Abstract
The multivesicular body (MVB) pathway delivers membrane proteins to the lumen of the vacuole/lysosome for degradation. The resulting amino acids are transported to the cytoplasm for reuse in protein synthesis. Our study shows that this amino acid recycling system plays an essential role in the adaptation of cells to starvation. Cells respond to amino acid starvation by up-regulating both endocytosis and the MVB pathway, thereby providing amino acids through increased protein turnover. Our data suggest that increased Rsp5-dependent ubiquitination of membrane proteins and a drop in Ist1 levels, a negative regulator of ESCRT activity, cause this response. Furthermore, we found that TORC1 and a second, unknown nutrient-sensing system are responsible for the starvation-induced protein turnover. Together, the data indicate that protein synthesis and turnover are linked by a common regulatory system that ensures adaptation and survival under nutrient-stress conditions.
Keywords: ESCRT, TOR, starvation, MVB pathway, endocytosis
Introduction
All eukaryotic organisms are able to respond rapidly to changing nutrient conditions through a complex array of cellular signaling pathways. Eukaryotic cells rely on these pathways to maintain homeostasis, as they are incapable of substantial nutrient storage. Fluctuations in extracellular nutrient availability are mitigated by physiological responses, including limitation of non-essential protein synthesis, biosynthetic generation of deficient nutrients (e.g. amino acids), and conservation and reutilization of existing resources (reviewed in (1).
Central to eukaryotic nutrient sensing systems is the highly conserved target of rapamycin (TOR) kinase (mTOR in mammals) (reviewed in (2). This Ser/Thr kinase exists in two structurally and functionally distinct multiprotein complexes in the cell, termed TORC1 (TOR complex 1) and TORC2. Studies of TORC1 have revealed its role as a central regulator of several cellular processes, including translation, nutrient transport, ribosome biogenesis, cellular proliferation, and autophagy. The function of TORC2, whose activity is insensitive to rapamycin, is less understood and is involved in regulation of the actin cytoskeleton, endocytosis, and lipid biogenesis.
Amino acids stimulate TORC1 signaling, though the mechanisms remain unclear. Nutrient deprivation, or treatment with rapamycin, results in rapid TORC1 inactivation and the instigation of cellular starvation-response mechanisms. These include a block of general translation initiation, transcriptional upregulation of stress-response genes, and derepression of autophagy (3). A key step in inhibiting general translation is the phosphorylation and inactivation of translation initiation factor eIF2 by the kinase Gcn2. TORC1 inactivation induces Gcn2 activity, though Gcn2 can also be activated by the presence of uncharged t-RNAs (4).
In addition to transcriptional and translational alterations, starving cells degrade organelles and portions of cytoplasm to recycle existing resources. This process, known as autophagy, is highly conserved in eukaryotes and is necessary for survival during prolonged starvation (5).
The multivesicular body (MVB) pathway is the primary means of degrading transmembrane proteins in a cell, linking its function to processes such as nutrient uptake and signaling downregulation (6). MVB protein sorting results in the formation of late endosomes containing cargo-laden intralumenal vesicles. These MVBs fuse with the lysosome / vacuole and release the cargo to the compartment's interior for degradation. The minimum signal for MVB sorting is the covalent attachment of a single ubiquitin molecule to the cytoplasmic portion of a target protein by ubiquitin ligase machinery, often Rsp5 in yeast and Nedd4-related ligases in mammalian cells (7). Ubiquitinated cargo is sorted at the MVB by a set of protein complexes collectively termed ESCRTs (Endosomal Sorting Complex Required for Transport). The four ESCRTs, ESCRT-0, -I, -II, and -III are recruited sequentially from the cytosol to the endosomal membrane where these complexes are thought to recognize, concentrate, and sequester transmembrane cargoes for packaging into vesicles that bud into the lumen of the compartment (6). Additionally, the ESCRT machinery is utilized throughout the cell in other membrane fission events such as cytokinesis and retroviral budding (8).
MVB formation requires that the ESCRT machinery be released from the endosomal membrane in a reaction catalyzed by the ATPase Vps4. This energy-requiring step is regulated in part by Ist1, which has been shown to physically interact with the N-terminal substrate-recognition motif of Vps4 (9-12). Regulation of Vps4 by Ist1 seems to be two fold. Genetic data suggest that Ist1 is involved in the recruitment of Vps4 to the endosomal membrane and, as such, is supportive of Vps4 function. Conversely, in vitro data shows Ist1 as having a potent negative effect on Vps4 by inhibiting its assembly and ATPase activity (9). These observations suggest that Ist1 fills a unique regulatory niche within the framework of the MVB pathway.
This work presents evidence for a regulatory link connecting canonical cellular starvation-response/nutrient-sensing systems (TORC1- and Gcn2-mediated processes) with the MVB sorting and degradation pathway. Starvation induces the degradation of numerous plasma membrane proteins by increasing the efficiency of both endocytosis and MVB sorting. The latter seems to be mediated by the MVB factor Ist1, whose protein levels vary dramatically in response to changing nutrient conditions.
Results
Recycling of amino acids through the MVB pathway is important for survival during starvation
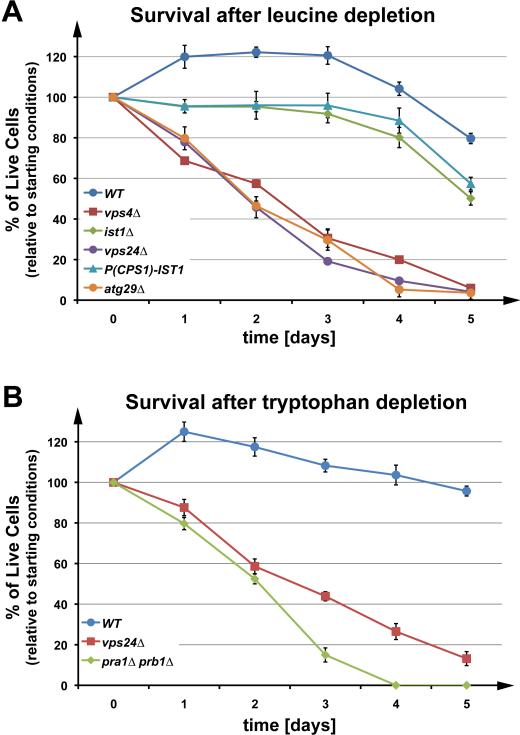
During our work with ESCRT mutants, we observed that these strains rapidly lost viability when kept in stationary phase on agar plates. Furthermore, previous studies have shown that diploid strains carrying mutations in the ESCRT machinery exhibited sporulation defects (13). These observations suggested that a block in the MVB pathway might affect starvation- response pathways. To test this idea further, we examined the survival rate of wild-type and ESCRT-mutant strains under starvation conditions. Note that the yeast strain SEY6210, referred to as ‘wild type’, contains several mutations that render the strain auxotrophic for leucine, tryptophan, histidine, lysine and uracil (Table 1). For most of our experiments, we used leucine-free medium to induce starvation conditions. Leucine is the most common amino acid found in proteins, and leucine synthesis is dependent on LEU2, which is mutated in SEY6210. The data shown in Figure 1 A demonstrated that lack of either Vps4 or Vps24, two proteins essential for a functional MVB pathway, dramatically decreased the survival rate during leucine-starvation conditions (see Fig. S1 A for a prolonged analysis of cell survival during starvation). This decreased survival rate was similar to that of the autophagy mutant atg29Δ. These data showed that half of the ESCRT mutant cells died within the first two days. In contrast, wild-type cells exhibited no loss of viability in the same time period, but instead showed a ~20 percent increase in cell number, possibly due to the completion of cytokinesis by cells in later stages of mitosis. This increase in cell number was not observed in cells deleted for IST1, a gene encoding a regulatory ESCRT factor not essential for MVB formation (9, 14). These data demonstrated a strong correlation between MVB pathway activity and the survival of yeast cells under starvation conditions.
Table 1.
| Strain or Plasmid | Descriptive Name | Genotype or Description | Reference or Source |
|---|---|---|---|
| Yeast Strains | |||
| SEY6210 | WT | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | (41) |
| MBY3 | vps4Δ | SEY6210, VPS4::TRP1 | (39) |
| MBY63 | ist1Δ | SEY6210, IST1::HIS3 | (9) |
| BWY102 | vps24Δ | SEY6210, VPS24::HIS3 | (42) |
| CJY13 | IST1-HA | SEY6210, IST1-HA::KanMX6 | This study |
| GOY23 | pra1Δ prb1Δ | SEY6210; prb1Δ::LEU2 pep4 Δ::LEU2 | (43) |
| MCY52 | npr1Δ | SEY6210, NPR1::KanMX6 | This study |
| CBY118 | FTR1-GFP | SEY6210.1, FTR1-GFP::HISMX | C. Burd, unpublished |
| MCY51 | FTR1-GFP ist1Δ | CBY118, IST1::URA3 | This study |
| EOY14 | CAN1-GFP | SEY6210, CAN1-GFP::TRP1 | This study |
| JKY4 | CAN1-GFP ist1Δ | EOY14, IST1::HIS3 | This study |
| TSY183 | FTR1-GFP rsp5-1 | SEY6210, RSP5::HIS3 + pDsRED415-rsp5L733S, FTR1-GFP::URA3 | (21) |
| MCY54 | FTR1-GFP npr1Δ | CBY118, NPR1::KanMX6 | This study |
| EOY9 | FTR1-HA | SEY6210, FTR1-HA::HIS5 | This study |
| EOY21 | FTR1-HA | MBY63, FTR1-HA::KanMX6 | This study |
| MCY29 | pdr5Δ | SEY6210, PDR5::HIS3 | This study |
| MCY30 | pdr5Δ vps4Δ | MBY3 (SEY6210, VPS4::TRP1), PDR5::HIS3 | This study |
| EOY77 | FTR1-GFP npr1Δ bit61Δ | MCY54 (SEY6210.1, FTR1-GFP::HISMX, NPR1::KanMX6), BIT61::TRP1 | This study |
| EOY78 | FTR1-GFP avo2Δ | CBY118 (6210.1, Ftr1-GFP::HISMX), AVO2::TRP1 | This study |
| EOY79 | FTR1-GFP npr1Δ avo2Δ | MCY54 (SEY6210.1, FTR1-GFP::HISMX, NPR1::KanMX6), AVO2::TRP1 | This study |
| EOY80 | FTR1-GFP bit61Δ | EOY75 (SEY6210, BIT61::HIS3), FTR1GFP::KanMX | This study |
| E. coli | |||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIqZ□M15 Tn10(tetr)] | Stratagene (La Jolla, CA) | |
| Plasmids | |||
| pMB241 | IST1-HA | URA3 Apr (pRS416) IST1-HA | (9) |
| pMB243 | IST1-GFP | URA3 Apr (pRS416) IST1-GFP | (9) |
| pMB409 | P(SNF7)-IST1-HA | URA3 Apr (pRS416) P(SNF7)-IST1-HA | This study |
| pMB327 | P(CPS1)-IST1-HA | URA3 Apr (pRS416) P(CPS1)-IST1-HA | This study |
| pJK6 | P(CPS1)-FUR4-GFP | LEU2 Apr (pRS415) P(CPS1)-FUR4-GFP | This study |
| pJK12 | P(SNF7)-FUR4-GFP | TRP1 Apr (pRS414) P(SNF7)-FUR4-GFP | This study |
| pJK19 | P(CUP1)-FUR4-GFP | URA3 Apr (pRS416) P(CUP1-1)-FUR4-GFP | This study |
| pMB277 | P(GAL4)-IST1-HA | URA3 Apr (pRS416) P(GAL1)-IST1-HA | (9) |
| pCJ50 | DID2-HA | HIS3 Apr (pRS423) DID2-HA | This study |
| pCD2 | GST-IST1 | Apr Kanr (pGEX-KG) GST-IST1 | (9) |
| pAH32 | GST-DID2(CT) | Apr (pGEX-KG) GST-DID2(113-204) | (12) |
| pMB63 | GST-VPS4E233Q | Apr (pGEX-KG) GST-vps4(E233Q) | (17) |
| pCJ51 | VPS4 | HIS3 Apr (pRS423) VPS4 | This study |
| pRS316GFP-Aut7 | GFP-ATG8 | URA3 Apr (pRS316) GFP-ATG8 | (31) |
| pJV40U | CRH2-GFP (internal in-frame fusion) | URA3 Apr (pRS416) CRH2-GFP | (44) |
| pMC47 | ist1(S244A)-HA | URA3 Apr (pRS416) ist1(S244A)-HA | This study |
| pMB66 | vps4(E233Q) | HIS3 Apr (pRS413) vps4(E233Q) | (17) |
| pCJ54 | P(SNF7)-HA-DID2 | URA3 Apr (pRS416) P(SNF7)-HA-DID2 | This study |
| pCJ55 | P(SNF7)-HA-IST1 | URA3 Apr (pRS416) P(SNF7)-HA-IST1 | This study |
| pCJ56 | P(SNF7)-HA-VPS60 | URA3 Apr (pRS416) P(SNF7)-HA-VPS60 | This study |
| pCJ57 | P(SNF7)-HA-VTA1 | URA3 Apr (pRS416) P(SNF7)-HA-VTA1 | This study |
Figure 1.
The MVB pathway is instrumental in maintaining cellular viability during starvation. Survival of auxotrophic wild-type and mutant strains in YNB (-Leu) (A) or YNB (-Trp) (B) over five days. The data show the average of three parallel experiments (+/- standard deviation).
Deletion of two key vacuolar peptidases caused a loss in viability during starvation similar to that observed with ESCRT mutants (pra1Δ prb1Δ, Fig. 1 B). This result is consistent with the idea that the diminished survival of ESCRT mutants during starvation was due to the lack of protein degradation via the MVB pathway. Finally, as a control we compared the survival rate of wild-type and vps4Δ cells in YNB complete medium after the cells reached stationary phase (Fig. S1 B). Under these naturally achieved starvation conditions, both strains showed survival rates similar to those observed following leucine depletion, indicating that the survival phenotype of the ESCRT mutants is not specific to leucine starvation.
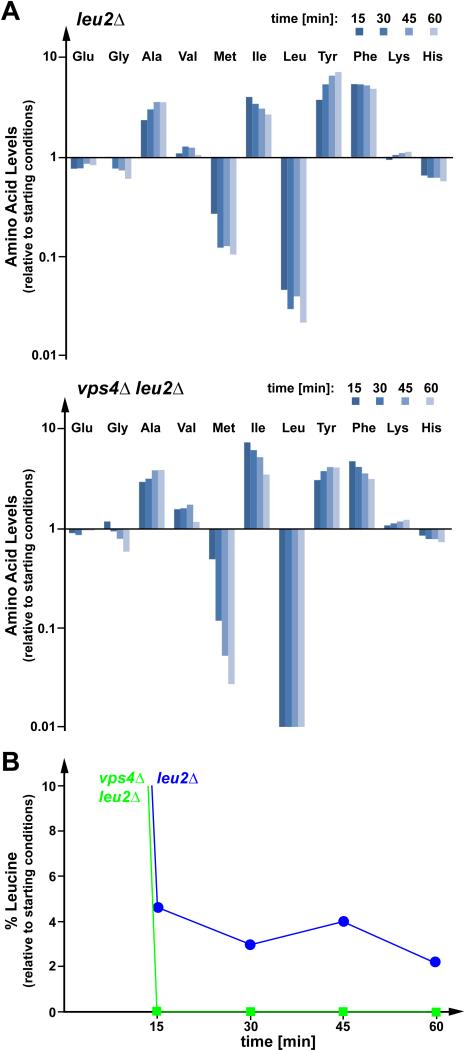
The MVB pathway delivers membrane proteins into the lumen of the vacuole for degradation. The resulting amino acids are pumped into the cytoplasm by vacuolar transporters where they are reused for protein synthesis (15). This amino acid recycling pathway might play an important role during starvation conditions. To test this hypothesis, wild-type (for ESCRT function) or vps4Δ strains, both leucine auxotrophs (leu2Δ), were transferred into growth medium lacking leucine, and the cellular concentrations of various free amino acids were determined at different time points. Overall, the starting amino acid levels and the response to leucine starvation was very similar between wild-type and the MVB mutant strain vps4Δ (Fig. 2 A, Table S1). The levels of alanine, isoleucine, tyrosine and phenylalanine rapidly increased following leucine starvation, which is possibly caused by a broad translational shutdown combined with the continued uptake of these amino acids from the medium. The major difference between the two strains was the extent to which the leucine levels dropped. Wild-type cells maintained 3-5% of the initial leucine concentration, while that of vps4Δ cells decreased to non-detectable amounts of leucine (Fig. 2 B). This result was consistently observed in multiple independent experiments (Fig. S2 A). The detection limits of the amino acid analyses are at least 10-fold less than the values measured in starved wild-type cells (<0.3%). These data indicated that the MVB pathway plays a role in maintaining minimal levels of amino acids during starvation conditions. Interestingly, we observed a dramatic decrease in methionine levels in both strains, though methionine was not limited in the growth medium. Similar decreases in methionine levels were observed in cells starved for lysine (Fig. 4 A) or histidine (data not shown), suggesting that this drop in free methionine might be part of the starvation response and connected to a block in general translation initiation.
Figure 2.
The MVB pathway is important for maintaining amino acid levels during starvation. (A) Analysis of free cellular amino acid content in leucine auxotrophic wild-type (leu2Δ) and vps4Δ (vps4Δleu2Δ) cells following a shift from YNB Complete Synthetic Medium (CSM) to YNB (-Leu). Charts indicate changes in cellular amino acid content relative to starting conditions. (B) Graphical representation of changes in cellular leucine levels relative to starting point in wild-type and vps4Δ cells as shown in (A).
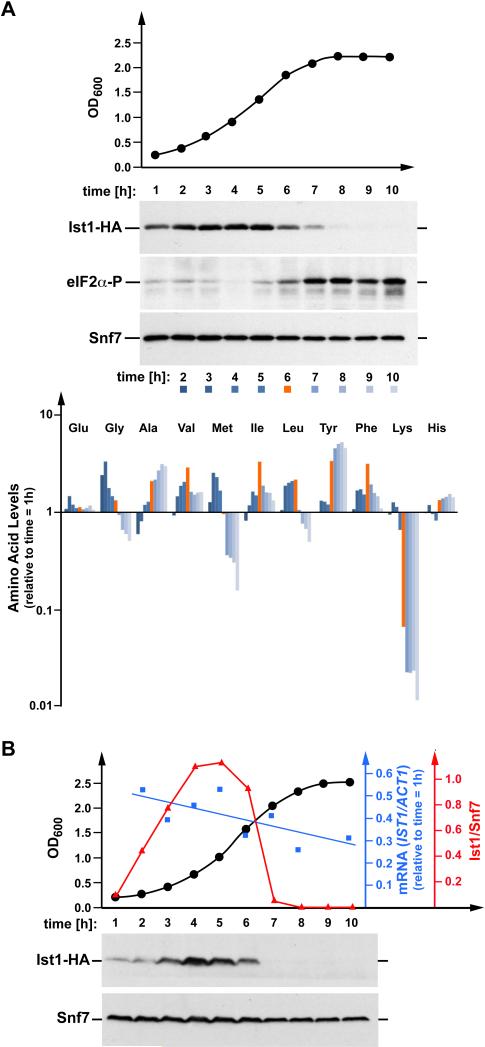
Figure 4.
Ist1 protein levels mirror the translational activity of the cell. (A) Time-course samples analyzed by Western blot to detect HA-tagged Ist1 protein levels (expressed from pMB241) and levels of phospho-eIF2α in MBY63 grown in YNB (-Ura) medium. Snf7 is a loading control. The same samples were analyzed for free amino acid content to track changes relative to starting conditions throughout growth. Red bar indicates the transitional point at hour six, wherein cellular free lysine levels drop dramatically, Ist1 protein levels begin to decline, and phosphorylation of eIF2α increases. (B) Western blot analysis and quantitative RT-PCR performed on samples collected over a 10 hour period encompassing the entire yeast growth curve (black trendline shows OD600 of culture). The strain contains a genomically integrated HA-tag downstream of the native IST1 locus. Snf7 is a loading control. Blue trendline shows IST1 mRNA levels relative to actin. Red line represents densitometric quantitation of Ist1-HA protein levels from the blot shown (relative to Snf7).
Ist1 protein levels regulate the MVB pathway
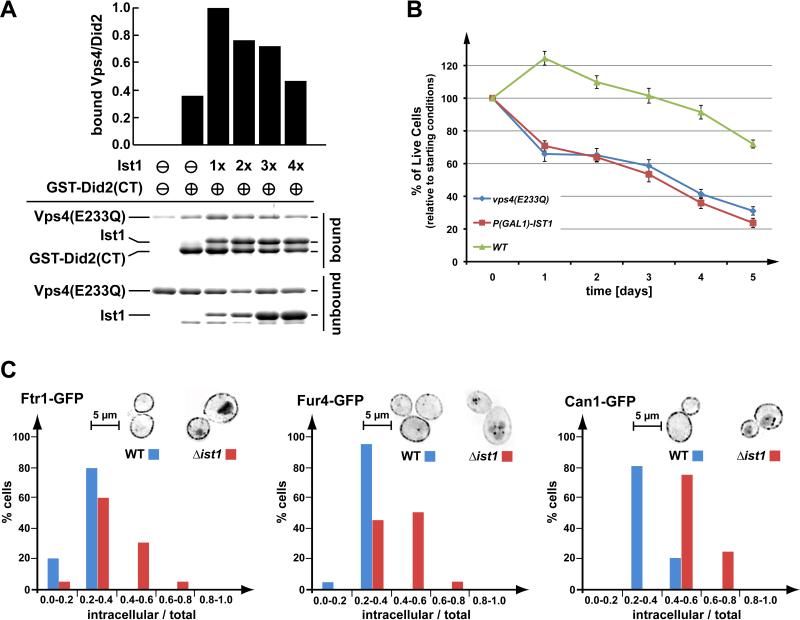
Ist1 has been shown to function in the recruitment of Vps4 to ESCRT-III, suggesting a supportive role for Ist1 in MVB trafficking (9, 12). However, overexpression of Ist1 results in reduced recruitment of Vps4 to ESCRT-III and a defect in MVB trafficking (9), suggesting that Ist1 can have both a positive and negative effect on Vps4 activity, depending on Ist1 protein levels. To test this idea, we preformed an in vitro ‘GST-pull-down’ experiment using purified recombinant proteins. Did2 is an important factor in the recruitment of Vps4 due to its interactions with Ist1, Vps4 and ESCRT-III (12). Therefore, we used the binding of Vps4 to Did2 as readout for recruitment efficiency. For the assay, the C-terminal half of Did2 (GST-Did2(CT), amino acids 113-204), which contains the Vps4 and Ist1 interaction regions, was immobilized on glutathione-sepharose beads. These beads were then incubated with an equal amount of the ATP-locked form of Vps4 (Vps4E233Q) in the presence of various concentrations of Ist1. The results indicated that, consistent with previously published data (12), the addition of an approximate equimolar amount of Ist1 increased the recruitment of Vps4 to Did2 (Fig. 3 A), possibly through the formation of a trimeric Vps4-Ist1-Did2 complex. In contrast, increasing amounts of Ist1 caused a reduction of Did2-associated Vps4 (Fig. 3 A) suggesting that at higher concentrations, Ist1 might bind to Did2 and Vps4 independently, resulting in the formation of Did2-Ist1 and Ist1-Vps4 complexes that inhibit the formation of the trimeric complex. This effect of Ist1 on Vps4 recruitment to GSTDid2 was consistently observed in other independent pulldown assays, although the Ist1 concentration necessary to affect the Vps4-Did2 interaction varied (Fig. S2 B). These variations are likely due to the tendency of Ist1 to oligomerize/aggregate (16), thereby changing the concentration of soluble Ist1 protein in the in vitro assay. These in vitro observations are consistent with the altered endosomal recruitment of Vps4 when Ist1 protein levels are elevated in vivo (9).
Figure 3.
Regulation of Vps4 by Ist1. (A) In vitro binding studies using recombinant Vps4(E233Q), Ist1 and GST-Did2(CT) (amino acids 113-204). GST-Did2(CT) was immobilized on GSH-sepharose and an approximately equimolar amount of Vps4(E233Q) was added in the presence of ATP. The Ist1 concentrations were varied from zero to approximately four times the molar amount of Vps4(E233Q). Bound and unbound fractions were analyzed by SDS-PAGE and coomassie staining, and the amount of bound Vps4(E233Q) was determined by densitometric scanning. The obtained values were normalized according to the amount of bound GST-Did2(CT), then plotted relative to the negative control (no GST-Did2(CT) = 0) and the maximum signal (1xIst1 = 1). (B) Survival of wild-type yeast, yeast expressing a dominant-negative version of Vps4 (vps4E233Q, pMB66), and yeast with GAL1-driven overexpression of Ist1 over a period of five days. The data show the average of three parallel experiments (+/- standard deviation). (C) Steady state localization of three permeases, Ftr1-GFP (iron), Fur4-GFP (uracil), and Can1-GFP (arginine) after extended exponential phase growth in WT and ist1Δ cells. Images were inverted for better visualization (brightest GFP signal appears black). The total and internal (excluding plasma membrane) GFP signal was quantified from 20 cells of each microscopy set. The graph shows the percentage of cells with a particular range of internal-to-total GFP signal. Analysis using the Kolmogorov-Smirnov test indicated that all discussed differences are statistically relevant.
Additional support for this regulatory model comes from the observation that artificially high cellular levels of Ist1 induced by using a GAL1 promoter (pGAL1-Ist1-HA), resulted in loss of viability during leucine starvation (Fig. 3 B). This drop in viability was similar to the drop observed in cells expressing the dominant-negative mutant vps4(E233Q), which renders the MVB pathway nonfunctional (17). In comparison to the data shown in Figure 1 A, this viability assay was performed using a different yeast strain (W303) and a different carbon source (galactose).
Analysis of cell-surface protein trafficking indicated that Ist1 protein levels in exponentially growing cells have a negative regulatory effect on the MVB pathway. GFP-tagged versions of the plasma membrane proteins Ftr1 (iron transporter), Fur4 (uracil transporter) and Can1 (arginine transporter) showed obvious vacuolar staining when expressed in cells deleted for IST1, whereas no vacuolar signal was observed in wild-type cells (Fig. 3 C). These observations suggested that loss of Ist1 caused heightened downregulation of plasma membrane proteins, likely due to increased MVB pathway activity in these mutant cells.
We analyzed Ist1 protein levels in growing cells by Western blot using a functional HA-tagged version of Ist1. The data showed that Ist1-HA levels change dramatically with growth conditions (Fig. 4 A). Ist1 levels increased at low cell densities, peaked during exponential growth and then fell to barely detectable levels as cells entered stationary phase. In contrast, the concentration of the control protein Snf7 (ESCRT-III subunit) showed only minor changes during the different growth phases of the cells.
The drop in Ist1 levels coincided with the end of the growth phase and an increase in phosphorylated translation-initiation factor eIF2 (time = 6h, Fig. 4 A). Phosphorylation of eIF2 at the α-subunit has been shown to block general protein translation in response to amino acid starvation (1), suggesting that starvation might have caused the decrease in Ist1 levels. Consistent with this idea, free amino acid analysis of the same samples revealed a sharp drop in cellular lysine levels after six hours of growth, which occurred concurrently with a marked decline in Ist1 levels (red-marked time point in Fig. 4 A). The yeast strain used for the experiment is a lysine-auxotroph, and after approximately six hours of growth, the lysine provided by the medium was depleted (Fig. 4 A, bottom panel), causing the induction of starvation response pathways. The resulting changes in the levels of other tested amino acids were similar to those observed during leucine starvation (Fig. 2 A).
To identify the mechanism responsible for regulating Ist1 protein levels, the growth-dependent Ist1 expression analysis was repeated using a strain containing a genomically integrated HA-tag immediately downstream from the IST1 locus. This strain was grown in YNB Complete Synthetic Medium (CSM). Samples were taken every hour and analyzed by Western blot for the presence of Ist1-HA, phospho-eIF2α and Snf7. Furthermore, quantitative RT-PCR was performed to determine the amount of IST1-HA mRNA relative to the control mRNA of the actin gene ACT1 (Fig. 4 B). The results showed growth-dependent levels of Ist1-HA, eIF2α-P and Snf7 similar to the patterns observed in the experiment of Figure 4 A, which used plasmid-encoded IST1-HA. In contrast to the Ist1 protein levels, the relative amount of IST1-HA mRNA exhibited only minor changes during growth of the yeast culture, indicating that Ist1 levels are not transcriptionally controlled; rather, they are likely regulated at the level of protein translation and/or degradation.
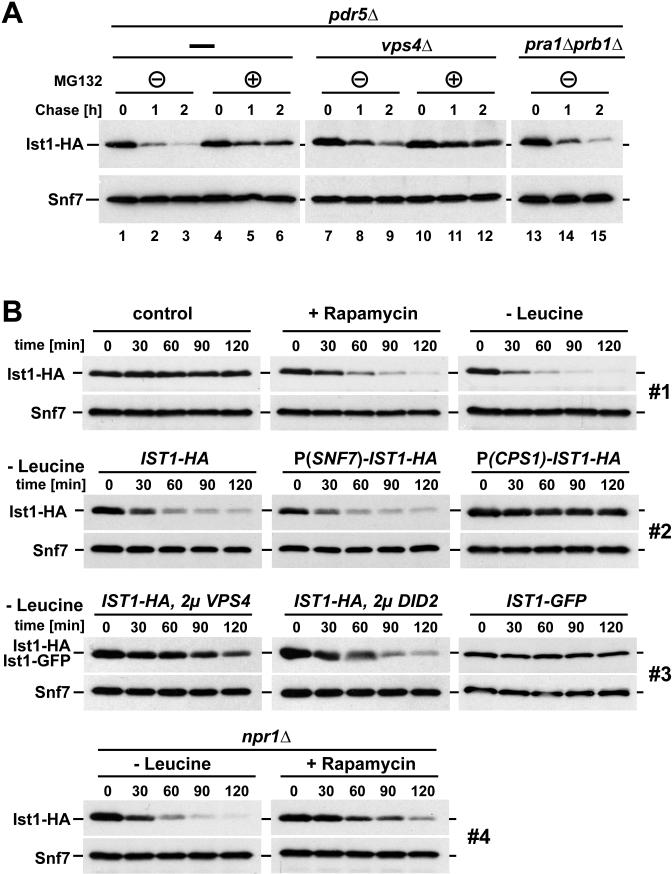
The rate of Ist1 degradation was determined by adding the translational inhibitor cycloheximide to cells and monitoring Ist1 levels over time by Western blot. The strains used for these experiments were deleted for the gene encoding the multidrug transporter Pdr5 in order to enhance the effect of the proteasomal inhibitor MG132. The results showed that in cells treated with cycloheximide, Ist1-HA was almost completely degraded within two hours; whereas, the control protein Snf7 (ESCRT-III subunit) remained stable during the same time period (Fig. 5 A, lanes 1-3). The addition of the proteasomal inhibitor MG132 partially stabilized Ist1-HA (Fig. 5 A, lanes 4-6). In contrast, no change in Ist1-HA turnover was observed in cells lacking Pra1 and Prb1, two proteases that play a key role in vacuolar protein degradation (Fig. 5 A, lanes 13-15). These results indicated that Ist1-HA is rapidly degraded, and that the proteasome, rather than the MVB pathway, mediates the degradation.
Figure 5.
Ist1 is degraded by the proteasome, and its levels drop dramatically during starvation conditions. (A) Ist1-HA protein levels (expressed from pMB241) of exponentially growing cells (OD600 = 0.7) were analyzed at intervals following cycloheximide addition in pdr5Δ, pdr5Δvps4Δ, and pra1Δprb1Δ cells in the presence or absence of the proteasomal inhibitor MG132. Snf7 is a loading control. (B) Western blot detection of Ist1 protein degradation of under different conditions. All treatments and collections were performed on exponentially growing cells. Individual sets (1-4) were obtained with parallel sample collection, preparation, and film exposure to ensure comparability. Snf7 serves as a loading control. P(SNF7)-IST1-HA and P(CPS1)-IST1-HA refer to non-endogenous promoters driving IST1-HA expression. 2μ VPS4 and 2μ DID2 indicate strains overexpressing the corresponding proteins Vps4 and Did2, respectively, using a high-copy vector.
We observed a decreased rate of Ist1-HA degradation in cells deleted for VPS4 (Fig. 5 A, lanes 7-9), a mutation that causes the accumulation of Ist1 at the endosome with ESCRT-III (9). This stabilization effect was even more pronounced in vps4Δ cells that were treated with MG132 (Fig. 5 A, lanes 10-12). These results suggested that predominantly the soluble pool of Ist1, not the membrane associated pool, is targeted for degradation by the proteasome.
The observed drop in Ist1 levels as cells enter stationary phase (Fig. 4) suggested that starvation conditions might result in the loss of Ist1 protein. To test this idea, we followed yeast Ist1-HA levels by Western blot during acute starvation. Starvation conditions were induced by either transferring cells to growth medium lacking leucine or by adding rapamycin, a drug that blocks the kinase activity of the TORC1 complex. In contrast to the non-treated control samples, both leucine starvation and rapamycin addition resulted in the rapid disappearance of Ist1 (Fig. 5 B, set 1). This rapid drop in Ist1-HA levels was also observed when Ist1-HA was expressed from the constitutive SNF7 promoter (the promoter driving expression of the control protein Snf7), further support that Ist1 levels are not transcriptionally regulated (Fig. 5 B, set 2). In contrast, a fusion of IST1-HA to the 5’ UTR region of CPS1 resulted in stable Ist1-HA protein levels during leucine-starvation (Fig. 5 B, set 2). The expression of the vacuolar hydrolase Cps1 is upregulated during starvation conditions, indicating that Cps1 belongs to a set of proteins that is efficiently translated during these stress conditions (18). Together, the results suggested that the observed drop in Ist1 levels during starvation is mainly caused by general translational repression in combination with the intrinsically rapid turnover rate of the protein. We predicted that the starvation-induced drop in Ist1 levels increases the efficiency of the MVB pathway and thus increases the recycling of amino acids required for the stress-response pathways. Consistent with this idea, we observed diminished fitness in starving cells that maintain artificially high levels of Ist1 due to the CPS1 promoter fusion (P(CPS1)-IST1, Fig. 1 A).
C-terminally GFP-tagged Ist1 (Ist1-GFP) is a non-functional fusion protein; it does, however, localize properly to MVBs (Dimaano et al., 2008). Ist1-GFP did not exhibit a starvation-induced drop in protein levels (Fig. 5 B, set 3). This result suggested that the large, C-terminal GFP domain might interfere with the degradation of Ist1. Interestingly, the C-terminal region of Ist1 contains a cluster of lysine residues that could be targeted for ubiquitination. Rsp5 is the principle ubiquitin ligase acting in endocytosis and the MVB pathway. However, starvation-induced degradation of Ist1 was found to be independent of Rsp5 function (Fig. S3 A). Similarly, mutation of a reported Ist1 C-terminal phosphorylation site (19), showed no effect on the rapid degradation of the protein during starvation (Ist1(S244A), Fig. S3 A). Finally, microscopy of Ist1-GFP-expressing cells showed no obvious change in Ist1 localization during acute starvation (Fig. S3 B). Therefore we found that neither Rsp5, nor phosphorylation of S244 mediates the degradation of Ist1. Also, the degradation is not attributable to a change in Ist1 localization following starvation.
Because the lysine-rich C-terminus of Ist1 contains the Vps4-interaction domain, we tested whether high levels of Vps4 would also stabilize the Ist1 protein. The experiment demonstrated that overexpression of Vps4 (expressed from a 2μ high copy vector) was indeed able to inhibit the starvation-induced degradation of Ist1-HA. In contrast, overexpression of Did2, which binds to the N-terminal region of Ist1, had only a minor effect on the Ist1 degradation rate (Fig. 5 B, set 3). In summary, our analysis suggested that soluble, non-Vps4-associated Ist1 was targeted for proteasomal degradation. This degradation, coupled with decreased translation, is the most likely cause for the rapid loss of Ist1 observed following the induction of starvation-response pathways.
To test if protein stability of Vps4 regulators other then Ist1 were affected by starvation, we constructed N-terminal HA-fusions of VPS60, VTA1, DID2 and IST1. All of these fusion proteins were expressed under the control of the SNF7 promoter. These constructs were expressed in wild-type cells and protein levels were followed by Western blot using anti-HA antibodies during one hour of leucine starvation (Fig. S4). The results demonstrated that, similar to the data obtained with the C-terminal HA fusion, HA-Ist1 was rapidly degraded upon induction of starvation. The protein levels of the other HA-tagged regulatory factors also declined, but to a lesser extent than observed for HA-Ist1. The MVB pathway was found to be functional during the initial phase of starvation and thus the observed decline in the levels of Vps60, Did2 and Vta1 were not sufficient to cause MVB sorting defects. Importantly, our data indicated that after prolonged starvation, cells seem to discontinue protein turnover via the MVB pathway and shift to the autophagy pathway. This is discussed in more detail later in this publication. This suggests that the loss of ESCRT regulatory factors might be inconsequential at later time points in starvation.
Starvation induces downregulation of plasma membrane proteins
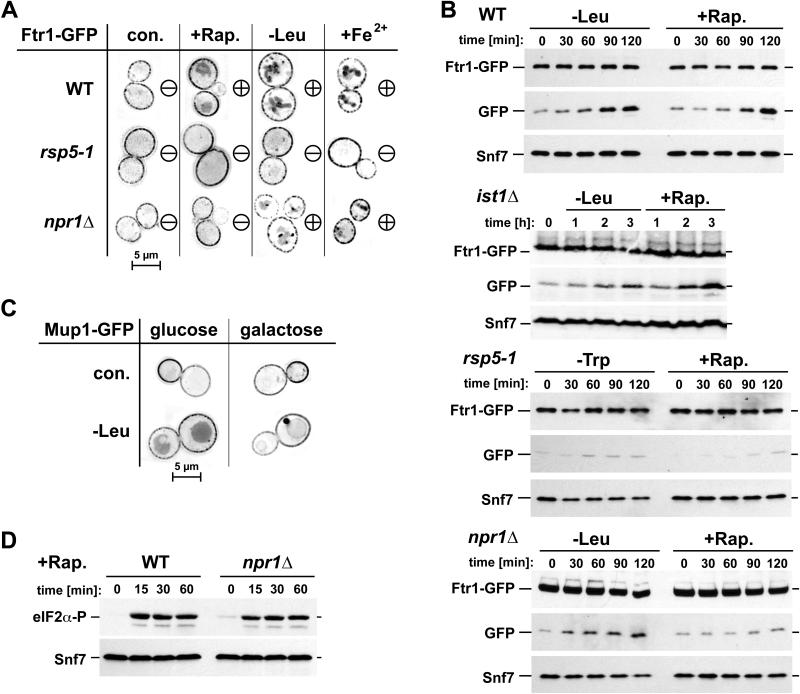
We investigated the effect of starvation on Ftr1-GFP trafficking using fluorescence microscopy. Within 45 minutes of either leucine starvation or rapamycin addition, we observed the relocalization of Ftr1-GFP from the plasma membrane to vacuole-proximal compartments, likely late endosomes, and to the vacuolar lumen (Fig. 6 A). A similar Ftr1-GFP relocalization was observed when cells were treated with high concentrations of iron, which causes substrate-dependent downregulation and has been described previously (20, 21). Consistent with the microscopy data, Western blot analysis showed the emergence of a smaller GFP protein band (~25 kD) following starvation, which is likely an intermediate in the vacuolar degradation of Ftr1-GFP (Fig. 6 B). Quantitative analysis of the Western blot data indicated a loss of 10-30 percent of Ftr1-GFP within two hours of starvation (data not shown). These results suggested that starvation induced the rapid endocytosis of Ftr1-GFP and its delivery to the vacuole for degradation. Cells deleted for IST1 still showed increased Ftr1-GFP turnover with starvation (Fig. 6 B), suggesting that the starvation-response not only upregulates the MVB pathway via Ist1 degradation, but also increases the efficiency of endocytosis through an unknown mechanism.
Figure 6.
Starvation conditions cause downregulation of Ftr1 and Mup1. (A) Localization of Ftr1-GFP in exponentially growing cells with no treatment (con.) or after 45 minutes of rapamycin treatment (+Rap.), leucine starvation (-Leu), or iron treatment (+Fe2+, 0.5 mM) in wild-type cells, cells expressing an rsp5 mutant allele (rsp5-1), or npr1Δ cells. ‘Minus’ (-) designates minimal internalization of Ftr1, ‘plus’ (+) designates heightened internalization and accompanying late endosomal/vacuolar staining. (B) Time-course Western blot analysis of Ftr1-GFP stability with amino acid starvation (-Leu, -Trp) or rapamycin treatment (+Rap.) in WT, ist1Δ, cells expressing defective Rsp5 (rsp5-1, this strain is a leucine-autotroph and thus tryptophan-depletion is used for starvation), or npr1Δ cells. Blots show full length Ftr1-GFP, free GFP (cleaved from chimera in vacuole), and Snf7 as a loading control. (C) Localization of Mup1-GFP in cells overexpressing IST1-HA from a galactoseinducable promoter (Ist1 levels are normal when grown in glucose). Cells with no treatment (con.) or after 45 min leucine starvation (-Leu) were analyzed by fluorescence microscopy. (D) Western blot analysis of eIF2α phosphorylation upon rapamycin-treatment of wild-type and npr1Δ cells using a phospho-specific antibody.
The starvation-induced internalization was not limited to Ftr1 but was also observed with the arginine transporter Can1 (Fig. S5 A) and the methionine transporter Mup1 (Fig. 6 C). Furthermore, previous studies have shown starvation-induced downregulation of Fur4 (uracil transporter, (22), Tat2 (tryptophan transporter, (23), Mal61 (maltose transporter, (24), Bap2 (amino acid transporter, (25) and Hxt17 (glucose transporter, (26). These data suggested that amino acid starvation might cause a broad endocytic response that results in rapid internalization and degradation of multiple plasma membrane proteins. However, starvation does not induce non-specific, bulk-flow endocytosis, as rapamycin treatment did not increase the internalization of the GPI-anchored protein Crh2-GFP (Fig. S5 B). Furthermore, we observed that a pool of Ftr1, Can1 and Fur4 remained at the plasma membrane even after 16 hours of starvation (Fig. S5 C), indicating that the rapid downregulation of membrane proteins is limited to the initial phase of the starvation response. It is possible that protein turnover via the MVB pathway ceases after prolonged starvation and amino acid recycling is mediated predominantly by autophagy.
Starvation-induced downregulation of plasma membrane proteins occurred via the MVB pathway and required the function of the ESCRT machinery including Vps4. During starvation, overexpression of Ist1, which has been shown to interfere with Vps4 activity (9), resulted in the accumulation of endocytosed Mup1-GFP in an aberrant endosomal compartment adjacent to the vacuole (Fig. 6 C). Furthermore, rapid degradation of Ftr1 under starvation conditions was dependent on the function of Rsp5, the same ubiquitin-ligase associated with downregulation of Ftr1 in high iron concentrations (21). Both microscopy and Western blot analysis indicated that cells expressing the rsp5-1 mutant allele (defective in ubiquitin conjugation) are deficient in the starvation-induced degradation of Ftr1-GFP, irrespective of the type of starvation (Fig. 6 A and B). This Rsp5-dependence suggested that starvation conditions might simply upregulate the same machinery that normally regulates Ftr1 endocytosis.
Addition of rapamycin or depletion of leucine resulted in similarly increased protein internalization (Fig. 6 A and B), suggesting that TORC1 might be the key regulator for the endocytic effects observed during both treatments. The TORC1 downstream kinase Npr1 has been implicated in the regulation of membrane protein trafficking (27, 28). Therefore, we tested whether Npr1 was required for Ftr1 internalization during acute starvation by analyzing Ftr1-GFP trafficking in a NPR1 deletion strain. The result showed that rapamycin-induced endocytosis of Ftr1 was impaired in npr1Δ cells. However, leucine-starved npr1Δ cells showed Ftr1 internalization being only slightly delayed when compared to wild-type cells (Fig. 6 A and B, see Fig. S5 D for quantification of Fig. 6 A). Similar results were obtained with Can1-GFP expressed in npr1Δ (Fig. S5 B). These findings indicated the presence of at least two systems that regulate the nutrient-dependent degradation of Ftr1 and Can1. One system is regulated by TORC1 and its downstream kinase Npr1. A second system is induced by amino acid deficiency and causes downregulation in a TORC1-independent manner. Deletion of NPR1 had no effect on rapamycin-induced phosphorylation of eIF2α (Fig. 6 D) and showed only a slight delay in the starvation-induced drop in Ist1 levels (Fig. 5 B, set 4), suggesting that Ist1 does not have a primary role in Npr1-dependent regulation of protein trafficking.
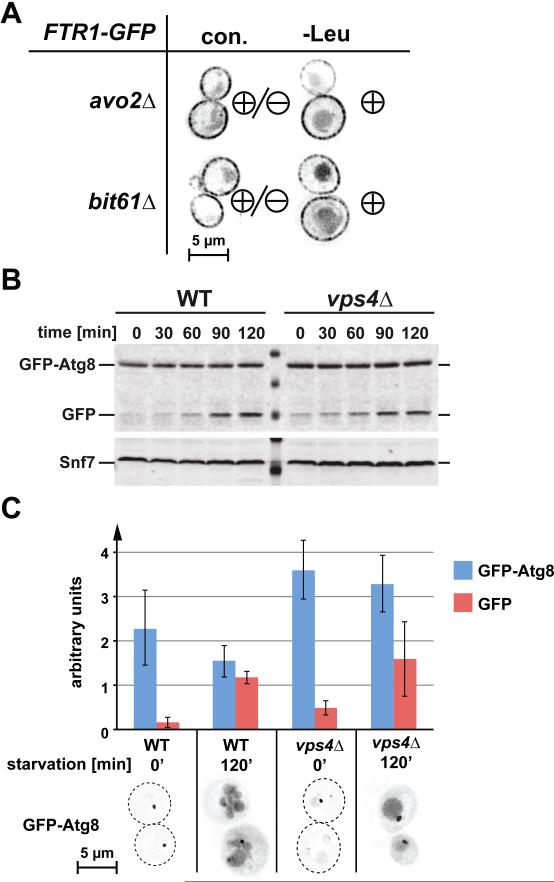
An obvious candidate for the TORC1-independent regulation of the endocytic pathway is the rapamycin-insensitive TORC2 complex. Because deletion of the Tor2 kinase is lethal, we analyzed the starvation response in cells lacking the non-essential TORC2 subunits Avo2 or Bit61. Deletion of either AVO2 or BIT61 increased Ftr1 downregulation even under rich medium conditions (Fig. 7 A). Furthermore, leucine-starvation increased Ftr1 downregulation in both mutant strains (Fig. 7 A). These data suggested that TORC2 does not play an important role for the downregulation of plasma membrane proteins during starvation.
Figure 7.
TORC2 does not mediate starvation-induced downregulation of Ftr1; autophagy is not inhibited in ESCRT mutants. (A) Localization of Ftr1-GFP in the TORC2-deletion mutants avo2Δ and bit61Δ under normal conditions (con.) and following 1 hr. of leucine starvation. ‘Minus’ (-) designates minimal internalization of Ftr1, ‘plus’ (+) designates heightened internalization and accompanying late endosomal/vacuolar staining. (B) Wild-type and vps4Δ cells expressing GFPAtg8 were starved for leucine at time=0 following 24 hours of continuous growth. Samples were taken at indicated time points and analyzed by anti-GFP Western blot. (C) Fluorescence microscopy of the cells used for the Western blot analysis shown in B. The amount of GFP-Atg8 and its GFP-containing degradation product were determined based on the Western blot shown in B and two other Western blots performed in parallel experiments (amounts are shown relative to loading control Snf7, +/- standard deviation).
Autophagy is not blocked in yeast ESCRT mutants
Recent studies in mammalian cells showed that loss of ESCRT function impairs fusion of autophagosomes with lysosomes, thereby impeding degradation of autophagic cargo (29, 30). To test if the observed starvation-response defect in yeast ESCRT mutant strains (Fig. 1 A) is caused by a block in autophagy, we analyzed the starvation-induced expression and degradation of GFP-tagged Atg8 in wild-type and vps4Δ cells. Atg8 is highly expressed under starvation conditions, where it functions in the formation of autophagosomes and is subsequently degraded in the lysosome. Therefore GFP-Atg8 is a useful tool to study both induction and functionality of the autophagy pathway (31). For our analysis, we grew wild-type and vps4Δ cells expressing GFP-Atg8 continuously for one day to ensure steady-state conditions, then induced starvation by transferring the cells into medium lacking leucine. We analyzed samples at time zero and after two hours of starvation by fluorescence microscopy and anti-GFP Western blot (Fig. 7 B and C). The results showed that within two hours of starvation both strains accumulated a stable intermediate of GFP-Atg8 degradation in the vacuole. This observation indicated that the efficiency of autophagy induction and execution was similar in both strains. However, vps4Δ cells showed increased levels of GFP-Atg8 and the stable intermediate before starvation, suggesting that ESCRT mutants might exhibit a low-level autophagic response under normal growth conditions.
Taken together, the data demonstrated that, unlike mammalian cells, yeast does not require MVBs for a functional autophagy pathway. Therefore, the observed phenotypes in the starvation response of ESCRT mutants are likely a direct consequence of a defective MVB pathway.
Discussion
The role of the MVB pathway in the degradation of membrane proteins is well established (6). The MVB pathway delivers these proteins into the lumen of vacuoles/lysosomes where they are degraded by hydrolases. Resident transporters at the vacuolar membrane pump the resulting amino acids into the cytoplasm for reuse in protein synthesis. Here we demonstrate that this recycling pathway provides a vital source of amino acids for the cell during starvation conditions.
Our studies indicated that yeast cells have a very limited storage capacity for amino acids, and, as a consequence, amino acid levels drop rapidly during starvation (<15 min in the case of leucine). Furthermore, we found that the MVB pathway plays an important role in maintaining minimal amino acid levels during starvation, which explains the rapid loss of viability of ESCRT mutants when starved for amino acids (Fig. 1). Similarly, studies in mammalian cells have shown a role for the proteasomal degradation pathway in maintaining amino acid levels during acute starvation (32). Thus, both of the major protein degradation systems, the proteasome and the MVB pathway, provide an immediate source of amino acids when limitations occur. These amino acids allow for synthesis of starvation-response proteins, which are vital for the cell to adapt to stress conditions. Such proteins include vacuolar hydrolases and components of the autophagy pathway. Full induction of autophagy requires hours, during which requisite autophagic proteins must be synthesized (33, 34). Therefore, during acute starvation, the MVB pathway and the proteasome serve to maintain translational capability until the major starvation response system, autophagy, is fully active.
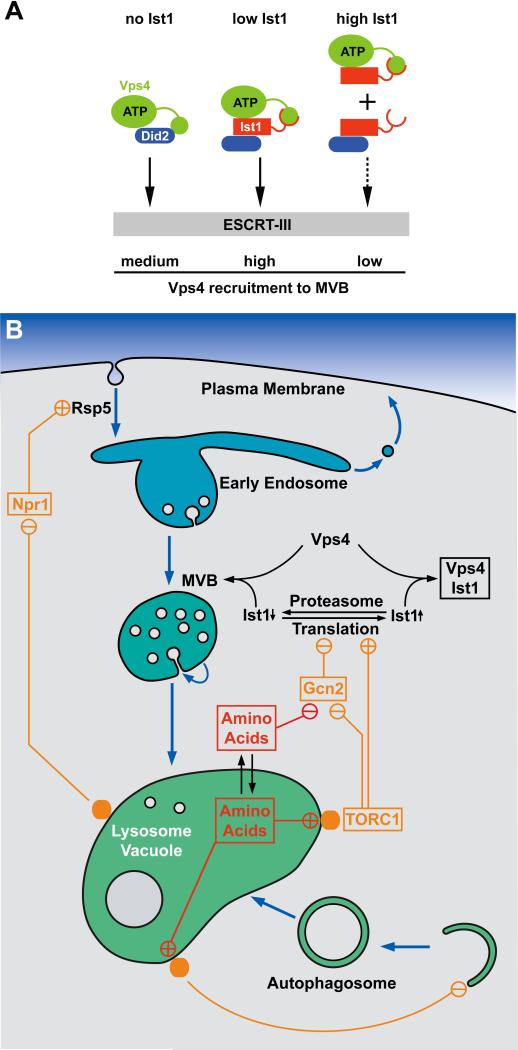
Our data suggested that the activity of the MVB pathway might be modulated according to metabolic conditions through changes in the cellular concentration of Ist1, a protein that binds to the essential ESCRT factor Vps4. Exponentially growing cells have high Ist1 levels; whereas, cells approaching stationary phase show a dramatic decrease in Ist1 levels (Fig. 4). For lack of proper tools, we don't have direct evidence that Ist1 levels regulate Vps4 kinetics in vivo. Our in vitro studies, however, are consistent with a model in which low cellular Ist1 concentrations promote efficient Vps4 recruitment to endosomes by binding simultaneously to both Vps4 and the ESCRT-III protein Did2 (Vps4-Ist1-Did2), whereas high Ist1 levels promote the formation of binary Vps4-Ist1 and Ist1-Did2 complexes that impair Vps4 recruitment (Fig. 8 A). Loss of Ist1 results in intermediate Vps4 recruitment efficiency. Based on the propensity of Ist1 to oligomerize in vitro (16), an alternate model could be proposed in which cellular Ist1 levels determine the oligomeric state of the protein. Low levels might result in primarily monomeric Ist1, which promotes recruitment of Vps4 to MVBs; whereas, high Ist1 levels could cause self-oligomerization and thus result in the inhibition of Vps4 recruitment.
Figure 8.
Regulation of translation, autophagy and the MVB pathway by nutrient-sensing systems. (A) Model for the regulation of Vps4 recruitment to ESCRT-III by cellular Ist1 levels. (B) Model for the concerted regulation of translation initiation, autophagy, the MVB pathway and endocytosis by amino acid levels.
Exponentially growing ist1Δ cells exhibit increased turnover of plasma membrane proteins compared to wild-type cells (Fig. 3 C), suggesting that the high Ist1 concentration in rapidly-growing cells suppresses MVB-dependent degradation of membrane proteins. Consistent with this idea, a recent study has shown that deletion of IST1 resulted in an increase in the size and number of MVB vesicles, suggestive of an increased flux of cargo through the MVB pathway (35). Therefore, an important function of Ist1 might be to suppress membrane protein degradation during the exponential growth phase of the cell, a phase in which cells focus on protein production and cell expansion. Inducing starvation in exponentially growing cells resulted in a rapid drop of Ist1 levels (Fig. 5), which is proposed to increase MVB pathway throughput and, in turn, membrane protein turnover.
The variation in Ist1 levels is not transcriptionally generated, but is the result of changes in the translational output of the cell combined with a rapid turnover rate of Ist1 (Fig. 4, Fig. 5). As such, Ist1 levels are a direct readout of the metabolic state of the cell, providing a connection between the MVB pathway and the TOR and Gcn2 regulatory systems that govern translation initiation. This connection is predicted to result in an inverse coupling of protein synthesis and protein degradation via the MVB pathway: high levels of protein synthesis result in reduced MVB pathway throughput, and conversely, low levels of protein synthesis during starvation result in heightened protein degradation via the MVB pathway (Fig. 8 B).
Starvation conditions or addition of rapamycin have been shown to induce endocytosis and subsequent degradation of numerous plasma membrane transporters, indicating that the TOR signaling pathway is involved in the regulation of membrane protein turnover (22-26). Similarly, we observed that starvation caused rapid internalization and degradation of the iron-transporter Ftr1, the arginine-transporter Can1 and the methionine transporter Mup1. We found that this downregulation is not solely caused by the degradation of Ist1 and the predicted subsequent upregulation of the MVB pathway (Fig. 6), but also seems to involve increased internalization of the transporters through separate mechanisms. Taken together, these observations suggest that starvation induces both endocytosis and MVB sorting of a large set of plasma membrane proteins, leading to increased amino acid recycling. Importantly, the general amino acid transporter Gap1 has been shown to be stabilized at the plasma membrane during starvation (28), further support that starvation does not increase bulk flow uptake of plasma membrane proteins, but enhances endocytosis of a specific set of proteins that are not essential for survival under the given stress conditions.
Under normal growth conditions, Ftr1 degradation is regulated in response to levels of available iron. High iron concentrations result in the ubiquitination of Ftr1 by Rsp5 and the subsequent degradation of Ftr1 via the MVB pathway (21). Starvation-induced degradation of Ftr1 was also found to require Rsp5, suggesting that similar mechanisms are involved in both starvation- and iron-induced Ftr1 downregulation. Npr1 is a downstream effector of TOR, and we found this kinase to be important for the rapamycin-induced downregulation of Ftr1 and other cell-surface proteins. A recent proteomics study identified Rsp5 as a target of Npr1 (36), suggesting that TOR regulates the activity of Rsp5 via Npr1. However, deletion of NPR1, or genes encoding subunits of the TORC2 complex, did not block the rapid downregulation of Ftr1 during leucine starvation, suggesting that an additional, TOR-independent pathway also regulates membrane protein degradation.
Long-term starvation induces autophagy, which, in mammalian cells, is impaired in ESCRT mutants (29, 30). Our analysis indicated that yeast ESCRT mutants are not defective in autophagy induction or autophagosome fusion with the vacuole. Moreover, ESCRT mutants showed slightly induced autophagy even in rich medium, indicating that loss of the MVB pathway possibly decreases steady-state TOR activity. Long-term starvation did not cause a complete loss of the plasma membrane proteins Ftr1, Can1 and Fur4, suggesting that the rapid degradation of plasma membrane proteins observed during the first 1-2 hours of starvation discontinues after some time, and the remaining pool of plasma membrane proteins is not further degraded. This observation fits to the model that the MVB pathway plays an important role for the initial phase of the starvation response, but is eventually replaced by autophagy, the long-term starvation response.
In summary, we propose a model in which MVB-dependent protein degradation is regulated by the growth phase and metabolic state of a cell (Fig. 8 B). Ist1 seems to be involved in this regulation: its levels are regulated by the translational output of the cell, and these varying levels impact the activity of Vps4. High translational output in rapidly growing cells results in high Ist1 protein levels, which impair the function of Vps4 in the MVB pathway. Under these conditions, degradation of membrane proteins is low. Decreased translational activity, as observed in non-growing or starving cells, causes a drop in Ist1 levels due to its intrinsically rapid degradation. This drop increases Vps4 recruitment to MVBs, thereby increasing protein degradation. Additionally, starvation causes increased endocytosis of a large number of plasma membrane proteins, and their subsequent delivery to the MVB pathway.
TORC1 plays an important role in regulating both endocytosis and, via Ist1, the MVB pathway. The TORC1 complex localizes to the vacuolar membrane, which has led to speculation that TORC1 is regulated by the vacuolar amino acid pool rather than the cytoplasmic pool (37). The result of starvation-induced upregulation of endocytosis and vacuolar degradation is a pool of free amino acids that is important for the adaptive synthesis of stress-response proteins. Thus the MVB pathway helps to maintain translation during acute starvation until autophagy, the long-term starvation-response pathway, is fully active.
Materials and methods
Antibodies
HA (hemagglutinin) specific antibody was purchased from Covance (Princeton, NJ). GFP-specific monoclonal antibody was purchased from Roche Applied Science (Indianapolis, IN). Antiserum specific to phospho(S51)-eIF2α was purchased from Abcam (Cambridge, MA). Antisera against Snf7 has been characterized previously (17).
Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. To maintain plasmids, yeast were grown in appropriate complete synthetic dropout medium. Wild type cells as well as those containing genomic integrations or deletions were grown in either rich YPD medium (yeast extract-peptone-dextrose) or complete synthetic media YNB (Yeast Nitrogen Base) with 2% Glucose + Drop-out Mix Synthetic from US Biological (Swampscott, MA) containing the following components in [mg/L]: Adenine [10], Ala [40], Arg [40], Asn [40], Asp [40], Cys [40], Gln [40], Glu [40], Gly [40], His [40], Myo-Inositol [40], Ile [40], Leu [200], Lys [40], Met [40], Para-Aminobenzoic Acid [4], Phe [40], Pro [40], Ser [40], Thr [40], Trp [40], Tyr [40], Uracil [40], Val [40].
DNA Manipulations
Plasmids used in this study are listed in Table 1. All plasmids were constructed using standard cloning techniques. The pRS4XX shuttle vectors used in this study have been described previously (38). Plasmid-based GFP fusions used pEGFP-C1 obtained from Clontech Laboratories Inc. (Palo Alto, CA).
Protein Purification
Proteins used in pulldown assays were expressed in Escherichia coli. Vps4(E233Q) (pMB63), GST-Did2(CT) (pAH32) and Ist1 (pCD2) were purified previously reported (9, 12, 39).
Methods
Survival assays used exponentially growing cells diluted uniformly into YNB (-Leu) composed as detailed above. Cells were grown at 30°C with agitation. Samples were collected daily, diluted 1:106 then plated on YPD (Yeast Extract Peptone Dextrose). Colonies were counted following two days of growth at 30°C; viability was gauged as number of colonies with respect to starting point. Free amino acid analysis of yeast was performed using a Hitachi L8800 Analyzer (Hitachi, Schaumburg, IL). For amino acid analysis of cells starved for leucine, exponentially growing cultures were spun down, washed twice in YNB (-Leu) and resuspended in YNB (-Leu). For each time point, 5 ODV of cells was harvested, washed 3X on ice with Milli-Q water and resuspended in 10% TCA. Cells were lysed with glass beads and vortexing, cell debris was pelleted, and soluble material was spun hard at 4°C for 10 minutes. Supernatants were kept overnight at -20°C, spun hard again at 4°C, and diluted for analysis. Quantitative RT-PCR was performed using standard techniques with actin mRNA as a control. Sub-cellular fractionation was performed as previously described (40). Western blot samples tracking transmembrane protein degradation (Fig. 6 B) were initially TCA precipitated, acetone washed, then lysed with glass beads in SDS-PAGE sample buffer (2% SDS, 0.1 M Tris, pH 6.8, 10% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol) containing 6 M Urea. All other Western blot samples were collected and lysed with glass beads in the presence of SDS-PAGE sample buffer. Western blots were performed using standard techniques. Rapamycin was used at a final concentration of 200 ng/mL. Cycloheximide was used at a final concentration of 25 mg/L. Fluorescence microscopy was performed on a deconvolution microscope (DeltaVision, Applied Precision, Issaquah, WA). GST-pulldown assays were performed using recombinant proteins as previously described (12).
Supplementary Material
Supporting Information
Figure S1. Survival rate of wild-type and vps4Δ strains during amino acid starvation. The graphs show the average and standard deviation from three parallel cultures. (A) Exponentially growing leucine-auxotrophic strains were depleted for leucine and the survival rate was determined. (B) Cells were grown in YNB complete to stationary phase (~14 hours) and the survival rate during stationary phase was determined.
Figure S2. Function and regulation of Vps4. (A) Free amino acid analysis of WT and vps4Δ cells following one hour of leucine starvation. Values are represented as change relative to starting conditions. (B) In vitro binding studies using recombinant Vps4(E233Q), Ist1 and GSTDid2(CT) (amino acids 113-204). GST-Did2(CT) was immobilized on GSH-sepharose and approximately equimolar amounts of Vps4(E233Q) was added in the presence of ATP. The Ist1 concentrations were varied from zero to approximately four times the molar amount of Vps4(E233Q). Bound fractions were analyzed by SDS-PAGE and coomassie staining, and the amount of bound Vps4(E233Q) relative to GSTDid2(CT) was determined by densitometric scanning. The obtained values were plotted relative to the negative control (no GST-Did2(CT) = 0) and the maximum signal (2xIst1 = 1).
Figure S3. Ist1 protein levels and sub-cellular localization. (A) Western blot analysis of Ist1-HA following leucine starvation in strains containing functional Rsp5 (RSP5) or an Rsp5 mutant deficient in ubiquitin conjugation (rsp5-1) (panel 1), and cells expressing either wild-type Ist1 or Ist1 containing a point mutation in a reported C-terminal phosphorylation site, Ist1(S244A)-HA (panel 2). (B) Ist1-GFP localization under normal conditions (con.) and after 20 minutes of leucine starvation.
Figure S4. Protein levels of HA-Ist1, HA-Did2, HA-Vta1 and HA-Vps60 during starvation conditions. (A) Cells expressing N-terminal HA-tagged ESCRT proteins grown in complete medium were shifted to leucine-starvation conditions (t=0) and the effect of starvation on levels of HA-tagged proteins were determined by Western blot (Snf7 serves as a control). (B) Quantification of the Western blots shown in A. Protein amounts are relative to starting conditions and to levels of Snf7, the stable control protein.
Figure S5. Downregulation of Can1, Crh2, and Ftr1 during starvation. (A) Localization of Can1-GFP in untreated (con.), rapamycin-treated, or leucine-starved wild-type or npr1Δ cells. Cells grown to exponential phase in YNB CSM, then treated with rapamycin or transferred to YNB (-Leu). (B) Localization of Crh2-GFP in cells before (con.) and after 1 hr. of rapamycin-treatment, as well as under normal conditions in ist1Δ cells. (C) Localization of Can1-GFP, Ftr1-GFP and Fur4-GFP after 16 hours of starvation (stationary phase). (D) Quantification of the microscopy analysis of wild-type and npr1Δ cells shown in Fig. 6A. Total and intracellular GFP signal was determined for 50 cells from each strain. The graph shows the distribution of cells with a particular intracellular-to-total ratio. Analysis using the Kolmogorov-Smirnov test indicated that all discussed differences are statistically relevant.
Acknowledgements
Special thanks to Inge Stijleman and Philip Bernard for assistance with the RT-PCR experiments. Kelsey Fulkerson aided in strain construction. The yeast strain CBY118 was a gift from Chris Burd. This work has been supported by NIH Grants R01 GM074171.
Abbreviations used in this paper
- MVB
multivesicular body
- ESCRT
endosomal sorting complex required for transport
- TORC1
target of rapamycin complex one
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Proud CG. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009;19(6):260–267. doi: 10.1016/j.tcb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. Integration of general amino acid control and TOR regulatory pathways in nitrogen assimilation in yeast. J Biol Chem. 2010 doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, Neufeld TP. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans. 2009;37(Pt 1):232–236. doi: 10.1042/BST0370232. [DOI] [PubMed] [Google Scholar]
- 6.Piper RC, Katzmann DJ. Biogenesis and Function of Multivesicular Bodies. Annu Rev Cell Dev Biol. 2007 doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belgareh-Touze N, Leon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem Soc Trans. 2008;36(Pt 5):791–796. doi: 10.1042/BST0360791. [DOI] [PubMed] [Google Scholar]
- 8.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 9.Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates vps4 localization and assembly. Mol Biol Cell. 2008;19(2):465–474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20(5):1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20(5):1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010 doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163(1):47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19(2):475–484. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekito T, Fujiki Y, Ohsumi Y, Kakinuma Y. Novel families of vacuolar amino acid transporters. IUBMB Life. 2008;60(8):519–525. doi: 10.1002/iub.92. [DOI] [PubMed] [Google Scholar]
- 16.Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16(7):754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. Embo J. 1998;17(11):2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordallo J, Suarez-Rendueles P. Control of Saccharomyces cerevisiae carboxypeptidase S (CPS1) gene expression under nutrient limitation. Yeast. 1993;9(4):339–349. doi: 10.1002/yea.320090404. [DOI] [PubMed] [Google Scholar]
- 19.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4(3):310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Felice MR, De Domenico I, Li L, Ward DM, Bartok B, Musci G, Kaplan J. Post-transcriptional regulation of the yeast high affinity iron transport system. J Biol Chem. 2005;280(23):22181–22190. doi: 10.1074/jbc.M414663200. [DOI] [PubMed] [Google Scholar]
- 21.Strochlic TI, Schmiedekamp BC, Lee J, Katzmann DJ, Burd CG. Opposing activities of the Snx3-retromer complex and ESCRT proteins mediate regulated cargo sorting at a common endosome. Mol Biol Cell. 2008;19(11):4694–4706. doi: 10.1091/mbc.E08-03-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galan JM, Volland C, Urban-Grimal D, Haguenauer-Tsapis R. The yeast plasma membrane uracil permease is stabilized against stress induced degradation by a point mutation in a cyclin-like “destruction box”. Biochem Biophys Res Commun. 1994;201(2):769–775. doi: 10.1006/bbrc.1994.1767. [DOI] [PubMed] [Google Scholar]
- 23.Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146(6):1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penalver E, Lucero P, Moreno E, Lagunas R. Catabolite inactivation of the maltose transporter in nitrogen-starved yeast could be due to the stimulation of general protein turnover. FEMS Microbiol Lett. 1998;166(2):317–324. doi: 10.1111/j.1574-6968.1998.tb13907.x. [DOI] [PubMed] [Google Scholar]
- 25.Omura F, Kodama Y, Ashikari T. The N-terminal domain of the yeast permease Bap2p plays a role in its degradation. Biochem Biophys Res Commun. 2001;287(5):1045–1050. doi: 10.1006/bbrc.2001.5697. [DOI] [PubMed] [Google Scholar]
- 26.Krampe S, Boles E. Starvation-induced degradation of yeast hexose transporter Hxt7p is dependent on endocytosis, autophagy and the terminal sequences of the permease. FEBS Lett. 2002;513(2-3):193–196. doi: 10.1016/s0014-5793(02)02297-4. [DOI] [PubMed] [Google Scholar]
- 27.Kamura T, Burian D, Khalili H, Schmidt SL, Sato S, Liu WJ, Conrad MN, Conaway RC, Conaway JW, Shilatifard A. Cloning and characterization of ELL-associated proteins EAP45 and EAP20: A role for yeast EAP-like proteins in regulation of gene expression by glucose. J Biol Chem. 2001;5:5. doi: 10.1074/jbc.M010142200. [DOI] [PubMed] [Google Scholar]
- 28.De Craene JO, Soetens O, Andre B. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J Biol Chem. 2001;276(47):43939–43948. doi: 10.1074/jbc.M102944200. [DOI] [PubMed] [Google Scholar]
- 29.Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17(18):1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 30.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17(20):1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 31.Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15(5):2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310(5756):1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 33.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147(2):435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson DP, West M, Henry R, Odorizzi G. Regulators of vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell. 2010;21(6):1023–1032. doi: 10.1091/mbc.E09-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturgill TW, Cohen A, Diefenbacher M, Trautwein M, Martin DE, Hall MN. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7(10):1819–1830. doi: 10.1128/EC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 39.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16(8):1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 41.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8(11):4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3(2):283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 43.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Pena JM, Cid VJ, Arroyo J, Nombela C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol Cell Biol. 2000;20(9):3245–3255. doi: 10.1128/mcb.20.9.3245-3255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Figure S1. Survival rate of wild-type and vps4Δ strains during amino acid starvation. The graphs show the average and standard deviation from three parallel cultures. (A) Exponentially growing leucine-auxotrophic strains were depleted for leucine and the survival rate was determined. (B) Cells were grown in YNB complete to stationary phase (~14 hours) and the survival rate during stationary phase was determined.
Figure S2. Function and regulation of Vps4. (A) Free amino acid analysis of WT and vps4Δ cells following one hour of leucine starvation. Values are represented as change relative to starting conditions. (B) In vitro binding studies using recombinant Vps4(E233Q), Ist1 and GSTDid2(CT) (amino acids 113-204). GST-Did2(CT) was immobilized on GSH-sepharose and approximately equimolar amounts of Vps4(E233Q) was added in the presence of ATP. The Ist1 concentrations were varied from zero to approximately four times the molar amount of Vps4(E233Q). Bound fractions were analyzed by SDS-PAGE and coomassie staining, and the amount of bound Vps4(E233Q) relative to GSTDid2(CT) was determined by densitometric scanning. The obtained values were plotted relative to the negative control (no GST-Did2(CT) = 0) and the maximum signal (2xIst1 = 1).
Figure S3. Ist1 protein levels and sub-cellular localization. (A) Western blot analysis of Ist1-HA following leucine starvation in strains containing functional Rsp5 (RSP5) or an Rsp5 mutant deficient in ubiquitin conjugation (rsp5-1) (panel 1), and cells expressing either wild-type Ist1 or Ist1 containing a point mutation in a reported C-terminal phosphorylation site, Ist1(S244A)-HA (panel 2). (B) Ist1-GFP localization under normal conditions (con.) and after 20 minutes of leucine starvation.
Figure S4. Protein levels of HA-Ist1, HA-Did2, HA-Vta1 and HA-Vps60 during starvation conditions. (A) Cells expressing N-terminal HA-tagged ESCRT proteins grown in complete medium were shifted to leucine-starvation conditions (t=0) and the effect of starvation on levels of HA-tagged proteins were determined by Western blot (Snf7 serves as a control). (B) Quantification of the Western blots shown in A. Protein amounts are relative to starting conditions and to levels of Snf7, the stable control protein.
Figure S5. Downregulation of Can1, Crh2, and Ftr1 during starvation. (A) Localization of Can1-GFP in untreated (con.), rapamycin-treated, or leucine-starved wild-type or npr1Δ cells. Cells grown to exponential phase in YNB CSM, then treated with rapamycin or transferred to YNB (-Leu). (B) Localization of Crh2-GFP in cells before (con.) and after 1 hr. of rapamycin-treatment, as well as under normal conditions in ist1Δ cells. (C) Localization of Can1-GFP, Ftr1-GFP and Fur4-GFP after 16 hours of starvation (stationary phase). (D) Quantification of the microscopy analysis of wild-type and npr1Δ cells shown in Fig. 6A. Total and intracellular GFP signal was determined for 50 cells from each strain. The graph shows the distribution of cells with a particular intracellular-to-total ratio. Analysis using the Kolmogorov-Smirnov test indicated that all discussed differences are statistically relevant.