Abstract
Though loss of function in CBP/p300, a family of CREB-binding proteins, has been causally associated with a variety of human neurological disorders, such as Rubinstein-Taybi syndrome, Huntington’s disease and drug addiction, the role of EP300 interacting inhibitor of differentiation 1 (EID1), a CBP/p300 inhibitory protein, in modulating neurological functions remains completely unknown. Through the examination of EID1 expression and cellular distribution, we discovered that there is a significant increase of EID1 nuclear translocation in the cortical neurons of Alzheimer’s disease (AD) patient brains compared to that of control brains. To study the potential effects of EID1 on neurological functions associated with learning and memory, we generated a transgenic mouse model with a neuron-specific expression of human EID1 gene in the brain. Overexpression of EID1 led to an increase in its nuclear localization in neurons mimicking that seen in human AD brains. The transgenic mice had a disrupted neurofilament organization and increase of astrogliosis in the cortex and hippocampus. Furthermore, we demonstrated that overexpression of EID1 reduced hippocampal long-term potentiation and impaired spatial learning and memory function in the transgenic mice. Our results indicated that the negative effects of extra nuclear EID1 in transgenic mouse brains are likely due to its inhibitory function on CBP/p300 mediated histone and p53 acetylation, thus affecting the expression of downstream genes involved in the maintenance of neuronal structure and function. Together, our data raise the possibility that alteration of EID1 expression, particularly the increase of EID1 nuclear localization that inhibits CBP/p300 activity in neuronal cells, may play an important role in AD pathogenesis.
Keywords: EP300 interacting inhibitor of differentiation 1, CBP/p300, histone, p53, Acetylation, Alzheimer’s disease
Introduction
EID1 (EP300 interacting inhibitor of differentiation 1), also known as CRI1, was originally identified as a pRb-interacting protein negatively regulates myogenesis (MacLellan et al., 2000; Miyake et al., 2000). EID1 inhibits myogenic differentiation by blocking the histone acetyltransferase (HAT) activity of p300/CREB (cAMP response element binding protein) binding protein, a transcriptional co-activator, required for MyoD dependent transactivation. Its physical interaction with pRb brings EID1 to MDM2, an E3 ubiquitin ligase, leading to its proteasomal degradation up on cell cycle exit. In contrast, its interaction with the RET finger protein (RFP, TRIM 27), an upstream modulator of Rb, increases the stability of EID1protein thus inhibits Rb mediated transcription activation (Krutzfeldt et al., 2005). A newly identified EID1 binding partner, Necdin, antagonizes the repressive effects of EID1 whereby promotes myoblast differentiation. This interaction also stabilizes EID1 and promotes its relocalization to the cytoplasm (Bush and Wevrick, 2008). EID1 can also be recruited and bridged by other transcriptional inhibitors, such as small heterodimer partner (SHP), which directly binds and inhibits a large set of nuclear receptors, or bind directly to nuclear receptors, such as orphan nuclear receptor, SF1, to accomplish transcriptional inhibition (Bavner et al., 2002; Macchiarulo et al., 2006; Park et al., 2007). It is suggested that the mechanism of these transcriptional inhibitions involves EID1 antagonism of the p300/CBP (CREB binding protein)-dependent co-activator functions (Bavner et al., 2002; Chen et al., 2005; Park et al., 2007).
Transcriptional co-activator CBP and p300 have high degree of homology and similar pattern of expression. They both stimulate CREB-dependent gene expression by interacting with and integrating a variety of signal-responsive transcription factors, modifying transcription factors and histone by acetylation (Vo and Goodman, 2001). Their co-activator functions regulate many physiological processes including cell growth, differentiation and apoptosis. Therefore, CBP/p300 promoted signaling mechanism is important for long-term memory and neuronal survival (Maggirwar et al., 2000; Yukawa et al., 1999; Alarcon et al., 2004). Mutations and decreased levels of CBP/p300 are associated with a number of neurological disorders including Rubinstein-Taybi Syndrome (RTS) and Spinocerebellar ataxia type 1 (SCA1) (Roelfsema et al., 2005; Tsirigotis et al., 2006). Loss of function in CBP/p300 has been directly associated with familial Alzheimer’s disease (FAD) (Francis et al., 2007; Marambaud et al., 2003) and critical loss of CBP/p300 histone acetylase activity has been linked to neuronal apoptosis (Rouaux et al., 2003). FAD related studies have demonstrated that mutations in APP and PS1 alter CBP/CREB function. In a cell culture model, Vitolo et al. demonstrated a decrease of PKA activity, CREB activation and LTP in hippocampal neurons by Aβ peptide (Vitolo et al., 2002). In a transgenic (Tg) mouse model, mutations of APP and PS1 were associate with CBP/CREB dysfunction (Gong et al., 2004). Treatments by rolipram that augment cAMP/CREB signaling improves LTP and memory function in the mouse model. In a conditional knockout mouse model, in which PS1/2 is selectively inactivated in excitatory neurons, there is a decrease of CBP and a reduction of transcription of CBP/CREB genes. Consequently, the conditional knockout mice have LTP and memory deficiency as they are aging (Saura et al., 2004).
EID1 has been identified as an inhibitor of p300 by suppression of the acetyltransferase activity during myogenesis. Its function in other tissues or cell type has not been elucidated. EID1 transcripts are ubiquitously detected in human tissues with greatest expression in cardiac and skeletal muscle and brain (MacLellan et al., 2000; Miyake et al., 2000), and has exceptionally high level of expression in mouse brain (Bavner et al., 2002). We were intrigued by this expression pattern especially since EID1 is reported to be degraded upon cell cycle exit in skeletal muscle, thus expected to show similar behaviors in neuronal differentiation and low levels of expression in terminally differentiated neurons. We set out to investigate the expression pattern and functions of EID1 in the brain with relation to its roles in memory loss and neurodegeneration in Alzheimer’s disease (AD). Surprisingly, we found that EID1 is highly expressed in neurons and barely detectable in astrocytes of mouse and human brains. More importantly, we showed an increase of EID1 nuclear translocation in the neurons of AD patient brains compared with that of normal brains. To our knowledge, this is the very first piece of evidence that links EID1 with the neurodegenerative diseases of the central nervous system (CNS). To further investigate the roles of EID1 in memory and LTP, we generated an EID1 transgenic mouse model that exhibits neuron-specific overexpression of human EID1 gene in the brain. The Tg mice showed clear nuclear translocation of EID1 in the brain mimicking that seen in human AD brains. Here, we also present evidence indicating that overexpression of EID1 reduces LTP and impairs spatial learning and memory function. The negative effects of extra nuclear EID1 in Tg mouse brains is likely due to its inhibitory function on the acetyltrasferase activity of CBP/p300, thus affects expression of downstream genes involved in the maintenance of neuronal structure and function. Together, our data raise the possibility that alteration of EID1 expression maybe causally related to pathogenesis of AD.
Materials and Methods
Cell culture and qRT-PCR analysis
Human embryonal teratocarcinoma Atera2/D1 (NT2) cells (Stratagene, La Jolla, CA) and human HEK 293 cells (Graham et al., 1977) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Bethesda, MD) supplemented with 10% fetal calf serum (GCS, Wisent, Inc. St. Bruno, PQ). NT2 cells were differentiated into neurons and astrocytes with all trans-retinoic acid (RA, Sigma, Oakville, ON) according to the method of Pleasure and Lee (Pleasure and Lee, 1993) as described previously (Sodja et al., 2002). RNA extraction, first strand cDNA synthesis, and qRT-PCR analysis were performed as described previously (Shen et al., 2006). To detect the expression level of the EID1 transcript in differentiated and undifferentiated NT2 cells, equal amounts of cDNA (2 ng each) were used with the primers: EID1F 5’ CTCAGTGGTGCCGGCTACA 3’ and EID1R 5’ CAGGGCTGGTTCTCTTGTTCTC 3’.
Generation of EID1 transgenic mice
The full-length human EID1 cDNA was amplified by RT-PCR from human brain mRNA with a forward primer carrying a HindIII restriction site, followed by the Kozak sequence of mouse EID1 cDNA and the 5’ coding region of human EID1 cDNA (EID1TgF 5’ GCCATCAAGCTTTTTTTCCACCTTGGTCGAGTCGCTTCCACCATGTCGGAAATGGC TGAGTTGTCC 3’). The reverse primer contains an EcoRV site and a HA-tag coding sequence on the 3’ end before the stop codon of human EID1 cDNA (EID1TgR 5’ CATGGCGATATCTCAGGCGTAATCCGGGACATCGTACTCTCTATCAATAATCTCAT CACAGCCGAGTTCTTC 3’). The EID1 PCR products were cut with HindIII and EcoRV and cloned into PDGF-SV40 vector cut with HindIII/EcoRV to form a transgene expression vector (Li et al., 2004). Linear PDGF-EID1-SV40 DNA fragments were excised by SalI and NdeI digestion, purified by agarose gel electrophoresis, and recovered using QIAquick kit (QIAGEN, Valencia, CA). Transgenic mice were generated by microinjecting a linealized SalI–NdeI restriction fragment into 0.5-day FVB/N pre-nuclear mouse embryos by standard methods and maintained in the FVB/N background. Four founder mouse lines have been generated. All transgenic mice were bred to FVB/N mice to produce F1 offspring. Genotyping to screen EID1 transgenic mice was performed from mouse tail DNA using the following primers: EID1GTF 5’CTC TTC ACT GCG ATG TTC TCC 3’ and EID1GTR 5’ GTC CTC GCT CTC GAA GTC TG 3’. Over expression of EID1 transcript in the Tg mice was detected by RT-PCR from cortex total RNA using the following primers: EID1RTF 5’ GTCGGAAATGGCTGAGTTGT 3’ and EID1RTR 5’ CTTCTTCAAGAGCGGCTGAT 3’. After characterization, we have been focusing on the higher EID1 expression line to study the effects of EID1 overexpression on neuronal functions.
Brain tissues and immunohistochemical (IHC) analysis
Human AD and age-matched control brain slices from mid-temporal gyrus were generous gifts from Dr CK Combs and as described previously (Austin and Combs, 2010). Cortex and hippocampus of FVB control and EID1 Tg mice, as well as the hippocampus of the pPDGF-APPSw, Ind mimicking human AD transgenic mice (Jackson Laboratory, Bar Harbor, ME) were used in this study. Animal handling and tissue harvesting were as described by Li et al. (Li et al., 2004). IHC staining of brain tissue sections was according to the method described previously (Gan et al., 2008). The sections were stained with the following primary antibodies: rabbit anti-EID1 (1:300, GeneTex Inc, Irvine, CA, USA), mouse anti-GFAP (1:200), mouse anti-NeuN (1:600), mouse anti-MAP-2 (1:400), mouse anti-CBP (1:200) were all obtained from Millipore (Billerica, MA, USA). The antigens were detected with specific fluorescein-conjugated secondary antibodies (JacksonImmunoResearch Laboratories Inc., West Grove, PA, USA). Nuclei were counterstained with DAPI. All images were captured with a Zeiss LSM 510 META confocal system and analyzed with Photoshop software. For Nissl staining, five 6 µm sections per mouse and four mice per group were analyzed. After deparaffinization in xylene for 10 min, followed by 100%, 95% and 70% ethanol (3 min each), the sections were rinsed in deionized water and stained with 0.1% cresyl violet for 10 min. After rinsing with deionized water, they were dehydrated and mounted with Permount (Daigger, IL USA). Images were collected and analyzed with a Nikon fluorescent microscope 851 (Nikon Corporation, Tokyo). The numbers of normal neurons from the CA1 and cortex areas in each image were counted and analyzed by blinded investigators as previously described (Gan et al., 2008).
Hippocampal slice preparation and recording of potentials
Horizontal hippocampal slices (400 µm) including the EC, subiculum and hippocampus were cut using a Vibratome (Leica VT1000S) from 2.5–3.5 months old mice as described previously (Deng et al., 2006). After being deeply anesthetized with isoflurane, mice were decapitated, and their brains were dissected out in ice-cold saline solution that contained 130 mM NaCl, 24 mM NaHCO3, 3.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, 5.0 mM MgCl2, and 10 mM glucose, saturated with 95% O2 and 5% CO2, pH 7.4. Slices were initially incubated in the preceding solution at 35°C for 40 min for recovery and then kept at room temperature (~24°C) until use. All animal procedures conformed to the guidelines approved by the University of North Dakota Animal Care and Use Committee.
Whole-cell patch-clamp recordings using a Multiclamp 700B amplifier in voltage-clamp mode from in vitro hippocampal slices were used for our experiments. Cells in the slices were visually identified with infrared video microscopy and differential interference contrast optics (Deng et al., 2006). Recording electrodes were filled with the solution containing 100 mM Cs-gluconate, 0.6 mM EGTA, 5 mM MgCl2, 8 mM NaCl, 2 mM ATP2Na, 0.3 mM GTPNa, 40 mM HEPES and 1 mM QX-314, pH 7.3. The extracellular solution comprised 130 mM NaCl, 24 mM NaHCO3, 3.5 mM KCl, 1.25 mM NaH2PO4, 2.5 mM CaCl2, 1.5 mM MgCl2 and 10 mM glucose, saturated with 95% O2 and 5% CO2, pH 7.4. Bicuculline (10 µM) was included in the extracellular solution to block GABAA receptors. We recorded from the CA1 pyramidal neurons. The holding potential was at −60 mV. AMPA receptor-mediated EPSCs were evoked by placing a stimulation electrode in the stratum radiatum of the CA1 region to stimulate the Schaffer collateral fibers. Under these conditions, the recorded currents were completely blocked by application of DNQX (10 µM) or GYKI 52466 (100 µM) at the end of experiments, confirming that they were mediated by AMPA receptors. LTP was induced by presynaptic stimulation at 0.33 Hz paired with postsynaptic (CA1 pyramidal neurons) depolarization from −60 to −10 mV for 5 min. Series resistance was rigorously monitored by the delivery of 5 mV voltage steps after each evoked current. Experiments were discontinued if the series resistance changed by >10%. Data were filtered at 2 kHz, digitized at 10 kHz, acquired on-line and analyzed off-line using pCLAMP 9 software (Molecular Devices, Sunnyvale, CA).
Morris water maze test
The water maze consisted of a circular pool, 1.2 m in diameter and 0.6 m in height, filled to a level of 35 cm with water and maintained at a temperature of 22–23°C (Morris, 1984; Xu et al., 2004). Pool water was made opaque by the addition of 100 ml of nontoxic white tempera paint to reduce reflection. A Plexiglas escape platform (12 cm in diameter) was positioned 1 cm below the water surface and can be placed at various locations throughout the pool. Water maze performance was recorded with a video camera suspended above the maze and interfaced with a video tracking system (HVS Imaging, Hampton, UK). FVB control (n=10) and EID Tg mice (n=10) (15–19 months old) were tattooed with a black mark to allow video tracking. One week prior to learning and memory assay, mice were habituated to the water maze for swimming. On each spatial memory measurement, mice were placed into the pool from 4 quasi-random start points and allowed a maximum of 60 seconds to escape to the platform where the mouse was remain for 15 seconds. Mice that fail to escape were led to the platform. The mean escape latencies, swim distances, and swim speeds were statistically analyzed with the software (HVS Imaging, Hampton, UK).
Western blotting and Co-immunoprecipitation
Mouse tissues were frozen in liquid nitrogen and homogenized in RIPA buffer in an electronic homogenizer and then kept on ice for 45 min. The samples were centrifuged at 14,000xg for 10 min at 4 °C to collect the supernatant for total cellular proteins. Western blotting analyses were performed as previously described (Liu et al., 1998). Nuclear and cytoplasmic fractionation was performed using NE-PER nuclear and cytoplasmic extraction reagent (Thermo Scientific, Rockford, IL, USA) according to the manufacturer’s instruction. The blots were probed with the following primary antibodies: mouse polyclonal anti-EID1 (1:2000, Abnova, Taiwan, Corporation, Taipei, Taiwan), mouse monoclonal anti-GFP, anti-p300 and anti-βIII tubulin (1:2000, 1:500, 1:1000, respectively, all from Millipore, Temecula, CA); mouse monoclonal anti-β-actin (1:5000, Sigma, Oakville, ON); rabbit polyclonal anti-CBP, anti-acetyl His3, anti-acetyl p53 (1:500, 1:5000, 1:1000, respectively, all from Millipore, Temecula, CA) and anti-His3 (1:10,000 Novus, CO USA); mouse monoclonal anti-p53 (1:200, GenePharma Co., Ltd , Shanghai, PRC). The antigens were detected using horseradish peroxidase-conjugated secondary antibodies: anti-mouse IgG (1:5000 v/v), anti-rabbit IgG (1:5000 v/v, both from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) or antigoat IgG (1:5000, Sigma, Oakville, ON). The antigen-antibody complexes were visualized by enhanced chemiluminescence using an ECL Plus detection kit (Amersham Phamacia Biotech, Baie d’Urfe, QC).
For the co-immunoprecipitation assay, human cDNA encoding full length EID1 protein was cloned in the pEGFP-N1 vector (Clontech, Palo Alto, CA, USA) to produce an EGFP-tagged EID1 construct. This construct was transiently transfected into HEK-293 cells and total cellular proteins were extracted as described above. The immunoprecipitation procedure was as described previously (Liu et al., 2004) and precipitated complexes were boiled in protein loading buffer and separated by SDS-PAGE. The presence of CBP, p300, and EID1-EGFP in the complex was revealed by Western blotting as described above.
Results
EID1 is highly expressed in neuronal cells and translocated into the nuclei of neuronal cells of AD brains
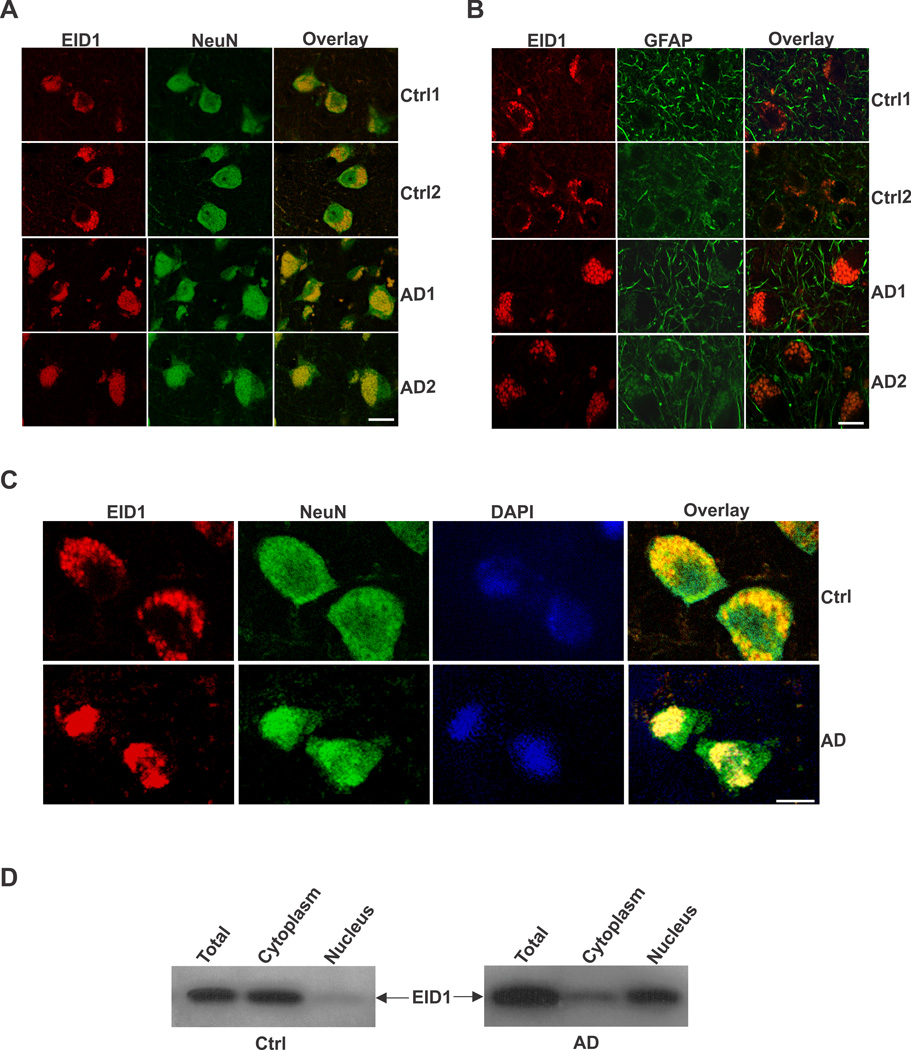
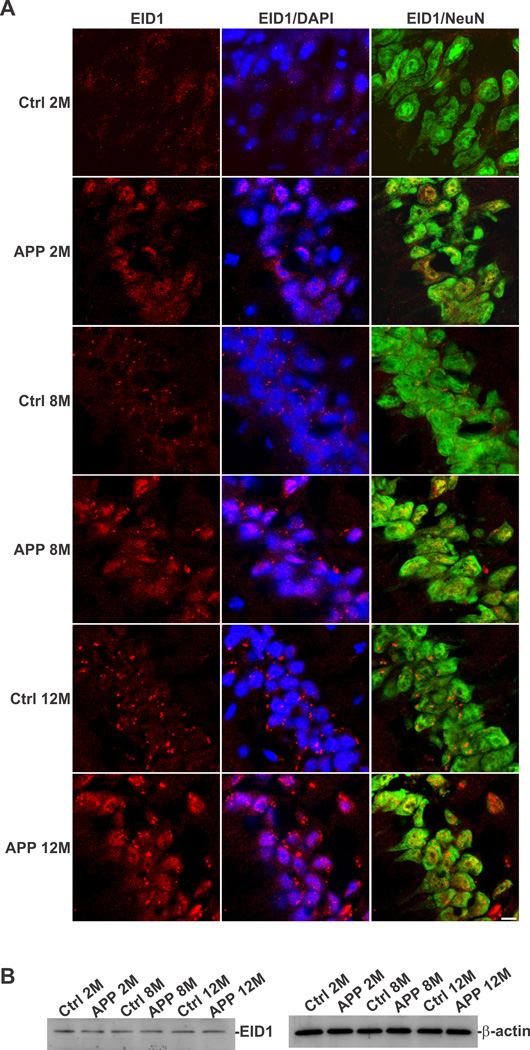
In order to determine the cellular distribution of EID1 in the brain, we carried out a double immunohistochemical (IHC) staining of EID1 with neuronal (NeuN) and glial (GFAP) markers. We discovered that this gene is primarily expressed in neuronal cells, as demonstrated by its co-localization with NeuN and is expressed to a much less degree, in glial cells in mouse brain (Fig. 1 A and B). EID1 is also highly expressed in cultured NT2 neurons and barely detectable in undifferentiated neuronal progenitor cells and astrocytes (Fig. 1C and D). Similarly, we detected EID1 protein distributed in the cytoplasm of neurons (Fig. 2A, Ctrl1 and Ctrl2), but not in glial cells (Fig. 2B, Ctrl1 and Ctrl2) of human brains. Interestingly, we noticed a significant increase in complete or partial nuclear translocation of EID1 in the cortical neurons of AD patients compared with that of control human brains (Fig. 2A, AD1 and AD2). This observation was further confirmed by triple staining of AD and control cortical neurons with anti-EID1, anti-NeuN and nuclear DAPI staining (Fig. 2C). This nuclear translocation is confirmed by nuclear and cytoplasmic fractionation of control and AD brain tissues, followed by Western blotting analysis (Fig. 2D). Furthermore, EID1 became partially aggregated and the IHC staining appeared more punctuated during aging (compare Fig. 3 Ctrl 2M with Ctrl 8M and Ctrl 12M), but nuclear translocation was not observed in these control mice. In contrast, EID1 nuclear translocation already occurred in 2 month old APP transgenic mice (Fig. 3 APP 2M) and both EID1 aggregation and nuclear translocation were progressively pronounced in the aged APP transgenic mice (Fig. 3 APP 8M and APP 12M). We did not observe changes in abundance of EID1 protein in aged or APP Tg mice as compared to younger or control mice (Fig. 3B). Our results together suggest that the increased EID1 nuclear translocation in neuronal cells may play an important role in the pathogenesis of AD and precedes Aβ plaque formation (Gan et al., 2008).
Figure 1. EID1 is highly enriched in neurons and much less in astrocytes.
A. Double staining of the Cornu Ammonis 3 (CA3) region of mouse hippocampus with anti-EID1 and anti-NeuN antibodies demonstrating that EID1 is expressed in neurons. B. Double staining of similar brain region (CA3) with anti-EID1 and anti-GFAP antibodies showing that there is no or less expression of EID1 in GFAP-positive cells. Rhodomine-conjugated anti-rabbit IgG (for EID1) and FITC-conjugated anti-mouse IgG (for NeuN and GFAP) were used to detect the specific immunostaining. Scale bar = 100 µM. C. Changes in mRNA levels of EID1 during RA-induced differentiation of NT2 cells were determined by qRT-PCR. The samples were measured against the cDNA of undifferentiated NT2 cells as a control, set at 100%. Percentage of each sample was calculated by 100× 2−ΔCt, where ΔCt is the cycle number difference between test sample and the control sample. undiff – undifferentiated NT2 cells (control), NT2A – NT2 astrocytes, NT2N – NT2 neurons. The experiments were performed in triplicate. D. Western blotting analysis of the three cell types shown in C indicating that EID1 protein is highly enriched in neuronal cells. β-actin was used as a loading control.
Figure 2. Subcellular localization of EID1 protein in control and AD brains.
The middle temporal gyrus of control and AD human brains was stained with anti-EID1 and anti-NeuN (A), or anti-EID1 and anti-GFAP (B) antibodies. Rhodomine-conjugated anti-rabbit IgG (for EID1) and FITC-conjugated anti-mouse IgG (for NeuN and GFAP) were used to detect the specific immunostaining. Scale bar = 30 µM. C. The middle temporal gyrus of control and AD human brains was stained with anti-EID1 and anti-NeuN antibodies. The nuclei were counterstained with DAPI. Scale bar = 10 µM. D. Western blotting analysis of fractionated EID1 protein from control and AD brains illustrating the increase of nuclear localization of this protein in AD brain.
Figure 3. EID1 aggregation and nuclear translocation in aged and APP transgenic mice.
The Cornu Ammonis 1 (CA1) region of the hippocampus from control and AD-like transgenic mice (pPDGF-APPSw,Ind), 2–12 months of age, was stained with anti-EID1 and anti-NeuN antibodies. The nuclei were counterstained with DAPI. Rhodomine-conjugated anti-rabbit IgG (for EID1) and FITC-conjugated anti-mouse IgG (for NeuN) were used to detect the specific immunostaining. Scale bar = 20 µM. B. Western blotting analysis EID1 from control and APP transgenic mouse brains, 2–12 months of age, demonstrating that the levels of this protein do not change significantly. β-actin was used as a loading control.
Overexpression of EID1 in mouse brain reduces hippocampal long-term potentiation and impairs spatial learning and memory function in the EID1 transgenic mice
The increased nuclear translocation in the neurons of AD patients and mutant APP AD transgenic mice further prompted us to develop EID1 transgenic mice to study the effects of EID1 on the potential pathogenesis of AD. Since the expression of EID1 in the brain is largely in neuronal cells, we generated neuron-specific EID1 overexpression Tg mice under the control of a PDGFB promoter (Fig. 4A). Three independent founder mouse lines were generated, all of which developed normally without gross abnormality. However, we found that the aging mice exhibit static and stereotypy behaviors. We focused on the higher expresser line and age-matched wild-type mice to study the potential role of EID1 in AD pathogenesis that lead to the increase of neurodegeneration, decrease of synaptic function and alteration of neurological function. We first confirmed the expression of exogenous EID1 in the cortex by RT-PCR with specific primers derived from human EID1 cDNA sequence that was introduced into the mouse brains (Fig. 4B). IHC analysis of the hippocampus sections from the control and Tg mice indicated that EID1 is translocated into the nuclei of neuronal cells in Tg mice (Fig. 4C). Nuclear and cytoplasmic fractionation of control and EID1 Tg brain tissues, followed by Western blotting analysis confirmed that overexpression of EID1 led to its nuclear translocation in neurons of the Tg mice (Fig. 4D), resembling the incidence observed in human AD brains (Fig. 2).
Figure 4. Characterization of EID1 transgenic mice.
A. A representative agarose gel image showing the identification of pPDGFB-EID1 transgenic mice by PCR amplification of mouse tail genomic DNA. In: lane P – Tg positive, lane N – Tg negative, lane M - molecular weight marker. B. RT-PCR characterization of EID1 expression in the brain (cortex) of different pPDGFB-CRI1 transgenic mice (lanes P) compared with non-transgenic littermate mice (lanes N). In: lane C - plasmid control, lane M - molecular weight marker. C. Nuclear localization of EID1 in the neurons of EID1 Tg mice. The Cornu Ammonis 3 (CA3) region of control and Tg mouse hippocampus (5 month old) was double stained with anti-EID1 and anti-GFAP antibodies. The nuclei were counterstained with DAPI. Rhodomine-conjugated anti-rabbit IgG (for EID1) and FITC-conjugated anti-mouse IgG (for GFAP) were used to detect the specific immunostaining. Scale bar = 50 µM. D. Western blotting analysis of fractionated EID1 protein from control and EID1 Tg mouse brains illustrating the increase of nuclear localization of this protein in Tg mice. Top left panel: 5 month old control mouse (Ctrl1); top right panel: 5 month old Tg mouse (Tg1); lower left panel: 19 month old control mouse (Ctrl2); lower right panel: 19 month old Tg mouse (Tg2).
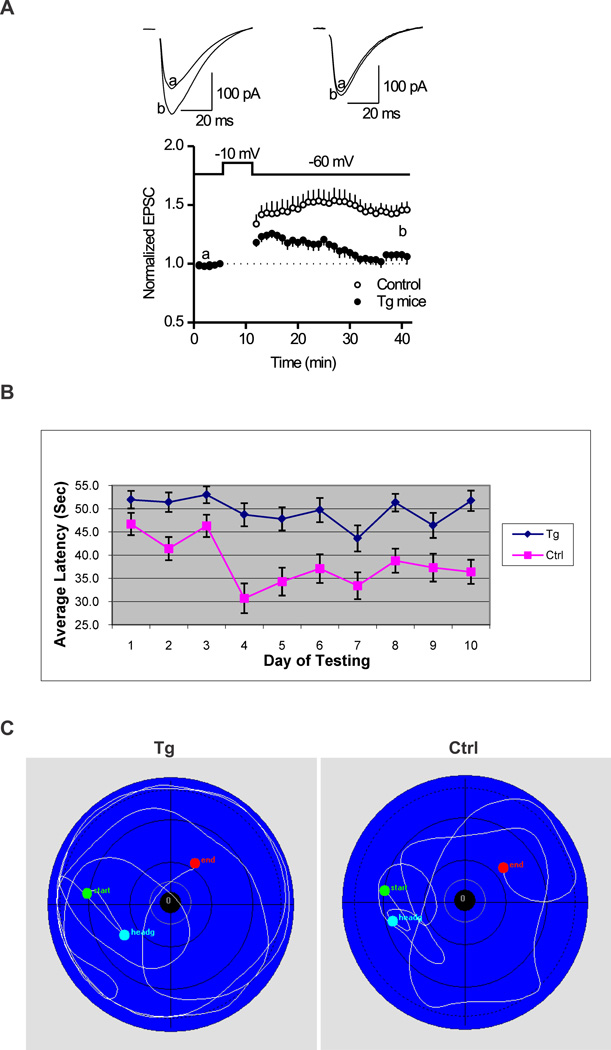
Long-term potentiation (LTP), the activity-dependent change in strength of neuronal connections, is a form of long-lasting synaptic plasticity that has been proposed as a cellular mechanism of learning and memory. To determine whether the transgenic mice overexpressing EID1 exhibit defect in synaptic plasticity, we measured and characterized the LTP in the CA1 region of hippocampal slices from 3 month EID1 mice and their wild-type littermates. We demonstrated that the amplitude of EPSC was significantly smaller in the EID1 Tg mice than that of age-matched control mice (Fig. 5A). At the time point of 30 min after inducing of LTP, the amplitude of EPSC is 145.9 ± 7.2% of baseline for control mice, and 106.1 ± 6.8% of baseline for Tg mice. The EID1 Tg mice failed to induce LTP. These results indicated that overexpression of EID1 inhibits long-lasting hippocampal synaptic plasticity.
Figure 5. EID1 Tg mice showed reduced hippocampal LTP and impaired spatial learning and memory function.
A. Hippocampal slices were prepared from three month old EID1 Tg and age matched control littermates as described in the Materials and Methods. At the time point of 30 min after induction of LTP, the amplitude of EPSC is 145.9 ± 7.2% of baseline for control mice (p < 0.001, vs. baseline, n=12 cells from 3 mice), and 106.1 ± 6.8% of baseline for Tg mice (p = 0.12, vs. baseline, n=11 cells from 3 mice). Note EID1 mice failed to induce LTP. B. Morris Water Maze test showing average latency to platform. EID1 Tg and control mice (15–19 month old, 8 each) were subjected to Morris Water Maze test for 10 days, 6–8 rounds/day. The escape latencies, swim distances, and swim speeds were statistically analyzed with the HVS Imaging software and plotted with Excel. C. Swimming path of EID1 Tg and control mice.
We further used the hidden-platform version of the Morris water maze to test the spatial memory and learning function in the EID1 Tg and age-matched control mice. As shown in Figure 5B and C, the escape latency and distance were significantly increased in EID1 Tg mice compared with that of control mice, while no difference was observed between the two groups in the cued testing. The average latency of control mice was significantly lower in day ten than day one, whereas the Tg mice showed no difference, suggesting a learning deficiency of the Tg mice. We also noticed that the time of EID1 mice spent in the target quadrant is significant less than that of wild-type control mice (Fig. 5C). Motor function analysis showed that both EID1 mice and wild-type mice have similar motor coordination either on a constant or accelerated speed (data not shown). Together, we conclude that the transgenic mice overexpressing EID1 in neurons have a significant impairment in spatial memory and learning function, which is not due to the deficits in visual and motor coordination.
EID1 overexpression alters neurofilament structure and global cellular organization in the hippocampus and cortex
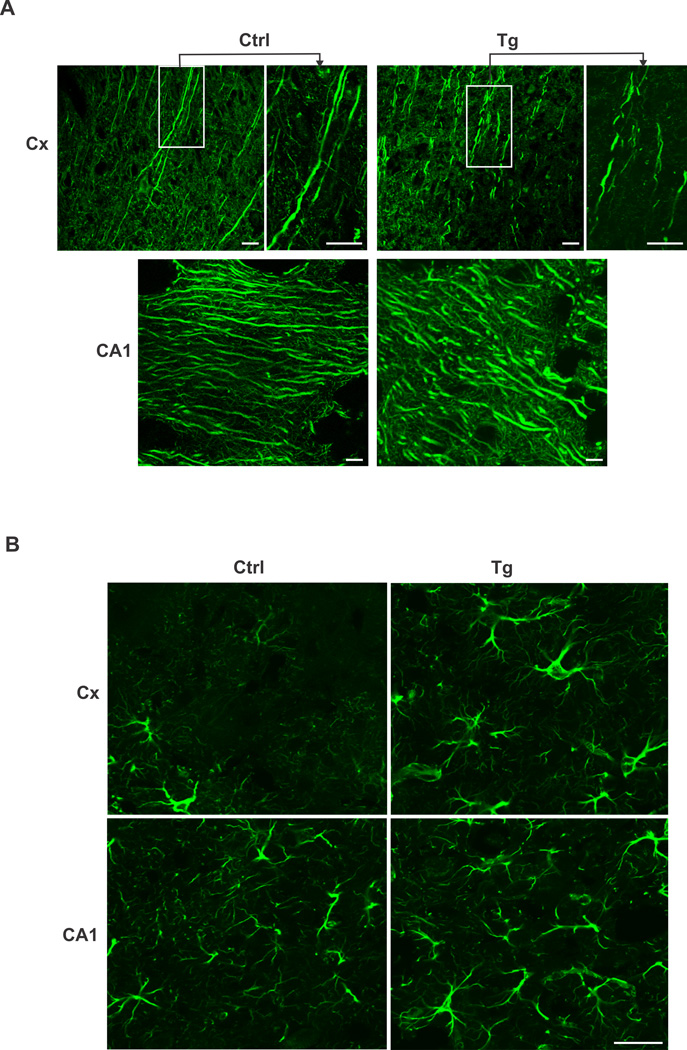
We carried out Nissl and IHC staining to determine whether overexpression of EID1 results in structural, morphological and functional alterations in the hippocampus and cortex. Nissl staining of the brain sections from control and EID1 Tg mice did not reveal significant difference in neuronal cell numbers in the cortex and hippocampus (data not shown), suggesting that the low EPSCs and failed LTP induction of the EID1 Tg mice as reflected on impaired memory and learning abilities was not caused by neuronal loss. This led us to speculate that the impairments in the Tg mice brains might be due to structural changes affecting neuronal connections or transmission. We therefore examined the brain sections of the control and Tg mice stained with anti-Map2 antibody (Fig. 6A). Higher magnification slices revealed a marked disruption of the integrity of neurofilament structure in the EID1 Tg mice (Fig. 6A top panel). Furthermore, we also observed a dramatic increase of astrogliosis in the cortex and hippocampus of aging EID1 Tg mice as indicated by abnormal increase in the number of astrocytes due to the destruction of nearby neurons (Fig. 6B). Collectively, these results document a marked increase of neuronal structural disruption, synaptic alteration and inflammatory response in the EID1 Tg mice that recapitulate the major histological and pathological characteristics of human AD.
Figure 6. Structural and cellular organization changes in EID1 Tg mouse brains.
A. disrupted neurofilamental structure in EID1 Tg mouse brains. Hippocampus (CA1) and cortex (Cx) sections from EID1 Tg and control mice were stained with anti-MAP2 antibody. FITC-conjugated anti-mouse IgG was used to detect the specific immunostaining. Scale bar = 10 µM. B. Increase of astrogliosis in EID1 Tg mouse brains. Hippocampus (CA1) and cortex (Cx) sections from EID1 Tg and control mice were stained with anti-GFAP antibody. FITC-conjugated anti-mouse IgG was used to detect the specific immunostaining. Scale bar = 50 µM.
EID1 physically interact with CBP and P300 in the nuclei
Loss of function in CBP/p300 has been directly associated with AD and EID1 has been characterized as a CBP/p300 inhibitor during myogenesis. To understand the role of EID1 in memory dysfunction in the brain, we focused on the interaction of EID1 with CBP/P300. We first confirmed the physical interactions of EID1 with both p300 and CBP by co-immunoprecipitation from cultured cells and from the brain tissues of Tg mice overexpressing human EID1 protein (Fig. 7 A, B and C). In another set of experiments, IHC was also used to study the interaction of EID1 with CBP (Fig. 7D). In the hippocampal neurons of control 19 month-old mice, CBP and EID1 were well co-localized in both the cytoplasm and nuclei. In the age matched EID1 Tg mice CBP and EID1 were completely co-localized in the nuclei, suggesting that the interaction of these proteins in the Tg mice brains may be involved in chromatin modification and/or transcription regulation in the nuclei. Next we performed Western analysis of CBP and p300 in control and EID1 Tg mice (Fig. 7E). Our results showed that the levels of these proteins were not altered in EID1 Tg mice, suggesting that it is the interaction of EID1 with CBP/p300 that is responsible for the memory and learning deficit observed in the EID1 Tg mice.
Figure 7. EID1 physically interacts with p300 and CBP.
Total cellular proteins were extracted from HEK-293 cells transfected with EGFP-tagged EID1 construct (A, B), or from the hippocampus of EID1 Tg mouse brain (C), and immunoprecipitated with antip300 (A, C) or anti-EGFP (B) antibodies. The presence of p300 in the complex from A and C was detected by anti-p300 antibody (A and C, top panels) and the presence of EID1 in the complex was detected by anti-EGFP antibody (A, bottom panel). The presence of EID1 in the complex from B and C was detected by anti-EID1 antibody (B, top panel, C bottom panel) and the presence of CBP in the complex was detected by anti-CBP antibody (B, bottom panel). In: lane1 - total cellular proteins extracted from untransfected cells (A and B) or brain tissue (C), lane 2 - total cellular proteins extracted from cells transfected with EID1-EGFP, lane 3 - mock immunoprecipitation without primary antibodies, lane 4 - proteins immunoprecipitated with anti-p300 (A, C) or anti- EGFP antibody (B). D. Co-localization of EID1 and CBP. The hippocampus of EID1 Tg mouse was stained with anti-EID1and anti-CBP antibodies. Rhodomine-conjugated anti-rabbit IgG (for EID1) and FITC -conjugated anti-mouse IgG (CBP) were used to detect the specific immunostaining. The nuclei were counterstained with DAPI. Scale bar = 50 µM. E. Western blotting analysis p300 and CBP from control and EID1 Tg mouse brains demonstrating that the levels of these proteins do not change significantly in Tg mice as compared to control mice. β-actin was used as a loading control.
Overexpression of EID1 inhibits CBP-mediated acetylation of histone and p53 proteins, thus affecting the expression of their target genes
During skeletal muscle differentiation, EID1 interacts with p300 and thus suppresses the acetyltransferase activity of p300. Since EID1 is translocated to the nuclei in neurons of the Tg mice brains, it is likely that this translocation will enable EID1 to inhibit the acetyltransferase activity of CBP/p300, thus decreasing the acetylation of the transcriptional regulatory proteins, such as histone and p53 in the nuclei. Since Necdin is linked to cytoplasmic localization of EID1, we first investigated Necdin expression by Western blot analysis in hippocampus of of EID1 Tg and control mice. We did not find changes in Necdin protein levels in EID1 Tg mice, suggesting that increase in nuclear localization of EID1 is not due to decreased expression of Necdin. Next, we examined the amounts of acetylated H2, H3 and H4 and p53 in hippocampus of the EID1 Tg and control mice. Western blotting of EID1Tg and control littermate populations revealed that approximately 20% of the EID1 Tg mice showed no significant changes in acetylated proteins examined; however, 40% of the Tg animals showed decreased amounts of acetylated H3 (Fig. 8A, top panel) and the other 40% had reduced acetylated p53 (Fig. 8B top panel). We rarely detected simultaneous decreases of both acetylated H3 and p53 in the same EID1 Tg mice, and no changes were detected for H2 and H4 protein acetylation (data not shown). Decreased histone acetylation has been linked to reduced level or loss of several key functional and structural proteins in peripheral nerve system and in cancer cells, including synaptophysin, E-cadherin, and βIII-tubulin (Akasaka et al., 2009; Ekici et al., 2008; Liu et al., 2008). We therefore performed Western analysis of these proteins from EID1 Tg and control animals. While we found no changes in synaptophysin or E-cadherin in the hippocampus of EID1 Tg mice, we did detect a decrease in βIII-tubulin protein levels, where there was a decrease in acetylated H3 (Fig. 8A, second panel from the top). It has been recently discovered that acetylated p53 promotes neurite outgrowth and axon regeneration through upregulation of its target genes, such as, coronin, actin binding protein, 1B (CORO1B), growth associated protein 43 (GAP43) and Ras-associated protein 13 (RAB13) following neuronal injury (Gaub et al., 2010; Tedeschi and Di, 2009; Tedeschi et al., 2009). Although we found decreased acetylated p53 in the Tg mice hippocampus, Western blot analysis of EID1 Tg and control animals did not reveal any changes in these proteins (data not show). We did once again, find reduced βIII-tubulin levels in these Tg mice hippocampus (Fig. 8B, second panel from the top). These data indicate that overexpression of EID1 indeed reduces CBP/p300-mediated acetylation of H3 and p53 and subsequently alters the levels of certain structural protein in neurons, thus offering partial explanation on the observation of disrupted neurofilament structure in the EID1 Tg mice brains (Fig. 6A).
Figure 8. EID1 mediated decrease of His3 and p53 acetylation leading to decreased βIII-tubulin expression.
Total cellular proteins were extracted from the hippocampus of control and EID1 Tg mice and separated by SDS-PAGE (50 µg/lane). The levels of acetylated his3, corresponding βIII-tubulin and total his3 (A), or acetylated p53, corresponding βIII-tubulin and total p53 (B) in each sample were detected by Western analysis. No significant changes were found in total his3 and p53. β-actin was used as a loading control.
Discussion
Neurodegenerative diseases are often associated with impaired learning and memory, eventually leading to dementia. The role of histone acetylation in regulation of genes associated with cognition has attracted great interest in recent years (Fischer et al., 2007; Francis et al., 2009; Peleg et al., 2010; Sharma, 2010). Studies on the use of histone deacetylase (HDAC) inhibitors to enhance memory storage and potentially treat mental retardation and neurodegenerative diseases have rapidly emerged from the literature (Fischer et al., 2010). Not only is CBP/p300 a key family of acetyltrasferases, CBP has also been shown to be the transcriptional co-activator regulating gene expression induced by certain HDAC inhibitors (Vecsey et al., 2007). While memory deficit caused by reduced histone acetylation has been unequivocally linked to CBP ablation in neurons (Valor et al., 2011), the role of its acetyltransferase inhibitor, EID1, has never been investigated in the brain. The present study opened a new window to the understanding of EID1 function in learning and memory. The analysis of neuron-specific overexpression of EID1 in a novel mouse model did not only provide new insights of the regulation of nuclear protein acetylation, it also contributed to the mechanism of CBP/p300 inhibition linked to neurodegenerative diseases.
EID1 nuclear translocation/overexpression did not interfere with neuronal viability
Earlier studies on EID1 suggested that the cellular distribution of the protein is mainly cytoplasmic and sometime it can be distributed throughout the whole cells (Bavner et al., 2002; Wen and Ao, 2001). Our results also showed that EID1 is mainly located in the cytoplasm of neurons in control human and mouse brains. It was translocated into the nuclei under stressed or pathological conditions such as the cases seen in AD patients and APP Tg mice brains (Fig. 2 and 3). We were able to recreate this phenomenon by generating EID1 Tg mice with neuron-specific overexpression of human EID1. This model allowed us to study the biological significance of this nuclear translocation. It is unclear at the present whether EID1 has a specific function in the cytoplasm under normal physiological condition. Its nuclear translocation appeared to be consistent with its proposed functions within the nucleus, including antagonism of CBP/p300-dependent coactivator functions, the direct inhibition of the HAT activity of CBP/p300 and interaction with histones (Bavner et al., 2002; MacLellan et al., 2000; Miyake et al., 2000). Necdin is mainly present in the cytoplasm of mature neurons and was reported to sequester EID1 in the cytoplasm, rendering EID1 less active in transcriptional repression because of its retention in the cytoplasm (Lee et al., 2005; Bush and Wevrick, 2008). However, we did not detect decreased necdin levels in the Tg mice brains, suggesting that a different pathway mediating the nuclear translocation of EID1 under these pathological conditions.
There was a significant spatial learning and memory deficit in the EID1 Tg mice; however, we were surprised to discover that there was no significant cell loss in the cortex and hippocampus of the EID1 Tg mice brains. Instead, we found severe structural disruption of neurofilaments in these regions. It is possible that only specific transcriptional programs were affected by EID1-mediated transcriptional suppression and these affected genes were not involved in neuronal apoptosis. This is in agreement with the findings by Valor and colleagues that loss of CBP and subsequent decrease in histone acetylation had a very modest impact on the expression of a number of immediate early genes and did not at all affect neuronal viability (Valor et al., 2011). Both lines of evidence suggest that sustained low levels of histone acetylation do not necessarily cause neuronal cell death. However, this may not apply to diseased conditions where neurodegeneration has already initiated. In these cases apoptosis is specifically accompanied by further decrease of histone acetylation, due to caspase-6 cleavage of CBP/p300, compromising neuronal survival (Rouaux et al., 2003).
EID1 inhibits the acetylation of selected transcriptional regulators which may control a specific set of structural proteins
Despite lack of neurodegeneration in the brains of the EID1 Tg mice, they suffer significant spatial learning and memory deficit, linking the function of EID1 to memory. EID1 binds p300 and our Co-IP experiment showed that it also binds CBP. Earlier studies have indicated that its interaction with CBP/p300 inhibits CBP/p300 HAT activity (MacLellan et al., 2000; Miyake et al., 2000). Increased amounts of EID1 in the nuclei of hippocampal neurons significantly reduced the acetylation of histone 3 or p53, but not other histones. The molecular mechanism of selective inhibition of histone and non-histone targets is unknown, but it seems to be consistent with the observations that training or environmental enrichment induced acetylation of specific histones. For example, environmental enrichment induced hippocampal and cortical acetylation of Histone 3 and 4 in CK-25 Tg mice (Fischer et al., 2007). During learning, aged mice exhibited specific H4 lysine 12 deregulation (Peleg et al., 2010). Whereas ablation of CBP in forebrain principal neurons preferentially reduced H2A and H2B acetylation (Valor et al., 2011). It appears that memory formation does not affect all histones and restricted CBP knockout or inhibition of CBP/p300 function by overexpression of their inhibitor, EID1, cause random deregulation of histone/non-histone protein acetylation.
The reduced acetylation of p53 in the hippocampus of some EID1Tg mice is intriguing because of its crucial role in eliciting neuronal cell death during development and in adult organisms after exposure to a range of stressor and/or DNA damage. CBP/p300 acetylates most lysine residues located in the last COOH-terminal portion of p53 (Gu and Roeder, 1997). It has been discovered recently that the non-apoptotic role of p53 in neuronal cells heavily involves the state of acetylation of p53 protein. CBP/p300 mediated increase of p53 acetylation was enriched during neuronal outgrowth and maturation (Tedeschi and Di, 2009). Enhanced p53 acetylated at specific sites was shown to promote neurite outgrowth and maturation in cultured neurons without affecting cell survival. Further investigation has identified Coronin 1b, Rab 13 and GAP43 as the p53 target genes involved in pro-neurite and axon-outgrowth in PC 12 cells (Di et al., 2006; Tedeschi et al., 2009). We did not however find any changes in protein levels of these p53 target genes in the EID1 Tg mice. It is possible that the involvement of these proteins only represents molecular pathways occurs during neuronal injury in PC12 cell lines. There may be different set of immediate downstream targets affected by reduced p53 acetylation in the neurons of EDI1 Tg mice.
Decreased histone/p53 acetylation may affect microtubule stability: implication in neuronal structure and plasticity
Although decreased histone and p53 acetylation may deregulate the transcriptions of different sets of immediate early genes, the affected pathways appear to converge on microtubule structure. We found that the levels of βIII-tubulin were decreased in mice where His 3 or p53 acetylation was reduced, offering a plausible explanation as why there was disrupted neurofilamental structure in the EID1 Tg mice brains (Fig. 6A). Microtubule stability is crucial for elongated cell such as neurons to function. They provide structure and network for the transport of cargo vesicles and disruption of this network leads to reduced synaptic plasticity and memory function. It has been reported that nicotinamice, a HDAC inhibitor restored cognition in AD Tg mice through increasing the levels of microtubule-stability associated proteins (Green et al., 2008). It is not clear currently how p53 de-acetylation leads to decreased βIII-tubulin level. There is direct evidence that reduced histone acetylation induced loss of βIII-tubulin in cancer cells (Akasaka et al., 2009). Our finding is also consistent with other studies reporting that histone deacetylation or increased histone acetylation have selective effect on gene expression (Peleg et al., 2010; Vecsey et al., 2007). Overall, our EID1 Tg mice provide a good model to study CBP/p300 mediated synaptic plasticity and memory enhancement associated with neurodegenerative diseases. Future genome wide analysis of gene expression will identify molecular networks affected by EID1 translocation and overexpression. This will undoubtedly shed light on the molecular mechanisms of intrinsic inhibition of the acetylation of histone or non-histone proteins in neurons.
Highlights.
We found EID1 is translocated into the nuclei in neurons of AD brains.
We developed a EID1 transgenic mouse model.
EID1 Overexpression and nuclear translocation reduced LTP and impaired memory.
EID1 inhibits CBP/p300 acetyltrasferase activity and disrupts neuronal structure.
Acknowledgements
This study was supported in part by NIH NS45829, and HL75034 Grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akasaka K, Maesawa C, Shibazaki M, Maeda F, Takahashi K, Akasaka T, Masuda T. Loss of class III beta-tubulin induced by histone deacetylation is associated with chemosensitivity to paclitaxel in malignant melanoma cells. J Invest Dermatol. 2009;129:1516–1526. doi: 10.1038/jid.2008.406. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Austin SA, Combs CK. Amyloid precursor protein mediates monocyte adhesion in AD tissue and apoE(−)/(−) mice. Neurobiol Aging. 2010;31:1854–1866. doi: 10.1016/j.neurobiolaging.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavner A, Johansson L, Toresson G, Gustafsson JA, Treuter E. A transcriptional inhibitor targeted by the atypical orphan nuclear receptor SHP. EMBO Rep. 2002;3:478–484. doi: 10.1093/embo-reports/kvf087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JR, Wevrick R. The Prader-Willi syndrome protein necdin interacts with the E1A-like inhibitor of differentiation EID-1 and promotes myoblast differentiation. Differentiation. 2008;76:994–1005. doi: 10.1111/j.1432-0436.2008.00281.x. [DOI] [PubMed] [Google Scholar]
- Chen WY, Juan LJ, Chung BC. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol Cell Biol. 2005;25:10442–10453. doi: 10.1128/MCB.25.23.10442-10453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng PY, Porter JE, Shin HS, Lei S. Thyrotropin-releasing hormone increases GABA release in rat hippocampus. J Physiol. 2006;577:497–511. doi: 10.1113/jphysiol.2006.118141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di GS, Knights CD, Rao M, Yakovlev A, Beers J, Catania J, Avantaggiati ML, Faden AI. The tumor suppressor protein p53 is required for neurite outgrowth and axon regeneration. EMBO J. 2006;25:4084–4096. doi: 10.1038/sj.emboj.7601292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekici M, Schmitz F, Hohl M, Seigel GM, Thiel G. Chromatin structure and expression of synapsin I and synaptophysin in retinal precursor cells. Neurochem Int. 2008;53:165–172. doi: 10.1016/j.neuint.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Francis YI, Diss JK, Kariti M, Stephanou A, Latchman DS. p300 activation by Presenilin 1 but not by its M146L mutant. Neurosci Lett. 2007;413:137–140. doi: 10.1016/j.neulet.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Francis YI, Fa M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, Arancio O. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer's disease. J Alzheimers Dis. 2009;18:131–139. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Qiao S, Lan X, Chi L, Luo C, Lien L, Yan LQ, Liu R. Neurogenic responses to amyloid-beta plaques in the brain of Alzheimer's disease-like transgenic (pPDGF-APPSw,Ind) mice. Neurobiol Dis. 2008;29:71–80. doi: 10.1016/j.nbd.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di GS. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 2010;17:1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- Gong B, Vitolo OV, Trinchese F, Liu S, Shelanski M, Arancio O. Persistent improvement in synaptic and cognitive functions in an Alzheimer mouse model after rolipram treatment. J Clin Invest. 2004;114:1624–1634. doi: 10.1172/JCI22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, Sellers WR, Mittnacht S. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, O'Neill MA, Wevrick R. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- Li B, Ryder J, Su Y, Moore SA, Jr, Liu F, Solenberg P, Brune K, Fox N, Ni B, Liu R, Zhou Y. Overexpression of GSK3betaS9A resulted in tau hyperphosphorylation and morphology reminiscent of pretangle-like neurons in the brain of PDGSK3beta transgenic mice. Transgenic Res. 2004;13:385–396. doi: 10.1023/b:trag.0000040039.44899.6f. [DOI] [PubMed] [Google Scholar]
- Liu QY, Lei JX, Leblanc J, Sodja C, Ly D, Charlebois C, Walker PR, Yamada T, Hirohashi S, Sikorska M. Regulation of DNaseY activity by actinin-alpha4 during apoptosis. Cell Death Differ. 2004;11:645–654. doi: 10.1038/sj.cdd.4401401. [DOI] [PubMed] [Google Scholar]
- Liu QY, Pandey S, Singh RK, Lin W, Ribecco M, Borowy-Borowski H, Smith B, Leblanc J, Walker PR, Sikorska M. DNaseY: a rat DNaseI-like gene coding for a constitutively expressed chromatin-bound endonuclease. Biochemistry. 1998;37:10134–10143. doi: 10.1021/bi9800597. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hong Y, Zhao Y, Ismail TM, Wong Y, Eu KW. Histone H3 (lys-9) deacetylation is associated with transcriptional silencing of E-cadherin in colorectal cancer cell lines. Cancer Invest. 2008;26:575–582. doi: 10.1080/07357900701837168. [DOI] [PubMed] [Google Scholar]
- Macchiarulo A, Rizzo G, Costantino G, Fiorucci S, Pellicciari R. Unveiling hidden features of orphan nuclear receptors: the case of the small heterodimer partner (SHP) J Mol Graph Model. 2006;24:362–372. doi: 10.1016/j.jmgm.2005.09.016. [DOI] [PubMed] [Google Scholar]
- MacLellan WR, Xiao G, Abdellatif M, Schneider MD. A novel Rb- and p300- binding protein inhibits transactivation by MyoD. Mol Cell Biol. 2000;20:8903–8915. doi: 10.1128/mcb.20.23.8903-8915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggirwar SB, Ramirez S, Tong N, Gelbard HA, Dewhurst S. Functional interplay between nuclear factor-kappaB and c-Jun integrated by coactivator p300 determines the survival of nerve growth factor-dependent PC12 cells. J Neurochem. 2000;74:527–539. doi: 10.1046/j.1471-4159.2000.740527.x. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, Robakis NK. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Miyake S, Sellers WR, Safran M, Li X, Zhao W, Grossman SR, Gan J, DeCaprio JA, Adams PD, Kaelin WG., Jr Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol Cell Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Park YY, Park KC, Shong M, Lee SJ, Lee YH, Choi HS. EID-1 interacts with orphan nuclear receptor SF-1 and represses its transactivation. Mol Cells. 2007;24:372–377. [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, gis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Lee VM. NTera 2 cells: a human cell line which displays characteristics expected of a human committed neuronal progenitor cell. J Neurosci Res. 1993;35:585–602. doi: 10.1002/jnr.490350603. [DOI] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyurek Y, Bartholdi D, Niedrist D, Papadia F, Bacino CA, den Dunnen JT, van Ommen GJ, Breuning MH, Hennekam RC, Peters DJ. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ, III, Kandel ER, Duff K, Kirkwood A, Shen J. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Sharma SK. Protein acetylation in synaptic plasticity and memory. Neurosci Biobehav Rev. 2010;34:1234–1240. doi: 10.1016/j.neubiorev.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Shen H, Sikorska M, Leblanc J, Walker PR, Liu QY. Oxidative stress regulated expression of ubiquitin Carboxyl-terminal Hydrolase-L1: role in cell survival. Apoptosis. 2006;11:1049–1059. doi: 10.1007/s10495-006-6303-8. [DOI] [PubMed] [Google Scholar]
- Sodja C, Fang H, Dasgupta T, Ribecco M, Walker PR, Sikorska M. Identification of functional dopamine receptors in human teratocarcinoma NT2 cells. Brain Res Mol Brain Res. 2002;99:83–91. doi: 10.1016/s0169-328x(01)00324-2. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Di GS. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10:576–583. doi: 10.1038/embor.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Nguyen T, Puttagunta R, Gaub P, Di GS. A p53-CBP/p300 transcription module is required for GAP-43 expression, axon outgrowth, and regeneration. Cell Death Differ. 2009;16:543–554. doi: 10.1038/cdd.2008.175. [DOI] [PubMed] [Google Scholar]
- Tsirigotis M, Tang MY, Beyers M, Zhang M, Woulfe J, Gray DA. Delayed spinocerebellar ataxia in transgenic mice expressing mutant ubiquitin. Neuropathol Appl Neurobiol. 2006;32:26–39. doi: 10.1111/j.1365-2990.2005.00694.x. [DOI] [PubMed] [Google Scholar]
- Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in Forebrain Principal Neurons Causes Modest Memory and Transcriptional Defects and a Dramatic Reduction of Histone Acetylation But Does Not Affect Cell Viability. J Neurosci. 2011;31:1652–1663. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo OV, Sant'Angelo A, Costanzo V, Battaglia F, Arancio O, Shelanski M. Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci U S A. 2002;99:13217–13221. doi: 10.1073/pnas.172504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wen H, Ao S. Identification and characterization of a novel human cDNA encoding a 21 kDa pRb-associated protein. Gene. 2001;263:85–92. doi: 10.1016/s0378-1119(00)00585-0. [DOI] [PubMed] [Google Scholar]
- Xu W, Chi L, Row BW, Xu R, Ke Y, Xu B, Luo C, Kheirandish L, Gozal D, Liu R. Increased oxidative stress is associated with chronic intermittent hypoxia-mediated brain cortical neuronal cell apoptosis in a mouse model of sleep apnea. Neuroscience. 2004;126:313–323. doi: 10.1016/j.neuroscience.2004.03.055. [DOI] [PubMed] [Google Scholar]
- Yukawa K, Tanaka T, Tsuji S, Akira S. Regulation of transcription factor C/ATF by the cAMP signal activation in hippocampal neurons, and molecular interaction of C/ATF with signal integrator CBP/p300. Brain Res Mol Brain Res. 1999;69:124–134. doi: 10.1016/s0169-328x(99)00086-8. [DOI] [PubMed] [Google Scholar]