Abstract
The role of estrogen receptor alpha (ER) in breast cancer development and as a primary clinical marker for breast cancer prognosis is well documented. In this study, we identified the oncogenic protein TWIST1 (Twist), which is over-expressed in high-grade breast cancers, as a potential negative regulator of ER expression. Functional characterization of ER regulation by Twist was carried out using Twist low (MCF-7, T-47D) and Twist high (Hs 578T, MDA-MB-231, MCF-7/Twist) expressing cell lines. All Twist high cell lines exhibited low ER transcript and protein levels. By chromatin immunoprecipitation and promoter assays, we demonstrated that Twist could directly bind to E-boxes in the ER promoter and significantly down-regulate ER promoter activity in vitro. Functionally, Twist over-expression caused estrogen independent proliferation of breast cells and promoted hormone resistance to the selective estrogen receptor modulator (SERM) tamoxifen and selective estrogen receptor down-regulator (SERD) fulvestrant. Importantly, this effect was reversible on down-regulating Twist. Additionally, orthotopic tumors generated in mice using MCF-7/Twist cells were resistant to tamoxifen. These tumors had high vascular volume and permeability surface area as determined by magnetic resonance imaging (MRI). Mechanistically, Twist recruited DNA methyltransferase 3B (DNMT3B) to the ER promoter leading to a significantly higher degree of ER promoter methylation compared to parental cells. Furthermore, we demonstrated by co-immunoprecipitation that Twist interacted with histone deacetylase 1 (HDAC1) at the ER promoter, causing histone deacetylation and chromatin condensation, further reducing ER transcript levels. Functional re-expression of ER was achieved using demethylating agent 5-azacytidine and histone deacetylase inhibitor valproic acid. Finally, an inverse relationship was observed between Twist and ER expression in human breast tumors. In summary, the regulation of ER by Twist could be an underlying mechanism for loss of ER activity observed in breast tumors and may contribute to the generation of hormone resistant ER negative breast cancer.
Keywords: Breast cancer, estrogen receptor, hormone resistance, TWIST1, Twist, transcriptional regulation, chromatin immunoprecipitation, co-immunoprecipitation, histone deacetylation, histone deacetylase 1 (HDAC1), methylation, DNA methyltransferase 3B (DNMT3B)
Introduction
The role of estrogen receptor alpha (ER) in normal breast development and in breast cancer is well established (Osborne 1998). For breast cancer treatment, ER status is of paramount importance for the selection of appropriate hormonal therapy and also as a prognostic marker. At presentation, over 75% of breast tumors are ER positive and are treated with targeted anti-estrogen therapy, including selective estrogen receptor modulators (SERMs) such as tamoxifen and raloxifene (Herynk and Fuqua 2007, Osborne et al 2000), and selective estrogen receptor down-regulators (SERDs) such as fulvestrant (Faslodex, ICI 182,780)(Osborne et al 2004). Recently, aromatase inhibitors (AIs) (i.e. letrozole and anastrazole) that directly target the production of estrogen from testosterone are increasingly being used in the clinic (Osborne and Tripathy 2005, Osborne and Schiff 2005). They are more effective and demonstrate fewer side effects when compared to tamoxifen, which is the most commonly prescribed anti-estrogen drug (Herynk and Fuqua 2007). ER negative tumors, on the other hand, are refractory to anti-estrogen therapy from the onset and are associated with a poor clinical prognosis (Herynk and Fuqua 2007). Additionally, a majority of ER positive breast tumors become hormone resistant through various mechanisms and relapse within five years (Herynk and Fuqua 2007, Schiff et al 2005). Epigenetic silencing through promoter hypermethylation of ER accounts for a major portion of ER gene silencing (Ottaviano et al 1994, Yan et al 2001). Mutations within the ER gene are observed in about 1% of primary breast tumors but it is unclear how these contribute to the regulation of ER expression (Fuqua et al 2000, Herynk and Fuqua 2004, Murphy et al 1997, Roodi et al 1995). In addition to these regulatory mechanisms, ER is also regulated transcriptionally (Angeloni et al 2004, deConinck et al 1995). Changes in proliferation and apoptotic stimuli including ER and cyclin D1 signaling which contributes towards tumor homeostasis (Herynk and Fuqua 2007) and activation of classical signaling pathways including HER-2 and EGFR (Herynk and Fuqua 2007), MAPK (Oh et al 2001), and PI3K/AKT (Campbell et al 2001) have also been implicated in hormone resistance.
TWIST1 (Twist) is a basic helix-loop-helix transcription factor involved in the negative regulation of cellular determination (Rose and Malcolm 1997) and in the differentiation of several lineages including myogenesis and osteogenesis (Bialek et al 2004, Hebrok et al 1997). Twist is over-expressed in breast cancers (Mironchik et al 2005), promotes chromosomal instability (Vesuna et al 2006), regulates the tumor suppressor protein E-cadherin (CDH1) (Vesuna et al 2008) and p53 (Stasinopoulos et al 2005), and promotes the generation of breast cancer stem cells (Vesuna et al 2009). However, its mechanistic role in breast cancer progression is still not completely understood.
In this study, we identified ER as a target of Twist by demonstrating that Twist binds to the ER promoter and down-regulates its transcription. This is accompanied by de novo methylation of the ER promoter caused by recruitment of DNA methyltransferase 3B (DNMT3B). Twist additionally mobilizes histone deacetylase 1 (HDAC1) which suppresses ER expression due to the formation of a repressive chromatin structure caused by deacetylation of the ER promoter. Both these effects can be partially reversed by using methylation inhibiting reagent 5-azacytidine and histone deacetylase inhibitor valproic acid. Functionally, the loss of ER in breast cancer cells over-expressing Twist results in hormonal independence and resistance to anti-estrogens tamoxifen and fulvestrant. Moreover, Twist over-expressing breast cancer cells are able to form orthotopic tumors in severe combined immunodeficient (SCID) mice in the absence of estrogen supplementation. Furthermore, these tumors have high vascular volume and high vascular permeability. Finally, we show an inverse correlation between Twist and ER expression in human breast tumors. Taken together, these data suggest a mechanistic link between the up-regulation of Twist and loss of ER leading to the progression of ER negative and hormone resistant breast cancer.
Results
ER is down-regulated in Twist over-expressing breast cancer cell lines
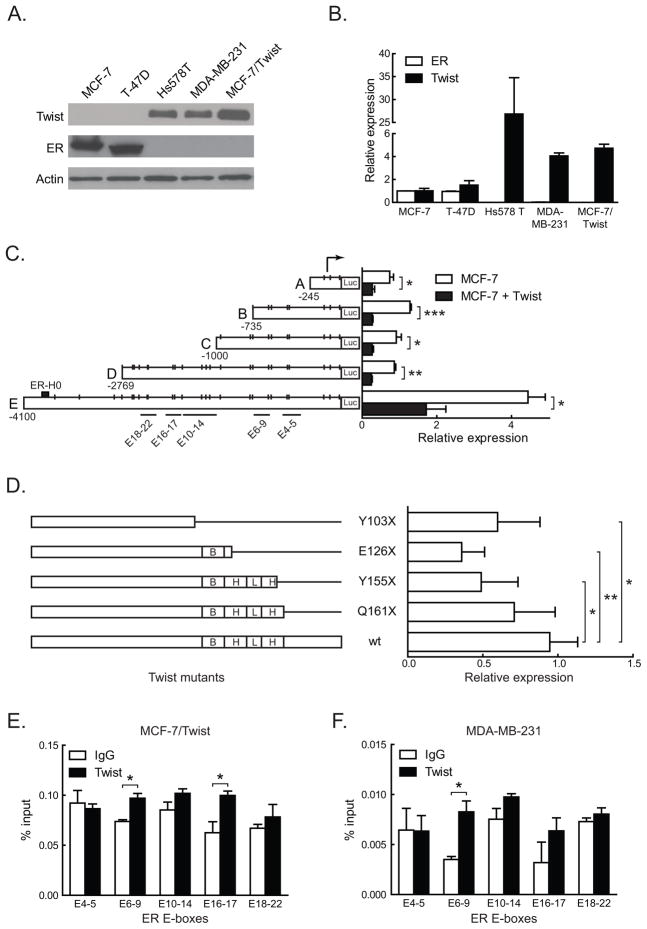
To characterize the role of Twist in breast cancer biogenesis, we initially analyzed the MCF-7/Twist cell line (Mironchik et al 2005) for differential gene expression using microarray analysis (Affymetrix, Santa Clara, CA), and identified the ER transcript which was down-regulated by 13 fold. To confirm this finding, breast cancer cell lines were evaluated for Twist and ER expression by immunoblotting and qRT-PCR. As shown in Figure 1A–B, there was an inverse correlation between Twist and ER protein and mRNA levels within the cell lines tested.
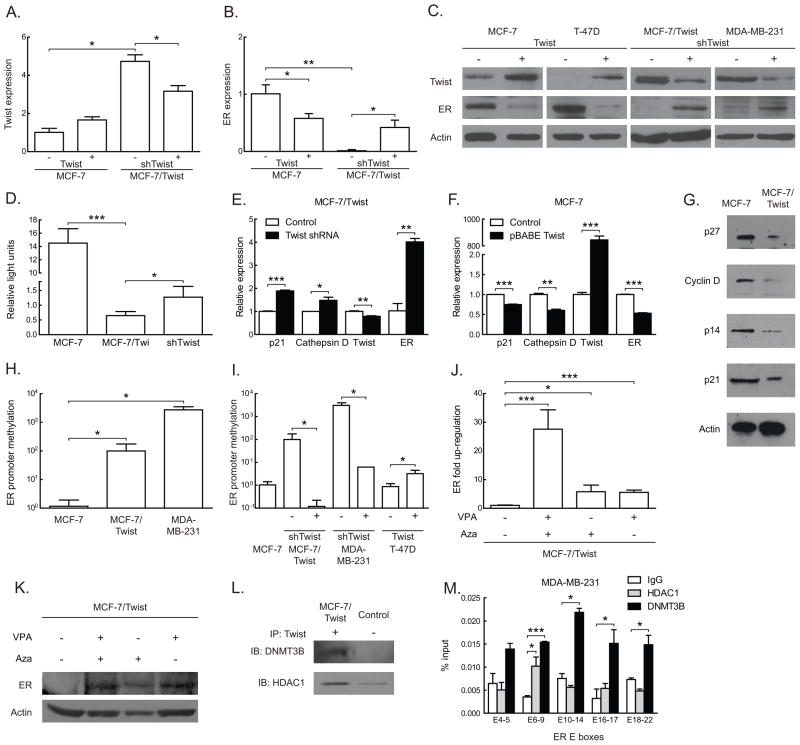
Figure 1. Twist regulation of ER by direct promoter binding.
A. Total proteins from three tumorigenic (MCF-7, T-47D, and Hs 578T) and two metastatic (MDA-MB-231 and MCF-7/Twist) breast cancer cell lines were immunoblotted for Twist and ER. Antibodies against Twist were made in-house, while ER (Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (Sigma-Aldrich) were obtained commercially.
B. Histogram depicting relative expression of Twist and ER mRNA from cell lines analyzed by qRT-PCR. The primers used were 5′-GGACAAGCTGAGCAAGATTCAGA-3′ and 5′-TCTGGAGGACCTGGTAGAGGAA-3′ for Twist and 5′-GACAGGGAGCTGGTTCACAT-3′ and 5′-AGGATCTCTAGCCAGGCACA-3′ for ER. Values for Twist and ER were normalized to values in MCF-7. Experiments were repeated thrice in duplicates. Error bars depict S.D.
C. Schematic representation of ER promoter constructs showing the location of putative Twist binding E-boxes, denoted by short vertical lines. Areas spanned by chromatin immunoprecipitation (ChIP) amplicons are denoted by lines with E-box numbers. Promoter reporter assay results of transient Twist transfections in MCF-7 cells are displayed in the histogram on the right. Experiments were repeated five times in duplicates. Error bars depict S.D.
D. Schematic displaying Twist wild-type (wt) and mutant constructs. Stop codons, the DNA binding basic domain (B), and the helix-loop-helix (HLH) domain are indicated. Promoter activities of the constructs are displayed on the adjacent histogram. All luciferase activities were normalized to the activity of wild type Twist. Experiments were repeated twice in duplicates. Error bars depict S.E.M.
E–F. Chromatin immunoprecipitation using Twist antibody was carried out using MCF-7/Twist and MDA-MB-231 cells and analyzed using E-box specific primers by quantitative real-time PCR. Histograms depict the amplification from each primer set as a percentage of input chromatin compared to IgG negative control. A rabbit monoclonal antibody against histone H3 was used as positive control.
Twist represses ER promoter activity in breast cancer cells
To functionally confirm the regulatory role of Twist in ER down-regulation, we carried out promoter-reporter assays in breast cancer cell lines. The 4 kb ER promoter has 26 canonical E-box sequences (CANNTG) (Murre et al 1989) to which Twist can potentially bind (Figure 1C). Transient transfections with Twist plasmids were carried out for promoter reporter assays in MCF-7 (Figure 1C) and MCF-7/Twist cells (data not shown). Twist repressed the full-length ER promoter by 2.5 fold, while the other deletion constructs were repressed from 2.5 to 3 fold.
In order to confirm the role of the bHLH regions of Twist in binding the ER promoter, we used the full-length ER promoter and Twist bHLH deletion mutants to assay for ER promoter repression. As seen in Figure 1D, none of the Twist mutants demonstrated repression comparable to wild-type Twist, except for the deletion mutant Q161X, which was downstream of the bHLH domain.
Twist binds directly to E-boxes within the ER promoter
To address if Twist binds directly to the ER promoter, we carried out chromatin immunoprecipitation (ChIP) assays using MCF-7/Twist and MDA-MB-231 cell lines (Figure 1E–F). MDA-MB-231 is an ER negative breast cancer cell line with high levels of endogenous Twist. Chromatin immunoprecipitation was carried out as per instructions (Cell Signaling, Danvers, MA) using antibodies against Twist. Normal rabbit IgG was used as a negative control, while Histone H3 was used as a positive control antibody. Five pairs of primers were designed to span most of the ER promoter and the 26 putative Twist binding E-boxes (Figure 1C). Maximum chromatin immunoprecipitation binding was seen in the areas of E-box 6–9 in MCF-7/Twist and MDA-MB-231 cells. Significant binding was also seen in the area of E-boxes 16–17 in MCF-7/Twist cells. These results indicate that Twist binds directly or as part of a complex to the endogenous ER promoter.
Twist facilitates estrogen independence in breast cells
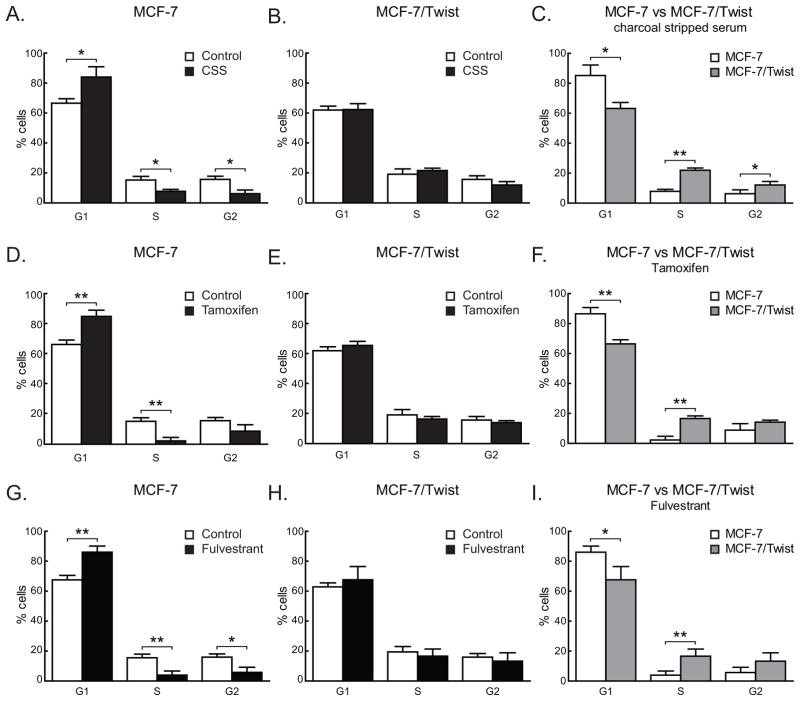
As we observed increased Twist expression in ER negative cell lines, we sought to determine if Twist promoted hormone independence by repressing ER expression. To confirm this observation, MCF-7 (ER positive) and MCF-7/Twist (ER negative) cells were grown for three days in estrogen depleted media containing 5% charcoal stripped serum (CSS) and cell cycle distribution was analyzed by flow cytometry (Figure 2). Proliferation of MCF-7 cells was significantly reduced in CSS (S=5.4%) compared to untreated cells (S=15.6%, P<0.05)(Figure 2A) but not in MCF-7/Twist cells (S=22.0% vs. 19.5%, P>0.05) (Figure 2B). Moreover, the percentages of cells in all three phases of the cell cycle was significantly altered between MCF-7 and MCF-7/Twist cells treated with CSS - G1=85.2% vs. 63.2%, P<0.05; S=5.4% vs. 22.0%, P<0.005; and G2=6.3% vs. 12.1%, P<0.05 (Figure 2C). The difference was insignificant in untreated controls of both MCF-7 and MCF-7/Twist. These results support our earlier data indicating that the down-regulation of ER by Twist in MCF-7 cells leads to estrogen independent growth.
Figure 2. Cell cycle profiles of MCF-7 and MCF-7/Twist cells.
A–C. Histograms depicting cell cycle phases for MCF-7 and MCF-7/Twist cells grown in the absence of estrogen (charcoal stripped serum).
D–F. Histograms displaying cell cycle profiles of MCF-7 and MCF-7/Twist cells grown in estrogen-free media followed by tamoxifen treatment.
G–I. Histograms depicting results of cell cycle analysis of MCF-7 and MCF-7/Twist cells following Fulvestrant treatment.
Cell cycle stage values were calculated by the Dean-Jett-Fox model (Fox 1980). Experiments were independently repeated four times.
Twist promotes hormone resistance in breast cancer cells
To investigate if the loss of ER brought about by Twist caused hormone resistance in breast cells, we treated MCF-7 and MCF-7/Twist cells with the selective estrogen receptor modulator (SERM) tamoxifen and the selective estrogen receptor down-regulator (SERD) fulvestrant. As seen in Figure 2D, MCF-7 cells were significantly arrested in presence of tamoxifen (S=2.3%) compared to untreated cells (S=15.6%, P<0.005). On the other hand, MCF-7/Twist cells were largely unaffected by tamoxifen treatment (S=16.6% vs. 19.5%, P>0.05) (Figure 2E). Also, G1 and S phases of the cell cycle were significantly altered in MCF-7 and MCF-7/Twist cells treated with tamoxifen (G1=86.6% vs. 66.5%, P<0.005; S=2.3% vs. 16.6%, P<0.005; G2=8.9% vs. 14.2%, P>0.05) (Figure 2F).
Treatment with fulvestrant exhibited comparable results to those of tamoxifen. MCF-7 cell growth was significantly affected by treatment (S=3.9%) compared to untreated controls (S=15.6%, P<0.005) (Figure 2G) while MCF-7/Twist cells were unaffected by the treatment (S=16.6% vs. 19.5%, P>0.05) (Figure 2H). Similarly, MCF-7 cells were significantly affected by the fulvestrant treatment compared to MCF-7/Twist cells (G1=86% vs. 67.7%, P<0.005; S=3.9% vs. 16.6%, P<0.0005; G2=5.8% vs. 13.3%, P<0.05) (Figure 2I). There were no significant differences in cell cycle phases of untreated MCF-7 and MCF-7/Twist (data not shown).
Hormone resistance is reversed by down-regulation of Twist in breast cancer cells
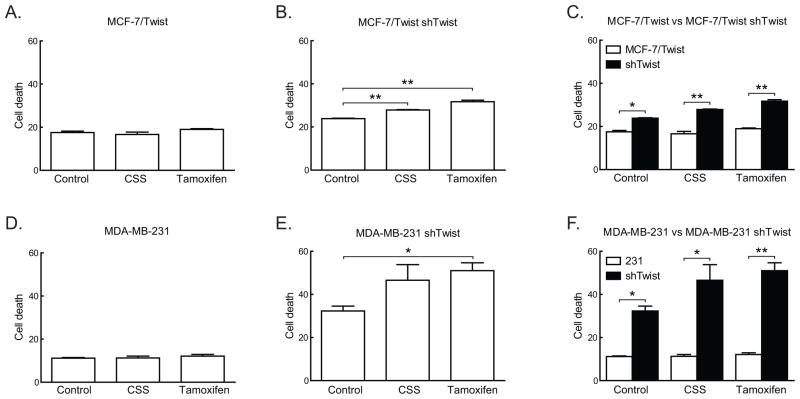
In order to study if the loss of Twist caused a reversion to the estrogen dependent hormone sensitive phenotype, we down-regulated Twist expression in MCF-7/Twist and MDA-MB-231 cells using a lentiviral delivered shRNA based approach. As seen in Figure 3A-C, MCF-7/Twist shTwist cells showed significantly higher cell death following growth in CSS or on exposure to tamoxifen compared to parental MCF-7/Twist cells, which remain unaffected. Similar effects were also seen in MDA-MD-231 shTwist cells (Figure 3D–F). This would indicate that ER independence and hormone resistance exhibited by MCF-7/Twist and MDA-MB-231 cells is at least partially due to Twist expression and can be reversed by down-regulation of Twist.
Figure 3. Effect of Twist down-regulation on cell cycle.
A–C. Histograms depicting increased cell death in Twist down-regulated MCF-7/Twist cells in the absence of estrogen (charcoal stripped serum) and presence of tamoxifen.
D–F. Histograms displaying increasing cell death in MDA-MB-231 shTwist cells in the absence of estrogen (charcoal stripped serum) and in the presence of tamoxifen.
Twist promotes growth of breast tumors in the absence of estrogen
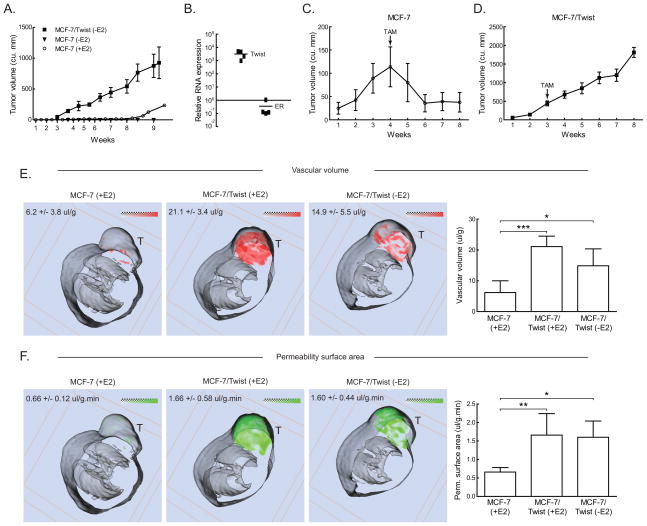
In order to strengthen our finding that over-expression of Twist induced estrogen independence in vivo, we orthotopically injected MCF-7/Twist cells into the mammary fat pad of SCID mice, which were not supplemented with estrogen. As seen in Figure 4A, MCF-7/Twist xenografts produced large tumors (greater >250 mm3) within four to five weeks of incubation. These results confirmed that MCF-7/Twist cells are estrogen independent in vivo. In order to confirm that the expression of Twist and ER in tumors was similar to that of MCF-7/Twist cells, we isolated RNA from four tumors and performed qRT-PCR using Twist and ER primers. As seen in Figure 4B, expression of Twist was inversely correlated with levels of ER transcripts.
Figure 4. In vivo growth characteristics of MCF-7 and MCF-7/Twist cells.
A. Line chart showing growth of 1 × 106 MCF-7 cells (without E2, solid triangles; with E2, hollow circles) and MCF-7/Twist cells (without E2, solid squares) orthotopically implanted into female SCID mice and allowed to grow for the time indicated. Tumors were measured weekly.
B. MCF-7/Twist tumors from mice (n=4) were excised and Twist and ER transcript levels determined by qRT-PCR. The graph depicts relative differences in Twist and ER transcript levels.
C, D. Growth of MCF-7 and MCF-7/Twist tumors over eight weeks treated with tamoxifen. tamoxifen pellet implantation is indicated by an arrow.
E, F. Representative false color coded MRI generated 3-D transverse slices of MCF-7 and MCF- 7/Twist xenografts in the mammary fat pad. Red and green represent the distributions of vascular volume (VV) and vascular permeability surface area product (PS), respectively. Gray-scale images represent the mouse body; while tumors are seen on top and indicated by “T”. Averaged values from all mice are indicated in the figures as well as displayed on the histograms at the right. Images depicted are a representative sample of five mice for MCF-7(+E2), six mice for MCF-7/Twist(−E2), and six mice for MCF-7/Twist (+E2).
Next, we injected mice (n=10) with MCF-7/Twist and MCF-7 cells in the presence of estrogen (17β-estradiol pellet implanted in the back). After 3–4 weeks of growth, all mice were implanted with a tamoxifen pellet. As seen in Figure 4C–D, MCF-7 tumors regressed to pre-treatment levels, while MCF-7/Twist tumors were unaffected by tamoxifen.
Twist increases vascular volume and vascular permeability of breast tumors in mice
Functional magnetic resonance imaging (fMRI) was used to non-invasively analyze the vascular volume (VV) and permeability-surface (PS) area product values in vivo. Figure 4E–F display representative false color-coded MRI generated 3-D transverse slices of xenograft tumors using MCF-7 and MCF-7/Twist cells in mice. The average tumor VV in MCF-7 (+estrogen)(E2) and MCF-7/Twist (−E2) xenografts was 6.2 and 14.9 μl/g respectively (Figure 4E). The average tumor PS in MCF-7 (+E2) and MCF-7/Twist (−E2) xenografts was 0.66 and 1.60 μl/g·min respectively (Figure 4F). Both results were significant according to the Scheffe test (F=15.9 and 7.04 respectively). VV and PS values in MCF-7 vector control xenografts were comparable to those in MCF-7 xenografts (data not shown), and were consistent with the previous report (36). VV and PS in MCF-7/Twist (+E2) xenografts were 21.1 ul/g and 1.66 ul/g.min respectively. These values were significantly higher than those in MCF-7 (+E2) controls (F=5.48 and 6.23 respectively). There was no significant difference between estrogen supplemented and non-supplemented MCF-7/Twist xenografts for VV and PS (F=3.00 and 0.03 respectively).
Twist down-regulation causes re-expression of ER
To investigate the mechanism of ER regulation by Twist, we transiently down-regulated Twist in MCF-7/Twist cells and up-regulated Twist in MCF-7 and T-47D. As seen in Figure 5A-B, down-regulation of Twist in MCF-7/Twist cells caused a significant drop in mRNA levels of Twist accompanied by an increase in ER expression. Transient expression of Twist, on the other hand, caused a significant drop in ER protein in MCF-7 and T-47D cells (Figure 5C). We also demonstrated that Her-2/neu protein levels were low in MCF-7/Twist cells, which indicates that the effect of Twist on ER is not mediated by Her-2/neu (Supplementary Figure 1). Furthermore, we determined that the reactivation of ER in the Twist down-regulated clones was functionally active. For this purpose, the ERE-luc construct was used as a functional reporter system (kind gift of Nancy Davidson) for the in vitro studies. As seen in Figure 5D, MCF-7/Twist cells show a significant drop in the activation of the reporter indicating the lack of ER functionality in these cells. Importantly, the re-expression of ER by down-regulating Twist in MCF-7/Twist cells increased reporter activity, an indication of functional ER proteins.
Figure 5. Functional effects of Twist on ER expression, promoter methylation, and histone deacetylation.
A,B. Histograms depicting changes in Twist and ER expression after Twist and shTwist mediated up- and down-regulation in MCF-7 and MCF-7/Twist cells respectively. Transcript levels were estimated by qRT-PCR and are derived from three independent experiments in duplicates. Error bars depict S.D.
C. A panel displaying immunoblots of Twist up- and down-regulated cell lines scored for Twist and ER.
D. Histogram depicting changes in relative binding of ER to ERE luciferase plasmid in MCF-7, MCF-7/Twist, and shTwist mediated Twist down-regulated MCF-7/Twist cells. Experiments were repeated thrice in duplicates. Error bars depict S.D.
E,F. Histogram of qPCR results displaying the effect of (E) Twist down-regulation on ER downstream target genes in MCF-7/Twist cells; (F) Twist up-regulation on downstream ER target genes in MCF-7 cells.
G. Immunoblots of ER downstream targets that are dysregulated in MCF-7/Twist cells compared to parental MCF-7 cells. Actin was used as a loading control.
H. Basal ER promoter methylation levels of MCF-7 (low Twist, high ER), MCF-7/Twist and MDA-MB-231 (high Twist, low ER). Experiments were repeated twice in duplicates. Error bars depict S.D.
I. Histogram displaying changes in ER promoter methylation in cell lines after Twist up- and down-regulation. Experiments were repeated twice in duplicates. Error bars depict S.D.
J. Histogram showing increase in ER expression in MCF-7/Twist cells treated with 1μM demethylating agent 5-azacytidine (AZA) and 10 μM HDAC inhibitor valproic acid (VPA) and assayed by qRT-PCR. Experiments were repeated twice in duplicates. Error bars depict S.D.
K. Immunoblots of ER re-expression in MCF-7/Twist cells after treatment by AZA, VPA, and in combination.
L. Immunoblots of co-immunoprecipitation of MCF-7/Twist lysates. Twist antibodies were used for the co-immunoprecipitation and immunoblots were probed with HDAC1 and DNMT3B.
M. Histogram of chromatin immunoprecipitation results of MDA-MB-231 cells using HDAC1 and DNMT3B antibodies. E-boxes covered in the ER promoter are indicated on the horizontal axis. Results are displayed compared to negative control IgG and are calculated as a percentage of total input DNA. Bars depict S.D.
To further characterize the functionality of Twist-mediated ER loss in breast cancer cells, we performed quantitative real-time PCR and immunoblotting for downstream targets of ER such as Cyclin D1, p21, p27 (Foster and Wimalasena 1996), p14 (Cho et al 2006), and Cathepsin D (Morisset et al 1986) in MCF-7, MCF-7/Twist, and MCF-7/Twist shTwist cells. As seen in Figure 5E-F, p21 and Cathepsin D were up-regulated when Twist was down-regulated in MCF-7/Twist cells and were down-regulated when Twist was up-regulated in MCF-7 cells. This would indicate that regulation of ER by Twist is functionally relevant. Furthermore, we also probed for intrinsic levels of ER target genes in MCF-7 and MCF-7/Twist cells. As seen in Figure 5G, all the ER targets genes were down-regulated in MCF-7/Twist cells. This suggests that over-expression of Twist, in this system, can repress ER levels, which in turns affects the expression of its downstream target genes.
Twist induces hyper-methylation of the ER promoter
A common mechanism of ER gene silencing is that of promoter hypermethylation and occurs in 5–49% of patient samples (Ottaviano et al 1994, Yan et al 2001). Indeed, we did find a significant increase in ER promoter methylation in MCF-7/Twist cells as observed by MS-qPCR analysis (Figure 5H). Subsequently, we used cell lines with transiently up- and down-regulated Twist to validate our earlier observations. As seen in Figure 5I, Twist over-expression in T-47D cells caused an increase in ER promoter methylation. On the other hand, Twist down-regulation in MCF-7/Twist and MDA-MB-231 caused a significant decrease in ER promoter methylation. In order to reverse the Twist induced methylation of the ER promoter, we treated MCF-7/Twist cells with the demethylating agent 5-azacytidine. We found a significant increase in ER transcript and protein levels as seen by qRT-PCR and immunoblotting (Figure 5J–K). To decipher the mechanistic cause of the increased methylation of ER brought about by Twist, we analyzed for recruitment of DNA methyltransferases. The de novo methyltransferase DNMT3B was co-immunoprecipitated by Twist from MCF-7/Twist lysates (Figure 5L). Other methyltransferases such as DNMT1 and DNMT3A were not co-immunoprecipitated by Twist (data not shown).
Twist causes histone deacetylation of the ER promoter
Regulation of genes via methylation is accompanied, in some cases, by an increase in histone deacetylase (HDAC) activity (Cameron et al 1999). Mechanistically, we determined that Twist recruited the histone deacetylase HDAC1 (Figure 5L) to the ER promoter leading to deacetylation, which resulted in lowered expression of ER. In order to functionally study the role of HDACs in the regulation of ER, we treated MCF-7/Twist cells with the HDAC inhibitor valproic acid (VPA). As seen in Figure 5K, we found a significant increase in ER expression in these cells when treated with the inhibitor. Use of AZA and VPA in combination was able to rescue ER to a higher degree as compared to either of them alone.
Twist recruits HDAC1 and DNMT3B to the ER promoter
In order to confirm our co-immunoprecipitation results, we carried out ChIP experiments on MDA-MB-231 cells using HDAC1 and DNMT3B antibodies. As seen in Figure 5M, DNMT3B was recruited to all the E-box sites of the ER promoter, while HDAC1 was significantly recruited to E-box 6–9. Taken together, the largest enrichment of DNMT3B and HDAC1 binding was in the E-box 6–9 regions. This would point to an important role of E-box 6–9 region in the regulation of ER by Twist via the interaction with HDAC1 and DNMT3B.
Twist and ER are inversely correlated in breast cancer patients
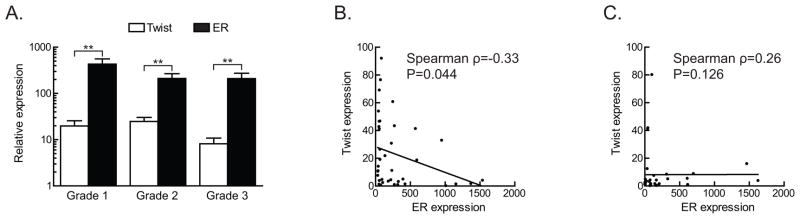
To confirm the inverse correlation between Twist and ER expression seen in breast cancer cell lines, Twist and ER mRNA levels in human breast tumors were quantified by qRT-PCR. A total of 73 primary breast cancers (grade 1, n=16; grade 2, n=22; grade 3, n=35) and four normal breast samples were analyzed (Figure 6). We examined correlation in normalized (to normal breast tissue) expression between TWIST and ER genes, overall, and within each grade, using the nonparametric, Spearman rank test. Twist expression levels were significantly different from ER levels (Figure 6A). We also observed an increase in Twist expression levels with increasing tumor grade while ER expression levels showed a decrease with increasing tumor grade for grades 1 and 2. An inverse relation was observed with increased Twist expression associated with decreased ER expression, which was statistically significant in grades 1 and 2 (Figure 6B) but not in grade 3 (Figure 6C). Overall, these results suggest that Twist expression inversely correlates with ER expression in human breast cancers of grades 1 and 2.
Figure 6. Correlation between Twist and ER mRNA levels in human breast tumors.
A. Histogram depicting Twist and ER transcript levels in various grades of breast cancer from 73 patient samples (Grade 1, n=16; grade 2, n=22; grade 3, n=35). Error bars display S.D.
B. Scatter plot of results of qPCR from samples of grades 1 and 2 breast cancer.
C. Scatter plot of qPCR results from grade 3 samples.
Discussion
The regulation of ER has been a topic of interest for numerous investigators given its importance in the development of breast tumor formation. The classification of breast tumors as either ER positive or negative has a significant impact on the selection of appropriate chemotherapeutic regimens and a loss of ER is correlated with poor prognosis, in part due to the exclusion of targeted anti-hormonal therapy. It is well established that a combination of genetic, epigenetic, and transcriptional controls regulate ER expression. However, the ontogeny of tumor progression leading to the formation of the ER negative and/or estrogen resistant state is still not clearly understood.
In the present study, we have demonstrated that the transcriptional regulation of ER can be modulated by Twist, a protein that has been shown to be up-regulated in high-grade breast cancer phenotypes (Mironchik et al 2005). This finding was corroborated using ER negative breast cancer cell line MDA-MB-231, which has intrinsically high levels of Twist expression and is very aggressive in mice. On the other hand, the ER positive cell line MCF-7 has very low levels of Twist and is less tumorigenic in SCID mice. These results indicated that the up-regulation of Twist in combination with the down-regulation of ER could be a potential mechanism for the acquisition of an aggressive breast cancer phenotype. However, alternative mechanisms probably exist to silence ER expression in the absence of Twist in breast cell lines like MCF-10A and MDA-MB-468 (Pilat et al 1998). The importance of this finding is that loss of ER in breast cancer patients has a poor prognosis (Herynk and Fuqua 2007, Schiff et al 2005) and this loss of ER expression brought about by Twist may have implications for ER negative and/or ER resistant breast cancers.
Twist exerts its negative regulatory activity at the transcriptional level by binding to E-boxes within promoter sequences. Overexpression of Twist showed a consistent three to four folds repression in promoter assays indicating that Twist is a transcriptional repressor of ER expression. In vivo, each of the 26 putative Twist binding sites may work independently or in some concerted fashion to down-regulate the expression of ER. We confirmed the specificity of Twist binding to the ER promoter by employing Twist bHLH deletion mutants in promoter assays. This indicated that Twist binding to the ER promoter is abrogated once the functional helix-loop-helix domain is excised from Twist. Chromatin immunoprecipitation using the Twist positive cell lines MCF-7/Twist and MDA-MB-231 confirmed that Twist binds in vivo to the ER promoter. The binding of Twist to the ER promoter in MDA-MB-231 cells provided further evidence that the down-regulation of ER by Twist is a possible functional mechanism for the generation of ER negativity in Twist positive breast cells.
A functional hallmark of ER negative cells is the ability to proliferate without estrogen stimulation and the propensity for hormone resistance (Herynk and Fuqua 2007, Schiff et al 2005). Significantly, Twist cells were able to grow without estrogen both in vitro, and in vivo. They were also resistant to the SERM tamoxifen and SERD fulvestrant in vitro without significant G1 arrest compared to the Twist negative parental MCF-7 cells. Moreover, Twist tumors were unaffected by tamoxifen treatment. These experiments strongly indicated that Twist expression caused hormone independence in breast cancer cells possibly leading to hormone resistance.
Vascular volume and permeability surface area product of tumors has been utilized as a surrogate marker for angiogenesis in tumors (Bikfalvi and Bicknell 2002, Mironchik et al 2005). We observed that the intratumoral distribution pattern of VV and PS in MCF-7/Twist xenografts (without estrogen) was quite different from those in MCF-7/Twist (with estrogen). In the absence of estrogen, VV and PS were high in the periphery of the tumors while in the presence of estrogen they were increased both at the periphery and in the central region of the tumor. This could indicate that Twist drives vasculogenesis independent of estrogen signaling. This would indicate that tumors expressing Twist are more likely to be aggressive, invasive, and ultimately more metastatic due to well-developed vasculature. This meshes in with earlier work showing that Twist is up-regulated in breast cancers and is a down-regulator of E-cadherin (Mironchik et al 2005, Vesuna et al 2008). On the other hand, the higher VV and PS in the estrogen supplemented tumors could possibly be ascribed to increased VEGF expression directly driven by estrogen (Sengupta et al 2003).
Mechanistically, we demonstrated that Twist interacts with DNMT3B and HDAC1 at the ER promoter. The recruitment of DNMT3B leads to de novo hypermethylation while HDAC1 would cause histone deacetylation, eventually leading to transcriptionally silenced chromatin (Figure 7). We were able to reverse this state using the methylation inhibitor 5-azacytidine and HDAC inhibitor valproic acid. Thus, we have presented the first evidence indicating that Twist can regulate ER expression by inducing promoter methylation and chromatin remodeling, in addition to transcriptional repression.
Figure 7. A model for the regulation of ER by Twist.
A proposed model depicting the normal, active state of transcription with unmethylated promoter DNA and acetylated chromatin leading to an ER+, hormone sensitive state. On binding to its target E-boxes, Twist recruits HDAC1 that deacetylates histone proteins leading to compaction of chromatin. Twist (with HDAC1) also interacts with DNMT3B at the E-boxes that causes de-novo methylation of the promoter area. Together with other, as yet unknown co-factors, Twist binding eventually leads to a repressive state of transcription ultimately culminating in the shutting down of ER transcription.
Next, we directly targeted Twist by a lentivirus mediated down-regulation strategy. We saw a larger change in expression of ER than Twist. However, the expression of ER was only partially functional (~10%) as compared to MCF-7 as seen by ERE binding assays. This could be attributed to factors such as promoter methylation and chromatin deacetylation, which we have determined earlier. The reactivation of ER by down-regulating Twist expression may be of some relevance in patients as even 1–10% of breast tumor cells producing ER are clinically responsive to tamoxifen treatment (Harvey et al 1999). These data indicated that re-expression of ER is possible by the use of methylation inhibitors in Twist over-expressing cells. The fact that an oncogene Twist is contributing to these epigenetic changes is a novel discovery and would indicate that use of epigenetic therapy might be beneficial for patients with either estrogen refractory disease or ER negative breast cancer.
Twist expression is increased in high-grade human breast tumors and a high percentage of these are ER negative (Mironchik et al 2005). We have observed similar results in breast cancer patient microarrays that are available from other studies that show a significant negative correlation between Twist and ER expression. The over-expression of Twist may provide a mechanistic link between development of aggressive breast cancer and loss of ER expression and may provide a means to elucidate the ontogeny of ER negative and/or estrogen resistant breast cancers (Figure 7). In summary, our results demonstrate an alternative mechanistic explanation for the loss of ER expression in breast tumors, which supplements promoter methylation and de-acetylation. The loss of ER expression brought about by Twist has important implications with respect to breast cancer treatment in that Twist over-expressing cells can proliferate in the absence of estrogen as well as in the presence of the anti-estrogen tamoxifen and fulvestrant. It may be possible to reverse the down-regulation of ER by treating ER negative tumors with a combination of Twist shRNA, methylation inhibitors, and HDAC inhibitors leading to a rescue of ER expression and reversing the ER negative phenotype. This could contribute towards clearing the roadblock of hormone independence and resistance to anti-hormonal treatment that currently exist in the treatment of ER negative breast tumors.
Materials & Methods
Cell culture
MCF-7, MDA-MB-231, and T-47D cell lines were obtained from American Type Tissue Collection (Manassas, VA) and maintained as instructed. Production of the stable Twist overexpressing cell line MCF-7/Twist has been described earlier (Mironchik et al 2005, Vesuna et al 2009). For experiments involving hormone independence, and anti-estrogen resistance, MCF-7 and MCF-7/Twist cells were plated at 500,000 cells per 100 mm dish in complete media. The following day, media was replaced by phenol-red free (PRF) minimal essential media (MEM) containing 5% charcoal stripped serum (CSS) and the cells were allowed to grow for two days. On the third day, media was replaced with complete MEM (control), and PRF-MEM/5% CSS (hormone independence). For anti-estrogen treatment, PRF-MEM/5% CSS media was supplemented with 1 μM 4-hydroxytamoxifen (tamoxifen) or 1 nM fulvestrant. Three days following drug treatment, the cells were harvested and fixed in 70% ethanol overnight. Cell cycle analysis was performed by laser scanning cytometry on a FacScan I (BD Biosciences, San Jose, CA). Independent experiments were repeated four times. Data was analyzed using FlowJo (Tree Star Inc., Ashland, OR) and ModFit LT 2.0 software (Verity House Software, Topsham, ME).
Promoter analysis
Cloning of the ER promoter and the Twist deletion constructs has been described elsewhere (deGraffenried et al 2002) (El Ghouzzi et al 2000). The ER promoter constructs were transiently transfected (TransIT-LT1, Mirus Bio Corporation, Madison, WI) along with the Twist expression constructs in the ER positive breast cancer cell line MCF-7. An enhanced green fluorescent protein expression plasmid pEGFP-N1 (Clontech, Mountain View, CA) was used to determine transfection efficiency (Vesuna et al 2005).
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described (Cell Signaling), with primers covering the entire region of the ER promoter. Primer sequences are in supplementary data.
Methylation assays
MCF-7, MCF-7/Twist and MDA-MB-231 cells were grown to 70% confluence and DNA extracted according to the manufacturers protocol (Qiagen, Valencia, CA). Subsequently, the DNA was processed for bisulfite treatment (Zymo Research, Orange, CA). Bisulfite treated DNA was PCR amplified using primers that amplify a 110 bp region around the Not I site of the CpG island in the first ER exon (O’Doherty et al 2002). Primers sequences are in supplementary data files.
Methylation inhibitor and HDAC inhibitor treatment
For demethylation treatment, cells were grown to 40–50% confluence in 6-well plates and treated with 1 μM 5-azacytidine (AZA) for three days. At the end of this period, DNA was isolated and processed for demethylation studies by MS-qPCR. Total proteins were extracted and immunoblotted to analyze the effect of demethylation on ER expression. Cells were treated with histone deacetylase (HDAC) inhibitor valproic acid (VPA) at 10 μM concentration for three days and processed similarly.
Animal studies
Mice were anesthetized with acepromazine (62.5 mg/kg) and xylazine (6.5 mg/kg) or ketamine (100 mg/kg) and xylazine (10 mg/kg) in saline administered i.p. for xenograft implantation procedures. Mice were orthotopically injected in the breast with 2×106 MCF-7/Twist or control MCF-7 cells in 100 μl sterile complete media in the second mammary fat pad. A total of 15 female mice were injected with MCF-7/Twist cells and five female mice were injected with control MCF-7 cells. Estradiol pellets were 90 day slow release (0.18 mg/pellet, Innovative Research of America, Sarasota, FL) and tamoxifen pellets were 60 day slow release (5 mg/pellet, IRA). All animal experiments were done under Institutional Animal Care and Use Committee (IACUC) guidelines established at the Johns Hopkins University School of Medicine.
Measurement of Vascular Volume (VV) and Permeability-surface Area Products (PS)
The measurement of VV and PS has been described in detail elsewhere (Bhujwalla et al 2001, Mironchik et al 2005). Three-dimensional images were drawn for all the 17 mice (MCF-7 (+E2) n=5, MCF-7/Twist (+E2) n=6, MCF-7/Twist (−E2) n=6), and the images presented are representative of each group. The parameters were 8 slices, 1 mm slice thickness, FOV = 32 mm, 8 scans, 0.25 mm in plane spatial resolution.
Twist and ER mRNA expression levels in human breast cancers
Frozen breast cancer samples controlled for adequate tumor content (over 80%) by laser capture dissection were obtained from the University Medical Center, Utrecht, The Netherlands. Total RNA was isolated using Trizol, reverse transcribed using SuperScript III (Invitrogen, Carlsbad, CA), and quantitative real-time PCR amplified. Expression values were normalized with the 36B4 gene.
Statistical analysis
Data was analyzed by independent, two sided Student’s T-test. Statistics with respect to vascular volume (VV) and permeability surface area (PS) were performed by Scheffe’s test. We examined correlation in normalized (to normal breast tissue) expression between TWIST and ER genes, overall, and within each grade, using the non-parametric, Spearman rank test. For all analysis, P values below 0.05 were considered significant. In all figures, (*) denotes P<0.05, (**) denotes P<0.005, and (***) denotes P<0.0005.
Supplementary Material
Immunoblot displaying Her-2/neu expression levels in MCF-7 and MCF-7/Twist cells.
Acknowledgments
We thank the UMC Utrecht Biobank and Mrs. M. van Blokland for their help in obtaining the human breast cancer RNA. We also thank Flonne Wildes and Gary Cromwell for helping with mice, Yelena Mironchik for helping with cell culture, and Paul T. Winnard Jr. and Balaji Krishnamachary for critically reading the manuscript. We also thank Kayleen Bailey for help with methylation experiments. ER promoter and ERE-luciferase constructs were kindly provided by Professor Nancy Davidson (Johns Hopkins University, MD). Twist mutant constructs were kindly provided by Professor Jacky Bonaventure (INSERM, France). This work was supported by the National Institutes of Health [1RO1CA140226 to V.R.].
Footnotes
Conflicts of Interest
None
References
- Angeloni SV, Martin MB, Garcia-Morales P, Castro-Galache MD, Ferragut JA, Saceda M. Regulation of estrogen receptor-alpha expression by the tumor suppressor gene p53 in MCF-7 cells. J Endocrinol. 2004;180:497–504. doi: 10.1677/joe.0.1800497. [DOI] [PubMed] [Google Scholar]
- Bhujwalla ZM, Artemov D, Natarajan K, Ackerstaff E, Solaiyappan M. Vascular differences detected by MRI for metastatic versus nonmetastatic breast and prostate cancer xenografts. Neoplasia. 2001;3:143–153. doi: 10.1038/sj.neo.7900129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Bikfalvi A, Bicknell R. Recent advances in angiogenesis, anti-angiogenesis and vascular targeting. Trends Pharmacol Sci. 2002;23:576–582. doi: 10.1016/s0165-6147(02)02109-0. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–9824. doi: 10.1074/jbc.M010840200. [DOI] [PubMed] [Google Scholar]
- Cho EY, Choi YL, Chae SW, Sohn JH, Ahn GH. Relationship between p53-associated proteins and estrogen receptor status in ovarian serous neoplasms. Int J Gynecol Cancer. 2006;16:1000–1006. doi: 10.1111/j.1525-1438.2006.00553.x. [DOI] [PubMed] [Google Scholar]
- deConinck EC, McPherson LA, Weigel RJ. Transcriptional regulation of estrogen receptor in breast carcinomas. Mol Cell Biol. 1995;15:2191–2196. doi: 10.1128/mcb.15.4.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGraffenried LA, Hilsenbeck SG, Fuqua SA. Sp1 is essential for estrogen receptor alpha gene transcription. J Steroid Biochem Mol Biol. 2002;82:7–18. doi: 10.1016/s0960-0760(02)00151-6. [DOI] [PubMed] [Google Scholar]
- El Ghouzzi V, Legeai-Mallet L, Aresta S, Benoist C, Munnich A, de Gunzburg J, et al. Saethre-Chotzen mutations cause TWIST protein degradation or impaired nuclear location. Hum Mol Genet. 2000;9:813–819. doi: 10.1093/hmg/9.5.813. [DOI] [PubMed] [Google Scholar]
- Foster JS, Wimalasena J. Estrogen regulates activity of cyclin-dependent kinases and retinoblastoma protein phosphorylation in breast cancer cells. Mol Endocrinol. 1996;10:488–498. doi: 10.1210/mend.10.5.8732680. [DOI] [PubMed] [Google Scholar]
- Fox MH. A model for the computer analysis of synchronous DNA distributions obtained by flow cytometry. Cytometry. 1980;1:71–77. doi: 10.1002/cyto.990010114. [DOI] [PubMed] [Google Scholar]
- Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, et al. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60:4026–4029. [PubMed] [Google Scholar]
- Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Fuchtbauer A, Fuchtbauer EM. Repression of muscle-specific gene activation by the murine Twist protein. Exp Cell Res. 1997;232:295–303. doi: 10.1006/excr.1997.3541. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA. Estrogen receptors in resistance to hormone therapy. Adv Exp Med Biol. 2007;608:130–143. doi: 10.1007/978-0-387-74039-3_10. [DOI] [PubMed] [Google Scholar]
- Mironchik Y, Winnard PT, Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, et al. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801–10809. doi: 10.1158/0008-5472.CAN-05-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisset M, Capony F, Rochefort H. Processing and estrogen regulation of the 52-kilodalton protein inside MCF7 breast cancer cells. Endocrinology. 1986;119:2773–2782. doi: 10.1210/endo-119-6-2773. [DOI] [PubMed] [Google Scholar]
- Murphy LC, Dotzlaw H, Leygue E, Douglas D, Coutts A, Watson PH. Estrogen receptor variants and mutations. J Steroid Biochem Mol Biol. 1997;62:363–372. doi: 10.1016/s0960-0760(97)00084-8. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- O’Doherty AM, Church SW, Russell SE, Nelson J, Hickey I. Methylation status of oestrogen receptor-alpha gene promoter sequences in human ovarian epithelial cell lines. Br J Cancer. 2002;86:282–284. doi: 10.1038/sj.bjc.6600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El-Ashry D. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol. 2001;15:1344–1359. doi: 10.1210/mend.15.8.0678. [DOI] [PubMed] [Google Scholar]
- Osborne C, Tripathy D. Aromatase inhibitors: rationale and use in breast cancer. Annu Rev Med. 2005;56:103–116. doi: 10.1146/annurev.med.56.062804.103324. [DOI] [PubMed] [Google Scholar]
- Osborne CK. Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat. 1998;51:227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Zhao H, Fuqua SA. Selective estrogen receptor modulators: structure, function, and clinical use. J Clin Oncol. 2000;18:3172–3186. doi: 10.1200/JCO.2000.18.17.3172. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Wakeling A, Nicholson RI. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action. Br J Cancer. 2004;90(Suppl 1):S2–6. doi: 10.1038/sj.bjc.6601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK, Schiff R. Aromatase inhibitors: future directions. J Steroid Biochem Mol Biol. 2005;95:183–187. doi: 10.1016/j.jsbmb.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- Pilat MJ, Schwab ED, Yao KL, Pienta KJ. Examination of the DNA methylation properties in nontumorigenic and tumorigenic breast epithelial cell lines. Anticancer Res. 1998;18:2575–2582. [PubMed] [Google Scholar]
- Roodi N, Bailey LR, Kao WY, Verrier CS, Yee CJ, Dupont WD, et al. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. J Natl Cancer Inst. 1995;87:446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- Rose CS, Malcolm S. A TWIST in development. Trends Genet. 1997;13:384–387. doi: 10.1016/s0168-9525(97)01296-1. [DOI] [PubMed] [Google Scholar]
- Schiff R, Massarweh SA, Shou J, Bharwani L, Arpino G, Rimawi M, et al. Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol. 2005;56(Suppl 1):10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- Sengupta K, Banerjee S, Saxena N, Banerjee SK. Estradiol-induced vascular endothelial growth factor-A expression in breast tumor cells is biphasic and regulated by estrogen receptor-alpha dependent pathway. Int J Oncol. 2003;22:609–614. [PubMed] [Google Scholar]
- Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P, Jr, Raman V. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J Biol Chem. 2005;280:2294–2299. doi: 10.1074/jbc.M411018200. [DOI] [PubMed] [Google Scholar]
- Vesuna F, Winnard P, Jr, Raman V. Enhanced green fluorescent protein as an alternative control reporter to Renilla luciferase. Anal Biochem. 2005;342:345–347. doi: 10.1016/j.ab.2005.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna F, Winnard P, Jr, Glackin C, Raman V. Twist overexpression promotes chromosomal instability in the breast cancer cell line MCF-7. Cancer Genet Cytogenet. 2006;167:189–191. doi: 10.1016/j.cancergencyto.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna F, van Diest P, Chen JH, Raman V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun. 2008;367:235–241. doi: 10.1016/j.bbrc.2007.11.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesuna F, Lisok A, Kimble B, Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–1328. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang X, Davidson NE. Role of DNA methylation and histone acetylation in steroid receptor expression in breast cancer. J Mammary Gland Biol Neoplasia. 2001;6:183–192. doi: 10.1023/a:1011308707512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblot displaying Her-2/neu expression levels in MCF-7 and MCF-7/Twist cells.