Abstract
Mutations of the Matrix metalloproteinase-20 (MMP20, enamelysin) gene cause autosomal recessive amelogenesis imperfecta and Mmp20 ablated mice also have malformed dental enamel. Here we show that Mmp20 null mouse secretory stage ameloblasts maintained a columnar shape and were present as a single layer of cells. However, the null maturation stage ameloblasts covered extraneous nodules of ectopic calcified material formed at the enamel surface. Remarkably, nodule formation occurs in null mouse enamel when MMP20 is normally no longer expressed. The malformed enamel in Mmp20 null teeth was loosely attached to the dentin and the entire enamel layer tended to separate from the dentin indicative of a faulty DEJ. The enamel rod pattern was also altered in Mmp20 null mice. Each enamel rod is formed by a single ameloblast and is a mineralized record of the migration path of the ameloblast that formed it. The Mmp20 null mouse enamel rods were grossly malformed or were absent indicating that the ameloblasts do not migrate properly when backing away from the DEJ. Thus, MMP20 is required for ameloblast cell movement necessary to form the decussating enamel rod patterns, for the prevention of ectopic mineral formation, and to maintain a functional DEJ.
Keywords: MMP20, enamelysin, enamel, enamel organ, ameloblast
Introduction
Enamel development is stage specific. The two predominant stages are the secretory and maturation stages. During the secretory stage, MMP20 is expressed while a protein scaffold is formed and while the mineral precipitates as carbonated hydroxyapatite in long thin ribbons that grow out to form the full thickness of the enamel layer. The secretory stage is when the tall columnar ameloblasts of the enamel organ begin moving in rows to from the rod and inter-rod enamel. During the maturation stage, MMP20 expression ends and the ameloblasts shorten and cease movement. This is when kallikrein-4 (KLK4) assists in removal of the protein scaffold, which allows the enamel ribbons to grow further in width and thickness into large hexagonal crystals (1, 2). The crystals eventually press against one another and interlock as the enamel matures into its final hardened form (3).
MMPs are a family of proteinases that cleave virtually all extracellular matrix proteins. MMPs play critical roles in reproduction, development, morphogenesis, wound healing, tissue repair, regeneration, remodeling, and cell migration (reviewed in (4)). MMP20 is required for healthy dental enamel development. People and mice with homozygous MMP20 mutations have soft discolored enamel that may be hypoplastic and abrades easily from the dentin surface (5–11). Although, the expression of MMP20 during the secretory stage is necessary to cleave enamel matrix proteins (7, 12–22), MMP20 expression is also required to maintain a normal enamel organ morphology during the maturation stage of development when MMP20 is no longer expressed (6, 7). In the absence of MMP20, ectopic calcified nodules appear on the maturation stage enamel surface which disrupts the continuity of the almost linear ameloblast layer.
Here we demonstrate using the Mmp20 null mouse that: (1) calcified nodules form during the maturation stage of enamel development; (2) the maturation stage ameloblasts completely cover these nodules, (3) the enamel is only loosely connected to the underlying dentin; and (4) the enamel rod pattern is severely malformed or nonexistent. These results are consistent with MMP20 playing a key role in cell movement, formation of a strong DEJ, and prevention of ectopic mineral formation.
Materials and Methods
All animals used in this study were housed in Association for Assessment and Accreditation of Laboratory Animal Care approved facilities and all operations were performed in accord with protocols approved by the Forsyth Institute Animal Care and Use Committee.
Histology
Incisors were from adult C57BL/6 mice. Mice were anaesthetized with chloral hydrate and fixed by intravascular perfusion with 1% or 2.5% glutaraldehyde buffered with 0.08M sodium cacodylate containing 0.05% CaCl2, pH 7.2. The hemimandibles were removed and most jaws were decalcified at 4°C in 4.13% disodium ethylenediaminetetraacetic acid (EDTA), post-fixed in osmium tetroxide reduced with potassium ferrocyanide, dehydrated in graded alcohols, and embedded in LR White acrylic resin (London Resin, Berkshire, UK) as previously described (23). Jaws fixed with 2.5% glutaraldehyde were either decalcified as above or left undecalcified. All of these samples were treated with reduced osmium and processed for embedding in Epon 812 substitute. Semi-thin sections of incisor segments were cut with glass knives or with a diamond histoknife and stained with toluidine blue for examination by light microscopy.
Scanning electron microscopy (SEM)
For SEM imaging of surfaces (Figures 1, 2 and 3B), the labial bone encasing incisors was removed and the enamel organ cells were gently removed from the enamel and discarded. The exposed enamel surfaces were lightly brushed with dry Kimwipes and the incisors were examined at ×50 magnification without any further processing in a Hitachi S-3000N variable pressure scanning electron microscope using the backscatter mode at 25 kV and 20 Pa pressure. In other experiments, whole erupted incisors (Figure 3A) were air-dried, fastened to stubs, sputter coated with palladium gold, and examined using a JEOL 6400 scanning electron microscope. SEM evaluations of incisor prism patterns (Figure 4) were performed on rehydrated freeze-dried mandibular jaws from adult wild type and Mmp20 null mice. Samples were washed briefly in diluted sodium hypochlorite solution, rinsed in deionized water, lightly etched with 0.1% nitric acid, re-rinsed, air dried and examined without coating in backscatter mode using a JEOL-JSM6460LV (JEOL Ltd., Japan) SEM operated at 20 kV.
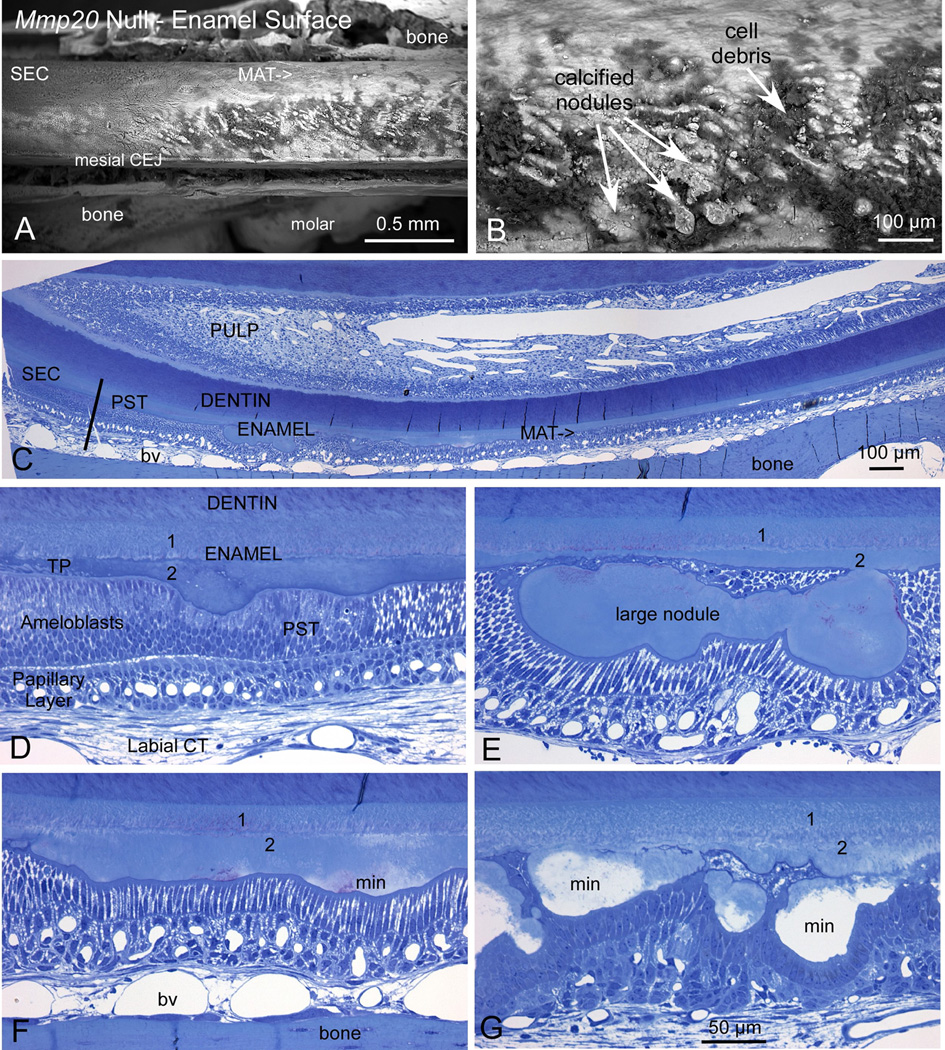
Fig. 1.
Mmp20 null mouse enamel was present in two distinct layers and had a rough surface due to the presence ectopic calcified nodules. Backscattering SEM images of the enamel surface from mandibular incisors of adult Mmp20 null mice (A, B) revealed malformed enamel. The enamel was especially malformed during the maturation stage (A; MAT) where numerous calcified nodules of irregular sizes and shapes were intermixed with cell debris (B). Semi-thin plastic demineralized histological sections (C–G) show that at the end of the secretory stage (C; SEC) the ameloblasts appear to undergo typical postsecretory transition (C, D; PST) into shorter cells typical of the maturation stage of amelogenesis. However, the enamel was thin and was composed of two layers (1, 2). Calcified nodules extend from the outermost layer nearest the ameloblasts to form a rough and uneven surface (2). Paradoxically, the innermost layer near the dentin (1) appeared relatively smooth and even (D–G). Both ameloblast and papillary layer cells were arrayed around the nodules to cover them, which distorted the spatial arrangement of the cells and, at times, made them appear folded (E, G). The nodules mineralized (F, G; min) during the maturation stage and at a somewhat faster pace than the inner two layers of enamel as observed by backscatter imaging (A, B) and the clear spaces left in the tissue following demineralization (G). The magnification bar in G applies equally to panels D–G. CEJ, cementoenamel junction; labial CT, labial connective tissue; bv, blood vessel; TP, Tomes’ Process.
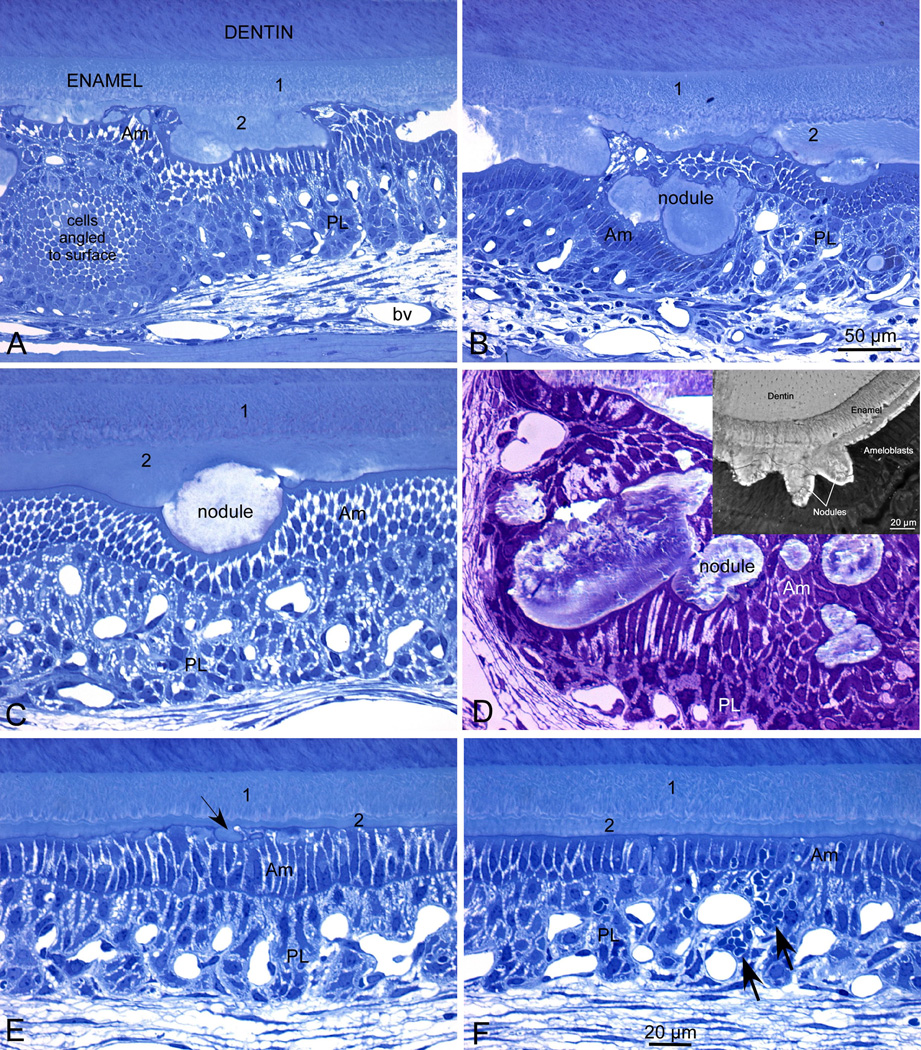
Fig. 2.
Mmp20 null maturation stage ameloblasts form a continuous covering over the nodules that project from the enamel surface. Semi-thin plastic demineralized histological sections (A–C, E, F) and a non-demineralized section (D) show maturation stage enamel organ from mandibular incisors of adult Mmp20 null mice. The inset in panel D shows backscatter imaging of the nodules at the enamel surface. Modulating ameloblasts (Am) and associated papillary layer cells (PL) formed a continuous covering over all nodules that projected from the enamel surface in Mmp20 null mice (A–D). This caused distortions in the normal spatial arrangement of the cells perhaps giving the illusion in some cases that isolated masses of cells were swirling and/or layered (A, B) or that nodules were embedded inside the cell layers of the enamel organ (B, D). In undemineralized sections (D), nodules appeared layered with a malformed structure. Nodule-free areas were sometimes encountered in the maturation stage (E, F). The enamel in these locations often appeared thin and ameloblasts appeared somewhat distorted with small pools of protein at their apices (E, arrow). Inflammatory cell infiltration into the enamel organ was sometimes observed in sections closest to the gingival margin of the tooth (F, arrows). 1 and 2 designate the two different enamel layers. The magnification bar in B applies also to panel A; the magnification bar in F applies equally to panels C–F.
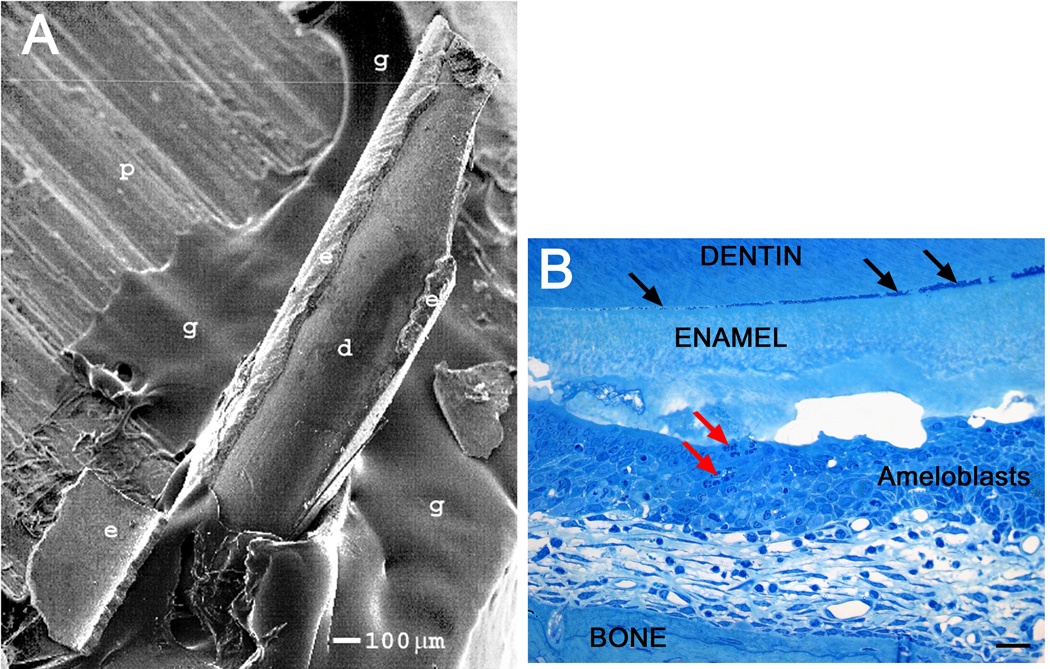
Fig. 3.
The dentin-enamel junction (DEJ) is weak in Mmp20 null enamel. SEM image of an incisor from an Mmp20 null mouse glued to a metal pedestal (A). Note that in the process of gluing the incisor to the pedestal a semi-conical piece of enamel fell off the dentin onto the pedestal at the base of the incisor. A semi-thin plastic demineralized histological section showing inflammatory cell infiltration into the ameloblast layer (B, red arrows). Debris appears to be accumulating at the DEJ where the enamel may already be separating from the underlying dentin (B, black arrows). d, dentin; e, enamel; g, glue; p, metal pedestal, panel B scale bar 20 µm.
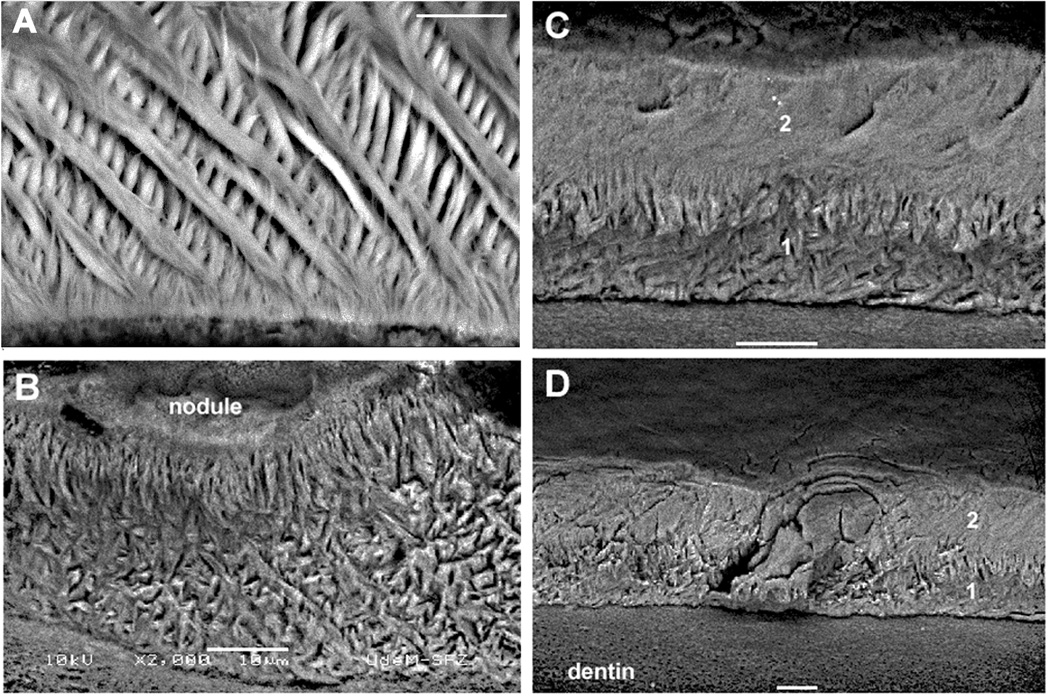
Fig. 4.
Enamel rod patterns of mandibular incisors from wild-type and Mmp20 null mice. The wild-type enamel had crisscrossing (decussating) rows of enamel rods (A). The Mmp20 null enamel may have a poorly organized rod pattern (B), no rod pattern with a poorly organized rod layer beneath (C), or virtually no rod pattern whatsoever (D). 1 and 2 designate the two different enamel layers. Note that relative to Figures 1 and 2, the enamel from these incisors are displayed upside down. All magnification bars are 10 µm in length.
Results
Mmp20 null mouse enamel appears histologically as two distinct layers and the enamel surface is marred by calcified nodules
Backscattering scanning electron imaging of mandibular incisors from adult Mmp20 null mice revealed that the maturation stage enamel surface was rough and contained numerous calcified nodules of irregular sizes and shapes (Fig. 1A, B). These nodules were intermixed with cell debris on the enamel surface. At the end of the secretory stage (Fig. 1C; SEC), ameloblasts appeared to undergo “true” postsecretory transition (Fig. 1C, D; PST) into shorter modulating cells typical of the maturation stage of amelogenesis. Strikingly, both modulating ameloblasts and papillary layer cells were arrayed around nodules which distorted the spatial arrangement of these cells and made them appear at times highly folded (Fig. 1E, G). Unlike normal mice where the enamel surface is flat and smooth, the Mmp20 null enamel surface was mostly irregular and undulating (Fig. 1D–G). Histological sections (Fig. 1D–G) showed that the abnormally thin enamel was comprised of two layers (1, 2) plus the mineralized nodules at the enamel surface. Interestingly, the inner enamel layer closest to the dentin appeared homogenous and did not vary greatly in thickness. However, the outer layer closest to the ameloblasts displayed large variations in thickness apparently due to the ameloblast layer accommodating the mineralized and mineralizing nodules (Fig. 1C–G). The nodules mineralized (Fig. 1 F, G; min) during the maturation stage at a somewhat faster pace than the inner two layers of enamel as observed by backscatter imaging (Fig. 1A, B) and the clear spaces remaining in the tissue following demineralization (Fig. 1G). This demonstrates that absence of MMP20 activity during the secretory stage causes ectopic mineralizations at the enamel surface during the maturation stage of enamel development.
The maturation stage ameloblast layer of Mmp20 null mice completely covers the calcified nodules
Rodent incisor maturation stage ameloblasts normally have a low columnar morphology consisting of single, almost linear, groupings of cells. However, maturation stage ameloblasts in Mmp20 null incisors have a markedly altered arrangement and morphology where the null ameloblasts (Am) and associated papillary cells (PL) form a continuous covering over extraneous nodules of calcified material formed at the enamel surface (Fig. 2A–C). This causes distortions in the normal spatial arrangement of the cells so that the cells can appear to swirl or to be layered (Fig. 2A, B). Although it is possible that the nodules are imbedded inside the cell layers of the enamel organ (Fig. 2B, D), in our experience the nodules located to the enamel surface (Fig. 2D inset) and the plane of sectioning made them appear within the cell layers. In undemineralized sections, nodules appear layered and to have abnormal structure (Fig. 2D). Nodule-free areas are also sometimes encountered in the maturation stage (Fig. 2E, F). The enamel in these locations often appears thin and the ameloblasts appear somewhat distorted with small pools of protein at their apices (Fig. 2E; arrow). Signs of inflammatory cell infiltration into the enamel organ are also sometimes seen in regions closest to the gingival margin of the tooth (Fig. 2F; arrows; Fig 3B red arrows). This shows that the ameloblast and papillary layers from Mmp20 null mice cover and promote the growth of the ectopic calcified nodules.
Mmp20 null mouse enamel is loosely connected to the underlying dentin
Enamel in Mmp20 null mice has almost no resistance to mechanical stress and is so frail that during processing of incisors for SEM analyses, large pieces of enamel often become dislodged from the tooth and fall off (Fig. 3A). Since the enamel does not crumble but instead dislodges in large sheets from the incisor, this suggests that the interface between enamel and underlying dentin in null mice is particularly weak. Prior to Mmp20 null mouse incisor eruption, a gap can be observed between the dentin and enamel that appears to accumulate debris (Fig. 3B black arrows). This shows that MMP20 is essential for formation of a mechanically strong DEJ typical of normal mice.
Mmp20 null mouse enamel has a disrupted or nonexistent rod pattern
Normal rodent incisor enamel has a decussating mineralized enamel rod pattern. Each rod is formed by one ameloblast and each rod preserves a complete record of the migratory path of the ameloblast that formed it. Figure 4A shows normal decussating enamel rods; rows of which can be observed to cross adjacent rows. In contrast, Mmp20 null mouse enamel has either a highly disorganized almost unrecognizable rod pattern (Fig. 4B, C) or it has virtually no rod pattern at all (Fig. 4D). This suggests that MMP20 is required for ameloblasts to move synchronously in rows that slide by each other to form the normal decussating enamel rod pattern.
Discussion
Here we show that enamel organ cells on the incisors of Mmp20 null mice: (1) form thinner than normal enamel that is arranged histologically in two distinct enamel layers; (2) form ectopic calcified nodules during the maturation stage that are covered by the ameloblast and associated papillary layers; (3) form enamel with a very weak DEJ; and (4) form enamel with a dysplastic or virtually nonexistent rod pattern.
In addition to the well defined roles for MMP20 in hydrolyzing newly secreted enamel matrix proteins and for maintaining normal enamel thickness (7, 12–22), MMP20 is also required to maintain a smooth outer enamel surface during the maturation stage of development when MMP20 is no longer expressed (7). If MMP20 is not present during earlier development, the enamel surface becomes lumpy and covered with numerous variably sized calcified nodules during later enamel development. Perhaps the earlier presence of MMP20 is crucial for definitive ameloblast progression from the secretory to maturation stage of enamel development. Deregulated developmental progression may promote the observed ectopic calcifications. This is supported by the observation that the Mmp20 null mouse secretory stage ameloblasts retract their Tomes’ processes as if preparing to enter the maturation stage, but later re-extend their Tomes’ processes as if resuming the secretory stage of development (24). Alternatively, it is possible that despite expression of KLK4 in the Mmp20 null mice, an abundance of un-reabsorbed enamel matrix protein at the ameloblast-enamel interface promotes ectopic nodule calcification. In either case, MMP20 is unexpectedly important for enamel maturation after it has normally ceased to be expressed.
The weak DEJ that allows the enamel to fall off in sheets was also interesting because it demonstrates that MMP20 activity is essential for establishing a strong bond between the dentin and forming enamel. Odontoblasts express MMP20 (13, 25) and this suggests that MMP20 may cleave important dentin protein(s), the cleavage products of which are essential for creating a strong DEJ. MMP20 does not cleave the most abundant dentin protein type I collagen (26), but it does cleave dentine sialophosphoprotein, which is the major non-collagen secretory product of odontoblasts responsible for dentin formation (27). MMP20 also cleaves aggrecan and cartilage oligomeric matrix protein (28), type V collagen (29), type XVIII collagen (30), fibronectin, type IV collagen, tenascin-C, laminin-1 and -5, but not type II collagen (26). Therefore, it is possible that cleavage of one or more of these proteins is essential for formation of the characteristically strong DEJ. Alternatively, since the KLK4 is expressed after the enamel has reached its full thickness; it may have limited access to the protein present in the deepest enamel layers. Thus, un-cleaved protein at the DEJ in Mmp20 null mice may interfere with the formation of a proper dentin-enamel interface. This could occur through a failure of the dentin crystals to properly seed the enamel crystals or could occur from a failure to produce a functional basement membrane at the apical end of the ameloblasts.
The Mmp20 null maturation stage ameloblasts maintain continuity by covering nodules projecting from the enamel surface. Perhaps, the normal complement of ameloblasts over the abnormally thin enamel causes cell buckling because too many ameloblasts are present in a given area. In this case, cell buckling would precede nodule formation. Conversely, the ameloblasts may simply cover the forming nodules and this may contribute to the observed distorted intercellular spatial relationships. In this case, nodule formation would be concurrent with formation of the irregular ameloblast layer (Fig. 1G; 2A, B). In any case, the distortion becomes progressively worse as enamel matures and may be a contributing factor for infiltration of inflammatory cells as ameloblasts approach the gingival margin where the tooth erupts (Figs. 2F and 3B). Ameloblast cell-cell contacts may also be affected in Mmp20 null mice. Cadherins are transmembrane proteins with extracellular domains that provide important adhesive contacts between neighboring cells. Previously it was demonstrated that ameloblasts express E-cadherin (31–37) and MMP20 cleaves the E-cadherin extracellular domain (24). Therefore in Mmp20 null mice, the distorted spatial relationships may also result, in part, from cell attachments that would normally not exist because of MMP20 activity. These normally cleaved attachments also likely interfere with the ability of rows of ameloblasts to slide by one another to form the characteristic decussating enamel rod pattern.
We have previously demonstrated that cadherins are essential for dental enamel development. We showed that conditional deletion of the cadherin stabilizing molecule, p120-catenin (p120), from epithelial tissues had a striking effect on mouse enamel development (31). Binding of p120 to the cadherin intracellular domain prevents cadherins from becoming internalized and degraded (38–40). When p120 was deleted from the enamel epithelium, E-cadherin was no longer immunolocalized to the ameloblasts, and the ameloblasts failed to properly attach to neighboring ameloblast, the stratum intermedium, or the enamel surface. Although, the general shape of the teeth was normal, the resulting dental enamel had no rod pattern and the malformed enamel easily abraded from the tooth surface (31). Therefore, stabilization of cadherins to the cell surface is essential for dental enamel formation. This also suggests that a balance exists between proteolytic processing of cadherins for normal ameloblast cell-cell contact versus a complete disruption of ameloblast function by an almost complete loss of ameloblast adhesive contacts. Thus, too many contacts may inhibit cell movement and too few may cause the ameloblast layer to fall apart.
Absence of cadherin hydrolysis by MMP20 may also be partially responsible for the poor or nonexistent rod pattern observed in the Mmp20 null enamel. Each enamel rod is formed by a single ameloblast (41). Therefore, each enamel rod is a mineralized record of the migratory path of the ameloblast that formed it (42). Since the Mmp20 null enamel rod pattern is dysplastic or nonexistent, this demonstrates that the null ameloblasts fail to migrate properly and implicates a role for MMP20 in modulation of ameloblast cell-cell contacts.
This study demonstrates that MMP20 is an important mediator necessary for maintaining a smooth enamel surface, a strong DEJ, and for establishing the decussating enamel rod pattern.
Acknowledgements
The authors gratefully acknowledge help provided by Micheline Fortin (LR White sectioning, decalcified samples) and Sylvia Zalzal (SEM) from the Université de Montréal, Line Mongeon (SEM) and Jeannie Mui (Epon embedding and sectioning, calcified samples) from McGill University, and Dr. Yuanyuan Hu (perfusions of mice with 2.5% glutaraldehyde) from the University of Michigan. This work was funded by NIDCR grant DE016276 (JDB) and by the CIHR (AN).
References
- 1.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit RevOral BiolMed. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 2.Smith CE. Cellular and chemical events during enamel maturation. Crit RevOral BiolMed. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 3.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roycik MD, Fang X, Sang QX. A fresh prospect of extracellular matrix hydrolytic enzymes and their substrates. Curr Pharm Des. 2009;15:1295–1308. doi: 10.2174/138161209787846676. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett JD, Beniash E, Lee DH, Smith CE. Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. JDentRes. 2004;83:909–913. doi: 10.1177/154405910408301204. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett JD, Skobe Z, Lee DH, Wright JT, Li Y, Kulkarni AB, Gibson CW. A developmental comparison of matrix metalloproteinase-20 and amelogenin null mouse enamel. Eur J Oral Sci. 2006;114 Suppl 1:18–23. doi: 10.1111/j.1600-0722.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- 7.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 8.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J MedGenet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, Piesco N, Hart TC. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papagerakis P, Lin HK, Lee KY, Hu Y, Simmer JP, Bartlett JD, Hu JC. Premature stop codon in MMP20 causing amelogenesis imperfecta. J Dent Res. 2008;87:56–59. doi: 10.1177/154405910808700109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SK, Seymen F, Kang HY, Lee KE, Gencay K, Tuna B, Kim JW. MMP20 hemopexin domain mutation in amelogenesis imperfecta. J Dent Res. 2010;89:46–50. doi: 10.1177/0022034509352844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett JD, Ryu OH, Xue J, Simmer JP, Margolis HC. Enamelysin mRNA displays a developmentally defined pattern of expression and encodes a protein which degrades amelogenin. ConnectTissue Res. 1998;39:101–109. doi: 10.3109/03008209809023916. [DOI] [PubMed] [Google Scholar]
- 13.Fukae M, Tanabe T, Uchida T, Lee SK, Ryu OH, Murakami C, Wakida K, Simmer JP, Yamada Y, Bartlett JD. Enamelysin (matrix metalloproteinase-20): localization in the developing tooth and effects of pH and calcium on amelogenin hydrolysis. JDentRes. 1998;77:1580–1588. doi: 10.1177/00220345980770080501. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Machule D, Gao C, DenBesten PK. Activation of recombinant bovine matrix metalloproteinase-20 and its hydrolysis of two amelogenin oligopeptides. Eur J Oral Sci. 1999;107:352–359. doi: 10.1046/j.0909-8836.1999.eos107506.x. [DOI] [PubMed] [Google Scholar]
- 15.Llano E, Pendas AM, Knauper V, Sorsa T, Salo T, Salido E, Murphy G, Simmer JP, Bartlett JD, Lopez-Otin C. Identification and structural and functional characterization of human enamelysin (MMP-20) Biochemistry. 1997;36:15101–15108. doi: 10.1021/bi972120y. [DOI] [PubMed] [Google Scholar]
- 16.Moradian-Oldak J, Jimenez I, Maltby D, Fincham AG. Controlled proteolysis of amelogenins reveals exposure of both carboxy- and amino-terminal regions. Biopolymers. 2001;58:606–616. doi: 10.1002/1097-0282(200106)58:7<606::AID-BIP1034>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 18.Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. EurJOral Sci. 2002;110:358–365. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 20.Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, Simmer JP. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- 21.Chun YH, Yamakoshi Y, Yamakoshi F, Fukae M, Hu JC, Bartlett JD, Simmer JP. Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res. 2010;89:785–790. doi: 10.1177/0022034510366903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamakoshi Y, Hu JC, Fukae M, Yamakoshi F, Simmer JP. How do enamelysin and kallikrein 4 process the 32-kDa enamelin? Eur J Oral Sci. 2006;114 Suppl 1:45–51. doi: 10.1111/j.1600-0722.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 23.Zalzal SF, Smith CE, Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27:352–359. doi: 10.1016/j.matbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Bartlett JD, Yamakoshi Y, Simmer JP, Nanci A, Smith CE. MMP20 Cleaves E-Cadherin and Influences Ameloblast Development. Cells Tissues Organs. 2011;294(2–4):222–226. doi: 10.1159/000324205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 26.Vaananen A, Srinivas R, Parikka M, Palosaari H, Bartlett JD, Iwata K, Grenman R, Stenman UH, Sorsa T, Salo T. Expression and regulation of MMP-20 in human tongue carcinoma cells. JDentRes. 2001;80:1884–1889. doi: 10.1177/00220345010800100501. [DOI] [PubMed] [Google Scholar]
- 27.Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem. 2006;281:38235–38243. doi: 10.1074/jbc.M607767200. [DOI] [PubMed] [Google Scholar]
- 28.Stracke JO, Fosang AJ, Last K, Mercuri FA, Pendas AM, Llano E, Perris R, Di Cesare PE, Murphy G, Knauper V. Matrix metalloproteinases 19 and 20 cleave aggrecan and cartilage oligomeric matrix protein (COMP) FEBS Lett. 2000;478:52–56. doi: 10.1016/s0014-5793(00)01819-6. [DOI] [PubMed] [Google Scholar]
- 29.Turk BE, Lee DH, Yamakoshi Y, Klingenhoff A, Reichenberger E, Wright JT, Simmer JP, Komisarof JA, Cantley LC, Bartlett JD. MMP-20 Is Predominately a Tooth-Specific Enzyme with a Deep Catalytic Pocket that Hydrolyzes Type V Collagen. Biochemistry. 2006;45:3863–3874. doi: 10.1021/bi052252o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaananen A, Tjaderhane L, Eklund L, Heljasvaara R, Pihlajaniemi T, Herva R, Ding Y, Bartlett JD, Salo T. Expression of collagen XVIII and MMP-20 in developing teeth and odontogenic tumors. Matrix Biol. 2004;23:153–161. doi: 10.1016/j.matbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Bartlett JD, Dobeck JM, Tye CE, Perez-Moreno M, Stokes N, Reynolds AB, Fuchs E, Skobe Z. Targeted p120-catenin ablation disrupts dental enamel development. PLoS One. 2010;5:e12703. doi: 10.1371/journal.pone.0012703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fausser JL, Schlepp O, Aberdam D, Meneguzzi G, Ruch JV, Lesot H. Localization of antigens associated with adherens junctions, desmosomes, and hemidesmosomes during murine molar morphogenesis. Differentiation. 1998;63:1–11. doi: 10.1046/j.1432-0436.1998.6310001.x. [DOI] [PubMed] [Google Scholar]
- 33.Heymann R, About I, Lendahl U, Franquin JC, Obrink B, Mitsiadis TA. E- and N-cadherin distribution in developing and functional human teeth under normal and pathological conditions. Am J Pathol. 2002;160:2123–2133. doi: 10.1016/S0002-9440(10)61161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obara N, Suzuki Y, Nagai Y, Takeda M. Expression of E- and P-cadherin during tooth morphogenesis and cytodifferentiation of ameloblasts. AnatEmbryol(Berl) 1998;197:469–475. doi: 10.1007/s004290050157. [DOI] [PubMed] [Google Scholar]
- 35.Palacios J, Benito N, Berraquero R, Pizarro A, Cano A, Gamallo C. Differential spatiotemporal expression of E- and P-cadherin during mouse tooth development. Int J DevBiol. 1995;39:663–666. [PubMed] [Google Scholar]
- 36.Sorkin BC, Wang MY, Dobeck JM, Albergo KL, Skobe Z. The cadherin-catenin complex is expressed alternately with the adenomatous polyposis coli protein during rat incisor amelogenesis. J HistochemCytochem. 2000;48:397–406. doi: 10.1177/002215540004800309. [DOI] [PubMed] [Google Scholar]
- 37.Terling C, Heymann R, Rozell B, Obrink B, Wroblewski J. Dynamic expression of E-cadherin in ameloblasts and cementoblasts in mice. EurJ Oral Sci. 1998;106 Suppl 1:137–142. doi: 10.1111/j.1600-0722.1998.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 38.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda M, Johnson E, Mandal SH, Lawson KR, Keim SA, Svoboda RA, Caplan S, Wahl JK, III, Wheelock MJ, Johnson KR. Expression of inappropriate cadherins by epithelial tumor cells promotes endocytosis and degradation of E-cadherin via competition for p120(ctn) Oncogene. 2006;25:4595–4604. doi: 10.1038/sj.onc.1209396. [DOI] [PubMed] [Google Scholar]
- 41.Skobe Z. SEM evidence that one ameloblast secretes one keyhole-shaped enamel rod in monkey teeth. Eur J Oral Sci. 2006;114 Suppl 1:338–342. doi: 10.1111/j.1600-0722.2006.00305.x. [DOI] [PubMed] [Google Scholar]
- 42.Cox B. A multi-scale, discrete-cell simulation of organogenesis: Application to the effects of strain stimulus on collective cell behavior during ameloblast migration. J Theor Biol. 2010;262:58–72. doi: 10.1016/j.jtbi.2009.09.010. [DOI] [PubMed] [Google Scholar]