Abstract
RTKs, the second largest family of membrane receptors, exert control over cell proliferation, differentiation and migration. In recent years, our understanding of RTK structure and activation in health and disease has skyrocketed. Here we describe experimental approaches used to interrogate RTKs, and we review the quantitative biophysical frameworks and structural considerations that shape our understanding of RTK function. We discuss current knowledge about RTK interactions, focusing on the role of different domains in RTK homodimerization, and on the importance and challenges in RTK heterodimerization studies. We also review our understanding of pathogenic RTK mutations, and the underlying physical-chemical causes for the pathologies.
Introduction
RTKs, the second largest family of membrane receptors, transduce biochemical signals upon lateral dimerization in the membrane plane (1–3). RTKs are single-pass membrane proteins, shown schematically in Figure 1. The N-terminal extracellular (EC) domains, usually several hundred amino acids long, vary between families and contain characteristic arrays of structural motifs. The single transmembrane TM domain is followed by a 40 to 80 amino acid juxtamembrane (JM) region and a ~250 amino acid kinase domain, homologous to soluble tyrosine kinases (1;4;5).
Figure 1.
A schematic of RTK architecture, showing the extracellular (EC) domain, the transmembrane (TM) domain and the tyrosine kinase domain. The kinase domain is linked to the TM domain via a 40–80 amino acid juxtamembrane (JM) domain. RTKs transduce biochemical signals via lateral dimerization in the membrane. While the monomers (left) are inactive, the dimers (right) are phosphorylated and active.
RTK dimerization controls and ensures the close contact of the two kinase domains in the dimer. Upon dimerization each kinase domain catalyzes the phosphorylation of critical tyrosine residues in the activation loop of the neighboring kinase (6). This is followed by the phosphorylation of additional tyrosine residues in the juxtamembrane and catalytic domains, which serve as binding sites for docking proteins. Upon recruitment and/or phosphorylation, these docking proteins initiate intracellular signaling cascades that control vital cellular processes (7–9).
RTKs, with a single exception (ErbB2), are ligand-binding proteins. The ligands play intricate roles in the activation process (10), but are not always essential for RTK dimerization and activation. Their contribution to RTK activation is believed to be both thermodynamic (stabilizing RTK dimers) and structural (inducing RTK structural changes which enhance RTK activity).
There are two requirements for successful activation of an RTK. The first requirement is the close approach of the two kinase domains in the dimer. This is accomplished via dimerization. The dimerization efficiency, and therefore the time the kinase domains spend in close proximity of each other, is controlled by the dimerization constant. Dimerization is modulated by ligand binding, and thus the proximity of the kinase domains is also regulated by the ligand concentration and the ligand binding constant. The second requirement for successful activation of an RTK is that the structure of the kinase domain dimer is suited for the transfer of a PO4 group from a bound ATP molecule to a particular tyrosine (9;11). The phosphorylation-competent dimer structure is established via specific receptor-receptor and receptor-ligand interactions. Structural constraints in the extracellular and TM domains, imposed upon dimerization and ligand binding, propagate into the kinase domain and control RTK phosphorylation and activity.
Physical - chemical models of RTK activation
We and others have proposed that RTK activation can be described with physical-chemical models which account for dimerization, ligand binding and phosphorylation (12–16). Such models have been shown to give an adequate description of RTK phosphorylation data, despite their simplicity. Usually the models do not take explicitly into account all interactions that regulate RTK activation in cellular membranes. Rather, all these events contribute to apparent constants describing different steps in RTK activation. Here we provide a brief overview of such models.
A. Dimerization
Ligand-independent lateral dimerization, i. e., the lateral association of two RTK monomers into an unliganded dimer, is the first critical step in RTK activation. It is controlled by the dimerization constant K1 and is given by the following reaction scheme:
| (1) |
The dimerization constant K1 is defined as K1 = [d]/[M]2, with [M] and [d] being the monomer and dimer concentrations, respectively. The total receptor concentration is [T] = 2[d] + [M]. The free energy of dimerization is calculated as ΔG = −RT ln K1.
Unliganded RTK dimers can be phosphorylated or not. If the unliganded dimers are phosphorylated, equation (1) accounts for the so-called “basal or constitutive activation”. If the unliganded RTK dimers are not phosphorylated (inactive), equation (1) accounts for the so-called “pre-dimerization”. These two terms are vaguely defined in biology, but they have the same physical-chemical basis, i.e. equation (1).
B. Ligand binding
The next step in RTK activation is ligand binding, which is believed to stabilize RTK dimers and likely alter their structure (discussed below). The processes of dimerization and ligand binding are coupled, and can be described by the following reaction scheme:
| (2) |
where D denotes the liganded dimer (see also Figure 2).
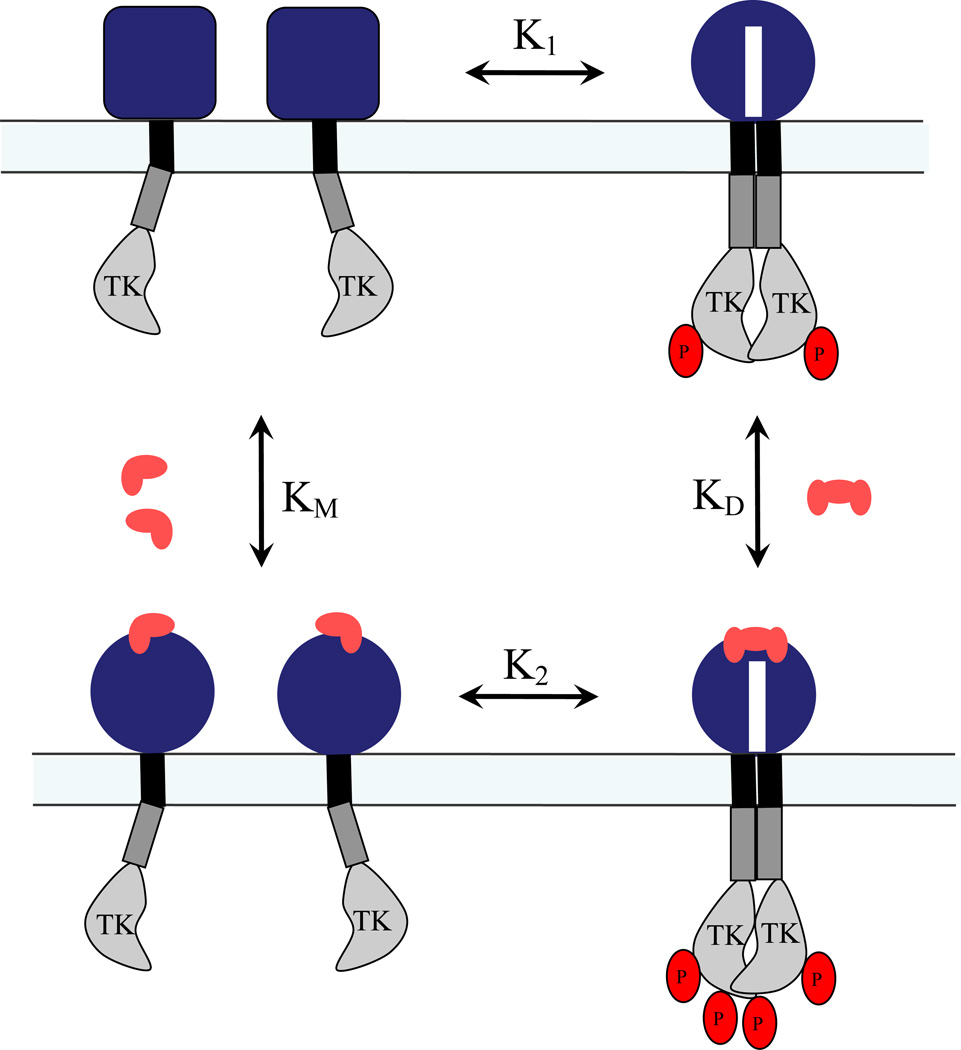
Figure 2.
A schematic representation of the activation model given by equation (2). Two monomers interact laterally to form dimers, both in the absence and presence of bound ligand (dimerization constants K1 and K2, respectively). The ligand binds to both monomers (association constant KM) and dimers (association constant KD). The receptors in the unliganded and liganded dimeric states are phosphorylated, but the probabilities for phosphorylation may be different (Φd for the unliganded state and ΦD for the liganded state).
For the sake of simplicity, sometimes it is assumed that the ligand binds preferentially to the unliganded dimers, rather than the monomers (14). In this case, scheme (2) is reduced to the following two coupled reactions:
| (3) |
In scheme (3), the dimerization and the ligand binding constants are defined as K1=[d]/[M]2 and KD=[D]/[d][L]2, respectively. The dimerization reaction is driven, in part, by ligand binding: Ligand binding depletes the unliganded dimers by converting them to liganded dimers, thus promoting the dimerization of the monomers.
All three schemes, (1) through (3), as well as more complex models accounting for negative cooperativity in ligand binding, have been used in the literature to rationalize experimental data and to gain insights into how dimerization and ligand binding regulate RTK activation (12–16). While these models are simple and do not capture the full complexity of RTK signaling, they are very useful in comparative studies, such as studies of the effects of sequence variations and pathogenic mutations on different steps in RTK activation, as discussed below.
C. Phosphorylation
In the dimer, the two receptors autophosphorylate each other, with each kinase domain acting as an enzyme which facilitates the transfer of a phosphate group to the neighboring kinase. One way to account for the efficiency of phosphorylation is to define receptor phosphorylation probabilities, Φd and ΦD, within the unliganded and liganded dimers, respectively (17). The concentration of phosphorylated receptors is then calculated according to:
| (4) |
Note that Φd = 0 corresponds to inactive unliganded dimers and describes predimerization, while Φd ≠ 0 describes active unliganded dimers and accounts for basal or constitutive activation.
Experimental characterization of RTKs
A. Measurements of RTK dimerization
To characterize RTK dimerization in the context of equation (1), one needs to measure directly two of the following three parameters over a range of RTK concentrations: (i) concentrations of RTK dimers, (ii) concentrations of RTK monomers, and (iii) total RTK concentrations. Quantitative measurements of these concentrations, and thus calculations of dimerization constants for full-length RTKs is a challenge. However, some widely used biochemical methods allow us to estimate and compare dimerization propensities.
One such method is chemical cross-linking (18–21). In this method the cells are incubated with a cross-linker, lysed, and analyzed using Western blotting. Since only RTKs in very close proximity can be cross-linked, the presence of cross-linked bands on Western blots suggests that dimers exist on the cell surface. Furthermore, higher cross-linking correlates with higher dimerization. While the method is useful and well established, it should be kept in mind that the cross-linker is non-specific, such that it cross-links all proteins in close proximity. As a result, gels of cross-linked proteins are usually smeared, and very difficult to accurately quantify. Furthermore, a limitation of this experimental approach is that the cross-linking propensities depend not only on close proximity, but also on structure. Thus, a structural change can alter the probability for RTK cross-linking, despite the fact that RTK dimerization is not affected.
Another method used in the literature for assessment of RTK dimerization is immunoprecipitation, followed by SDS-PAGE (22). Disulfide-linked RTK dimers have been observed using this technique under oxidizing conditions without cross-linking. In this case, however, the assay reports on interactions occurring within the immunoprecipitates. These might be different from interactions in the native plasma membrane, which imposes structural constraints on RTK dimers. In particular, the presence of unpaired cysteines in the receptors may lead to the formation of non-native disulfide bonds in the immunoprecipitate and give a false positive for dimer formation.
A powerful method used to study dimerization is Förster resonance energy transfer (FRET) (23–27). FRET involves the non-radiative transfer from a fluorescent donor to a fluorescent acceptor (28–32), and is manifested in a decrease in donor fluorescence and an increase in acceptor fluorescence (30–32) upon dimerization. The efficiency of energy transfer E is inversely proportional to the sixth power of the distance, r, between the donor and acceptor. The transfer efficiency E is a function of r and Ro, the characteristic Förster radius for the donor and acceptor pair:
| (5) |
Typical donor/acceptor pairs, such as the widely used fluorescent proteins, have Ro of 50–60 Å. Thus, if the two fluorophores are closer than 50 Å in a dimer, FRET will occur.
FRET can be measured in the native cellular environment. In these experiments the genes encoding the RTKs are modified by attaching sequences encoding fluorescent proteins, usually at RTK’s C-termini. While most in-cell FRET experiments measure the sensitized acceptor emission, sophisticated FRET techniques have been developed to assess donor quenching (33–38). When measuring interactions in membranes using FRET, however, special care needs to be exercised since FRET occurs even if there are no specific interactions, due to random co-localization of donors and acceptors (39–42). Furthermore, challenges in data interpretation arise because the cellular environment is highly heterogeneous, and supramolecular organizations in clusters/domains introduce additional heterogeneities affecting the measured FRET efficiency and complicating data analysis.
To overcome the above challenges, we have established plasma membrane-derived vesicles (43–48) as a model system for studies of RTK interactions in mammalian membranes via FRET (Figure 3). Plasma membrane-derived vesicles are produced using either a mechanical method, by breaking the plasma membrane, or using chemical methods, by disrupting the cytoskeleton in a direct or indirect (apoptotic) way (49–52). Plasma membrane-derived vesicles are a simplified model of the cell membrane because there is no cytoskeleton and no TM potential (43). Yet, plasma membrane-derived vesicles possess complex features that are characteristic of native cellular membranes (43–48). Their lipid composition is similar to the one in the native membrane (43), and they maintain the plasma membrane asymmetry (44–46). The membranes of the vesicles contain various membrane proteins and mimic the natural crowded membrane environment.
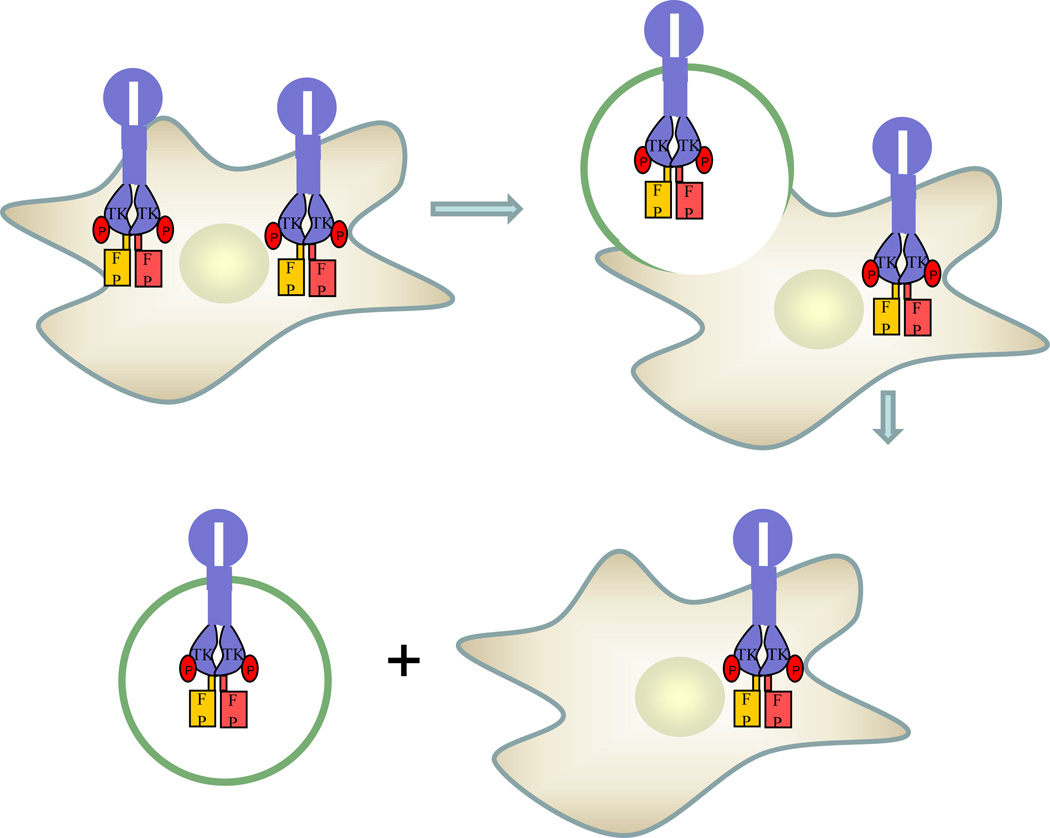
Figure 3.
Overview of RTK dimerization measurements in plasma membrane-derived vesicles. Cells are transfected with genes encoding RTKs fused to fluorescent proteins, and then vesiculated using established protocols. The vesicles are imaged in a confocal microscope, acquiring donor, acceptor and FRET images for each vesicle. The QI-FRET method, described in (53–55), is used to determine the donor and acceptor concentration, and the FRET efficiency in each vesicle. This information is then used to determine the concentrations of monomers and dimers in each vesicle, the dimerization constant K1 and the free energy of dimerization.
We have demonstrated that quantitative measurements of RTK dimerization in single vesicles are feasible with the QI-FRET method (53). The QI-FRET method, discussed in (53–55), yields the unknown donor and acceptor concentrations, and the FRET efficiency (i.e. the three parameters that are sufficient to calculate free energies of dimerization) (38). Because the experiments utilize transient transfection, expression levels vary from cell to cell, and vesicles with a wide range of receptor concentrations (i.e., number of receptors per unit membrane area) can be produced in a single transfection experiment. Thus, a wide protein concentration range is sampled, and the dimerization energetics are measured for different receptor concentrations, to obtain dimerization curves.
Using this methodology, we have measured the energetics of FGFR3 TM domain dimerization in the plasma membrane of CHO cells, as well as the dimerization energetics of a construct containing both FGFR3 EC and TM domains (55). The two dimerization free energies were determined as ΔG(TM) = −RTlnKD(TM) = −4.2 ± 0.2 kcal/mole and ΔG(EC+TM) = −3.3 ± 0.1 kcal/mole, respectively.
B. Characterization of ligand binding
The binding of ligands to full-length receptors on the surface of living cells is usually measured by quantifying bound radiolabeled or fluorescently-labeled ligands. Binding of ligands to isolated RTK extracellular domains can be characterized using isothermal titration calorimetry, ultracentrifugation, and surface plasmon resonance (SPR), as described below.
The most popular method to probe interactions between ligands and cell surface receptors involves quantification of bound radiolabeled ligands. In these experiments, cells are incubated with radiolabled ligands either on ice or at ambient temperature (to minimize receptor cell uptake). The free ligands are removed, a process which usually requires extensive washing with concentrated salt solutions. Then the cells are lysed and the radioactivity of the receptor-bound ligands is measured. Binding curves can be generated by performing the measurements at different concentrations of ligands. This method has been used to investigate EGF binding affinity to EGFR (56;57), and the interactions between FGF1 and FGF receptors (58;59). Furthermore, the bound ligands can be crosslinked to the receptors, such that the ligand-receptor complexes remain intact when the cell lysates are run on SDS-PAGE gels (59).
Fluorescence-based methods are an alternative to the traditional radiolabeling approach since the fluorescence intensity of the bound ligand is easily measured in a standard spectrofluorometer (60). The receptor–ligand interactions on the cell surface can be further investigated using single molecule fluorescence techniques. These techniques have allowed the visualization of single bound EGF molecules, and have provided insight into the binding kinetics (61;62). Fluorescence correlation spectroscopy (FCS) with single-molecule sensitivity is also used to quantitatively analyze ligand-receptor interaction on surface of living cell. This technique has yielded the dissociation constant for EGF-EGFR interactions, via the analysis of fluorescence intensity fluctuations in a small confocal volume (63).
Ligand binding to the isolated RTK extracellular domains has also been characterized. For instance, isothermal titration calorimetry, ultracentrifugation and molecular modeling (64) has been used to characterize the energetics and kinetics of the interactions between FGF1, FGFR1 extracellular domain, and heparan sulfate in solution. Alternatively, the ligands or the extracellular domains can be attached to a surface, and the interactions can be analyzed using SPR. Such studies have provided valuable insights about the specificity in FGF-FGFR interactions (65–67).
C. RTK phosphorylation measurements
Methods to measure RTK phosphorylation are well established. Phosphorylation levels are monitored by either (1) in-vitro kinase assays, (2) quantification of bound antibodies that specifically recognize phosphorylated tyrosines, or (3) mass spectrometry identifying attached phosphate groups.
In vitro kinase assays often utilize radioactive phosphorous. In these assays cell lysates containing RTKs of interest are incubated with kinase buffer with ATP containing radiolabeled phosphates. The radioactivity of the receptors is quantified upon the transfer of the radioactive phosphate groups to the kinase domains (68). Alternatively, RTK phosphorylation can be quantified without the use of radioactivity by coupling it with the oxidation of NADH, and then following NADH oxidation using spectrophotometric assays (69).
RTK phosphorylation can be measured with the use of antibodies that recognize phosphorylated tyrosines. The amounts of such bound antibodies can be determined using traditional Western blots, ELISA, or in-cell Western blot techniques. These experiments usually involve secondary antibodies which interact with the primary ones, and are conjugated to reporter moieties, detectable using either chemiluminescence or fluorescence. Two different types of primary antibodies can be used: (1) antibodies recognizing all phospho-tyrosines, such as the 4G10 anti-phosphotyrosine antibody, capable of detecting over 30 tyrosine kinases, including ErbB2 (70), PDGFR (71), and FGFR3 (72), and (2) antibodies generated against specific phosphorylated sites, recognizing a particular phosphotyrosine together with the sequence around it (14;15). The latter are useful because RTK kinase domains normally contain more than one tyrosine residue, and the phosphorylation of the different tyrosines occurs sequentially (73;74).
In recent years, mass-spectrometry has become popular in studies of RTK phosphorylation (75–77), and we foresee that its usage will continue to increase. In this method, the phosphorylated RTKs are purified via immunoprecipitation and are digested with trypsin. The products of digestion (peptides) are analyzed by either Matrix-associated laser desorption ionisation time-of-flight Mass Spectrometry (MALDI-TOF MS) or Electrospray Ionization Mass Spectrometry (ESI-MS) (78). By comparing the masses of the peptides with a “theoretical digest”, the presence of phosphate groups can be detected. This method has been used to identify the phosphorylation sites of PDGFR (79) and EGFR (80).
Structural requirements for RTK activation
As discussed above, RTK activation requires precise orientation and positioning of the catalytic domains with respect to each other, such that the phosphate group can be successfully transferred from ATP to the neighboring receptor. For EGFR, this entails the formation of an asymmetric kinase dimer in which the C-lobe of one kinase contacts the N-lobe of the second kinase and positions the activation loop of the second kinase to catalyze phosphate group transfer (9). The existence of asymmetric dimers have been proposed for FGF receptors, too (81). It is not yet clear, however, whether such asymmetric dimers form for all RTKs, and whether the two kinases alternate over time and act both as catalysts and substrates.
The order of the phosphorylation of the different tyrosines is also controlled by the structure of the kinase domain. For instance, the order of tyrosine phosphorylation in FGFR1 kinase domain is strict, and kinetically controlled. It is limited by the rate of transfer of the phosphate group from ATP to the tyrosines (73), and strongly influenced by the kinase tertiary structure (73;74).
The structure and orientation of the kinase domains, on the other hand, are controlled by specific interactions between the TM domains, as well as interactions between the EC domains and the ligands, as discussed below.
Structural constraints imposed by the TM domain dimers
The TM domain structure has been shown to control the orientation of the kinase domains in the dimer, such that successful phosphorylation can occur (11). In particular, rotation of the dimer interface has been shown to induce periodic oscillations in kinase activity (11). Therefore, the structure of the TM domain dimer is an important determinant in RTK signal transduction. About 20 years ago, a five-residue sequence motif (the so-called P0–P4 motif) was proposed to mediate dimerization of RTK TM domains (82). The characteristics of the P0-P4 motif are: P0 exhibits a small side chain, such as Gly, Ala, Ser, Thr, or Pro; P3 requires an aliphatic side chain, i.e. Ala, Val, Leu, or Ile; and P4 must be a Gly or Ala residue. The P0-P4 motif is similar to the GxxxG motif (83;84), shown to be important for Glycophorin A (GpA) dimerization. Later, the TM sequences of ErbB1, ErbB2, and ErbB4 were noticed to have at least two such GxxxG-like motifs (85), and the different GxxxG-like motifs were proposed to mediate either homo or hetero-dimerization (90;91). Alternatively, based on calculations of dimerization free energies of alternate ErbB2 TM domain structures, it was proposed that some RTK TM domains may form two alternative homodimer structures utilizing different GxxxG-like motifs, a phosphorylation-competent structure and an inactive structure (17;86;87).
The solved crystal structure of the ErbB2 TM dimer provided demonstration that dimerization occurs via the N terminal GxxxG dimerization motif, while the C terminal motif does not participate in the dimer interface (88). A similar GxxxG-like motif is involved in the dimerization of a different RTK, EphA1 (89). Yet, the GxxxG motif is not required for the dimerization of all RTKs. For instance, the dimer interfaces of ErbB3, EphA2, PDGFR and FGFR3 TM domains do not involve GxxxG-like motifs, despite the fact that their sequences contain several such motifs (92–96).
Structures of ligand-bound extracellular domain dimers
The solved crystal structures of isolated RTK extracellular domains reveal tight contacts between the two EC domains, and between the EC domains and ligands. The contacts between EGFR EC domains are mediated via a “dimerization arm”, exposed only upon ligand binding (97–99). A recent crystal structure of an EGFR drosophila variant reveals very strong interactions between the two EC domains when only one ligand is bound to dimer (100).
In the FGF2 structure found to FGFR1, there are many contacts between each ligand and the two receptors, as well as direct receptor-receptor interactions (101–103). The structure further reveals a positively-charged lysine-rich “canyon” where heparin oligosaccharides bind, interacting with both the extracellular domains and the ligands, further stabilizing the dimer. Growth factors for families other than EGFRs and FGFRs, (such as KIT and VEGFR), are covalently linked homodimers that bring the extracellular domains together, which promotes additional interactions between the extracellular domains (104;105).
Structural changes mediated by ligands
The ligands of some receptors are believed to not only introduce a structural change in the extracellular domain, but also affect the conformation of the kinase domain, thus increasing RTK activation. For instance, the EGFR unliganded dimer is believed to be inactive because the asymmetric kinase dimer, required for activity (9), cannot form in the absence of ligand. Ligand binding likely induces a rotation in the EGFR dimer and ensures correct positioning of the two kinase domains for phosphorylation (106). The dimeric ligand of KIT, believed to stabilize the dimer by cross-linking the two monomers (104), also induces a structural change which enables receptor-receptor interactions. These interactions are weak as compared to ligand-mediated dimer stabilization and thus they do not drive dimerization by themselves. However, these interactions define the relative orientations of the two receptors, which likely helps to position the kinase domains in the correct orientation for productive phosphorylation.
RTK involvement in human disease
A. Pathogenic mutations in RTKs
Since RTKs play a key role in the regulation of cellular processes that are critical for cell growth, differentiation, and motility, defects in their activation lead to human pathologies. There are many pathogenic single amino acid mutations in RTKs that are usually (but not always) gain-of-function mutations. Because RTK activity is determined by RTK phosphorylation, studies of pathogenic mutations invariably involve experiments which assess the effect of these mutations on phosphorylation. The simplest way to address this question is to compare wild-type and mutant expression and phosphorylation, side-by-side on Western blots. If the expression of the wild-type and the mutant are the same, then the phosphorylation levels can be directly compared. Such direct comparisons have shed light on the molecular basis behind many pathologies (68;107–109).
Direct comparisons of phosphorylation on Western blots have also shed light on how different mutations in a particular RTK can give rise to different phenotypes. One such study compared the phosphorylation of two FGFR3 mutants, linked to achondroplasia (ACH) and thanatophoric dysplasia (TD) (109). ACH, the most common form of human dwarfism(110;111), is a relatively mild phenotype characterized by short stature (112).
On the other hand, TD is much more severe and always lethal in the neonatal period, characterized by severe shortening of the limbs, macrocephaly, and a narrow thorax with small ribs (112). The phosphorylation of the R248C and K650E mutants, associated with TD, has been shown to be higher than the phosphorylation of the G380R mutant linked to ACH (109). Thus, there is a link between higher phosphorylation and more severe phenotypes.
The effect of pathogenic mutations may be diverse, and may include processing defects, such as impeded trafficking and defective downregulation (111;113;114). In many cases, the expression of the mutant is different from the expression of the wild-type in cell lines that are widely used in biomedical research. If the expressions are different, side-by-side comparison of phosphorylation on Western blots is not very informative. To be able to carry out such a comparison, we need to obtain an absolute measure of the phosphorylation at a particular expression level. We have shown that measurements of “phosphorylated fractions” as a function of expression levels can serve this purpose (15;16). In these experiments we treat RTKs with their ligands and we measure phosphorylation over a very wide range of ligand concentrations, including very high ligand concentrations (14): At high ligand concentration all receptors that are exposed to ligand and capable of binding ligand are driven to their liganded dimeric state. At these levels, phosphorylation is saturated and is not further increased when more ligand is added, providing a measure of the maximum possible phosphorylation (14;16). Phosphorylated fractions are then determined as the ratio of measured phosphorylation at a particular ligand concentration over the maximum possible phosphorylation (14;16), and are independent of the specific conditions used in the Western blot experiments. Using this technique, we have compared the phosphorylation of wild-type FGFR3 and the pathogenic A391E mutant linked to Crouzon syndrome in the absence of ligand, demonstrating an increase in FGFR3 phosphorylation due to the A391E mutation, despite the fact that the expression of the wild-type and the mutant were different (15).
B. Physical-chemical causes for RTK-linked pathologies
Altered dimerization
Some pathogenic mutations stabilize RTK dimers, by promoting more intimate contacts between the two mutant receptors in the dimer (see Table 1 for examples). As a result, the dimerization propensity (i.e. K1 in schemes (1) through (3)) increases. The distribution between inactive monomers and active dimers shifts towards the active dimeric state, increasing the over-all RTK activity.
Table 1.
A list of characterized pathogenic RTK mutations, and the underlying physical-chemical cause for the pathologies. All RTKs are human, unless stated otherwise.
| RTK | Mutations | Location | Phenotype | Cause for pathology |
|---|---|---|---|---|
| FGFR1 | P252R | Extracellular domain | Pfeiffer syndrome | Increased ligand binding (145). |
| FGFR1 | Y372C | Extracellular domain | Osteoglophonic dysplasia | Increased dimerization (103). |
| FGFR2 | S252W, P253R | Extracellular domain | Apert syndrome | Increased ligand binding (146). |
| FGFR2 | D321A | Extracellular domain | Pfeiffer syndrome | Increased ligand binding (146). |
| FGFR2 | C278F, C342Y, | Extracellular domain | Crouzon syndrome and Pfeiffer Syndrome | Increased dimerization (22). |
| FGFR2 | W290G, S354C, Y340H C342Y | Extracellular domain | Crouzon syndrome | Increased dimerization (22;147). |
| FGFR2 | T341P | Extracellular domain | Pfeiffer syndrome | Increased dimerization (22). |
| FGFR3 | R248C, S249C | Extracellular domain | Thanatophoric dysplasia type I | Increased dimerization (148). |
| FGFR3 | P250R | Extracellular domain | Muenke syndrome | Increased ligand binding (145). |
| FGFR3 | G370C, S371C, Y373C | Transmembrane domain | Thanatophoric dysplasia type I | Increased dimerization(148;149). |
| FGFR3 | G375C | Transmembrane domain | Achondroplasia | Increased dimerization (149). |
| FGFR3 | G380R | Transmembrane domain | Achondroplasia | Structural change (14). |
| FGFR3 | A391E | Transmembrane domain | Crouzon syndrome with acanthosis nigricans | Increased dimerization (15). |
| FGFR3 | K650E | Kinase domain | Thanatophoric dysplasia type 2 | Structural change in the kinase domain (150). |
| FGFR4 | Y367C | Extracellular domain | Breast cancer | Increased dimerization (151). |
| EGFR | EGFRvIII. 6-273 deletion | Extracellular domain | Glioblastoma | Structural change (152). |
| EGFR | T263P, A289V, P596L, G598V | Extracellular domain | Glioblastoma | Increased dimerization (153). |
| EGFR | G719S | Kinase domain | Non-small-cell lung carcinoma | Structural change in the kinase domain (154). |
| EGFR | L858R | Kinase domain | Non-small-cell lung carcinoma | Structural change in the kinase domain and increased ligand binding (9;155). |
| EGFR | 752–759 deletion and L861Q | Kinase domain | Non-small-cell lung carcinoma | Structural change in the kinase domain (155). |
| EGFR | 959–1030 deletion | C-terminal tail | Glioblastoma | Structural change in the kinase domain (156). |
| ErbB2 (Rat) | V664E | Transmembrane domain | Oncogenic in rat | Increased dimerization (16). |
| KIT | 417–419 deletion and T417I | Extracellular domain | Acute myeloid leukemia (AML) | Increased dimerization (157). |
| KIT | V559D | Juxtamembrane domain | Gastrointestinal stromal tumor | Structural change in the juxtamembrane domain (158) (159). |
| KIT | V560G | Juxtamembrane domain | Mastocytosis | Increased dimerization (158;160). |
| KIT (Murine) | KΔ27.547–555 deletion and D560K | Juxtamembrane domain | Mastocytoma | Increased dimerization (161). |
| KIT | D816V | Kinase domain | Mastocytosis | Structural change and increased dimerization (under debate) (162–164). |
| PDGFRA | V561D | Juxtamembrane domain | Gastrointestinal stromal tumors | Increased dimerization (165). |
| PDGFRA | D842V | Kinase domain | Gastrointestinal stromal tumors | Increased dimerization (165). |
To test for enhanced dimerization due to pathogenic mutations, one needs to use a direct dimerization assay such as the one shown in Figure 3. In our lab we have used this assay to investigate the effect of two pathogenic FGFR3 mutations, A391E and G380R. While the A391E mutation linked to Crouzon syndrome increases FGFR3 dimerization, the G380R mutation linked to ACH does not (paper in preparation). This behavior correlates with the mutation-induced stabilization of the isolated FGFR3 TM domain dimers in lipid bilayers (95;115).
Receptor overactivation due to increased dimerization may occur even for wild-type receptors if the receptor is overexpressed. This mechanism can be understood using equation (1). As the total concentration of receptors increases, the equilibrium is shifted towards the active dimeric state.
Altered ligand binding
There are pathogenic RTK mutations that increase ligand binding (KM or KD in schemes (2) and (3)). Increased ligand binding leads to dimer overstabilization and enhanced phosphorylation. Techniques that assess ligand binding strengths are well established (and reviewed above), and can be used to compare the binding of ligands to wild-type and mutant receptors. Such direct binding measurements, for example, have demonstrated aberrant ligand binding to FGFR mutants implicated in human growth disorders (Table 1).
Structural changes in the kinase domains
Some pathogenic mutations induce structural changes in RTK dimers (Table 1), which affect the receptor phosphorylation probabilities, Φd and ΦD in equation (4). In this case the number of dimers is not increased, but the mutant dimers are more active than the wild-type dimers, leading to an over-all increase in activity. Presumably, this occurs because it is easier to phosphorylate critical tyrosines in the mutant dimers, as compared to the wild-type dimers (116). Such structural changes may originate outside the kinase domain and propagate throughout the whole structure (106).
High resolution structures of full-length RTKs are not available yet, due to experimental challenges in the expression of full-length RTKs in large quantities (117). Since the determination of full-length RTK dimer structures is challenging, studies of the effect of pathogenic mutations on dimerization, ligand binding, and phosphorylation may help us deduce possible structural changes in mutant RTK dimers. For example, the G380R mutation increases FGFR3 phosphorylation, but does not increase FGFR3 cross-linking or ligand binding (14). We have interpreted these findings as an indication for a structural change. In support of this view, a molecular model of the FGFR3 TM dimer structure suggests that the mutation induces a rotation in the TM dimer interface (14). On the other hand, the changes in the phosphorylation and dimerization propensities due to the A391E mutation linked to Crouzon syndrome are very similar, suggesting that the mutation likely affects FGFR3 dimerization propensity, but does not induce a significant structural change (15;95).
Multiple effects
Some mutations may affect multiple steps in the RTK activation process. An approach to understand the effect of pathogenic mutations on dimerization, ligand binding, or receptor phosphorylation probabilities is to model RTK activation using physical-chemical models such as the ones given by equations (1) through (4). By fitting the models to experimental data, we can determine if a mutation affects dimerization (i.e K1 or K2), ligand binding, (KL or KM) or phosphorylation (Φd or ΦD). In one example, Pike and colleagues measured EGF binding to EGFR while varying both the receptor and ligand concentrations (12;13). By fitting a model similar to the one given by equation (2) to their data, they were able to determine that an engineered mutation in the extracellular domain of EGFR, Y246D, decreases ligand binding, while an L680N mutation in the kinase domain of EGFR affects both EGFR dimerization and ligand binding. While quantitative measurements of RTK phosphorylation, dimerization, and ligand binding may be very tedious, their pay-off could be significant: The understanding of how a mutation affects RTK activity is the first step towards the development of new therapeutic strategies with high efficiencies and low toxicities.
Thermodynamics of RTK dimerization
In this last section, we overview recent findings that pertain to the thermodynamics of RTK dimerization as a regulator of RTK activity.
A. Contributions of RTK domains to RTK dimerization thermodynamics
Transmembrane (TM) domains
Biophysical studies of the isolated RTK TM domains have provided a direct assessment of the contribution of RTK TM domains to RTK dimerization energetics. All isolated RTK TM domains have been shown to dimerize in bacterial membranes, with the dimerization strength varying between families and within the families (118). Quantitative studies of dimerization in lipid vesicles have shown that the contribution of FGFR3, ErbB1 and EphA TM domains to dimerization is about −3 kcal/mole (119–123). FGFR3 TM domains has been further shown to drive ligand-independent dimerization in plasma membrane-derived vesicles in the absence of ligand in the presence of the EC domain (54). As a result, sequence changes in RTK TM domains affect RTK dimerization at zero ligand and at low ligand concentrations (124;125). At high ligand concentrations, however, the contribution of the TM domains to dimerization is overshadowed by the large contributions of the EC domains and ligands. Thus, TM domain sequence alterations do not affect RTK dimerization and activation significantly at high ligand concentrations (124;126).
Extracellular domains and ligands
Based on the solved dimeric structures of isolated extracellular domains bound to ligand, it can be expected that ligand binding stabilizes RTK dimers. Indeed, in all such structures, despite a great deal of structural diversity, we see tight contacts between the two EC domains in the dimer and between the EC domains and ligands (discussed above). However, this view was questioned recently for EGFR in a study of EGFR diffusion, as ligand binding per se did not appear to stabilize EGFR dimers (127).
The crystal structure of the EC domain dimer of a drosophila EGFR variant shows very intimate contacts between the two EC domains when only one ligand is bound (100). The comparison of this structure to the symmetric human EGFR structure in the presence of two bound ligands (97;128) suggests that the binding of the first ligand is strongly stabilizing, while the second ligand binding event decreases dimer stability. This new finding suggests that the mechanism of ligand-induced RTK dimer stabilization is unexpectedly complex.
In the absence of ligand, the EC domains have been shown to inhibit dimerization. The contribution of FGFR3 extracellular domain to dimerization has been demonstrated to be inhibitory in plasma membrane derived vesicles, and its magnitude has been measured as ΔΔG = 0.9 ± 0.2 kcal/mole (55). Thus, the EC domains play a dual thermodynamic role in RTK dimerization, inhibiting dimerization in the absence of ligand and stabilizing the dimers in the presence of ligand.
Catalytic domains
The cross-phosphorylation of the kinase domains implies the occurrence of contacts between them (7–9). These contacts are likely be stabilizing. Yet, the contribution of the kinase domains to RTK dimerization energetics has not been measured experimentally thus far. It is possible that this contribution depends on the phosphorylation state. The JM domain, a 40 to 80 amino acid-long sequence between the TM domain and the catalytic domain may be further contributing to the interactions, by stabilizing the active conformation of the kinase domain, and mediating direct receptor-receptor contacts (129;130).
B. RTK heterodimerization
Heterodimerization between RTKs is a means of signal amplification, as well as diversification (131). RTK heterodimers have been shown to enhance receptor activation and downsteam signaling, as compared to homodimers (131–133). For instance, the ErbB2/ErbB3 heterodimer is believed to be the most biologically active and the most pro-tumorigenic of all ErbB homo and heterodimers (134;135). Multiple studies, focusing primarily on the ErbB family of receptors, have demonstrated the importance of heterodimerization in normal function and in disease (136–140). For example, ErbB1/ErbB2, ErbB1/ErbB4, and ErbB2/ErbB4 heterodimers have been shown to play a role in cell transformation. Furthermore, ErbB3 is overexpressed in many tumors that overexpress ErbB2, including breast, bladder, and melanomas (131;140). Tumors that overexpress ErbB2 also exhibit elevated ErbB3 phosphorylation levels (134). Yet, our understanding of RTK heterodimerization is only rudimentary, in part due to a paucity of methods that provide quantitative information about RTK heterodimerization. Thus far, heterodimerization between the isolated ErbB TM domains has been studied in detergents using FRET (141) and in bacterial membranes using the genetic assay GALLEX (91). These studies have suggested that ErbB TM domains form both homodimers and heterodimers with various stabilities.
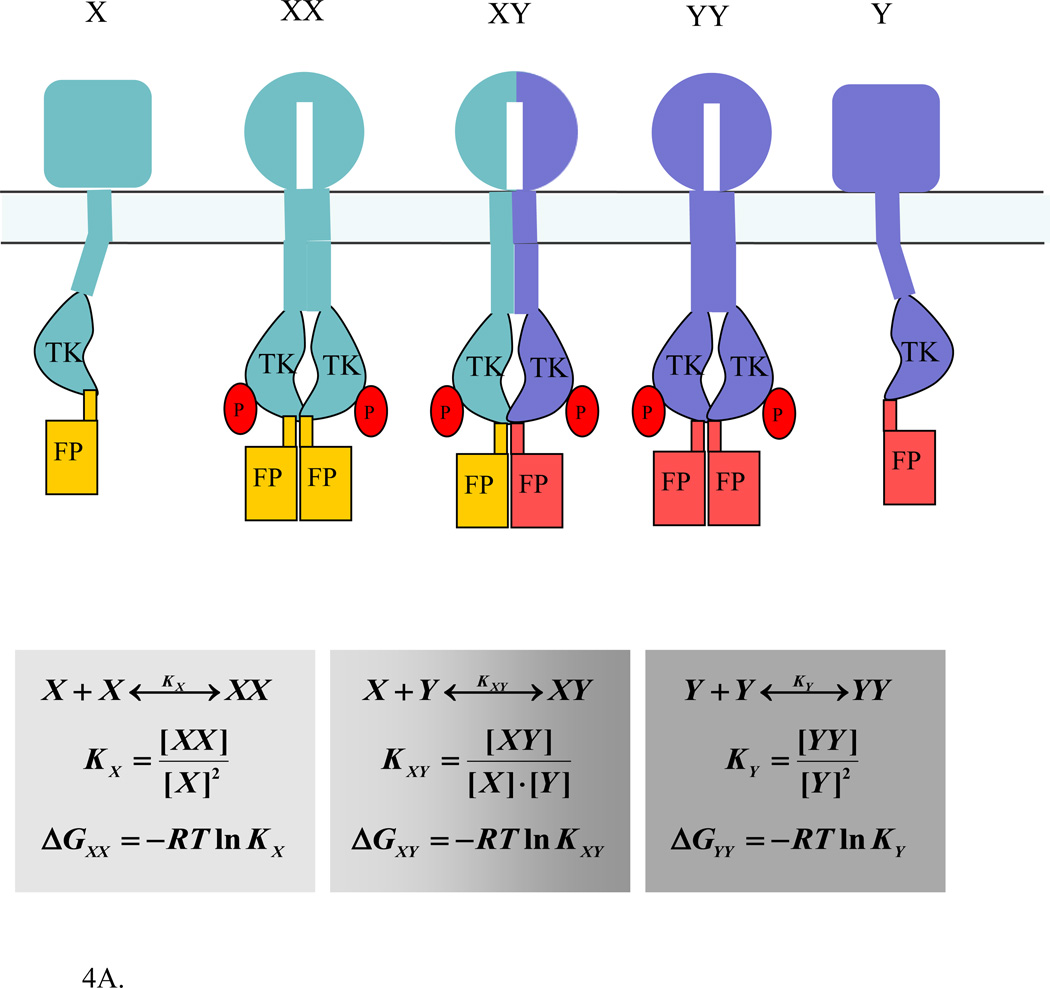
Germ-line mutations in FGFR receptors cause heterozygous disorders of bone development (112;142;143), as the homozygous conditions are lethal. Thus both wild-type and mutant receptors are co-expressed in cells, and questions arise if wild-type/mutant heterodimers form and what their activity is. The interpretation of heterodimerization studies, however, is very challenging because three different dimeric species usually exist: (1) wild-type homodimers, (2) mutant homodimers, and (3) wild-type/mutant heterodimers. These dimers are indistinguishable in many biochemical experiments, as wild-type and mutants are reactive to the same antibodies. One way to characterize heterodimerization is to use FRET, with the wild-type labeled with a donor and the mutant with an acceptor (or vice versa). In this case, the FRET efficiency can be measured easily. However, the heterodimerization propensities are challenging to quantify because RTKs also form homodimers. Figure 4A illustrates the challenge in such measurements, arising due to coupling between the homo and heterodimerization equilibria. The association constant of heterodimerization, Kxy, depends on the monomer concentrations of the two receptors, [X] and [Y]. The same monomer concentrations, however, appear in the equations describing the homodimerization constants Kx and Ky. Thus, homodimer stabilities need to be determined first, and then these are used to determine heterodimer stabilities (141;144).
Figure 4.
Characterization of RTK heterodimerization.
(A). FRET measurements of heterodimerization propensities between RTKs. One RTK is labeled with a donor, and the second RTK is labeled with the acceptor, such that the FRET efficiency can be measured. However, the calculation of the heterodimerization free energy is not trivial because the homo and heterodimerization equilibria are coupled. A detailed protocol for characterization of heterodimerization energetics using FRET is given in (144).
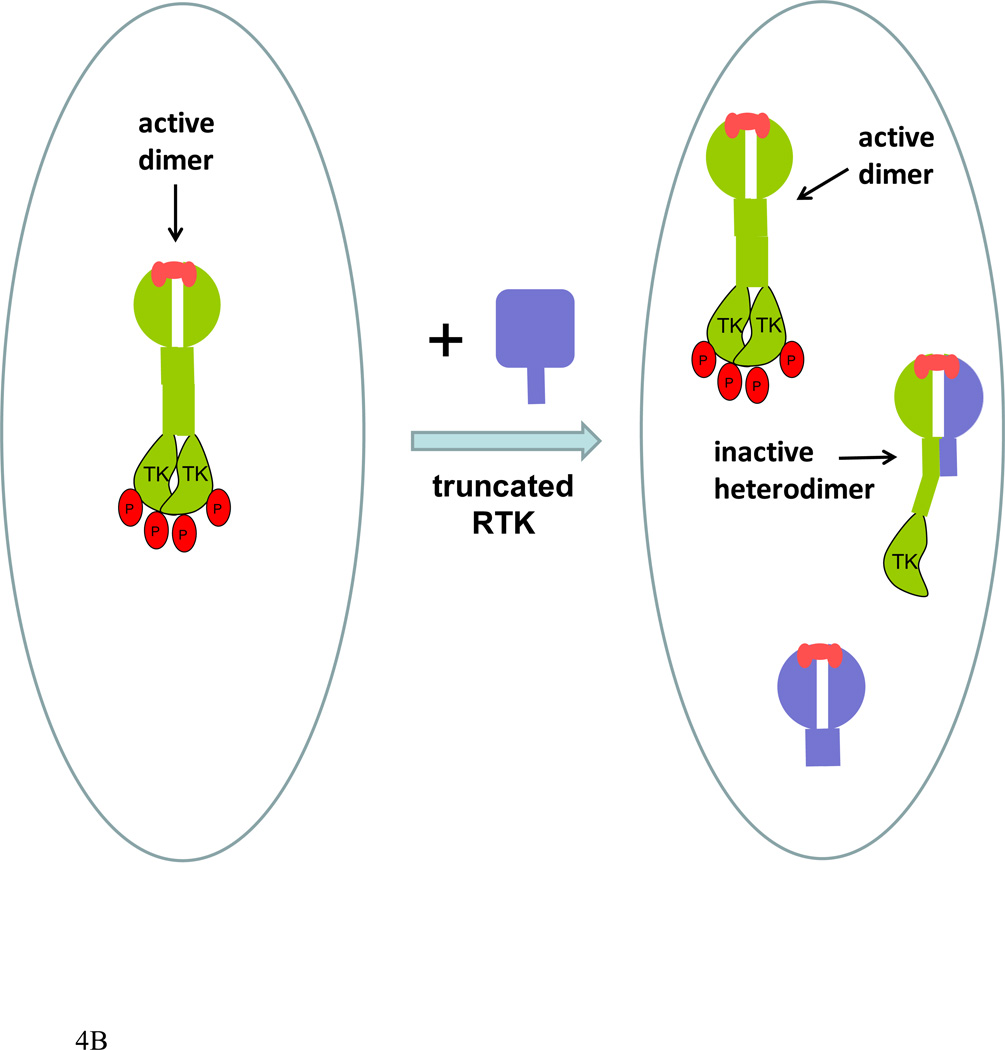
(B). An overview of an assay for RTK heterodimerization that does not require modification with fluorescent proteins. A stable cell line expressing full-length RTKs (green) is transiently transfected with a truncated version of a second RTK (blue) lacking the intracellular domain. The inactive full-length/truncated heterodimers deplete the pool of full-length receptors capable of forming homodimers and reduce their phosphorylation. The decrease in the phosphorylation of the full-length receptors is measured experimentally. This decrease is a reporter of heterodimerization strength between the RTKs that are stably and transiently expressed. Detailed description of the assay is given in (124).
We have demonstrated the feasibility of this approach by determining the propensity for heterodimer formation between the wild-type FGFR3 TM domain and its A391E mutant (144), linked to Crouzon syndrome. The free energy of heterodimerization was determined as −3.37 ± 0.25 kcal/mol (144). Comparison of this value to the homodimerization free energies for the wild-type, −2.8 ± 0.2 kcal/mol, and the mutant, −4.1 ± 0.2 kcal/mol, demonstrates that the heterodimer stability is the average of the two homodimer stabilities (144).
Another approach to investigate the formation of RTK heterodimers is to design an RTK construct that lacks the kinase domain, and then monitor the formation of heterodimers between this construct and full length RTKs (Figure 4B). The full-length/truncated heterodimers are inactive. They deplete the pool of full-length receptors capable of forming homodimers and ultimately reduce the concentration of active homodimers. Thus, the presence of the truncated receptors leads to a decrease in the phosphorylation of the full-length receptors if the full-length and the truncated receptors dimerize. This decrease in phosphorylation can be measured experimentally. Because the contributions of the kinase and the juxtamembrane domains to RTK dimerization may be non-negligible and different for different RTKs, this approach can only be used to assess the effect of pathogenic mutations in the extracellular and TM domains on heterodimerization (124). This is because the contributions of the intracellular domains are expected to be the same for the wild-type and mutant homodimers and heterodimers, but not for heterodimers of different RTKs. This approach has revealed that heterodimers of wild-type FGFR3 and the G380R mutant linked to ACH form with lower probability than wild-type FGFR3 homodimers in cellular membranes (124).
Final remark
As discussed in this review, quantitative biophysical frameworks, as well as sophisticated biochemical and biophysical experimental methods are now being used to unravel the physical-chemical principles behind RTK activation and their involvement in disease. The recent progress in the field has been impressive, and we look forward to future studies that will further deepen our understanding of RTK structure and function and pave the way for the development of novel targeted therapies for RTK-linked pathologies.
Highlights.
Physical-chemical models provide an adequate description of RTK activation.
Quantitative experimental methods shed new light on RTK function.
Methods are available to characterize RTK homo and heterodimerization.
Acknowledgement
Supported by NIH GM068619 and GM095930.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Fantl WJ, Johnson DE, Williams LT. Signaling by Receptor Tyrosine Kinases. Annu. Rev. Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- 2.Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L'Horte CGM, Knowles MA. Cell responses to FGFR3 signaling: growth, differentiation and apoptosis. Experim. Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends in Cell Biology. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 7.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XW, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 11.Bell CA, Tynan JA, Hart KC, Meyer AN, Robertson SC, Donoghue DJ. Rotational coupling of the transmembrane and kinase domains of the Neu receptor tyrosine kinase. Mol. Biol. Cell. 2000;11:3589–3599. doi: 10.1091/mbc.11.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macdonald-Obermann JL, Pike LJ. The Intracellular Juxtamembrane Domain of the Epidermal Growth Factor (EGF) Receptor Is Responsible for the Allosteric Regulation of EGF Binding. J. Biol. Chem. 2009;284:13570–13576. doi: 10.1074/jbc.M109.001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L, Horton WA, Hristova K. The physical basis behind achondroplasia, the most common form of human dwarfism. J. Biol. Chem. 2010;285:30103–30114. doi: 10.1074/jbc.M109.094086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Degnin C, Laederich MB, Horton AW, Hristova K. The A391E mutation enhances FGFR3 activation in the absence of ligand. Biochimica et Biophysica Acta-Biomembranes. 2011;1808:2045–2050. doi: 10.1016/j.bbamem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Hristova K. Pathogenic activation of receptor tyrosine kinases in mammalian membranes. J. Mol. Biol. 2008;384:1130–1142. doi: 10.1016/j.jmb.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Li E, Hristova K. Receptor Tyrosine Kinase transmembrane domains: function, dimer structure, and dimerization energetics. Cell Adhesion and Migration. 2010;4:249–254. doi: 10.4161/cam.4.2.10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanger BO, Stephens JE, Staros JV. High-Yield Trapping of Egf-Induced Receptor Dimers by Chemical Cross-Linking. FASEB J. 1989;3:71–75. doi: 10.1096/fasebj.3.1.2783412. [DOI] [PubMed] [Google Scholar]
- 19.Monsonego-Ornan E, Adar R, Feferman T, Segev O, Yayon A. The transmembrane mutation G380R in fibroblast growth factor receptor 3 uncouples ligand-mediated receptor activation from down-regulation. Mol. Cell. Biol. 2000;20:516–522. doi: 10.1128/mcb.20.2.516-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiner DB, Liu J, Cohen JA, Williams WV, Greene MI. A Point Mutation in the Neu Oncogene Mimics Ligand Induction of Receptor Aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 21.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta(2)-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson SC, Meyer AN, Hart KC, Galvin BD, Webster MK, Donoghue DJ. Activating mutations in the extracellular domain of the fibroblast growth factor receptor 2 function by disruption of the disulfide bond in the third immunoglobulin-like domain. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4567–4572. doi: 10.1073/pnas.95.8.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nature Cell Biology. 2003:S1–S7. [PubMed] [Google Scholar]
- 24.Grailhe R, Merola F, Ridard J, Couvignou S, Le Poupon C, Changeux JP, Laguitton-Pasquier H. Monitoring protein interactions in the living cell through the fluorescence decays of the cyan fluorescent protein. Chemphyschem. 2006;7:1442–1454. doi: 10.1002/cphc.200600057. [DOI] [PubMed] [Google Scholar]
- 25.Yerrapureddy A, Korte T, Hollmann S, Nordhoff M, Ahnert-Hilger G, Herrmann A, Veit M. Intracellular interaction between syntaxin and Munc 18-1 revealed by fluorescence resonance energy transfer. Molecular Membrane Biology. 2005;22:401–410. doi: 10.1080/09687860500224892. [DOI] [PubMed] [Google Scholar]
- 26.Raicu V, Jansma DB, Miller RJD, Friesen JD. Protein interaction quantified in vivo by spectrally resolved fluorescence resonance energy transfer. Biochem. J. 2005;385:265–277. doi: 10.1042/BJ20040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He LS, Bradrick TD, Karpova TS, Wu XL, Fox MH, Fischer R, McNally JG, Knutson JR, Grammer AC, Lipsky PE. Flow cytometric measurement of fluorescence (Forster) resonance energy transfer from cyan fluorescent protein to yellow fluorescent protein using single-laser excitation at 458 nm. Cytometry Part A. 2003;53A:39–54. doi: 10.1002/cyto.a.10037. [DOI] [PubMed] [Google Scholar]
- 28.Wu P, Brand L. Resonance energy transfer: Methods and applications. Anal. Biochem. 1994;218:1–13. doi: 10.1006/abio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 29.Kenworthy AK, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy tranfer. J. Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clegg RM. Fluorescence resonance energy transfer. Curr. Opin. Biotech. 1995;6:103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 32.Clegg RM. Fluorescence resonance energy transfer (FRET) In: Wang XF, Herman B, editors. Fluorescence Imaging Spectroscopy and Microscopy. New York: John Wiley; 1996. pp. 179–252. [Google Scholar]
- 33.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen HM, Puhl HL, Ikeda SR. Estimating protein-protein interaction affinity in living cells using quantitative Forster resonance energy transfer measurements. Journal of Biomedical Optics. 2007:12. doi: 10.1117/1.2799171. [DOI] [PubMed] [Google Scholar]
- 35.Zal T, Gascoigne NRJ. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zal T, Gascoigne NRJ. Using live FRET imaging to reveal early protein-protein interactions during T cell activation (vol 16, pg 418, 2004) Current Opinion in Immunology. 2004;16:674–683. [PubMed] [Google Scholar]
- 37.Hoppe A, Christensen K, Swanson JA. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys. J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merzlyakov M, Chen L, Hristova K. Studies of receptor tyrosine kinase transmembrane domain interactions: The EmEx-FRET method. J. Membr. Biol. 2007;215:93–103. doi: 10.1007/s00232-007-9009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schick S, Chen LR, Li E, Lin J, Koper I, Hristova K. Assembly of the M2 Tetramer Is Strongly Modulated by Lipid Chain Length. Biophys. J. 2010;99:1810–1817. doi: 10.1016/j.bpj.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You M, Li E, Wimley WC, Hristova K. FRET in liposomes: measurements of TM helix dimerization in the native bilayer environment. Analytical Biochemistry. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merzlyakov M, Hristova K. Forster Resonance Energy Transfer Measurements of Transmembrane Helix Dimerization Energetics. Methods in Enzymology: Fluorescence Spectroscopy. 2008;450:107–127. doi: 10.1016/S0076-6879(08)03406-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posokhov YO, Merzlyakov M, Hristova K, Ladokhin AS. A simple "proximity" correction for Forster resonance energy transfer efficiency determination in membranes using lifetime measurements. Analytical Biochemistry. 2008;380:134–136. doi: 10.1016/j.ab.2008.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott RE, Perkins RG, Zschunke MA, Hoerl BJ, Maercklein PB. Plasma-Membrane Vesiculation in 3T3-Cells and Sv3T3-Cells .1. Morphological and Biochemical Characterization. J. Cell Sci. 1979;35:229–243. doi: 10.1242/jcs.35.1.229. [DOI] [PubMed] [Google Scholar]
- 44.Holowka D, Baird B. Structural Studies on the Membrane-Bound Immunoglobulin E-Receptor Complex .1. Characterization of Large Plasma-Membrane Vesicles from Rat Basophilic Leukemia-Cells and Insertion of Amphipathic Fluorescent-Probes. Biochemistry. 1983;22:3466–3474. doi: 10.1021/bi00283a025. [DOI] [PubMed] [Google Scholar]
- 45.Holowka D, Baird B. Structural Studies on the Membrane-Bound Immunoglobulin E-Receptor Complex .2. Mapping of Distances Between Sites on Ige and the Membrane-Surface. Biochemistry. 1983;22:3475–3484. doi: 10.1021/bi00283a025. [DOI] [PubMed] [Google Scholar]
- 46.Holowka D, Baird B. Lactoperoxidase-Catalyzed Iodination of the Receptor for Immunoglobulin-e at the Cytoplasmic Side of the Plasma-Membrane. J. Biol. Chem. 1984;259:3720–3728. [PubMed] [Google Scholar]
- 47.Baumgart T, Hammond AT, Sengupta P, Hess ST, Holowka DA, Baird BA, Webb WW. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sengupta P, Hammond A, Holowka D, Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochimica et Biophysica Acta-Biomembranes. 2008;1778:20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen S, Ushiro H, Stoscheck C, Chinkers M. A Native 170,000 Epidermal Growth-Factor Receptor-Kinase Complex from Shed Plasma-Membrane Vesicles. J. Biol. Chem. 1982;257:1523–1531. [PubMed] [Google Scholar]
- 50.Scott RE, Maercklein PB. Plasma-Membrane Vesiculation in 3T3-Cells and Sv3T3 Cells .2. Factors Affecting the Process of Vesiculation. J. Cell Sci. 1979;35:245–252. doi: 10.1242/jcs.35.1.245. [DOI] [PubMed] [Google Scholar]
- 51.Pick H, Schmid EL, Tairi AP, Ilegems E, Hovius R, Vogel H. Investigating cellular signaling reactions in single attoliter vesicles. J. Am. Chem. Soc. 2005;127:2908–2912. doi: 10.1021/ja044605x. [DOI] [PubMed] [Google Scholar]
- 52.Hagmann J, Burger MM, Dagan D. Regulation of plasma membrane blebbing by the cytoskeleton. Journal of Cellular Biochemistry. 1999;73:488–499. [PubMed] [Google Scholar]
- 53.Li E, Placone J, Merzlyakov M, Hristova K. Quantitative measurements of protein interactions in a crowded cellular environment. Anal. Chem. 2008;80:5976–5985. doi: 10.1021/ac800616u. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Novicky L, Merzlyakov M, Hristov T, Hristova K. Measuring the Energetics of Membrane Protein Dimerization in Mammalian Membranes. J. Am. Chem. Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L, Placone J, Novicky L, Hristova K. The extracellular domain of fibroblast growth factor receptor 3 inhibits ligand-independent dimerization. Science Signaling. 2010;3:ra86. doi: 10.1126/scisignal.2001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ladda RL, Bullock LP, Gianopoulos T, Mccormick L. Radioreceptor Assay for Epidermal Growth-Factor. Analytical Biochemistry. 1979;93:286–294. doi: 10.1016/s0003-2697(79)80153-0. [DOI] [PubMed] [Google Scholar]
- 57.Guinivan P, Ladda RL. Decrease in Epidermal Growth-Factor Receptor Levels and Production of Material Enhancing Epidermal Growth-Factor Binding Accompany the Temperature-Dependent Changes from Normal to Transformed Phenotype. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:3377–3381. doi: 10.1073/pnas.76.7.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorensen V, Zhen Y, Zakrzewska M, Haugsten EM, Walchli S, Nilsen T, Olsnes S, Wiedlocha A. Phosphorylation of fibroblast growth factor (FGF) receptor 1 at Ser777 by p38 mitogen-activated protein kinase regulates translocation of exogenous FGF1 to the cytosol and nucleus. Mol. Cell. Biol. 2008;28:4129–4141. doi: 10.1128/MCB.02117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nugent MA, Edelman ER. Kinetics of Basic Fibroblast Growth-Factor Binding to Its Receptor and Heparan-Sulfate Proteoglycan - A Mechanism for Cooperativity. Biochemistry. 1992;31:8876–8883. doi: 10.1021/bi00152a026. [DOI] [PubMed] [Google Scholar]
- 60.Carraway KL, Cerione RA. Fluorescent-Labeled Growth-Factor Molecules Serve As Probes for Receptor-Binding and Endocytosis. Biochemistry. 1993;32:12039–12045. doi: 10.1021/bi00096a014. [DOI] [PubMed] [Google Scholar]
- 61.Uyemura T, Takagi H, Yanagida T, Sako Y. Single-molecule analysis of epidermal growth factor signaling that leads to ultrasensitive calcium response. Biophys. J. 2005;88:3720–3730. doi: 10.1529/biophysj.104.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teramura Y, Ichinose J, Takagi H, Nishida K, Yanagida T, Sako Y. Single-molecule analysis of epidermal growth factor binding on the surface of living cells. EMBO J. 2006;25:4215–4222. doi: 10.1038/sj.emboj.7601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pramanik A, Rigler R. Ligand-receptor interactions in the membrane of cultured cells monitored by fluorescence correlation spectroscopy. Biol. Chem. 2001;382:371–378. doi: 10.1515/BC.2001.045. [DOI] [PubMed] [Google Scholar]
- 64.Pantoliano MW, Horlick RA, Springer BA, Vandyk DE, Tobery T, Wetmore DR, Lear JD, Nahapetian AT, Bradley JD, Sisk WP. Multivalent Ligand-Receptor Binding Interactions in the Fibroblast Growth-Factor System Produce A Cooperative Growth-Factor and Heparin Mechanism for Receptor Dimerization. Biochemistry. 1994;33:10229–10248. doi: 10.1021/bi00200a003. [DOI] [PubMed] [Google Scholar]
- 65.Hoshikawa M, Yonamine A, Konishi M, Itoh N. FGF-18 is a neuron-derived glial cell growth factor expressed in the rat brain during early postnatal development. Molecular Brain Research. 2002;105:60–66. doi: 10.1016/s0169-328x(02)00393-5. [DOI] [PubMed] [Google Scholar]
- 66.Ohmachi S, Mikami T, Konishi M, Miyake A, Itoh N. Preferential neurotrophic activity of fibroblast growth factor-20 for dopaminergic neurons through fibroblast growth factor receptor-1c. J. Neuroscience Res. 2003;72:436–443. doi: 10.1002/jnr.10592. [DOI] [PubMed] [Google Scholar]
- 67.Olsen SK, Ibrahimi OA, Raucci A, Zhang FM, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:935–940. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- 69.Barker SC, Kassel DB, Weigl D, Huang XY, Luther MA, Knight WB. Characterization of Pp60(C-Src) Tyrosine Kinase-Activities Using A Continuous Assay - Autoactivation of the Enzyme Is An Intermolecular Autophosphorylation Process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 70.Hoffmann I, Eugene E, Nassif X, Couraud PO, Bourdoulous S. Activation of ErbB2 receptor tyrosine kinase supports invasion of endothelial cells by Neisseria meningitidis. J. Cell Biol. 2001;155:133–143. doi: 10.1083/jcb.200106148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou L, Takayama Y, Boucher P, Tallquist MD, Herz J. LRP1 Regulates Architecture of the Vascular Wall by Controlling PDGFR beta-Dependent Phosphatidylinositol 3-Kinase Activation. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0006922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Human Molecular Genetics. 2001;10:1255–1264. doi: 10.1093/hmg/10.12.1255. [DOI] [PubMed] [Google Scholar]
- 73.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Molecular Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 74.Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Science Signaling. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 76.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotech. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 77.Salomon AR, Ficarro SB, Brill LM, Brinker A, Phung QT, Ericson C, Sauer K, Brock A, Horn DM, Schultz PG, Peters EC. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:443–448. doi: 10.1073/pnas.2436191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watts JD, Affolter M, Krebs DL, Wange RL, Samelson LE, Aebersold R. Identification by Electrospray-Ionization Mass-Spectrometry of the Sites of Tyrosine Phosphorylation-Induced in Activated Jurkat T-Cells on the Protein-Tyrosine Kinase Zap-70. J. Biol. Chem. 1994;269:29520–29529. [PubMed] [Google Scholar]
- 79.Soskic V, Gorlach M, Poznanovic S, Boehmer FD, Godovac-Zimmermann J. Functional proteomics analysis of signal transduction pathways of the platelet-derived growth factor beta receptor. Biochemistry. 1999;38:1757–1764. doi: 10.1021/bi982093r. [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Molecular & Cellular Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Bae JH, Boggon TJ, Tome F, Mandiyan V, Lax I, Schlessinger J. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2866–2871. doi: 10.1073/pnas.0914157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sternberg MJE, Gullick WJ. A Sequence Motif in the Transmembrane Region of Growth Factor Receptors with Tyrosine Kinase Activity Mediates Dimerization. Protein Eng. 1990;3:245–248. doi: 10.1093/protein/3.4.245. [DOI] [PubMed] [Google Scholar]
- 83.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 84.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: The GxxxG motif occurs frequently and in association with b-branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 85.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 86.Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15937–15940. doi: 10.1073/pnas.252640799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landau M, Ben Tal N. Dynamic equilibrium between multiple active and inactive conformations explains regulation and oncogenic mutations in ErbB receptors. Biochimica et Biophysica Acta-Reviews on Cancer. 2008;1785:12–31. doi: 10.1016/j.bbcan.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 89.Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA, Artemenko EO, Efremov RG, Arseniev AS. Spatial Structure and pH-dependent Conformational Diversity of Dimeric Transmembrane Domain of the Receptor Tyrosine Kinase EphA1. J. Biol. Chem. 2008;283:29385–29395. doi: 10.1074/jbc.M803089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerber D, Sal-Man N, Shai Y. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J. Biol. Chem. 2004;279:21177–21182. doi: 10.1074/jbc.M400847200. [DOI] [PubMed] [Google Scholar]
- 91.Escher C, Cymer F, Schneider D. Two GxxxG-Like Motifs Facilitate Promiscuous Interactions of the Human ErbB Transmembrane Domains. J. Mol. Biol. 2009;389:10–16. doi: 10.1016/j.jmb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 92.Bocharov EV, Mayzel ML, Volynsky PE, Mineev KS, Tkach EN, Ermolyuk YS, Schulga AA, Efremov RG, Arseniev AS. Left-Handed Dimer of EphA2 Transmembrane Domain: Helix Packing Diversity among Receptor Tyrosine Kinases. Biophys. J. 2010;98:881–889. doi: 10.1016/j.bpj.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mineev KS, Khabibullina NF, Lyukmanova EN, Dolgikh DA, Kirpichnikov MP, Arseniev AS. Spatial structure and dimer-monomer equilibrium of the ErbB3 transmembrane domain in DPC micelles. Biochimica et Biophysica Acta-Biomembranes. 2011;1808:2081–2088. doi: 10.1016/j.bbamem.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 94.Bocharov EV, Mineev KS, Lesovoy D, Goncharuk MV, Goncharuk S, Bocharova OV, Volynsky PE, Efremov RG, Arseniev AS. Structural Aspects of Transmembrane Domain Interactions of Receptor Tyrosine Kinases. Biophysical Journal, Supplement. 2011;100 207a-1131-Pos. [Google Scholar]
- 95.Li E, You M, Hristova K. FGFR3 dimer stabilization due to a single amino acid pathogenic mutation. J. Mol. Biol. 2006;356:600–612. doi: 10.1016/j.jmb.2005.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oates J, King G, Dixon AM. Strong oligomerization behavior of PDGF beta receptor transmembrane domain and its regulation by the juxtamembrane regions. Biochimica et Biophysica Acta-Biomembranes. 2010;1798:605–615. doi: 10.1016/j.bbamem.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 97.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Molecular Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 98.Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 99.Bouyain S, Longo PA, Li SQ, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alvarado D, Klein DE, Lemmon MA. Structural Basis for Negative Cooperativity in Growth Factor Binding to an EGF Receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Plotnikov AN, Schlessinger J, Hubbard SR, Mohammadi M. Structural basis for FGF receptor dimerization and activation. Cell. 1999;98:641–650. doi: 10.1016/s0092-8674(00)80051-3. [DOI] [PubMed] [Google Scholar]
- 102.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligandreceptor specificity. Cell. 2000;101:413–424. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 103.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine & Growth Factor Reviews. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 104.Yuzawa S, Opatowsky Y, Zhang ZT, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 105.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 107.Harada D, Yamanaka Y, Ueda K, Tanaka H, Seino Y. FGFR3-related dwarfism and cell signaling. Journal of Bone and Mineral Metabolism. 2009;27:9–15. doi: 10.1007/s00774-008-0009-7. [DOI] [PubMed] [Google Scholar]
- 108.Li Y, Mangasarian K, Mansukhani A, Basilico C. Activation of FGF receptors by mutations in the transmembrane domain. Oncogene. 1997;14:1397–1406. doi: 10.1038/sj.onc.1200983. [DOI] [PubMed] [Google Scholar]
- 109.Naski MC, Wang Q, Xu JS, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat. Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 110.Ponseti IV. Skeletal Growth in Achondroplasia. Journal of Bone and Joint Surgery-American Volume A. 1970;52:701. [PubMed] [Google Scholar]
- 111.Horton WA, Hall JG, Hecht JT. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- 112.Vajo Z, Francomano CA, Wilkin DJ. The molecular and genetic basis of fibroblast growth factor receptor 3 disorders: The achondroplasia family of skeletal dysplasias, Muenke craniosynostosis, and Crouzon syndrome with acanthosis nigricans. Endocrine Reviews. 2000;21:23–39. doi: 10.1210/edrv.21.1.0387. [DOI] [PubMed] [Google Scholar]
- 113.Cho JY, Guo CS, Torello M, Lunstrum GP, Iwata T, Deng CX, Horton WA. Defective lysosomal targeting of activated fibroblast growth factor receptor 3 in achondroplasia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:609–614. doi: 10.1073/pnas.2237184100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bonaventure J, Gibbs L, Horne WC, Baron R. The localization of FGFR3 mutations causing thanatophoric dysplasia type I differentially affects phosphorylation, processing and ubiquitylation of the receptor. Febs Journal. 2007;274:3078–3093. doi: 10.1111/j.1742-4658.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- 115.You M, Li E, Hristova K. The achondroplasia mutation does not alter the dimerization energetics of FGFR3 transmembrane domain. Biochemistry. 2006;45:5551–5556. doi: 10.1021/bi060113g. [DOI] [PubMed] [Google Scholar]
- 116.Chen HB, Ma JH, Li WQ, Eliseenkova AV, Xu CF, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Molecular Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O'Connor S, Li E, Majors BS, He L, Placone J, Baycin D, Betenbaugh MJ, Hristova K. Increased Expression of the Integral Membrane Protein ErbB2 in Chinese Hamster Ovary Cells Expressing the Anti-apoptotic Gene Bcl-xL. Protein Expr. Purif. 2009;67:41–47. doi: 10.1016/j.pep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Finger C, Escher C, Schneider D. The Single Transmembrane Domains of Human Receptor Tyrosine Kinases Encode Self-Interactions. Science Signaling 2. 2009 doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- 119.Artemenko EO, Egorova NS, Arseniev AS, Feofanov AV. Transmembrane domain of EphA1 receptor forms dimers in membrane-like environment. Biochim. Biophys. Acta. 2008;1778:2361–2367. doi: 10.1016/j.bbamem.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 120.Li E, You M, Hristova K. SDS-PAGE and FRET suggest weak interactions between FGFR3 TM domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 121.Iwamoto T, You M, Li E, Spangler J, Tomich JM, Hristova K. Synthesis and initial characterization of FGFR3 transmembrane domain: Consequences of sequence modifications. Biochim. Biophys. Acta. 2005;1668:240–247. doi: 10.1016/j.bbamem.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 122.You M, Spangler J, Li E, Han X, Ghosh P, Hristova K. Effect of pathogenic cysteine mutations on FGFR3 transmembrane domain dimerization in detergents and lipid bilayers. Biochemistry. 2007;46:11039–11046. doi: 10.1021/bi700986n. [DOI] [PubMed] [Google Scholar]
- 123.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys. J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.He L, Wimley WC, Hristova K. FGFR3 heterodimerization in achondroplasia, the most common form of human dwarfism. J. Biol. Chem. 2011;286:13272–13281. doi: 10.1074/jbc.M110.205583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.He LJ, Shobnam N, Hristova K. Specific inhibition of a pathogenic receptor tyrosine kinase by its transmembrane domain. Biochimica et Biophysica Acta-Biomembranes. 2011;1808:253–259. doi: 10.1016/j.bbamem.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kashles O, Szapary D, Bellot F, Ullrich A, Schlessinger J, Schmidt A. Ligand-Induced Stimulation of Epidermal Growth-Factor Receptor Mutants with Altered Transmembrane Regions. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:9567–9571. doi: 10.1073/pnas.85.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464 doi: 10.1038/nature08827. 783-U163. [DOI] [PubMed] [Google Scholar]
- 128.Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TPJ, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An openand-shut case? Recent insights into the activation of EGF/ErbB receptors. Molecular Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 129.Brewer MR, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The Juxtamembrane Region of the EGF Receptor Functions as an Activation Domain. Molecular Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang XW, Kuriyan J. Mechanism for Activation of the EGF Receptor Catalytic Domain by the Juxtamembrane Segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Experim. Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 133.Tzahar E, Waterman H, Chen XM, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Difiore PP, Kraus MH. Cooperative Signaling of Erbb3 and Erbb2 in Neoplastic Transformation and Human Mammary Carcinomas. Oncogene. 1995;10:1813–1821. [PubMed] [Google Scholar]
- 135.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-Dependent Regulation of Her2/Neu Oncogenic Signaling by Heterodimerization with Her3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Grausporta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Riese DJ, Bermingham Y, vanRaaij TM, Buckley S, Plowman GD, Stern DF. Betacellulin activates the epidermal growth factor receptor and erbB-4, and induces cellular response patterns distinct from those stimulated by epidermal growth factor or neuregulin-beta. Oncogene. 1996;12:345–353. [PubMed] [Google Scholar]
- 138.Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE. ErbB receptor-induced activation of Stat transcription factors is mediated by Src tyrosine kinases. J. Biol. Chem. 1999;274:17209–17218. doi: 10.1074/jbc.274.24.17209. [DOI] [PubMed] [Google Scholar]
- 139.PinkasKramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 140.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nature Reviews Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 141.Duneau JP, Vegh AP, Sturgis JN. A dimerization hierarchy in the transmembrane domains of the HER receptor family. Biochemistry. 2007;46:2010–2019. doi: 10.1021/bi061436f. [DOI] [PubMed] [Google Scholar]
- 142.Mcintosh I, Bellus GA, Jabs EW. The pleiotropic effects of fibroblast growth factor receptors in mammalian development. Cell Structure and Function. 2000;25:85–96. doi: 10.1247/csf.25.85. [DOI] [PubMed] [Google Scholar]
- 143.Cunningham ML, Seto ML, Ratisoontorn C, Heike CL, Hing AV. Syndromic craniosynostosis: from history to hydrogen bonds. Orthodontics & Craniofacial Research. 2007;10:67–81. doi: 10.1111/j.1601-6343.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- 144.Merzlyakov M, You M, Li E, Hristova K. Transmembrane helix heterodimerization in lipids bilayers: probing the energetics behind autosomal dominant growth disorders. J. Mol. Biol. 2006;358:1–7. doi: 10.1016/j.jmb.2006.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ibrahimi OA, Zhang V, Eliseenkova AV, Linhardt RJ, Mohammadi M. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Human Molecular Genetics. 2004;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- 146.Ibrahimi OA, Zhang FM, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Human Molecular Genetics. 2004;13:2313–2324. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Galvin BD, Hart KC, Meyer AN, Webster MK, Donoghue DJ. Constitutive receptor activation by Crouzon syndrome mutations in fibroblast growth factor receptor (FGFR) 2 and FGFR2/Neu chimeras. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7894–7899. doi: 10.1073/pnas.93.15.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.d'Avis PY, Robertson SC, Meyer AN, Bardwell WM, Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by mutations responsible for the lethal skeletal dysplasia thanatophoric dysplasia type I. Cell Growth & Differentiation. 1998;9:71–78. [PubMed] [Google Scholar]
- 149.Adar R, Monsonego-Ornan E, David P, Yayon A. Differential activation of cysteine-substitution mutants of fibroblast growth factor receptor 3 is determined by cysteine localization. Journal of Bone and Mineral Research. 2002;17:860–868. doi: 10.1359/jbmr.2002.17.5.860. [DOI] [PubMed] [Google Scholar]
- 150.Webster MK, dAvis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol. Cell. Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roidl A, Foo P, Wong W, Mann C, Bechtold S, Berger HJ, Streit S, Ruhe JE, Hart S, Ullrich A, Ho HK. The FGFR4 Y367C mutant is a dominant oncogene in MDA-MB453 breast cancer cells. Oncogene. 2010;29:1543–1552. doi: 10.1038/onc.2009.432. [DOI] [PubMed] [Google Scholar]
- 152.Fernandes H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association - A study with an aberrant epidermal growth factor receptor (EGFRvIII/Delta EGFR) expressed in cancer cells. J. Biol. Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 153.Lee JC, Vivanco I, Beroukhim R, Huang JHY, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, Xu Q, Greulich H, Thomas RK, Paez JG, Peck TC, Linhart DJ, Glatt KA, Getz G, Onofrio R, Ziaugra L, Levine RL, Gabriel S, Kawaguchi T, O'Neill K, Khan H, Liau LM, Nelson SF, Rao PN, Mischel P, Pieper RO, Cloughesy T, Leahy DJ, Sellers WR, Sawyers CL, Meyerson M, Mellinghoff IK. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. Plos Medicine. 2006;3:2264–2273. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yun CH, Boggon TJ, Li YQ, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26:1567–1576. doi: 10.1038/sj.onc.1209957. [DOI] [PubMed] [Google Scholar]
- 156.Landau M, Fleishman SJ, Ben Tal N. A putative mechanism for downregulation of the catalytic activity of the EGF receptor via direct contact between its kinase and C-terminal domains. Structure. 2004;12:2265–2275. doi: 10.1016/j.str.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 157.Kohl TM, Schnittger S, Ellwart JW, Hiddemann W, Spiekermann K. KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood. 2005;105:3319–3321. doi: 10.1182/blood-2004-06-2068. [DOI] [PubMed] [Google Scholar]
- 158.Tarn C, Merkel E, Canutescu AA, Shen W, Skorobogatko Y, Heslin MJ, Eisenberg B, Birbe R, Patchefsky A, Dunbrack R, Arnoletti JP, von Mehren M, Godwin AK. Analysis of KIT mutations in sporadic and familial gastrointestinal stromal tumors: Therapeutic implications through protein modeling. Clinical Cancer Research. 2005;11:3668–3677. doi: 10.1158/1078-0432.CCR-04-2515. [DOI] [PubMed] [Google Scholar]
- 159.Ma YS, Cunningham ME, Wang XM, Ghosh I, Regan L, Longley BJ. Inhibition of spontaneous receptor phosphorylation by residues in a putative alpha-helix in the KIT intracellular juxtamembrane region. J. Biol. Chem. 1999;274:13399–13402. doi: 10.1074/jbc.274.19.13399. [DOI] [PubMed] [Google Scholar]
- 160.Kitayama H, Kanakura Y, Furitsu T, Tsujimura T, Oritani K, Ikeda H, Sugahara H, Mitsui H, Kanayama Y, Kitamura Y, Matsuzawa Y. Constitutively Activating Mutations of C-Kit Receptor Tyrosine Kinase Confer Factor-Independent Growth and Tumorigenicity of Factor-Dependent Hematopoietic-Cell Lines. Blood. 1995;85:790–798. [PubMed] [Google Scholar]
- 161.Casteran N, De Sepulveda P, Beslu N, Aoubala M, Letard S, Lecocq E, Rottapel R, Dubreuil P. Signal transduction by several KIT juxtamembrane domain mutations. Oncogene. 2003;22:4710–4722. doi: 10.1038/sj.onc.1206587. [DOI] [PubMed] [Google Scholar]
- 162.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield JH, Ashman LK, Kanayama Y, Matsuzawa Y, Kitamura Y, Kanakura Y. Identification of Mutations in the Coding Sequence of the Protooncogene C-Kit in A Human Mast-Cell Leukemia-Cell Line Causing Ligand-Independent Activation of C-Kit Product. Journal of Clinical Investigation. 1993;92:1736–1744. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Orfao A, Garcia-Montero AC, Sanchez L, Escribano L. Recent advances in the understanding of mastocytosis: the role of KIT mutations. British Journal of Haematology. 2007;138:12–30. doi: 10.1111/j.1365-2141.2007.06619.x. [DOI] [PubMed] [Google Scholar]
- 164.Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: A novel molecular approach to the treatment of KIT-positive malignancies. Journal of Clinical Oncology. 2002;20:1692–1703. doi: 10.1200/JCO.2002.20.6.1692. [DOI] [PubMed] [Google Scholar]
- 165.Tan PK, Wang J, Littler PLH, Wong KK, Sweetnam TA, Keefe W, Nash NR, Reding EC, Piu F, Brann MR, Schiffer HH. Monitoring interactions between receptor tyrosine kinases and their downstream effector proteins in living cells using bioluminescence resonance energy transfer. Molecular Pharmacology. 2007;72:1440–1446. doi: 10.1124/mol.107.039636. [DOI] [PubMed] [Google Scholar]