi: 10.1002/prot.340150206. [DOI] [PubMed] [Google Scholar]
- 41.Dembowski NJ, Kantrowitz ER. The use of alanine scanning mutagenesis to determine the role of the amino terminus of the regulatory chain in the heterotropic mechanism of Escherichia coli aspartate transcarbamoylase. Protein Eng. 1994;7:673–679. doi: 10.1093/protein/7.5.673. [DOI] [PubMed] [Google Scholar]
- 42.Sakash JB, Kantrowitz ER. The N-terminus of the regulatory chain of Escherichia coli aspartate transcarbamoylase is important for nucleotide binding and heterotropic effects. Biochemistry. 1998;37:281–288. doi: 10.1021/bi972102g. [DOI] [PubMed] [Google Scholar]
- 43.Winlund CC, Chamberlin MJ. Binding of cytidine triphosphate to aspartate transcarbamylase. Biochem. Biophys. Res. Commun. 1970;40:43–49. doi: 10.1016/0006-291x(70)91043-0. [DOI] [PubMed] [Google Scholar]
- 44.Mendes KR, Martinez JA, Kantrowitz ER. Asymmetric allosteric signaling in aspartate transcarbamoylase. ACS Chem Biol. 2010;5:499–506. doi: 10.1021/cb9003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipscomb WN. Aspartate Transcarbamoylase from Escherichia coli: Activity and Regulation. Adv. Enzymol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 46.Koshland DE, Hamadani K. Proteomics and models for enzyme cooperativity. J. Biol. Chem. 2002;277:46841–46844. doi: 10.1074/jbc.R200014200. [DOI] [PubMed] [Google Scholar]
- 47.Kantrowitz ER, Pastra-Landis SC, Lipscomb WN. E. coli Aspartate Transcarbamylase. Part I. Catalytic and Regulatory Functions. Trends Biochem. Sci. (Pers. Ed.) 1980;5:124–128. [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Kraulis PJ. MOLSCRIPT: A Program to Produce Both Detailed and Schematic Plots of Protein Structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]