Abstract
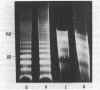
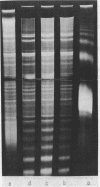
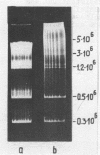
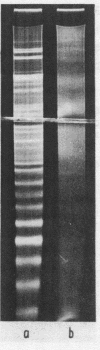
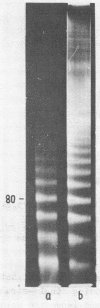
Chromatin DNA of rat thymus nuclei was cleaved by Serratia marcescens endonuclease. The fragments have been examined by polyacrylamide gel electrophoresis under denaturing conditions. The results obtained are interpreted to mean that the internucleosomal DNA is cleaved by the endonuclease into fragments which are multiples of 10 nucleotides. The 10 nucleotide periodicity in fragmentation of internucleosomal DNA is independent of the presence of histone H1 and is likely to be determined by the interaction of this DNA stretch with the histone core of nucleosomes. Such interaction implies a close association between the nucleosomes in the chromatin thread. Quasi-limit chromatin digest (50-55% of DNA hydrolysis) contains undegraded DNA fragments with length of up to 1000 nucleotides or more. A part of this resistant DNA consists of single-stranded fragments or contains single-stranded regions. These data may be accounted for by a very compact nucleosome packing in the resistant chromatin in which one of the DNA strands is more accessible to the endonuclease action.
Full text
PDF



















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballal N. R., Goldberg D. A., Busch H. Dissociation and reconstitution of chromatin without appreciable degradation of the proteins. Biochem Biophys Res Commun. 1975 Feb 17;62(4):972–982. doi: 10.1016/0006-291x(75)90418-0. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Mobbs J. D., Marshall A. J. Chromatin structure: a property of the higher structures of chromatin and in the time course of its formation during chromatin replication. Nucleic Acids Res. 1976 Dec;3(12):3293–3304. doi: 10.1093/nar/3.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley R. Histone propinquity using imidoesters. Biochem Biophys Res Commun. 1975 May 19;64(2):587–594. doi: 10.1016/0006-291x(75)90362-9. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Tatchell K., Van Holde K. E. On the occurrence of nucleosome phasing in chromatin. Cell. 1977 Nov;12(3):829–836. doi: 10.1016/0092-8674(77)90281-1. [DOI] [PubMed] [Google Scholar]
- Mandel R., Fasman G. D. Chromatin and nucleosome structure. Nucleic Acids Res. 1976 Aug;3(8):1839–1855. doi: 10.1093/nar/3.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle M., Roberts W. K. An extracellular nuclease from Serratia marcescens. I. Purification and some properties of the enzyme. J Biol Chem. 1969 Oct 10;244(19):5213–5218. [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Oliver D., Chalkley R. Asymmetric distribution of histone on DNA: a model for nucleohistone primary structure. Biochemistry. 1974 Dec 3;13(25):5093–5098. doi: 10.1021/bi00722a006. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Sokolenko A. A., Dianov G. L. Issledovanie DNK, sviazannoi s gistonami v khromatine. Mol Biol (Mosk) 1975 Sep-Oct;9(5):691–698. [PubMed] [Google Scholar]
- Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Elektroforeticheskoe izuchenie geterogennosti sub'edinits khromatina. Mol Biol (Mosk) 1977 Jul-Aug;11(4):781–789. [PubMed] [Google Scholar]
- Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Heterogeneity of chromatin subunits. FEBS Lett. 1977 Mar 1;74(2):229–233. doi: 10.1016/0014-5793(77)80852-1. [DOI] [PubMed] [Google Scholar]
- Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. Structure of chromatin subunits: an endonuclease Serratia marcescens study. Nucleic Acids Res. 1977 Sep;4(9):3267–3279. doi: 10.1093/nar/4.9.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- van Bruggen E. F., Arnberg A. C., van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. Electron microscopy of chromatin subunit particles. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1365–1370. doi: 10.1016/0006-291x(74)90348-9. [DOI] [PubMed] [Google Scholar]